Abstract
Human immunodeficiency virus type 1 (HIV-1) gene expression in astrocytes is restricted, resulting in a brief and limited synthesis of HIV-1 viral structural proteins. Impaired Rev function has been documented in these cells. However, the molecular mechanisms underlying the impaired Rev function are not fully understood. Using the astroglial cell line U87.MG as a model, we report here that HIV-1 gene expression down-regulated expression of Sam68, the 68-kDa Src-associated protein in mitosis, which was constitutively expressed at a lower level in astrocytes. Elevating the endogenous level of Sam68 expression considerably restored HIV-1 Rev function in astrocytes, as determined by a Rev-dependent reporter gene assay. However, elevation of Sam68 expression achieved only a modest increase in HIV-1 production, further supporting the notion that there are multiple cellular restrictions of HIV-1 gene expression in astrocytes. Mutagenesis analysis identified the region between amino acids 321 and 410 of Sam68 as being directly involved in the binding of Sam68 to Rev, while a double mutation in Rev, L78D and E79L, like those in the dominant-negative Rev mutant M10, eliminated Rev binding to Sam68. Moreover, subcellular fractionation and digital fluorescence microscopic imaging revealed that Sam68 expression promoted Rev nuclear export. Taken together, our studies demonstrate that a lower level of constitutive Sam68 expression, followed by further down-regulation by HIV-1 infection, contributes to impaired Rev function in astrocytes, and they suggest that Sam68 may play an important role in Rev nuclear export.
Microglia and astrocytes are the major target cells for HIV-1 infection in the central nervous system (14, 22, 49). Direct infection of astrocytes has been well documented in pediatric and, to a lesser extent, adult AIDS patients (1, 36-38, 46-49, 51). One of the unique features of HIV-1 infection of astrocytes is nonproductive viral replication, which is due to inefficient virus entry and restricted viral gene expression (5, 6, 8, 43, 47, 48). Astrocytes are therefore an excellent model for studying the molecular mechanisms of HIV-1 replication and for developing novel strategies for anti-HIV therapeutics.
Productive HIV-1 replication requires efficient Rev-dependent nuclear export of unspliced and singly spliced viral RNA for viral gene expression and replication of viral genomic RNA (7). HIV-1 Rev binds to an RRE RNA sequence present in all unspliced and incompletely spliced viral mRNA transcripts, and it transports these mRNAs from the nucleus into the cytoplasm (29, 30). This is believed to be the rate-limiting step in regulating expression of RRE-containing HIV-1 mRNAs. Several cellular factors have been identified as involved in the Rev-mediated nuclear export pathway; these include CRM1 (9, 11), eIF5-A (42), and Rip/Rab (3, 10). Whether these cellular factors contribute to restricted HIV-1 gene expression in astrocytes remains unknown.
Evidence has accumulated to suggest that the inability of astrocytes to sustain HIV-1 gene expression is a result of multiple postentry restrictions in the viral life cycle (for a review, see reference 4). One of the restrictions is inadequate Rev function, which prevents expression of viral structural components from these incompletely spliced and unspliced mRNA transcripts in astrocytes (20, 27, 34). However, the molecular mechanisms of Rev function impairment in astrocytes are largely unknown.
To characterize the molecular mechanisms underlying restricted HIV-1 gene expression in astrocytes, we attempted to systematically profile gene expression in HIV-1-infected and uninfected astrocytes by using the subtractive cDNA cloning strategy. In this paper, we report that HIV-1 infection significantly down-regulated constitutive expression of Sam68 in U87.MG astrocytic cells and that HIV-1 infection-induced down-regulation of Sam68 was specific to U87.MG cells, which expressed considerably lower levels of constitutive Sam68 protein. Expression of exogenous Sam68 was able to restore Rev function and, to some extent, HIV-1 production in U87.MG cells. Moreover, Sam68 was found to specifically interact with Rev and to promote Rev nuclear export.
MATERIALS AND METHODS
Abbreviations.
The abbreviations used in this paper are as follows: HIV-1, human immunodeficiency virus type 1; Sam68, Src-associated protein in mitosis; RRE, Rev-responsive element; HA, hemagglutinin; RTase, reverse transcriptase; LTR, long terminal repeat; MLV, Moloney murine leukemia virus envelope; VSV-G, vesicular stomatitis virus envelope; p.i., postinfection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PBMCs, peripheral blood mononuclear cells; CAT, chloramphenicol acetyltransferase; GFP, green fluorescence protein; RFP, red fluorescence protein; DAPI, 4′,6′-diamidino-2-phenylindole.
Cell cultures and transfections.
U87.MG, U138.MG, Jurkat, CEM, and 293T cells were purchased from the American Type Culture Collection and maintained in either Dulbecco's modified Eagle's medium (U87.MG, U138.MG, and 293T cells), or RPMI 1640 medium (Jurkat and CEM cells) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. U87.MG cell lines stably expressing exogenous Sam68 were obtained by transfection of a Sam68 expression cassette, pc3.Sam68 (see below), followed by G418 selection (200 μg/ml). Cell transfections were performed by the standard calcium phosphate precipitation method.
Plasmids.
HIVΔenv.GFP, HIVΔenv.CAT, MLV env, and VSV-G env expression plasmids have been described elsewhere (14). The sources of other plasmids used in the studies were as follows: HIVΔenv, M. Emerman (23); pSam68.HA, T. Ishidate (18); pSG5.Sam68, S. Courtneidge (25); pc.Rev and M10, B. Cullen (28); pGFP.Rev, R. Brack-Werner (27); pRRE-CAT, T. Hope (17); and pSV2-CAT, J. Roth (41). The standard PCR cloning technique was used to construct the following expression plasmids, with the templates, primers, and cloning backbones (with restriction enzyme sites underlined) shown in parentheses: pc3.Sam68 (pSG5.Sam68, 5′-GGGGTACCATGCAGCGCCGGGACGAC-3′ and 5′-GGAATTCTTAATAACGTCCATATGGGTG-3′, pcDNA3), pAs.Sam68 (pSG5.Sam68, 5′-TAATGAATTCATGCAGCGCCGGGACGAC-3′ and 5′-GGAATTCTTAATAACGTCCATATGGGTG-3′, pcDNA3), pRev.Flag (pc.Rev, 5′-GGAATTCTATGGCAGGAAGAAGCGGA-3′ and 5′-GGGGTACCCTATTCTTTAGTTCCTGACTC-3′, pFlag.CMV-1 from Eastman Kodak), pM10.Flag (pM10, with the same primers and backbone as those for pRev.Flag), pSam68.RFP (pSG5.Sam68, 5′-TCGAGCTCAAATGCAGCGCCGGGACGAC-3′ and 5′-GGAATTCTTAATAACGTCCATATGGGTG-3′, pDsRed1-C1 from Clontech [Palo Alto, Calif.]), pRev.RFP (pc.Rev, 5′-GGAATTCTATGGCAGGAAGAAGCGGA-3′ and 5′-GGGGTACCTTCTTTAGTTCCTGACTCCAA-3′, pDsRed1-C1), pGFP.M10 (pM10, with the same primers as those for pRev.RFP above, pGFP-C1 from Clontech). Sam68 in-frame deletion mutants pSam68ΔR1.HA, pSam68ΔX1.HA, and pSam68ΔR5.HA were made by digesting pSam68.HA with the existing unique restriction enzyme sites EcoRI, XhoI, and EcoRV, respectively, and religating the digested parental vector DNA. All recombinant plasmids were verified by sequencing, and functional activities of pRev.Flag, pRev.GFP, pM10.Flag, and pM10.GFP were confirmed by both an RRE-mediated reporter gene assay and an in trans complementation assay.
mRNA isolation and cDNA subtractive hybridization cloning.
