Abstract
Using reverse transcription-PCR and clonal sequencing of the dengue virus envelope gene derived from the plasma samples of six patients, we reported for the first time that dengue virus circulates as a population of closely related genomes. The extent of sequence diversity varied among patients, with the mean pairwise proportions of difference ranging from 0.21 to 1.67%. Genome-defective viruses were found in 5.8% of the total number of clones analyzed. Our findings on the quasispecies nature of dengue virus and the defective virus in vivo have implications with regard to the pathogenesis of dengue virus.
Dengue virus belongs to the genus Flavivirus of the family Flaviviridae. It consists of four serotypes, DEN-1 to DEN-4. While most dengue virus infections are present as asymptomatic or mild, self-limited dengue fever (DF), some patients may develop severe and potentially life-threatening dengue hemorrhagic fever (DHF)-dengue shock syndrome (5, 6, 10, 25). Epidemics of the four dengue viruses continue to be a major public health problem in tropical and subtropical areas. It has been estimated that approximately 100 million cases of DF and 250,000 cases of DHF occur annually worldwide (5, 15).
Due partly to the nonproofreading and thus error-prone nature of viral RNA polymerase, many RNA viruses exhibit a high degree of sequence variation, not only among isolates from different individuals but also among viruses within the same individual (13, 26). RNA viruses therefore exist as a population of closely related sequences known as quasispecies (3, 8, 21). Quasispecies are believed to play an important role in the survival and evolution of RNA viruses as well as in the pathogenesis of disease. Well-studied examples are human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) (2, 4, 13, 24, 26). Little is known about the extent of sequence variation of dengue virus in vivo and its relationship to disease severity. In this study, we investigated sequence variation in the envelope (E) gene of dengue viruses in plasma samples from six patients confirmed to have dengue virus type 3 during an outbreak in southern Taiwan in 1998 (7, 11, 23, 25). We report for the first time that dengue virus is present as quasispecies in plasma. In addition, a deletion and stop codons were found in 4 out of 69 clones analyzed, indicating the presence of genome-defective viruses in vivo.
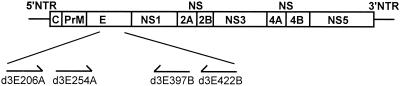
Dengue virus RNA was isolated from plasma samples from acutely ill patients collected within 7 days of the onset of illness with a QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) as described previously (23). The RNA eluates were subjected to reverse transcription (RT) by using a cDNA synthesis kit (Life Technologies, Rockville, Md.). Based on sequences available from GenBank, outer (d3E206A, d3E422B) and inner (d3E254A, d3E397B) primers were designed to amplify a 430-nucleotide region in the E gene (Fig. 1). This region covers all of domain III (which is presumably the receptor-binding domain), the connecting segment, and the hinge junction to domain II, according to the crystallographic model of the E protein of the tick-borne encephalitis virus (19). An aliquot of cDNA was subjected to first- and second-round PCR with the outer and inner primers, respectively. The PCR conditions were 95°C for 5 min, followed by 30 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 1 min, and then 72°C for 5 min. Each PCR product was cloned into the T/A cloning vector pCRII-TOPO, which was taken up by TOP10 competent cells by transformation (Invitrogen, San Diego, Calif.). To avoid the bias due to preferential amplification of certain templates in a single PCR, multiple clones derived from two separate PCRs were picked up and sequenced completely with a BigDye terminator cycle sequencing kit and a model ABI 373A automated sequencer (Applied Biosystems, Foster City, Calif.).
FIG. 1.
Schematic diagram of the dengue virus genome and the E region analyzed in this study. The relative positions of primers are shown. The primers and their sequences are as follows: d3E206A, 5′-TGGATGGTACATAGICAATGG-3′ (corresponding to genome positions 1543 to 1563 of dengue virus type 3 strain H87) (18); d3E422B, 5′-AGTCCCAGGCTGTGTCTCC-3′ (positions 2191 to 2173); d3E254A, 5′-ATCICAAGAGGGAGCAATGC-3′ (positions 1689 to 1710; I stands for deoxyinosine); and d3E397B, 5′-CGAGCTTCCCTTCCTGTACC-3′ (positions 2118 to 2099). NTR, nontranslated region; C, capsid; PrM, precursor membrane; NS, nonstructural.
