Abstract
Microtubule plus-end proteins CLIP-170 and EB1 dynamically track the tips of growing microtubules in vivo. Here we examine the association of these proteins with microtubules in vitro. CLIP-170 binds tubulin dimers and co-assembles into growing microtubules. EB1 binds tubulin dimers more weakly, so no co-assembly is observed. However, EB1 binds to CLIP-170, and forms a co-complex with CLIP-170 and tubulin that is recruited to growing microtubule plus ends. The interaction between CLIP-170 and EB1 is competitively inhibited by the related CAP-Gly protein p150Glued, which also localizes to microtubule plus ends in vivo. Based on these observations, we propose a model in which the formation of distinct plus-end complexes may differentially affect microtubule dynamics in vivo.
Keywords: Microtubule-binding proteins, EB1, CLIP-170, p150Glued
1. Introduction
Live cell imaging has revealed a group of proteins that specifically and dynamically track the tips of growing microtubules. These plus-end proteins are evolutionarily conserved and may regulate microtubule dynamics and microtubule-based transport (reviewed in [1]). While the in vivo behavior of these proteins has been examined, the mechanisms driving their plus-end localization remain unclear. Here we have examined the mechanisms of plus-end tracking for the proteins CLIP-170 and EB1.
CLIP-170 was originally identified as a linker between endosomes and microtubules[2] and EB1 was identified as a binding partner for the adenomatous polyposis coli protein (APC)[3]. GFP fusion proteins of each revealed that they track the plus-ends of growing microtubules in cells[4,5]. At the amino terminus of CLIP-170 are two tandem CAP-Gly microtubule binding domains[6]. A similar domain is found in the p150Glued subunit of the cytoplasmic dynein activator complex dynactin, which also shows plus-end localization in some cell types[7]. EB1 is part of a family of proteins that includes EB3 and RP1[8]; these proteins share a microtubule-binding domain that is structurally distinct from the CAP-Gly domain[9,10].
Proteins can show dynamic localization to microtubule plus-ends either through an association with a plus-end directed motor protein or by treadmilling on the microtubule end. While some proteins are actively transported to microtubule plus ends by kinesins[11], analysis of the dynamics of GFP fusion proteins has suggested that CLIP-170 and EB1 treadmill on the tips of growing microtubules by adding on at the plus-end and releasing just proximal to the end[4,12]. Several mechanisms could account for this treadmilling behavior. The end of a growing microtubule is biochemically and structurally distinct from that of a shrinking microtubule [13,14], and plus-end proteins may have a higher affinity for growing ends than for either shrinking ends or the microtubule lattice. Alternately, a plus-end protein could bind to tubulin dimers in the cytoplasm and co-polymerize with them, as suggested by Diamantopoulos et al. [15]. Here we have examined the plus end-targeting of CLIP-170 and EB1 in vitro, and provide evidence for distinct mechanisms for the plus end localization of these proteins.
2. Materials and Methods
Generation of Recombinant Proteins
Recombinant constructs of CLIP-170 (an in-frame T7 tag was fused to the N-terminus of the previously characterized H2 construct[16] generously provided by Holly Goodson, Notre Dame University) and p150Glued (residues 1–333)[17], which span the microtubule binding domains of these proteins, as well as full length EB1[17], were expressed as His-tagged fusion proteins in E. coli and purified by Ni2+-chromatography.
Microtubule Decoration Assays
Stable microtubules were polymerized from 15 μM tubulin dimers (Cytoskeleton, Denver, CO) in the presence of 1 mM GTP and 200 μM taxol, then centrifuged at 39,000xg and resuspended in warm PEM-50 buffer (100 mM PIPES, 1 mM EDTA, 1 mM MgCl2, pH 7.3) with 50 mM NaCl, 1 mM MgGTP, and 20 μM taxol. For assembly assays these microtubules were sheared to generate seeds[17]. For polarity marked microtubules, rhodamine-conjugated tubulin (Cytoskeleton) was polymerized at a 1:20 ratio to unlabeled tubulin to form brightly-labeled seeds, followed by polymerization in 15 μM tubulin at a ratio of 1:50 rhodamine-labeled to unlabeled tubulin dimers to generate more dimly labeled polymer.