U87.MG cells were infected with HIV-GFP reporter viruses, and infected cells were separated from uninfected cells by flow cytometry using GFP as a marker. mRNA was isolated from HIV-infected and uninfected cells by using the Micro-FastTrack 2.0 Kit (Invitrogen). cDNA subtractive hybridization was performed using the PCR-Select cDNA Subtraction Kit (Clontech) according to the manufacturer's instructions. After hybridization, differentially expressed transcripts were amplified by suppressive PCR, cloned, and sequenced. Sequence analysis was conducted by using the BLAST homology search program.
Preparation of whole-cell, cytoplasmic, and nuclear lysates.
Cells were washed twice with ice-cold phosphate-buffered saline and then harvested for whole-cell, cytoplasmic, or nuclear lysates by methods described previously (21). Briefly, cell pellets were resuspended in 2 volumes of whole-cell lysis buffer (10 mM NaHPO4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.2% sodium azide, 0.5% sodium deoxycholate, 0.004% sodium fluoride, and 1 mM sodium orthovanadate) and incubated on ice for 10 min. Whole-cell lysates were obtained by centrifugation and removal of the cell debris. To prepare cytoplasm and nuclear lysates, the cells were washed twice with phosphate-buffered saline and resuspended in cell lysis buffer (0.5% NP-40, 10 mM Tris-HCl [pH 8.0], 10 mM NaCl, 3 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol, 1 mM (each) phenylmethanesulfonyl fluoride and sodium orthovanadate, 2 mg of leupeptin/ml, and 2 mg of aprotinin/ml). After a 10-min incubation on ice, the cell suspension was spun at 500 × g at 4°C for 5 min. The supernatant was recovered as a cytoplasmic lysate, while pellets were saved as cell nuclei. Nuclei were then washed twice in the cell lysis buffer, followed by extraction for 10 min on ice with the whole-cell lysis buffer. Cell debris was removed by centrifugation at 12,000 × g at 4°C for 10 min. The resulting supernatant was saved as a nuclear lysate.
Northern and Western blot analyses.
Total cellular RNA was isolated by using Gibco/BRL's Trizol RNA isolation kit according to the manufacturer's instructions. After fractionation by electrophoresis, RNA was transferred to a Hybond-N nylon membrane (Amersham) and hybridized with 32P-radiolabeled DNA probes at 42°C overnight. Radiolabeled probes were prepared with the Random Primed DNA Labeling Kit (Boehringer-Mannheim), followed by removal of unincorporated radionucleotides with a Centri-Spin 40 column (Princeton Separation). For Western blot analysis, cell lysates (25 μg of protein) or immunoprecipitates with antibodies as indicated (1 mg of protein) were electrophoretically separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% acrylamide) gels and analyzed by immunoblotting using antibodies against either HA tag or Flag tag. The primary anti-Sam68, anti-HA, and anti-actin antibodies (Santa Cruz) and the primary anti-Flag antibody (M5; Sigma) were used at the dilutions recommended by the manufacturers. The blots were probed first with primary antibodies and then with appropriate horseradish peroxidase-conjugated secondary antibodies and were visualized with the ECL system (Amersham). Relative levels of protein or mRNA were determined by densitometric scanning of the blots using the housekeeping gene actin or GAPDH as an internal standard.
Preparation and infection of HIV-1 pseudotyped viruses and RTase assay.
HIV-1 viruses pseudotyped with different envelope proteins were prepared as previously described (14). Briefly, 293T cells (2 × 106 cells per 10-cm plate) were transfected with 20 μg of HIV-1 reporter plasmids (pHIV-GFP or pHIV-CAT) and 4 μg of pMLV-env, pVSV-G, or pcDNA3 by the calcium phosphate precipitation method. Cell culture supernatants were collected 48 h after medium change, filtered, and saved as virus stocks. Viral gene expression and viral replication were determined by measuring RTase activity, as previously described (15).
Digital fluorescence microscopic imaging analysis.
Fluorescence images were captured by using a digital video imaging microscope system consisting of a Zeiss Model Axiovert 200 M microscope with a 40× 1.4 UVF objective (Carl Zeiss) and an Axiovision video camera (Carl Zeiss). Images were analyzed on an IDE Imaging Workstation (Tegraf).
RESULTS
Down-regulation of Sam68 expression in U87.MG cells by HIV-1 infection.
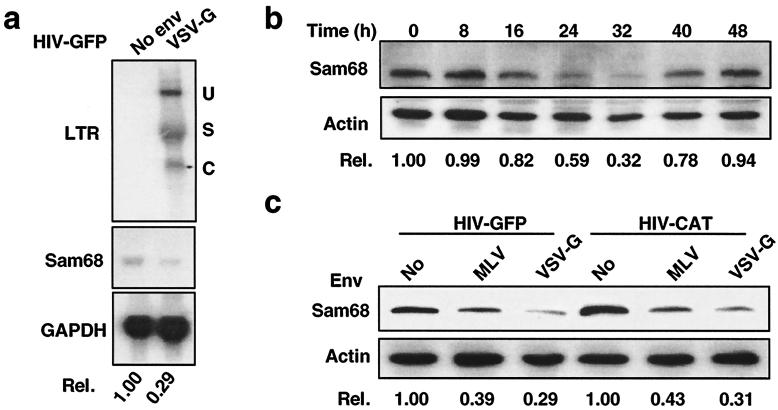
In order to identify cellular factors responsible for restricted HIV-1 gene expression in astrocytes, we utilized the subtractive cDNA cloning strategy to compare gene expression profiles between HIV-infected and uninfected astrocytes. We infected a well-characterized human astroglial cell line, U87.MG cells, with VSV-G-pseudotyped HIV-GFP reporter viruses (14). HIV-infected and uninfected cells were separated by flow cytometry for GFP expression. We then extracted mRNA, synthesized cDNA, and performed subtractive cDNA hybridization using the PCR-Select cDNA Subtraction Kit (Clontech). To confirm the reliability of subtractive cDNA cloning, Northern blot analysis was conducted using each of the isolated cDNA clones as a probe to hybridize against RNAs from HIV-infected and mock-infected U87.MG cells. Among the cDNA clones identified as affected by HIV-1 infection was the human Sam68 gene. Sam68 mRNA was down-regulated in U87.MG cells infected with 100 ng of p24gag of VSV-G-pseudotyped HIV-GFP viruses by 71% (Fig. 1a), which corresponded with the infection efficiency (about 69%) estimated by flow cytometry analysis for GFP expression.