The nucleotide sequences of 10 clones derived from dengue viruses from the plasma of a DF patient, ID17, were aligned with the program DNAMAN (version 4.15; Lynnon Biosoft, Quebec, Canada), and their 393-bp regions (excluding the sequences of primers) were compared. There were a total of 33 nucleotide substitutions, of which 30 were nonsilent and 3 were silent. To assess the extent of sequence variation, we determined the mean diversity, which was the number of substitutions divided by the total number of nucleotides sequenced (26). It was 0.84% for ID17 (Table 1). Among the 10 clones analyzed, some clones had more nucleotide substitutions whereas others had fewer. We therefore employed another method, pairwise comparison of each nucleotide sequence using the program MEGA, version 1.02 (Molecular Evolutionary Genetics Analysis, Pennsylvania State University, University Park), to assess the extent of sequence variation. The pairwise p-distances (proportions of difference in distances) thus determined ranged from 0.76 to 2.80% with a mean of 1.67% (Table 1).
TABLE 1.
Nucleotide sequence diversity of the E gene in dengue virus-infected patients
| Patient | Disease | No. of clones | No. of substitutionsa
|
No. of substitutions/total no. of nucleotides | Mean diversity (%)b | p-distance (%)c
|
||
|---|---|---|---|---|---|---|---|---|
| Nonsilent | Silent | Mean | Range | |||||
| ID3 | DF | 17 | 3 | 4 | 7/6,681 | 0.10 | 0.21 | 0-1.02 |
| ID7 | DF | 10 | 4 | 2 | 6/3,930 | 0.15 | 0.30 | 0-0.76 |
| ID8 | DF | 11 | 12 | 5 | 17/4,323 | 0.39 | 0.76 | 0.25-1.27 |
| ID17 | DF | 10 | 30 | 3 | 33/3,930 | 0.84 | 1.67 | 0.76-2.80 |
| ID19 | DHF | 10 | 3 | 2 | 5/3,930 | 0.13 | 0.25 | 0-1.02 |
| ID20 | DHF | 11 | 10 | 5 | 15/4,323 | 0.35 | 0.69 | 0-1.53 |
The nucleotide substitutions consist of nonsilent substitutions (those which change amino acids) and silent substitutions (those which do not change amino acids).
The mean diversity is the number of substitutions divided by the total number of nucleotides sequenced.
p-distances were calculated by pairwise comparison of nucleotide sequences between clones by the program MEGA.
To further examine the extent of sequence variation of dengue virus in vivo, RT and PCR were carried out for viral RNAs derived from the plasma of five other patients, including three DF and two DHF patients (25). Ten to seventeen clones from each sample were completely sequenced and analyzed, and the results are summarized in Table 1. There were more nonsilent substitutions than silent substitutions for most of the samples examined. The mean diversity ranged from 0.10 to 0.84%, indicating that the extent of sequence diversity varies among patients. Some patients, for example, ID3, had very homogeneous sequences (mean diversity, 0.10%) (Table 1). This was also revealed by the pairwise p-distance (mean, 0.21%) (Table 1). Overall, the extent of sequence variation determined by the mean diversity correlated with that determined by the mean pairwise p-distance (coefficient of correlation, r = 0.9998) (SPSS software, base 8.0; SPSS, Chicago, Ill.). These findings suggested that similar extents of sequence variation were seen in most of the clones analyzed and indicated that dengue virus exists as a population of closely related sequences in vivo.