For tip-localization experiments, microtubules were polymerized from 15 μM tubulin dimers and taxol-stabilized microtubules seeds in PEM-50 buffer, then plus-end proteins were added to final concentrations of 0.3 μM, 3.0 μM, or 9.0 μM monomer, as noted, and incubated two minutes at 37°C. For co-assembly experiments, plus-end proteins were pre-incubated with 15 μM tubulin dimers on ice for 5 minutes, mixed with microtubule seeds and 1 mM GTP to initiate polymerization, and incubated for 2 minutes at 37°C prior to fixation.
Microtubules were fixed with gluteraldehyde and pelleted onto coverslips as previously described[17], then labeled with primary antibodies to tubulin (Sigma, St. Louis, MO; Cytoskeleton), EB1 (BD Transduction Laboratories, San Jose, CA), T7 (Novagen, Madison, WI; Covance, Berkeley, CA), p150Glued (UP235 generated in our lab), and Alexa-conjugated secondary antibodies (Molecular Probes). Micrographs were acquired with consistent exposure times between comparable images; only linear image enhancement algorithms were applied. Fluorescence intensity linescans were performed on randomly selected microtubules with both ends visible in the frame using ImageJ (Rasband, W.S., ImageJ, NIH, Bethesda, MD). Pixel values along the microtubule were measured and the ratio of measured intensity values to the mean intensity value was calculated. To analyze distributions, microtubules were divided into three equal segments (end 1, middle and end 2) and the area under the curve of the fluorescence intensity plot was calculated for each segment.
Binding Assays
For microtubule binding assays, purified recombinant CLIP-170-H2 or EB1 at 3 μM (expressed as monomer concentration throughout) was incubated with increasing concentrations of microtubules for 30 min at 37°C in PEM-50 buffer with 1 mM MgGTP and 20 μM taxol. After centrifugation (39,000xg), supernatant and pellet fractions were analyzed by SDS-PAGE and ImageJ. The data were analyzed as saturation binding with ligand depletion[18], based on a 1:1 binding stoichiometry of tubulin dimers to CLIP-170 monomer and fit to the one site binding equation [y = (Bmax*x)/(Kd + x)], where x is the free ligand concentration, y is the fraction bound, Bmax is the maximal binding, and Kd is the concentration for half-maximal binding by nonlinear regression using Prism software (GraphPad, San Diego, CA). For comparison, binding data were also analyzed based upon a 2:1 binding stoichiometry and fit to either a one or two site model. The two site equation [y = (Bmax1*x)/(Kd1 + x) + (Bmax2*x)/(Kd2 + x)] represents a minimal model that assumes the two sites are independent, whereas the tandem sites in CLIP-170 are linked and therefore the actual fit is likely to be more complex. All binding studies were performed in triplicate.
For tubulin binding assays, purified recombinant H2 and EB1 were covalently linked to CH-Sepharose 4B beads at an initial concentration of 2–5 mg protein/ml beads[17]. Ten μl aliquots of beads were incubated for 90 min at 4°C with increasing concentrations of tubulin in PEM buffer with 0.1mM MgGTP. Tubulin dimers were centrifuged at 350,000xg for 10 min at 4°C immediately prior to incubation to remove oligomers and aggregates. Bound versus free fractions were separated by either low speed centrifugation or filtration through a PALL NanoSep device (300K cut-off) and analyzed by SDS-PAGE and densitometry using ImageJ; after subtracting background binding to blocked control beads, the percent maximal binding was plotted as a function of tubulin concentration and fit to the one site binding equation [y = (Bmax*x)/(Kd + x)] by nonlinear regression using Prism software. All binding experiments were performed in triplicate.
