Abstract
We describe the results of a study by electron microscopy and image processing of Gag protein shells—immature capsids—of Mason-Pfizer monkey virus assembled in Escherichia coli from two truncated forms of the Gag precursor: Δp4Gag, in which the C-terminal p4Gag was deleted, and Pro(−)CA.NC, in which the N-terminal peptides and proline 1 of the CA domain were deleted. Negative staining of capsids revealed small patches of holes forming a trigonal or hexagonal pattern most clearly visible on occasional tubular forms. The center-to-center spacing of holes in the network was 7.1 nm in Δp4Gag capsids and 7.4 nm in Pro(−)CA.NC capsids. Image processing of Δp4Gag tubes revealed a hexagonal network of holes formed by six subunits with a single subunit shared between rings. This organization suggests that the six subunits are contributed by three trimers of the truncated Gag precursor. Similar molecular organization was observed in negatively stained Pro(−)CA.NC capsids. Shadowed replicas of freeze-etched capsids produced by either construct confirmed the presence of a hexagonal network of holes with a similar center-to-center spacing. We conclude that the basic building block of the cage-like network is a trimer of the Δp4Gag or Pro(−)CA.NC domains. In addition, our results point to a key role of structurally constrained CA domain in the trimeric interaction of the Gag polyprotein.
Over the past few years there has been an explosion in structural information on retroviral proteins, specifically those of human immunodeficiency virus (reviewed in reference 54). Less progress has been made in understanding the process of assembly of viral proteins into the immature virus particle. Uncertainty about the overall shape and molecular organization of the major structural proteins still remains and has become a matter of debate.
In retroviruses, the shape-determining Gag polyprotein consists of several domains. Those common to all members of the family are MA (membrane-associated globular protein), CA (the major protein, which in mature particles forms the core shell), and NC (RNA-binding protein). In most cases a small C-terminal peptide completes the sequence. Lentiviruses and members of the human T-lymphotropic virus-bovine leukemia virus group (Deltaretrovirus) possess a Gag precursor with the above domains, while in the remaining genera (Betaretrovirus, Gammaretrovirus, and Spumavirus), the Gag precursor is substantially larger owing to the presence of one or more polypeptides intercalated between MA and CA. In Mason-Pfizer monkey virus (M-PMV), these are the pp24 phosphoprotein and the internal scaffold protein p12 (49). The C-terminal p4 peptide is rich in proline (52), like p6 in human immunodeficiency virus (HIV), and like p6 it may facilitate the final stages of budding of M-PMV (S. S. Rhee et al., personal communication).
The Gag polyprotein itself is capable of forming a spherical submembrane shell (23), often referred to as an immature capsid. In HIV, the molecular organization of the submembrane Gag shell has been studied in virus-like particles (VLP) by negative-stain electron microscopy and image processing (41, 43). This work revealed a hexagonal network of small rings and a stretch of short rods at the periphery of delipidized VLP. Subsequently, cryoelectron microscopy of HIV VLP confirmed the presence of a fringe of rods beneath the viral membrane (19). Similar images of rods were obtained by processing cryoimages of Moloney murine leukemia virus (MuLV) (60).
The length of the Gag polyprotein rods was estimated to be over 200 Å in HIV and at least 220 Å in MuLV. This is in agreement with the higher molecular weight of MuLV Gag protein, which has an additional protein, p12Gag, located between MA and CA. In contrast to negatively stained delipidized capsids (41, 43), cryoelectron microscopy of enveloped immature particles did not reveal evidence of hexagonal symmetry, although some evidence of local near-hexagonal arrays was reported (19, 60).