To investigate the extent and the distribution of sequence variation at the protein level, the deduced amino acid sequences of all clones from each patient were aligned and analyzed. The mean diversity of the amino acids ranged from 0.13 to 2.29%, and the mean p-distances ranged from 0.27 to 4.14% (Table 2). These results are generally in agreement with those at the nucleotide level in that the mean p-distances of amino acids correlated with those of the nucleotides (r = 0.997) (SPSS, base 8.0) (Table 1). Shown in Fig. 2 are the amino acid sequence alignments of viral E proteins from two patients, ID17 and ID20. For ID17, there were a total of 30 amino acid substitutions within the 131-amino-acid region analyzed (Fig. 2A). Some amino acid changes were conservative (such as threonine to serine and isoleucine to leucine) (Fig. 2A), whereas others were drastic (such as glutamic acid to valine or glycine) (Fig. 2A). Two in-frame stop codons were found at amino acid residues 290 (clone 7A) and 301 (clone 4A), indicating that genome-defective dengue viruses were present in the plasma. For ID20, there were 10 amino acid substitutions (Fig. 2B). Of note was a single nucleotide deletion at the third base of amino acid residue 353 (of clone 1B) that resulted in a frameshift and a premature stop codon 3 residues downstream (Fig. 2B). Among the 69 clones analyzed, there were 3 clones that had in-frame stop codons (clones ID17-4A, ID17-7A, and ID19-2B) and one clone (ID20-1B) with a deletion (Fig. 2 and data not shown). This corresponds to a frequency of defective viruses of 5.8%, based on the small fraction of the genome examined in this study.
TABLE 2.
Amino acid sequence diversity of the E protein in dengue virus-infected patients
| Patient | Disease | No. of clones | No. of substitutions/total no. of amino acids | Mean diversity (%)a | p-distance (%)b
|
|
|---|---|---|---|---|---|---|
| Mean | Range | |||||
| ID3 | DF | 17 | 3/2,227 | 0.13 | 0.27 | 0-1.53 |
| ID7 | DF | 10 | 4/1,310 | 0.31 | 0.61 | 0-1.53 |
| ID8 | DF | 11 | 12/1,441 | 0.83 | 1.67 | 0-3.05 |
| ID17 | DF | 10 | 30/1,310c | 2.29 | 4.14 | 1.54-7.63 |
| ID19 | DHF | 10 | 3/1,310d | 0.23 | 0.27 | 0-1.53 |
| ID20 | DHF | 11 | 10/1,441 | 0.69 | 1.39 | 0-3.05 |
The mean diversity is the number of substitutions divided by the total number of amino acids sequenced.
p-distances were calculated by pairwise comparison of amino acid sequences between clones by the program MEGA.
Two clones contained stop codons.
One clone contained a stop codon.
FIG. 2.
Alignment of deduced amino acid sequences of multiple clones from two patients, ID17 (A) and ID20 (B). The positions of amino acid residues and the corresponding domains within the E protein based on the tick-borne encephalitis virus model (19) are shown on top. A consensus sequence (CON) was generated for each sample. Dashes indicate sequence identity, asterisks indicate stop codons at those positions, and the underline indicates deletion at that position. Individual clone numbers are shown at the left following patient identification numbers, with the letters A and B indicating two separate PCRs.
Using the clonal sequencing analysis, we demonstrated that dengue virus is present as a quasispecies in vivo. This finding suggests that future analysis and interpretation of dengue viral sequences derived from clinical samples should take into consideration the quasispecies structure of dengue viruses in vivo, i.e., the simultaneous presence of multiple variant genomes. The possibility that the sequence variation observed was due to in vitro artifacts needs to be addressed. The error rate of RT was estimated to be around 10−4 on a complex template (20). The error rate of Taq polymerase was reported to be between 3.9 × 10−6 and 9.1 × 10−6/nucleotide/cycle under most PCR conditions (12-14). After 60 cycles of PCR amplification, the error frequencies were between 2.3 × 10−4 and 5.5 × 10−4, which, together with the RT errors, are lower than the mean diversity (0.10 to 0.84%) observed in our samples. We carried out a control experiment in which a recombinant E clone with a known sequence was transcribed in vitro with T7 polymerase, followed by RT, PCR, subcloning, and sequencing under identical conditions. Among the eight clones analyzed, only one substitution out of 3,144 bases sequenced was found. This corresponds to an error frequency of 0.03%, which was lower than the mean diversity observed in our samples. Considering the error frequencies and mean levels of diversity together, the sequence variation reported in this study is unlikely to be due to in vitro artifacts, though a small proportion of the substitutions might have been introduced by RT or Taq polymerase.