Column Chromatography
Purified tubulin, BSA, or the H2 fragment of CLIP-170 was covalently linked to CH Sepharose 4B beads[17]. Recombinant EB1 and p150Glued (1–333) at 5 μM each were incubated either singly or together with tubulin-bound, CLIP-170-bound or BSA-bound beads, as noted, for 1 hr in 50 mM Tris, 50 mM KCL, with 0.1% TritonX-100. The beads were washed and eluted with 2 M NaCl.
3. Results
Plus-End Proteins do not Preferentially Bind to Microtubule Ends In Vitro
To understand the mechanisms of microtubule plus-end specificity, we sought to reconstitute plus-end binding in vitro with proteins that track the ends of growing microtubules in vivo. We added full-length recombinant EB1 or a well-characterized recombinant fragment of CLIP-170 (H2)[16] that includes the N-terminal tandem CAP-Gly microtubule-binding domains (Fig. 1A) to microtubules polymerized in vitro. To test whether these proteins show a higher affinity for the newly polymerized end of the microtubule than for the microtubule lattice, we polymerized microtubules from purified tubulin and taxol-stabilized seeds in taxol-free buffer, added the recombinant proteins either prior to or immediately following a burst of polymerization; samples were then fixed after a two minute incubation. Both CLIP-170 and EB1 bound along the length of the microtubule with no enhancement at the ends, as shown by representative micrographs and fluorescent linescans (Fig. 1B,C).
Figure 1. Neither CLIP-170 nor EB1 preferentially localizes to newly polymerized microtubule plus ends in vitro.
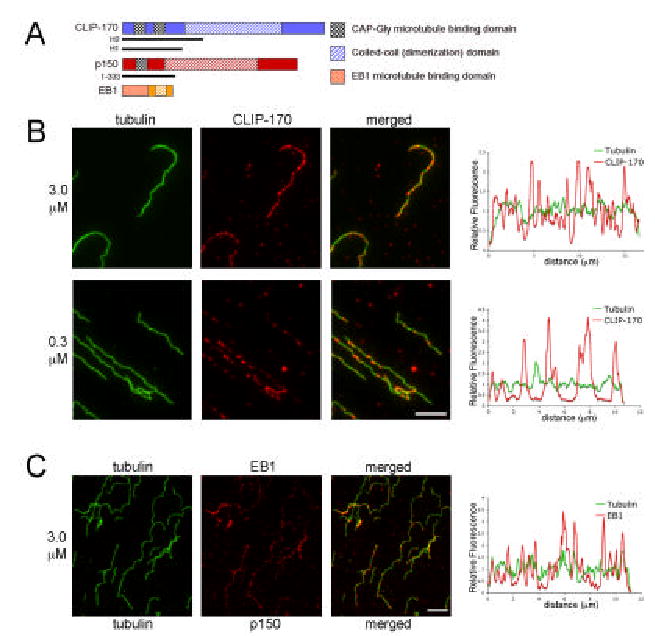
A. Schematic representations of the proteins CLIP-170, p150Glued and EB1, which show plus end specificity in vivo, and the constructs used in this study. The CLIP-170-H2 fragment used here contains the two N-terminal CAP-Gly microtubule-binding domains and part of the coiled-coil domain and the p150 (1–333) fragment includes the N-terminal CAP-Gly microtubule-binding domain; full-length EB1 was used. B. CLIP-170 (red) added to microtubules polymerized in vitro is not concentrated at microtubule ends, but rather labels the entire microtubule length (green). No end specificity was observed over a range of CLIP-170 concentrations including 3.0 μM (upper row) and (0.3 μM lower row). End to end fluorescence intensity linescans were performed on isolated microtubules to show the distribution of CLIP-170 labeling relative to tubulin labeling; no specificity for microtubule ends was observed. Scale=5μm. C. Recombinant EB1 (red) also did not localize specifically to newly polymerized microtubule ends (green) in either micrographs or linescans. Scale=5μm.