Retroviruses assemble either in the cytoplasm (Betaretrovirus and Spumavirus) or at the plasma membrane concomitant with the budding process. The latter are called C-type viruses, as proposed by Bernhard (6). However, a closer look at the morphology of retroviruses shows two different images in both sectioned and negatively stained virus particles (42). In lentiviruses the submembrane protein layer (Gag shell) of immature virus particles appears as a single dense layer in positively stained sections. In other retroviruses, such as mouse mammary tumor virus and murine leukemia virus, a pale intermediate layer separates the dense inner layer from the viral membrane. This layer was identified by cryoelectron microscopy as the less electron dense C terminus of MA and p12 (60). Analysis of thin sections of MuLV showed that the pale and dense submembrane layers in immature MuLV represent domains of the Gag precursor that stained differently with uranyl acetate (44).
Several successful attempts at in vitro assembly of retroviral spherical or tubular particles have been reported (8, 9, 24, 25, 26, 34, 35, 49, 50). The work by Rumlova-Klikova et al. (48) analyzed the assembly of M-PMV capsids from a variety of truncated and mutated Gag polyproteins in E. coli. The most attractive capsids for electron microscopic analysis were those assembled in E. coli by Pr78Gag missing the C-terminal p4 protein (Δp4Gag) and a construct comprised of CA.NC in which the N-terminal proline of CA was deleted [Pro(−)CA.NC]. Here, we describe our study of purified capsids using a variety of electron microscopy techniques: thin sectioning, freeze-etching and negative staining, followed by image processing. The results showed a regular hexagonal network of rings formed by six subunits of the Gag protein, suggesting that a fullerene-like organization of the submembrane Gag protein shell first proposed for HIV (41, 43) and amended by Forster et al. (16) might be present in other retroviruses.
MATERIALS AND METHODS
Construction of a vector for bacterial expression of M-PMV Δp4Gag gene and Pro(−)CA.NC.
The pG10GAG plasmid described previously (34) was used for the generation of the Δp4Gag expression vector named Δp4GAG. It was created by a HindIII deletion of gag sequences downstream from NC in pGI0GAG. The Pro(−)CA.NC construct was prepared as described before (48).
Expression of M-PMV Δp4Gag gene and liberation of inclusion bodies.
The colonies of E. coli BL21(DE3) transformed with Δp4Gag were resuspended in Luria-Bertani (LB) medium in the presence of 100 μg of ampicillin/ml to an optical density at 590 nm (OD590) of 0.1 and grown at 37°C. The cells were induced with 0.4 mM IPTG (isopropylthiogalactopyranoside) at an OD590 corresponding to 0.6. The cells harvested 4 h after induction were lysed with lysozyme and washed with detergent-containing buffers as described previously (34).
Preparation of capsids for electron microscopy.
Purified material released from cells was prepared for thin sectioning by conventional embedding in Araldite as described previously (32). For study by negative staining, Δp4Gag capsids were mixed 1:1 with 0.2 M glycine buffer (pH 10) containing 2 M NaCl and 10 mM EDTA (GNE-2) and incubated at 4°C. The aim of this treatment was to dissociate aggregates of capsids present in the starting material. Samples for negative staining were removed after 3, 6, and 24 h as well as after 7 and 14 days. Carbon-coated grids with adsorbed capsids were negatively stained with 4% sodium silicotungstate, pH 6.9 or 8.2, or 3% methylamine tungstate, pH 6.8. Valentine's modification of negative staining was used in some cases (55).
Dissociated Δp4Gag capsids were also gold immunolabeled with rabbit antibody against p27 (CA) followed by goat anti-rabbit antibody tagged by 5-nm colloidal gold (BioCell Int., Cardiff, United Kingdom) using the protocol described previously (41). In a control experiment, the specific antibody was replaced by a preimmune rabbit serum. All measurements were done on negatives of thin sections or negatively stained images using the Global Lab Image program (Data Translation, Marlboro, Mass.) or ScionImage software (Scion Corporation, Frederick, Md.).
For freeze-etching, the capsids were adsorbed to a freshly cleaved piece of mica (8 by 4 mm), sandwiched at right angles with a copper plate of the same size, and plunged in liquid nitrogen. The frozen sandwich was inserted into a double replica device as described before (39). A Balzers BAF 300 freeze-etch unit was used for fracturing, etching, shadowing with Pt/C, and carboning. Replicas were cleaned on sodium hypochlorite for several hours. A Philips CM12 electron microscope operating at 80 kV was used throughout this study.