The extent of sequence diversity observed in this study is similar to what has been reported for acute infection with HIV-1 and HCV, a member of Flaviviridae (4, 13, 26). In the case of HCV, the mean Hamming distances, which are the averages of the fractions of the amino acid differences taken for all sequence pairs derived from a single sample, ranged from 6 to 7% in hypervariable region 1 (HVR1) of E and ranged from less than 1 to 2% in the region outside HVR1 (4). The mean p-distances of amino acid sequences in our study (equivalent to the mean Hamming distances), ranging from 0.27 to 4.14%, were close to the mean Hamming distances of the region outside HVR1 of HCV and were lower than those in HVR1. This suggests that there is no hypervariable region like HVR1 of HCV identified in the 430-bp E region examined in this study. Consistent with this finding, analysis of the distribution of amino acid substitutions in different domains (the hinge junction, the connecting segment, and domain III) within this region of all clones reveals that there is no clustering of mutations in any particular domain examined (one-way analysis of variance, P = 0.734) (Fig. 2 and data not shown).
It should be noted that the sequence variation examined in this study was in the E gene, which is known to be the major determinant of cell tropism and the target of both humoral and cellular immune responses in many viruses. The E gene is therefore an area likely to be under positive selection. The difference in number of nonsynonymous nucleotide substitutions per site (dN) and the difference in number of synonymous nucleotide substitutions per site (dS) for each case were calculated with the MEGA program based on the method of Nei and Gojobori (17). The ratio of dN to dS was thus determined. With the exception of the mean dN/dS ratio of patient ID17, 2.13, the mean dN/dS ratios of five other patients (ranging from 0.48 to 0.85) were similar to those for the E region outside HVR1 in patients with acute HCV infection (ranging from 0.12 to 0.73) (4). The observation that the dN/dS ratios were generally lower than 1 suggests that strong positive selection for this E region is unlikely to have occurred in most cases of dengue virus infection, which is characterized by a short-lived resolving viremia (5, 10, 16, 22).
The relationship between the extent of sequence diversity and disease severity has been studied in several RNA viruses. For example, it was reported that a higher degree of sequence diversity of HIV-1 correlates with slower disease progression, suggesting an adaptive evolution in vivo (24). It has been reported recently that in the acute phase of HCV infection, progressing hepatitis was associated with genetic evolution whereas resolving hepatitis correlated with evolutionary stasis (4). By comparing the six samples in this study, we found that the levels of sequence diversity in the DHF patients were in the same range as those in the DF patients (Table 1). Recognizing as a caveat the small sample size in this study, studies of more cases are needed in the future to investigate the relationship between the extent of sequence diversity and disease severity in dengue virus infection.
Genome-defective dengue viruses containing either stop codons or a deletion, which have been confirmed by the sequencing of both strands, were identified in 5.8% of all clones analyzed. The frequency is similar to what has been reported for HCV, another member of Flaviviridae (13). There was evidence indicating that defective viruses can modulate viral replication in vitro, affect the clinical course of disease, or lead to the establishment of persistent infection (1, 3, 9). It should be noted that the 393-bp E region sequenced corresponds to 3.7% of the dengue virus genome. Although variation in the E gene may not be representative of the entire dengue virus genome, our finding that 5.8% of the clones in a small fraction of the genome were defective suggests that the frequency of defective viruses might be higher. Further analysis of more sequences of multiple regions would be required to confirm this. Interestingly, while the defective virus was found in only one of the four DF patients in this study, it was found in both of the DHF patients. Whether the higher frequencies of defective viruses seen in DHF patients result from higher levels of viral replication (16, 22) or whether the defective viruses contribute to the pathogenesis of DHF remains to be investigated.
Nucleotide sequence accession numbers
The sequences of the E genes of dengue viruses from the six patients studied here have been submitted to GenBank, and their accession numbers are AY053394 through AY053399.