Preferential plus-end localization was not dependent upon the concentration of the plus-end protein. Experiments were performed with 0.3, 3 and 9 μM recombinant proteins; at lower concentrations, less CLIP-170 or EB1 bound to the microtubules, but the bound protein was distributed along the length of the microtubule (Fig. 1B and data not shown).
CLIP-170 Binds to Tubulin Dimers and Co-Polymerizes In Vitro
While neither CLIP-170 nor EB1 was specific for newly formed microtubule ends, chemical crosslinking experiments have shown that a CLIP-170 N-terminal fragment binds to tubulin dimers, and therefore it has been proposed that CLIP-170 may co-assemble with tubulin at the plus-end [15]. We compared the relative affinity of CLIP-170 and EB1 for microtubules and tubulin. Using microtubule co-sedimentation assays, we measured the affinity of the H2 fragment of CLIP-170 for microtubules, and estimated a Kd = 1.3 ± 0.3 μM (Fig. 2A) based upon a 1:1 stoichiometry of tubulin dimer to CLIP-170 H2 polypeptide. Alternatively, Scheel et al. [16] have suggested that there is a 2:1 binding stoichiometry of tubulin dimers to CLIP-170-H2 polypeptide due to the presence of two tandem CAP-Gly domains. Fitting our data to a simple two site model, as shown in the inset to Fig. 2A, yields approximate Kds of 0.02 and 7.5 μM.
Figure 2. CLIP-170 binds to and co-polymerizes with tubulin, resulting in localization to growing microtubule plus ends.
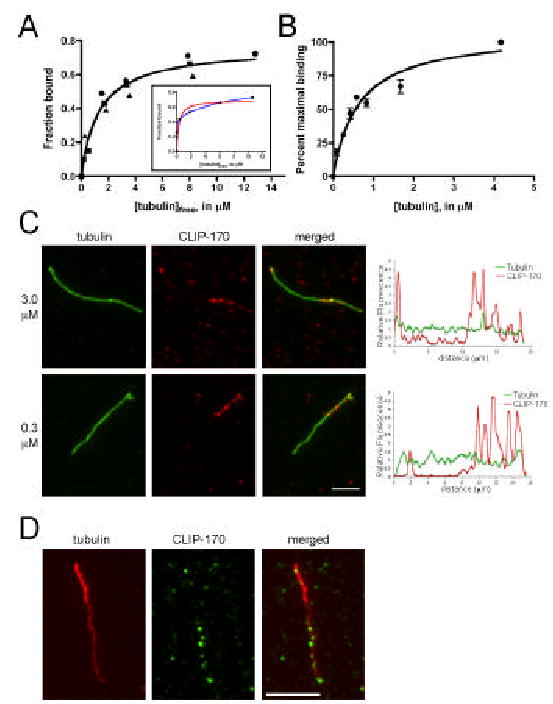
A. CLIP-170 bound to microtubules with a Kd of 1.3 ± 0.3 μM, assuming a 1:1 binding stoichiometry and a fit to a one site binding equation (see Methods); data from three independent experiments are shown (circles, squares, triangles). The inset shows the average data plotted assuming a binding stoichiometry of 2 tubulin dimers to each CLIP-170 polypeptide, with fits to either a one site (red line) or two site (blue line) binding model. B. GTP-tubulin bound to CLIP-170-beads with a Kd = 0.6 ± 0.1 μM. Error bars represent the standard error in the averages from three independent experiments. C. CLIP-170-H2 preincubated with tubulin dimers was added to microtubules. After a pulse of polymerization, CLIP-170 labeling (red) is concentrated at the end of the microtubule (green). Scale = 5μm. D. CLIP-170 (green) (3 μM) preincubated with tubulin dimers (15 μM ) labeled with a low concentration of rhodamine tubulin (1:50), was added to microtubule seeds (polymerized with 1:20 rhodamine tubulin for bright seeds) and allowed to polymerize, resulting in brighter rhodamine-tubulin labeling at the slow growing minus end in contrast to the brighter CLIP-170 labeling at the more rapidly growing microtubule plus end. Scale = 5μm.