FIG. 1.
Down-regulation of Sam68 expression by HIV infection in U87.MG cells. (a) Down-regulation of Sam68 mRNA by HIV-1 infection. U87.MG cells were infected with 100 ng of p24gag of HIV-GFP reporter viruses and were harvested for total RNA isolation at 32 h p.i., followed by Northern blotting. Probes were HIV-1 LTR (top), Sam68 cDNA (center), or GAPDH (bottom). U, unspliced RNA; S, singly spliced RNA; C, compeletely spliced RNA. (b) Down-regulation of Sam68 protein by HIV-1 infection. U87.MG cells were infected with 100 ng of p24gag of VSV-G-pseudotyped HIV-GFP viruses and were harvested at different time points p.i., as indicated, for whole-cell lysates, followed by Western blotting. (c) Viral envelope-independent Sam68 down-regulation. U87.MG cells were infected with 100 ng of p24gag of HIV reporter viruses pseudotyped with MLV or VSV-G, and Sam68 expression was analyzed by Western blotting. Quantitation of Sam68 mRNA and protein was performed as described in Materials and Methods.
To determine whether HIV-1 infection also led to a decrease in Sam68 protein expression, we infected U87.MG cells with VSV-G-pseudotyped HIV-GFP viruses, harvested cells at different time points p.i., and enriched HIV-infected cells for GFP expression by using flow cytometry. Western blot analysis showed that HIV-1 infection also resulted in a decrease in the Sam68 protein level, which began at 16 h, reached the lowest point at 32 h, and then recovered close to the initial level at 48 h (Fig. 1b, upper panel). In contrast, protein expression of the control housekeeping gene actin showed little or no change (Fig. 1b, lower panel). Moreover, Sam68 down-regulation in U87.MG cells was correlated with the amounts of VSV-G-pseudotyped HIV-GFP viruses used in the infection (data not shown). Based on these results, we used 24 to 32 h p.i. as the experimental time point throughout the studies unless stated otherwise.
To ensure that the observed effects of HIV-1 infection on Sam68 expression were not due to the VSV-G envelope and/or the HIV-GFP reporter virus used, we also tested MLV, as well as the HIV-CAT reporter virus (16). The results showed that MLV-pseudotyped HIV-GFP and HIV-CAT viruses down-regulated Sam68 expression by about 60%, while VSV-G-pseudotyped HIV-GFP and HIV-CAT viruses down-regulated Sam68 expression by about 70% (Fig. 1c), which corresponded with the infection efficiencies, 59% for MLV-pseudotyped viruses and 68% for VSV-G-pseudotyped viruses, at equal amounts of input virus (100 ng of p24gag). These results demonstrate that patterns of Sam68 down-regulation in U87.MG cells were basically identical, indicating that the observed down-regulation of Sam68 was due to postentry HIV-1 gene expression.
Rescue of HIV-1 Rev function in U87.MG cells by exogenous Sam68.
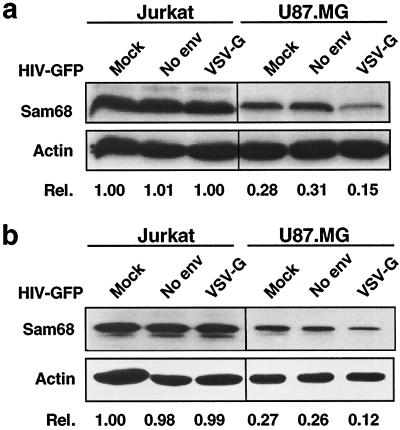
To investigate whether HIV-1 gene expression-induced down-regulation of Sam68 is specific to astrocytes, we then examined Sam68 expression in HIV-infected human T-lymphoid cell lines and PBMCs, which support productive viral replication. To obtain a comparable level of infection efficiency (about 60%), the amounts of viruses used for infection of each cell type were adjusted on the basis of GFP expression. The results showed that HIV-1 infection caused no detectable changes in Sam68 expression in Jurkat T lymphocytes (Fig. 2a), CEM cells, or PBMCs (data not shown). To ascertain that no changes in Sam68 expression were due to intracellular redistribution of Sam68 protein in these cells, as previously described for poliovirus (31), we determined Sam68 protein expression in the nucleus. Sam68 protein patterns in nuclei were similar to those in whole cells (Fig. 2b). In addition, we noticed that constitutive Sam68 expression in U87.MG cells was about 30% of that in all three cells supporting productive HIV-1 replication (e.g., Jurkat cells [Fig. 2a]). These results demonstrate that HIV-1 infection does not down-regulate Sam68 expression in cells supporting productive HIV-1 replication, indicating that the observed down-regulation of Sam68 was specific to astrocytes.
FIG. 2.
HIV-1 infection does not down-regulate Sam68 expression in HIV-permissive Jurkat cells. Cells were infected with HIV-GFP viruses (100 ng of p24gag) either containing no Env or containing VSV-G and were harvested 32 h after infection for whole-cell lysates (a) and nuclear lysates (b). Protein expression was analyzed by Western blotting. Mock infection with the culture medium was included.