Acknowledgments
We thank Shu-Mei Chang at Yuan's General Hospital in Kaoshiung, Taiwan, for kindly providing clinical samples and Day-Yu Chao, Tzu-Ling Sung, Yu-Chen Tsai, and Tsai-Yu Lin for statistical and technical assistance.
This work was supported by the National Health Research Institute (NHRI-CN-CL8903P) and in part by the National Science Council (NSC89-2320-B-002-206), Taipei, Taiwan, Republic of China.
REFERENCES
- 1.Barrett, A. D. T., and N. J. Dimmock. 1986. Defective interfering viruses and infections of animals. Curr. Top. Microbiol. Immunol. 128:55-84. [DOI] [PubMed] [Google Scholar]
- 2.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationship determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env gene. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 3.Domingo, E. J., J. J. Holland, and P. Ahlquist. 1988. RNA genetics, vol. 3. CRC Press Inc., Boca Raton, Fla.
- 4.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 5.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead, S. B. 1988. Pathogenesis of dengue: challenge to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 7.Harris, E., G. Roberts, L. Smith, J. Selle, L. D. Kramer, S. Valle, E. Sandoval, and A. Balmaseda. 1998. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 36:2634-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland, J. J., J. C. de la Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 9.Huang, A. S. 1988. Modulation of viral disease processes by defective interfering particles, p. 195-208. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. 3. CRC Press, Boca Raton, Fla. [Google Scholar]
- 10.Innis, B. L. 1995. Dengue and dengue hemorrhagic fever, p. 103-146. In J. S. Porterfield (ed.), Exotic viral infections—1995. Chapman & Hall, London, United Kingdom.
- 11.Lanciotti, R. S., C. H. Calisher, D. J. Gubler, G.-J. Chang, and A. V. Vorndam. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, M. J., B. Funsch, M. Wiese, and M. Roggendorf. 1995. Analysis of hepatitis C virus quasispecies populations by temperature gradient gel electrophoresis. J. Gen. Virol. 76:881-887. [DOI] [PubMed] [Google Scholar]
- 13.Martell, M., J. I. Esteban, J. Ouer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyerhans, A., R. Cheynier, J. Albert, S. Martina, S. Kwok, J. Sninsky, L. Morfeldt-Manson, B. Asjo, and S. Wain-Hobson. 1989. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell 58:901-910. [DOI] [PubMed] [Google Scholar]
- 15.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murgue, B., C. Roche, E. Chungue, and X. Deparis. 2000. Prospective study of the duration of magnitude of viremia in children hospitalized during the 1996-1997 dengue-2 outbreak in French Polynesia. J. Med. Virol. 60:432-438. [DOI] [PubMed] [Google Scholar]
- 17.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. E 3:418-426. [DOI] [PubMed] [Google Scholar]
- 18.Osatomi, K., and H. Sumiyoshi. 1990. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology 176:643-647. [DOI] [PubMed] [Google Scholar]
- 19.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Smith, D. B., J. McAllister, C. Casino, and P. Simmonds. 1997. Virus “quasispecies”: making a mountain out of a molehill? J. Gen. Virol. 78:1511-1519. [DOI] [PubMed] [Google Scholar]
- 21.Steinhauer, D. A., and J. J. Holland. 1987. Rapid evolution of RNA viruses. Annu. Rev. Microbiol. 41:409-433. [DOI] [PubMed] [Google Scholar]
- 22.Vaughn, D. W., S. Green, S. Kalayanarooj, B. L. Innis, S. Nimmannitya, S. Suntayakorn, T. P. Endy, B. Raengsakulrach, A. L. Rothman, F. A. Ennis, and A. Nisalak. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181: 2-9. [DOI] [PubMed] [Google Scholar]
- 23.Wang, W. K., C. N. Lee, C. L. Kao, Y. L. Lin, and C. C. King. 2000. Quantitative competitive reverse transcription-PCR for quantification of dengue virus RNA. J. Clin. Microbiol. 38:3306-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whestsell, M. R. Furtado, Y. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. 1997. Dengue hemorrhagic fever, diagnosis, treatment and control, 2nd ed. World Health Organization, Geneva, Switzerland.
- 26.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]