By incubating increasing concentrations of GTP-tubulin with a constant level of the H2 fragment of CLIP-170 covalently bound to Sepharose beads, we measured an approximate Kd of 0.6 ± 0.1 μM for the binding of CLIP-170 to unpolymerized tubulin, assuming a 1:1 stoichiometry of tubulin dimer to CLIP-170 (Fig. 2B). In contrast, the data for CLIP-170-tubulin binding cannot be fit to a two-site binding curve, suggesting that CLIP-170 binds to unpolymerized tubulin dimers with a 1:1 stoichiometry. Under our experimental conditions, EB1 bound weakly to both microtubules and tubulin dimers (see Fig. 3B below, and data not shown).
Figure 3. CLIP-170 binds to EB1, and recruits EB1 to growing microtubule plus ends.

A. Recombinant EB1 bound to a CLIP-170-H2 affinity matrix, but not to a control BSA matrix (L=load, F=flow-through, W=wash, E=eluate). B. CLIP-170 (middle panel) but not EB1 (left panel) bound to a tubulin-affinity matrix. Preincubation of EB1 with CLIP-170 resulted in the binding of the co-complex to the tubulin column (right panel; L=load, F=flow-through, W=wash, E=eluate). C. CLIP-170 (3 μM) and EB1 (3 μM) preincubated with tubulin dimers (15 μM) are recruited to growing microtubule ends, as shown by both micrographs (CLIP-170, green; EB1, red; tubulin, blue; scale = 5μm) and linescans.
As CLIP-170 binds directly to tubulin, co-polymerization may explain the robust plus-end specificity observed in vivo[15]. To test this, CLIP-170-H2 (3.0 μM) was pre-incubated with GTP-tubulin (15 μM) and then mixed with microtubule seeds and shifted to 37°C to allow a brief pulse of new polymerization. Under these conditions, CLIP-170-H2 is recruited to new polymer at the ends of microtubules (Fig. 2C); 66% of the microtubules scored (n=66) had concentrated CLIP-170-H2 labeling at one end. In contrast, only 6% of the microtubules scored (n=35) showed a concentration of CLIP-170-H2 labeling at one end if CLIP-170-H2 was not first pre-incubated with tubulin (as in Fig. 1B). We confirmed that the more heavily labeled end was the plus-end using polarity marked microtubules (Fig. 2D). Together these data suggest that CLIP-170 can bind to tubulin dimers and co-assemble with them into microtubules, resulting in the concentration of CLIP-170 at plus-ends, which are the site of most new microtubule polymerization. In contrast, preincubation of EB1 with tubulin did not result in the preferential localization of EB1 to the growing microtubule plus end (data not shown).
CLIP-170 Binds Directly to EB1 and Recruits EB1 to Microtubule Plus-Ends
As CLIP-170 and EB1 co-localize to dynamic microtubule plus ends in the cell, we tested whether these two proteins may interact directly, similar to our previous observation that EB1 binds to the related N-terminal CAP-Gly domain of p150Glued [17]. We found that recombinant EB1 was specifically retained on a CLIP-170-H2 column but not on a BSA control column (Fig. 3A). In the reciprocal experiment, CLIP-170 bound specifically to an EB1-affinity column (data not shown).
We then tested if CLIP-170 could recruit EB1 to tubulin. By itself, EB1 does not bind strongly to tubulin dimers bound to an affinity matrix (Fig. 3B, left panel), but when EB1 was applied to the tubulin column following preincubation with CLIP-170 both proteins bound specifically (Fig. 3B, right panel), suggesting that CLIP-170 can recruit EB1 to tubulin dimers.