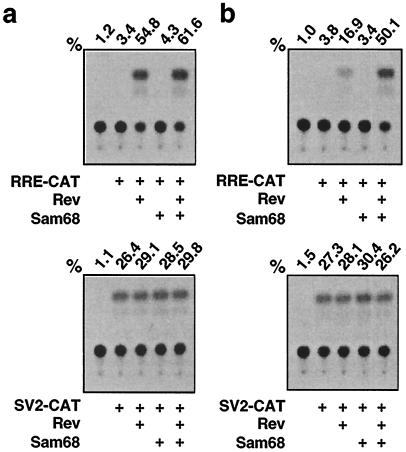
Since Sam68 has recently been shown to substitute for and synergize with HIV-1 Rev function (39), and our data indicate that HIV-1 gene expression down-regulates constitutive Sam68 expression in astrocytes, we decided to examine the effects of Sam68 expression on Rev function and HIV-1 production in astrocytes. To this end, we performed the Rev-dependent RRE-CAT reporter gene assay (17, 39). We rationalized that more exogenous Sam68 would be needed to activate Rev (RRE)-dependent gene expression in cells expressing a lower level of constitutive Sam68 protein, and vice versa. Thus, we transfected U87.MG cells with the RRE-CAT reporter plasmid (17) in combination with either a Rev expression plasmid or a Sam68 expression plasmid, or both. We also included 293T cells, which have been used in previous studies (39), and the Rev (RRE)-independent SV2-CAT reporter plasmid (41), as controls in these transfections. The results showed that Rev expression transactivated RRE-CAT expression by 53.6% in 293T cells (Fig. 3a, top) but increased RRE-CAT expression by only 15.9% in U87.MG cells (Fig. 3b, top), although both cell lines exhibited similar levels of Rev-independent SV2-CAT expression (Fig. 3, bottom). These results were consistent with the previous findings that Rev responsiveness is impaired in astrocytes (20, 27, 34). In contrast to the previous results (39), Sam68 expression alone exhibited little or no effect on RRE-CAT expression in 293T cells or U87.MG cells (Fig. 3a and b, top). Moreover, compared to Rev expression alone, coexpression of Sam68 and Rev increased RRE-CAT expression by 49.1% in U87.MG cells (Fig. 3b, top) but only 6.8% in 293T cells (Fig. 3a, top). The enhancement of Rev-dependent RRE-CAT activity by Sam68 expression in U87.MG cells was comparable to that by Rev expression alone in 293T cells. These results demonstrated that expression of exogenous Sam68 was able to relieve impaired Rev function in U87.MG cells, and they raised the possibility that a lower level of constitutive Sam68 expression followed by further down-regulation by HIV-1 infection may contribute to restricted HIV-1 gene expression in astrocytes.
FIG. 3.
Reconstitution of Rev function in U87.MG cells by expression of exogenous Sam68. 293T cells (a) and U87.MG cells (b) were transfected with 0.5 μg of RRE-CAT (top) or SV2-CAT (bottom) reporter DNA, in combination with 3 μg of the Rev expression plasmid pc.Rev and/or the Sam68 expression plasmid pc3.Sam68. pcDNA3 was used to equalize the total amounts of DNA transfected among all transfections. pCMVβGal was included to normalize variations of transfection efficiency among all transfections. CAT activity was determined by using cell lysates prepared 48 h posttransfection, as previously described (14, 15), and is expressed as a percentage of conversion. The CAT activity was determined to be in the linear range.
Enhancement of HIV-1 production in U87.MG cells by expression of exogenous Sam68.
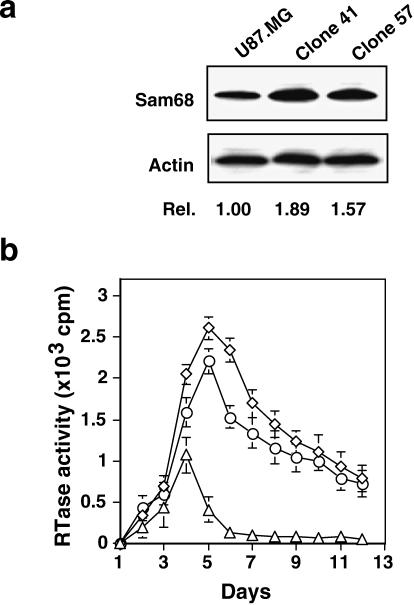
To investigate the possibility mentioned above, we elevated endogenous Sam68 expression in astrocytes and examined its effects on HIV-1 gene expression in these cells. We stably introduced a Sam68 expression cassette, pc3.Sam68, into U87.MG cells, and stable expression of Sam68 in these cells was analyzed by Western blot analysis. The results showed that stable introduction of exogenous Sam68 gene increased constitutive Sam68 expression in U87.MG cells by a range of 60 to 90% (data not shown). We then selected two representative clones, clones 41 and 57 (Fig. 4a), which exhibited 89 and 57% increases in Sam68 protein levels, respectively, and transfected them with HIV-1 NL4-3 proviral DNA. We monitored these cells for virus production for 12 consecutive days. The results showed that expression of exogenous Sam68 had little or no effect on the kinetics of virus production in U87.MG cells (48) but increased maximal virus production 2.7- and 2.3-fold in two different Sam68-expressing clones (Fig. 4b). The increase in virus production was consistent with the results obtained from the RRE-mediated reporter gene assay (Fig. 3b, top). Interestingly, we also detected considerable levels of virus production in these two stable U87.MG clones even at day 12 posttransfection, when there is little or no virus production in the parental U87.MG cells (Fig. 4b). The viruses collected from these two Sam68 stable cell clones at day 12 were able to infect and replicate in freshly isolated PBMCs (data not shown), suggesting that the progeny viruses were infectious. These results together demonstrated that despite the ability of Sam68 to fully restore Rev function in U87.MG cells, elevating Sam68 expression only modestly increased HIV-1 production in these cells, further supporting the notion that there are multiple restrictions of HIV-1 gene expression in astrocytes.
FIG. 4.
Augmented HIV-1 production in U87.MG cells by stable expression of exogenous Sam68. U87.MG cells were transfected with a pc3.Sam68 expression plasmid by the CaPO4 method. Stable clones were obtained by using 200 μg of G418/ml as the selection marker. (a) Sam68 expression in U87.MG cells and in two representative stable U87.MG clones, clones 41 and 57, determined by Western blotting. (b) HIV-1 production in U87.MG cells and the same two Sam68 stable clones. U87.MG cells (▵) and U87.MG stable clones 41 (◊) and 57 (○) were transfected with 1.5 μg of HIV-1 NL4-3 proviral DNA. A comparable level of the transfection efficiency was obtained, as confirmed by using peGFP (Clontech) as a marker. A 0.5-ml volume of culture medium was collected and processed at the time points indicated for the RTase activity assay (14).
Direct interaction of Sam68 with Rev.