Image processing.
The best electron micrographs were selected in terms of the quality of their optical Fourier transform. Micrographs presenting sharp spots and a first zero in the contrast transfer function with a frequency higher than 1/20 Å−1 resolution were chosen and digitized using a Leafscan 45 charge-coupled device microdensitometer with a step size corresponding to 5 Å on the specimen. Image processing was performed using the MRC image analysis package (13). Images were corrected twice for lattice distortion by real-space correlation analysis according to the protocol described by Henderson et al. (30). Considering defocus values used to record images (100 to 150 nm), no contrast transfer function correction was needed.
Projection maps were calculated from amplitude and phase data with or without symmetry and truncated at 30 Å. Determination of the space group was performed by using the Allspace program (56). Symmetrized images were computed in the real space using the Spider software system (17). Quasi-optical filtration (53) was used to obtain an initial raw filtered image (see Fig. 2A). The Fourier transform and power spectrum of the image were calculated. The resulting diffraction pattern was indexed to compute a Fourier filter mask that was applied to the Fourier transform.
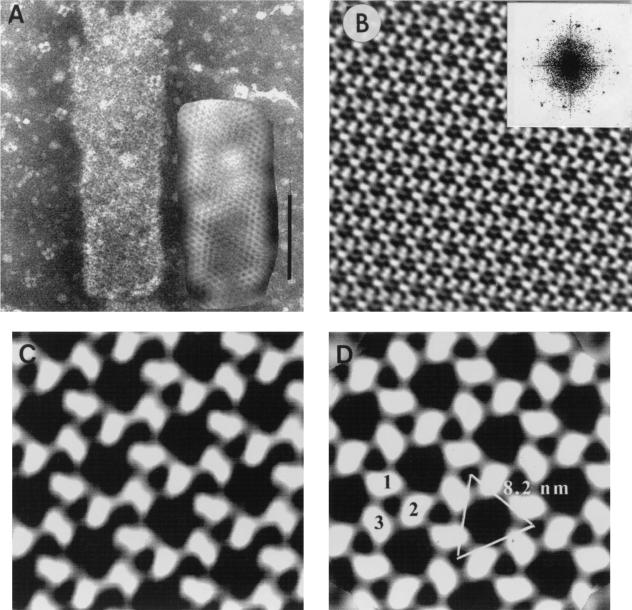
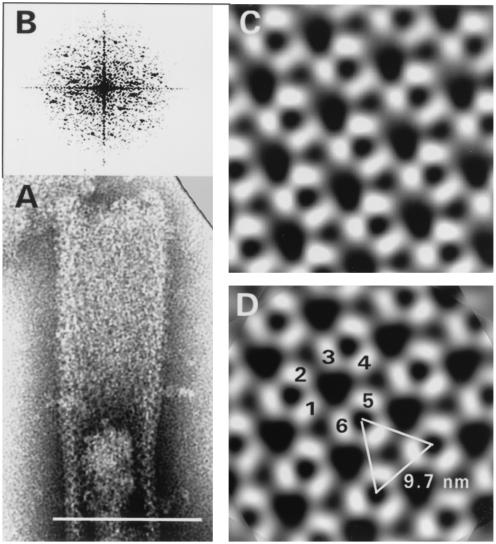
FIG. 2.
(A) Tubular form of Δp4Gag used for image processing. Quasi-optical filtration reveals hexagonal pattern of holes. Negative staining with sodium silicotungstate (pH 6.9). Bar, 100 nm. (B) Projection map of the crystal lattice shown in panel A after correction for distortion. Note well-visualized trimers in the upper left corner of the very regular hexagonal network resulting from processing. Inset: Power spectrum from tubular capsid shown in panel A. The first order is located at 8.2 nm, and the most distant spots are located at 2.8 nm. (C) Enlarged portion from panel B before symmetrization. (D) The same field as in panel C after threefold symmetrization. No correction for the contrast transfer function was carried out. Large holes are formed by three trimers (1, 2, and 3), each contributing two subunits.