We then tested whether CLIP-170 could recruit EB1 to microtubule plus-ends in vitro. EB1 and CLIP-170-H2 (each at 3μM) were pre-incubated together with GTP-tubulin (15 μM), added to microtubules and allowed to polymerize briefly. Under these conditions, both EB1 and CLIP-170-H2 show localization to the ends of microtubules (Fig. 3C). EB1 labeling was concentrated at one end of the microtubule in 55% of the microtubules scored (n=36) when EB1 and CLIP-170 were pre-incubated with tubulin. In contrast, EB1 labeling was concentrated at one end in only 4.5% of microtubules scored (n=44) when EB1 and CLIP-170 were added without pre-incubation. Together these data suggest that CLIP-170 can bind to EB1 and recruit it to a complex with tubulin dimers and this complex can copolymerize with growing microtubules.
CLIP-170 Competes with p150Glued for Binding to EB1
As both CLIP-170 and p150Glued bind to EB1 via their N-terminal CAP-Gly microtubule-binding domains, we tested whether these proteins compete for binding to EB1. EB1 is specifically retained on a CLIP-170-H2 column, but when EB1 and p150Glued are preincubated, the binding of EB1 to CLIP-170 is reduced, suggesting that p150Glued is competing for EB1 binding with CLIP-170 (Fig. 4A). This inhibition is not the result of p150Glued itself binding to CLIP-170, as this construct was not retained on the H2 column. Together these data suggest that EB1 can bind to either p150Glued or CLIP-170, resulting in distinct plus-end complexes, which may have differential functions.
Figure 4. Plus end proteins may form distinct complexes at the ends of growing microtubules.
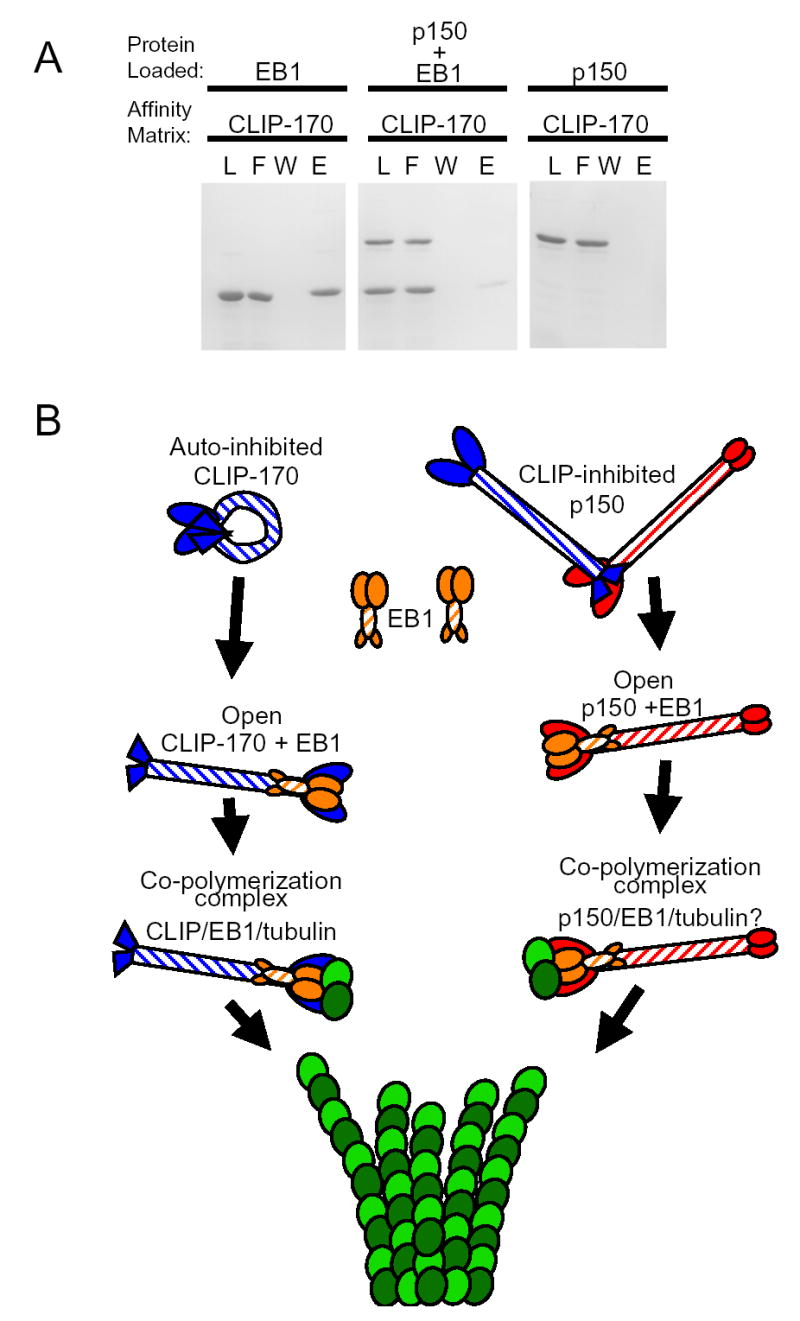
A. EB1 binds strongly to a CLIP-170-H2 affinity matrix (left panel), but preincubation of EB1 with p150Glued significantly reduced the observed binding (middle panel). p150Glued [1–333] does not bind to the CLIP-170 affinity matrix (right panel). The protein loaded and affinity matrix are as noted; L=load, F=flow-through, W=wash, E=eluate. B. Schematic illustrating a model for the interactions among microtubule plus end proteins. The C-terminal domain of CLIP-170 can bind to the N-terminal portion of the protein and hold it in an auto-inhibited confirmation. The C-terminus of CLIP-170 can also bind to the