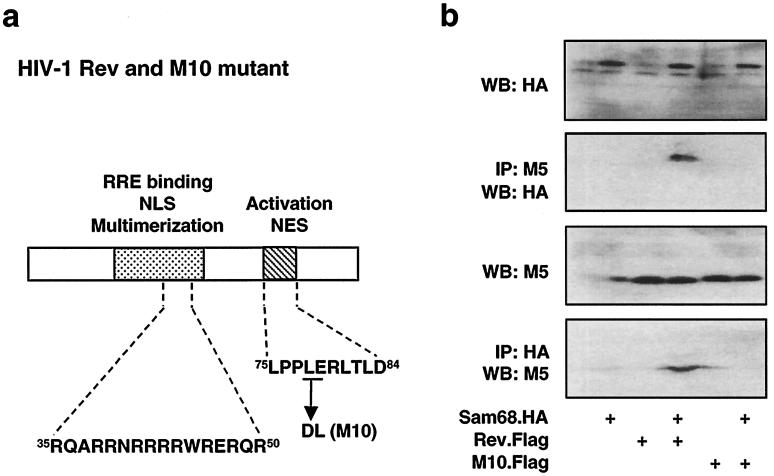
Our results demonstrated that Sam68 was able to restore Rev function in RRE-dependent gene expression (Fig. 3b, top) and accordingly improved HIV-1 gene expression (virus production) in U87.MG cells (Fig. 4b), suggesting that Sam68 may play an important role in Rev function. Thus, we decided to determine whether Sam68 interacted with Rev protein. We tagged Rev at the N terminus with a Flag epitope for immunodetection. We transfected 293T cells with expression plasmids for Sam68 (pSam68.HA), Rev (pRev.Flag), or both. Expression of Sam68 and Rev proteins was analyzed by Western blotting of cell lysates using antibodies HA and M5, which recognize the HA epitope and the Flag epitope, respectively (Fig. 5b, panels WB:HA and WB:M5). We performed immunoprecipitation with M5 followed by Western blotting with HA. The results showed that Sam68 bound to Rev (Fig. 5b, panel IP:M5/WB:HA), which was further confirmed by immunoprecipitation with HA followed by Western blotting with M5 (Fig. 5b, panel IP:HA/WB:M5). We also tagged M10, a well-characterized dominant-negative Rev mutant (28), with a Flag epitope and tested whether M10 retained the Sam68 binding function. Interestingly, two point mutations at amino acids 78 and 79 of Rev, as in M10 (Fig. 5a), abolished Rev binding to Sam68 (Fig. 5b, panels IP:M5/WB:HA and IP:HA/WB:M5). These results demonstrated that Sam68 bound to Rev and that these two amino acid residues within the nuclear export signal region of Rev were directly involved in Rev binding to Sam68.
FIG. 5.
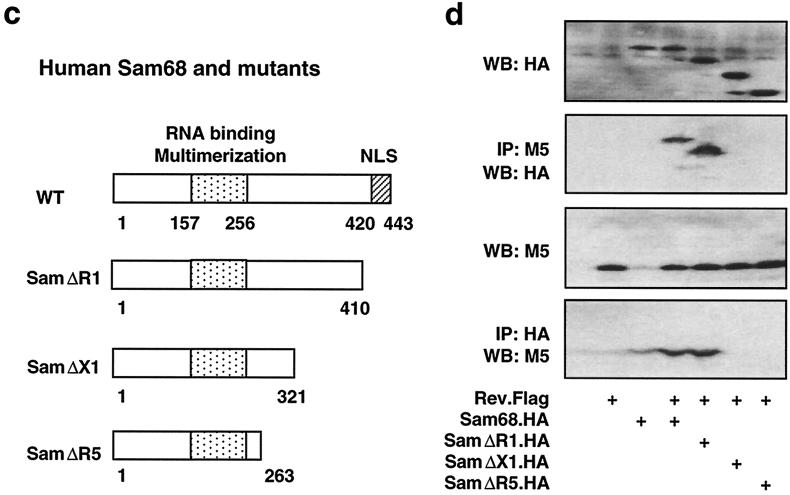
Direct interaction between Sam68 and Rev. 293T cells were transfected with 3 μg of different Rev and/or Sam68 expression plasmids, as indicated, and harvested 48 h after transfection for whole-cell lysates. pcDNA3 was used to equalize the total amount of DNA transfected among all transfections. Sam68 and Rev expression was analyzed by Western blotting (WB) of whole-cell lysates, and binding was detected by immunoprecipitation (IP) followed by Western blotting, as described in Materials and Methods. NLS, nuclear localization signal; NES, nuclear export signal.
We also made a series of Sam68 deletion mutants to identify the region(s) of Sam68 that was involved in its binding to Rev. Three C-terminal deletion mutants of Sam68 were constructed using the existing unique restriction sites (Fig. 5c), transfected into 293T cells in the presence of Rev, and analyzed for their binding to Rev by using a similar strategy. Expression of all Sam68 mutants (Fig. 5d, panel WB:HA) and Rev (Fig. 5d, panel WB:M5) was confirmed by Western blotting. Immunoprecipitation with M5 followed by Western blotting with HA showed that deletion of amino acids 410 to 433 of Sam68 (mutant designated SamΔR1) did not affect Sam68 binding to Rev (Fig. 5d, panel IP:M5/WB:HA). However, deletions of amino acids 321 to 433 (mutant designated SamΔX1) or amino acids 263 to 433 (mutant designated SamΔR5) of Sam68 abolished Sam68 binding to Rev (Fig. 5d, panel IP:M5/WB:HA). Conversely, immunoprecipitation with HA antibody followed by Western blotting with M5 antibody showed similar results (Fig. 5d, panel IP:HA/WB:M5). These data together demonstrated that the region between amino acids 321 and 410 of Sam68 is directly involved in Sam68 binding to Rev.
Rev nuclear export by Sam68 expression.
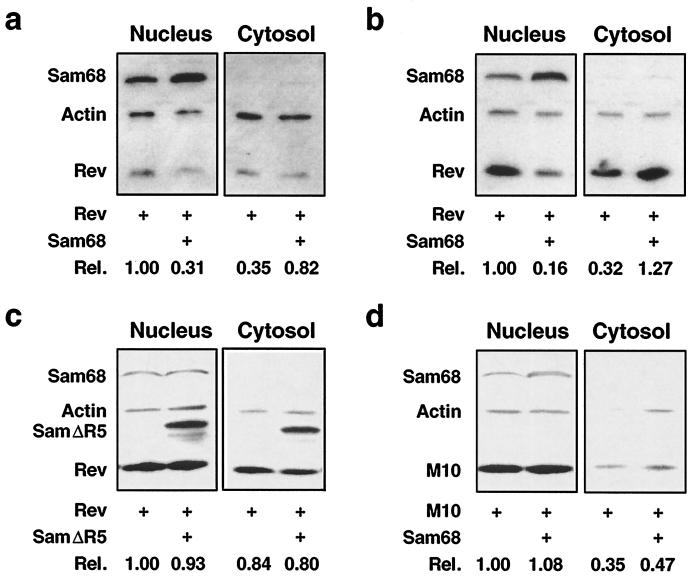
The primary function of Rev is to transport RRE-containing viral RNAs from the nucleus to the cytoplasm (19, 32, 40). Our findings that Sam68 was able to restore Rev function in astrocytes and that Sam68 directly interacted with Rev raised the possibility that Sam68 is involved in the dynamics of Rev protein between the nucleus and the cytoplasm. To address this possibility, we transfected U87.MG cells with Rev alone or together with Sam68 and determined the effects of Sam68 expression on Rev distribution between the nucleus and the cytoplasm by subcellular fractionation followed by Western blot analysis. The results showed that, as expected, Rev was mostly expressed in the nucleus (Fig. 6a). When coexpressed with Sam68, Rev protein levels were decreased by 69% in the nucleus and accordingly increased by 51% in the cytoplasm (Fig. 6a). Sam68 expression-induced Rev nuclear export appeared to be much more dramatic in 293T cells (Fig. 6b), which gave rise to a higher transfection efficiency (data not shown). More importantly, no detectable changes in Rev subcellular distribution were noted in 293T cells when Sam68 was replaced by the Sam68ΔR5 mutant (Fig. 6c) or when Rev was replaced by M10 (Fig. 6d). Taken together, these results demonstrate that Sam68 expression promoted Rev nuclear export by direct interaction of Sam68 with Rev.