N-terminal domain of p150Glued and may inhibit its binding to microtubules as well. EB1 can bind to the N-terminal domains of both CLIP-170 and p150Glued and may displace the C-terminal inhibitory domain of CLIP-170, thereby opening up the molecules. The open CLIP-170 or p150Glued in complex with EB1 can then bind to tubulin and co-polymerize into microtubules.
4. Discussion
In the cell, plus-end proteins dynamically localize to the growing ends of microtubules, but the mechanisms driving this localization are not well understood. Here we have reconstituted microtubule plus-end localization of CLIP-170 and EB1 in vitro. CLIP-170 binds to tubulin and co-polymerizes into growing microtubules, as previously proposed [15], resulting in preferential localization to the faster-growing plus end. In contrast, we have shown that in vitro, EB1 alone does not preferentially localize to microtubule plus ends. This observation is somewhat surprising, given the robust localization of EB1 to growing microtubule plus ends in vivo[5], but is consistent with our observations of relatively weak binding of EB1 to either tubulin or microtubules. EB1 has been shown to interact with multiple proteins in the cell, and the formation of a co-complex may be required to accurately target the protein in vivo.
Consistent with this hypothesis, we have shown here that EB1 binds directly to the CAP-Gly domain of CLIP-170, in parallel to the binding of EB1 to the CAP-Gly domain of p150Glued shown previously[17,19]. The EB1-CLIP-170 co-complex binds to tubulin dimers, and can be effectively recruited to the ends of growing microtubules in vitro.
A co-complex of EB1 and p150Glued may also specifically target microtubule plus ends in the cell; we have found in HeLa cells that the depletion of p150Glued by RNAi leads to a mislocalization of EB1 so that it is no longer as specific for microtubule plus ends (Levy et al., manuscript submitted). Overexpression of EB1 also leads to mislocalization along the length of the microtubule [17]. Finally, we have shown that CLIP-170 and p150Glued compete for binding to EB1, suggesting that there may be distinct plus-end complexes that differ functionally, thus providing mechanisms to specifically and sensitively regulate microtubule dynamics in the cell.