FIG. 6.
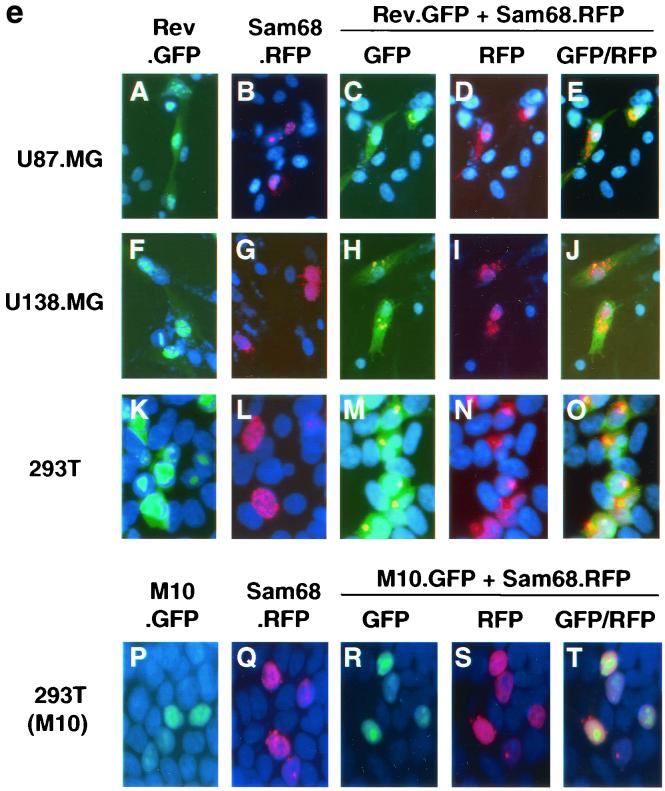
Rev nuclear export by Sam68 expression. (a through d) Rev nuclear export by subcellular fractionation. U87.MG cells (a) or 293T cells (b through d) were transfected with Rev, Sam68, M10, or Sam68ΔR5, or with different combinations, as indicated, and harvested 48 h after transfection for subcellular fractionation, as described elsewhere (21). pcDNA3 was used to equalize the total amount of DNA transfected among all transfections. Protein expression was detected by Western blotting using antibodies against Rev, Sam68, and actin, and levels were expressed as described above. (e) Rev nuclear export depicted by digital fluorescence microscopic imaging. U87.MG (panels A through E), U138.MG (panels F through J), and 293T (panels K through O) cells were transfected with Rev.GFP, Sam68.RFP, or both, as indicated. pM10.GFP was also included in 293T transfections (panels P through T). pcDNA3 was used to equalize the total amount of DNA transfected among all transfections. Cells were stained with 1 ng of DAPI/ml for the nuclei (blue fluorescence) 48 h after transfection and were then processed for digital fluorescence microscope imaging. In order to compare intracellular protein distribution among samples, images were taken with a constant exposure time with each fluorescent filter. Colocalization was analyzed by overlaying the images taken under the fluorescein isothiocyanate filter with those taken under the rhodamine filter and was identified by orange color.
To further ascertain the role of Sam68 in Rev nuclear export, we utilized GFP- or RFP (Clontech)-tagged Rev and Sam68, i.e., Rev.GFP and Sam68.RFP, and the digital fluorescence microscopic imaging technique to directly examine the relationship between Sam68 expression and intracellular localization of Rev. We transfected U87.MG cells, as well as another astroglial cell line, U138.MG cells, with Rev.GFP, Sam68.RFP, or both. We also included 293T cells as a nonastroglial control. We then fixed the cells in 2% paraformaldehyde and stained the cells with DAPI (blue fluorescence) to locate the nucleus. The results showed that when expressed alone, Rev, identified by green fluorescence, was mainly localized in the nucleus or nucleoli in all transfected cells (Fig. 6e, panel A), while Sam68, identified by red fluorescence, was also mainly localized in the nucleus (Fig. 6e, panel B). When coexpressed, Rev protein was significantly shifted into the cytoplasm (Fig. 6e, panel C), while Sam68 protein was localized mostly on or close to the nuclear membrane (Fig. 6e, panel D). Colocalization analysis showed that Rev and Sam68 proteins were colocalized (identified by orange color) more often on or close to the nuclear membrane (Fig. 6e, panel E). Comparable results were obtained with U138.MG cells (Fig. 6e, panels F through J), while more dramatic Rev cytoplasmic localization was noted in 293T cells (Fig. 6e, panels K through O), which was consistent with the previous data (Fig. 6a and b). To determine the specificity of Sam68-induced Rev nuclear export, we also transfected 293T cells with M10 tagged with GFP (M10.GFP), alone or in combination with Sam68.RFP. The results showed that when expressed independently, both M10 and Sam68 were localized in the nucleus, as expected (Fig. 6e, panels P and Q). Coexpression of Sam68 did not exhibit any detectable effects on the nuclear localization of M10 (Fig. 6e, panel R) or of Sam68 itself (Fig. 6e, panel S). Moreover, little or no colocalization of Sam68 and M10 was noted (Fig. 6e, panel T). Taken together, these results further demonstrated that Sam68 expression facilitated the export of Rev protein from the nucleus to the cytoplasm, and they suggest that a lower level of constitutive Sam68 expression followed by HIV-1 infection-induced down-regulation may impair Rev nuclear export and thereby Rev function in astrocytes.
DISCUSSION
It has been generally accepted that a block in Rev function contributes to the inability of astrocytes to support productive HIV-1 gene expression. Our results obtained from subtractive cDNA gene expression profiling showed that HIV-1 infection down-regulated Sam68 protein expression (Fig. 1). HIV-1 infection of cells supporting productive viral replication, e.g., Jurkat T lymphocytes, did not exhibit any detectable effects on Sam68 expression (Fig. 2), which may be due to a higher level of constitutive Sam68 expression in those cells and/or an unknown cell type dependence of Sam68 protein. The precise mechanisms of HIV-1 infection-induced Sam68 down-regulation in astrocytes are still under active investigation.
The differences in HIV-induced Sam68 down-regulation and constitutive Sam68 expression between cells supporting productive HIV-1 replication and astrocytes, along with the published results showing that Sam68 can function alone or with Rev in HIV-1 replication (39), prompted us to further characterize the roles of Sam68 protein in interaction between astrocytes and HIV-1 gene expression. We first asked whether Sam68 expression would relieve a Rev function block in astrocytes. The results showed that Sam68 expression restored Rev function almost to the full extent in the RRE-dependent reporter gene assay (Fig. 3b). Nevertheless, we obtained only an approximately threefold increase in HIV-1 viral production in astrocytes by stably introducing exogenous Sam68 gene expression (Fig. 4b), suggesting that other restrictions of HIV-1 gene expression exist in astrocytes. These may include transcriptional and translational suppression of viral genes (13, 26, 35), as well as improper posttranslational processing of viral proteins (44). In agreement with a previous report (27), we also observed that U87.MG and U138.MG astrocytes appeared to express more Rev in the cytoplasm (Fig. 6e, panels A and F). A recent study has further demonstrated that a defect in Rev nuclear import may also contribute to impaired Rev function in astrocytes (33).
In contrast with other published results (39), our results showed that Sam68 alone was not able to substitute for Rev in an RRE-mediated reporter gene assay (Fig. 3). Moreover, our results showed that only marginally synergistic effects between Sam68 and Rev were obtained in 293T cells (Fig. 3a), which was also confirmed in another recent study (45). This discrepancy can be attributed to the different levels of endogenous Sam68 expression in the cells that were used, as we demonstrated that Sam68 was able to function with Rev only in U87.MG cells, which expressed a lower level of constitutive Sam68 protein (Fig. 2a).
Sam68 was first identified as a tyrosine-phosphorylated protein in Src-transformed NIH 3T3 cells (12, 52) and has been shown to be involved in intracellular signal transduction and RNA metabolism (50). Sam68 has recently been demonstrated to preferentially bind to an RNA sequence, UAAA (24), which we located within stem I of RRE. Although these findings corroborated the previous report that Sam68 interacts with RRE in a specific manner (39), extensive studies have shown that only stems IIB and IID of RRE are required for Rev binding and thus Rev-mediated nuclear export of viral RNAs (2). Our results showed that Sam68 bound to Rev (Fig. 5) and promoted Rev nuclear export in the absence of RRE-containing RNAs (Fig. 6). Thus, Sam68 binding to RRE may not be necessary for the Sam68 function in Rev nuclear export. In addition, a very recent study has demonstrated that the dominant-negative Sam68ΔC mutant, in which the C-terminal nuclear localization signal has been deleted, inhibits Rev activity by causing perinuclear accumulation of unspliced RRE-containing RNAs (45), further suggesting that the effects of Sam68 and the dominant-negative Sam68 mutant on Rev function may be mediated by different mechanisms. However, the precise roles of Sam68 protein in the Rev nuclear export pathway and HIV-1 gene expression remain to be elucidated.
Acknowledgments
We thank M. Emerman, T. Ishidate, S. Courtneidge, R. Brack-Werner, B. Cullen, T. Hope, and J. Roth for materials and advice. We also thank H. Broxmeyer, J. Blum, S. Spinola, G. Alkhatib, A. Srivastava, and D. Gabuzda for critical reading of the manuscript.
This work was supported by grant R01NS39804 (to J.J.H.) and funds from the Department of Microbiology and Immunology, the Department of Medicine, and the Walther Oncology Center at the Indiana University School of Medicine.
REFERENCES
- 1.Bagasra, O., E. Lavi, L. Bobroski, K. Khalili, J. P. Pestaner, R. Tawadros, and R. J. Pomerantz. 1996. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS 10:573-585. [DOI] [PubMed] [Google Scholar]
- 2.Battiste, J. L., H. Mao, N. S. Rao, R. Tan, D. R. Muhandiram, L. E. Kay, A. D. Frankel, and J. R. Williamson. 1996. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273:1547-1551. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., R. A. Fridell, S. Madore, and B. R. Cullen. 1995. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 82:485-494. [DOI] [PubMed] [Google Scholar]
- 4.Brack-Werner, R. 1999. Astrocytes: HIV cellular reservoirs and important participants in neuropathogenesis. AIDS 13:1-22. [DOI] [PubMed] [Google Scholar]
- 5.Brack-Werner, R., A. Kleinschmidt, A. Ludvigsen, W. Mellert, M. Neumann, R. Herrmann, M. C. Khim, A. Burny, N. Muller-Lantzsch, D. Stavrou, et al. 1992. Infection of human brain cells by HIV-1: restricted virus production in chronically infected human glial cell lines. AIDS 6:273-285. [PubMed] [Google Scholar]
- 6.Chiodi, F., S. Fuerstenberg, M. Gidlund, B. Asjo, and E. M. Fenyo. 1987. Infection of brain-derived cells with the human immunodeficiency virus. J. Virol. 61:1244-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen, B. R. 1998. HIV-1 auxiliary proteins: making connections in a dying cell. Cell 93:685-692. [DOI] [PubMed] [Google Scholar]
- 8.Dewhurst, S., K. Sakai, X. H. Zhang, A. Wasiak, and D. J. Volsky. 1988. Establishment of human glial cell lines chronically infected with the human immunodeficiency virus. Virology 162:151-159. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 10.Fritz, C. C., M. L. Zapp, and M. R. Green. 1995. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 376:530-533. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 12.Fumagalli, S., N. F. Totty, J. J. Hsuan, and S. A. Courtneidge. 1994. A target for Src in mitosis. Nature 368:871-874. [DOI] [PubMed] [Google Scholar]
- 13.Gorry, P. R., J. L. Howard, M. J. Churchill, J. L. Anderson, A. Cunningham, D. Adrian, D. A. McPhee, and D. F. Purcell. 1999. Diminished production of human immunodeficiency virus type 1 in astrocytes results from inefficient translation of gag, env, and nef mRNAs despite efficient expression of Tat and Rev. J. Virol. 73:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, J., Y. Chen, M. Farzan, H. Choe, A. Ohagen, S. Gartner, J. Busciglio, X. Yang, W. Hofmann, W. Newman, C. R. Mackay, J. Sodroski, and D. Gabuzda. 1997. CCR3 and CCR5 are coreceptors for HIV-1 infection of microglia. Nature 385:645-649. [DOI] [PubMed] [Google Scholar]
- 15.He, J., C. M. deCastro, G. R. Vandenbark, J. Busciglio, and D. Gabuzda. 1997. Astrocyte apoptosis induced by HIV-1 transactivation of the c-kit protooncogene. Proc. Natl. Acad. Sci. USA 94:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishidate, T., S. Yoshihara, Y. Kawasaki, B. C. Roy, K. Toyoshima, and T. Akiyama. 1997. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 409:237-241. [DOI] [PubMed] [Google Scholar]