The binding of EB1 to the N-terminal domain of CLIP-170 may provide a mechanism for the co-regulation of the activity of plus-end proteins. It has recently been demonstrated that CLIP-170 folds to form an auto-inhibited conformation in which the C-terminal binds to and blocks the activity of the N-terminal portion of the protein [20]. These data suggest a model, illustrated by the schematic in Fig. 4B, in which EB1 binds to the N-terminal domains of CLIP-170, displacing the C-terminal inhibitory domain of CLIP-170 and opening up the molecule. The open CLIP-170 in complex with EB1 can then bind to tubulin and co-polymerize into microtubules. Structural studies have also suggested that the binding of EB1 to the CAP-Gly domain of p150Glued may activate this complex for microtubule binding [21].
Together these data suggest that plus-end proteins may utilize multiple mechanisms to localize to microtubule tips. There may also be redundant mechanisms in vivo, as EB1 may be localized via both CLIP-170-dependent and independent mechanisms[22]. One of the clearest functions of plus-end proteins is the regulation of microtubule dynamics. Both EB1 and CLIP-170 have been shown to affect the parameters of dynamic instability in cells and cell extracts. CLIP-170 promotes rescue events (change from shortening to growing states) and EB1 promotes rescues and decreases catastrophes (change from growing to shortening states)[12,22]. If EB1 and CLIP-170 co-polymerize with tubulin, then they are perfectly poised to affect the transitions between growth and shortening states. Further, both the cooperative and competitive interactions among these proteins that we have observed in vitro may function in vivo to provide microtubules with differential dynamics and/or targeting signals localized specifically to their plus ends.
References
- 1.Vaughan KT. Microtubule plus ends, motors, and traffic of Golgi membranes. Biochim Biophys Acta. 2005;1744:316–24. doi: 10.1016/j.bbamcr.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- 3.Su LK, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler KW. APC binds to the novel protein EB1. Cancer Res. 1995;55:2972–7. [PubMed] [Google Scholar]
- 4.Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–27. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- 5.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–8. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 6.Pierre P, Pepperkok R, Kreis TE. Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci. 1994;107 ( Pt 7):1909–20. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112 (Pt 10):1437–47. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- 8.Su LK, Qi Y. Characterization of human MAPRE genes and their proteins. Genomics. 2001;71:142–9. doi: 10.1006/geno.2000.6428. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–4. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Finley J, Liu ZJ, Qiu SH, Chen H, Luan CH, Carson M, Tsao J, Johnson D, et al. Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. J Biol Chem. 2002;277:48596–601. doi: 10.1074/jbc.M208512200. [DOI] [PubMed] [Google Scholar]
- 11.Maekawa H, Schiebel E. CLIP-170 family members: a motor-driven ride to microtubule plus ends. Dev Cell. 2004;6:746–8. doi: 10.1016/j.devcel.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–26. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–91. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller-Reichert T, Chrétien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (alpha,beta)methylenediphosphonate. Proc Natl Acad Sci U S A. 1998;95:3661–6. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamantopoulos GS, Perez F, Goodson HV, Batelier G, Melki R, Kreis TE, Rickard JE. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J Cell Biol. 1999;144:99–112. doi: 10.1083/jcb.144.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheel J, Pierre P, Rickard JE, Diamantopoulos GS, Valetti C, van der Goot FG, Häner M, Aebi U, Kreis TE. Purification and analysis of authentic CLIP-170 and recombinant fragments. J Biol Chem. 1999;274:25883–91. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- 17.Ligon LA, Shelly SS, Tokito M, Holzbaur EL. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell. 2003;14:1405–17. doi: 10.1091/mbc.E02-03-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol Pharmacol. 1995;47:1197–203. [PubMed] [Google Scholar]
- 19.Askham JM, Vaughan KT, Goodson HV, Morrison EE. Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol Biol Cell. 2002;13:3627–45. doi: 10.1091/mbc.E02-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lansbergen G, Komarova Y, Modesti M, Wyman C, Hoogenraad CC, Goodson HV, Lemaitre RP, Drechsel DN, Van Munster E, et al. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol. 2004;166:1003–14. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–60. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 22.Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol. 2002;159:589–99. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]