- 19.Kalland, K. H., A. M. Szilvay, K. A. Brokstad, W. Saetrevik, and G. Haukenes. 1994. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell. Biol. 14:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinschmidt, A., M. Neumann, C. Moller, V. Erfle, and R. Brack-Werner. 1994. Restricted expression of HIV-1 in human astrocytes: molecular basis for viral persistence in the CNS. Res. Virol. 145:147-153. [DOI] [PubMed] [Google Scholar]
- 21.Lawe, D. C., C. Hahn, and A. J. Wong. 1997. The Nck SH2/SH3 adaptor protein is present in the nucleus and associates with the nuclear protein SAM68. Oncogene 14:223-231. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. C., W. C. Hatch, W. Liu, Y. Kress, W. D. Lyman, and D. W. Dickson. 1993. Productive infection of human fetal microglia by HIV-1. Am. J. Pathol. 143:1032-1039. [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Q., S. J. Taylor, and D. Shalloway. 1997. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J. Biol. Chem. 272:27274-27280. [DOI] [PubMed] [Google Scholar]
- 25.Lock, P., S. Fumagalli, P. Polakis, F. McCormick, and S. A. Courtneidge. 1996. The human p62 cDNA encodes Sam68 and not the RasGAP-associated p62 protein. Cell 84:23-24. [DOI] [PubMed] [Google Scholar]
- 26.Ludvigsen, A., T. Werner, W. Gimbel, V. Erfle, and R. Brack-Werner. 1996. Down-modulation of HIV-1 LTR activity by an extra-LTR nef gene fragment. Virology 216:245-251. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, E., F. C. Silberstein, J. van Empel, V. Erfle, M. Neumann, and R. Brack-Werner. 1999. Diminished rev-mediated stimulation of human immunodeficiency virus type 1 protein synthesis is a hallmark of human astrocytes. J. Virol. 73:8279-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 29.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 30.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 31.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 33.Neumann, M., E. Afonina, F. Ceccherini-Silberstein, S. Schlicht, V. Erfle, G. N. Pavlakis, and R. Brack-Werner. 2001. Nucleocytoplasmic transport in human astrocytes: decreased nuclear uptake of the HIV Rev shuttle protein. J. Cell Sci. 114:1717-1729. [DOI] [PubMed] [Google Scholar]
- 34.Neumann, M., B. K. Felber, A. Kleinschmidt, B. Froese, V. Erfle, G. N. Pavlakis, and R. Brack-Werner. 1995. Restriction of human immunodeficiency virus type 1 production in a human astrocytoma cell line is associated with a cellular block in Rev function. J. Virol. 69:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niikura, M., G. Dornadula, H. Zhang, M. Mukhtar, D. Lingxun, K. Khalili, O. Bagasra, and R. J. Pomerantz. 1996. Mechanisms of transcriptional transactivation and restriction of human immunodeficiency virus type I replication in an astrocytic glial cell. Oncogene 13:313-322. [PubMed] [Google Scholar]
- 36.Nuovo, G. J., F. Galler, P. MacConnell, and A. Braun. 1994. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor-alpha RNA in the central nervous system. Am. J. Pathol. 144:659-666. [PMC free article] [PubMed] [Google Scholar]
- 37.Poland, S. D., G. P. Rice, and G. A. Dekaban. 1995. HIV-1 infection of human brain-derived microvascular endothelial cells in vitro. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:437-445. [DOI] [PubMed] [Google Scholar]
- 38.Ranki, A., M. Nyberg, V. Ovod, M. Haltia, I. Elovaara, R. Raininko, H. Haapasalo, and K. Krohn. 1995. Abundant expression of HIV Nef and Rev proteins in brain astrocytes in vivo is associated with dementia. AIDS 9:1001-1008. [DOI] [PubMed] [Google Scholar]
- 39.Reddy, T. R., W. Xu, J. K. Mau, C. D. Goodwin, M. Suhasini, H. Tang, K. Frimpong, D. W. Rose, and F. Wong-Staal. 1999. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 5:635-642. [DOI] [PubMed] [Google Scholar]
- 40.Richard, N., S. Iacampo, and A. Cochrane. 1994. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology 204:123-131. [DOI] [PubMed] [Google Scholar]
- 41.Ristea, S., M. Dobbelstein, and J. Roth. 2000. Rev protein of human immunodeficiency virus type 1 and cellular exportin 1 protein relocalize each other to a subnucleolar structure. AIDS Res. Hum. Retrovir. 16:857-865. [DOI] [PubMed] [Google Scholar]
- 42.Ruhl, M., M. Himmelspach, G. M. Bahr, F. Hammerschmid, H. Jaksche, B. Wolff, H. Aschauer, G. K. Farrington, H. Probst, D. Bevec, et al. 1993. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell Biol. 123:1309-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito, Y., L. R. Sharer, L. G. Epstein, J. Michaels, M. Mintz, M. Louder, K. Golding, T. A. Cvetkovich, and B. M. Blumberg. 1994. Overexpression of nef as a marker for restricted HIV-1 infection of astrocytes in postmortem pediatric central nervous tissues. Neurology 44:474-481. [DOI] [PubMed] [Google Scholar]
- 44.Shahabuddin, M., G. Bentsman, B. Volsky, I. Rodriguez, and D. J. Volsky. 1996. A mechanism of restricted human immunodeficiency virus type 1 expression in human glial cells. J. Virol. 70:7992-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soros, V. B., H. V. Carvajal, S. Richard, and A. W. Cochrane. 2001. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J. Virol. 75:8203-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi, K., S. L. Wesselingh, D. E. Griffin, J. C. McArthur, R. T. Johnson, and J. D. Glass. 1996. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann. Neurol. 39:705-711. [DOI] [PubMed] [Google Scholar]
- 47.Tornatore, C., R. Chandra, J. R. Berger, and E. O. Major. 1994. HIV-1 infection of subcortical astrocytes in the pediatric central nervous system. Neurology 44:481-487. [DOI] [PubMed] [Google Scholar]
- 48.Tornatore, C., K. Meyers, W. Atwood, K. Conant, and E. Major. 1994. Temporal patterns of human immunodeficiency virus type 1 transcripts in human fetal astrocytes. J. Virol. 68:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tornatore, C., A. Nath, K. Amemiya, and E. O. Major. 1991. Persistent human immunodeficiency virus type 1 infection in human fetal glial cells reactivated by T-cell factor(s) or by the cytokines tumor necrosis factor alpha and interleukin-1β. J. Virol. 65:6094-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vernet, C., and K. Artzt. 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13:479-484. [DOI] [PubMed] [Google Scholar]
- 51.Wiley, C. A., R. D. Schrier, J. A. Nelson, P. W. Lampert, and M. B. Oldstone. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc. Natl. Acad. Sci. USA 83:7089-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, G., O. Muller, R. Clark, L. Conroy, M. F. Moran, P. Polakis, and F. McCormick. 1992. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell 69:551-558. [DOI] [PubMed] [Google Scholar]