RESULTS
Electron microscopy and image processing of Δp4Gag capsids.
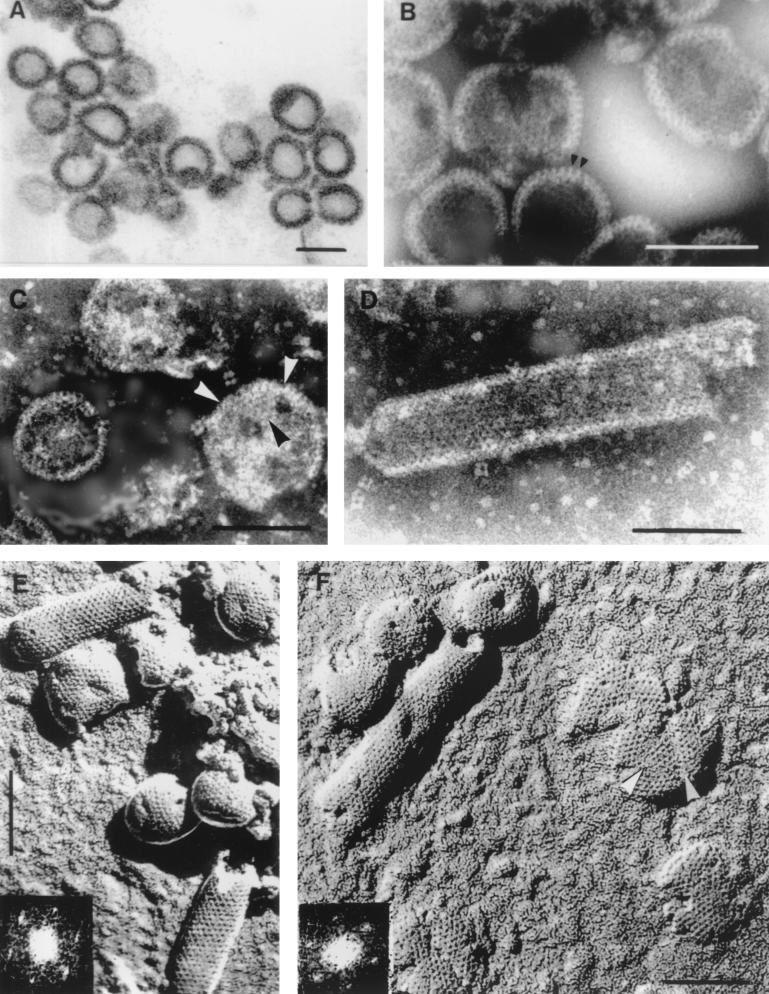
Thin sections of assembled structures from Δp4Gag-expressing cells revealed round or slightly distorted capsids (Fig. 1A) with an average diameter of 87 nm (standard deviation [SD] = 7, n = 96). For negative staining, aggregates or sheets of capsids were dissociated with GNE-2 buffer into individual round or tubular forms during the first 6 h, and this state did not change over a period of several weeks. A few capsids appeared damaged and showed small breaks or holes. Negative staining revealed peripheral projections about 7 nm apart (Fig. 1B) and small patches of regular arrays of holes 3 to 4 nm in diameter. The arrowheads in Fig. 1C point to an apparently triangular patch of holes similar to triangular patches observed on the inner surface of cytoplasmic membranes of Sf9 cells infected with recombinant baculovirus expressing HIV Gag protein. Triangular breaks were also described in HIV VLP (43), reminiscent of fine defects described in frozen-hydrated HIV VLP (19). A flat sheet with a triangular break is also shown in Fig. 1F. The Δp4Gag capsids were positively labeled with antibody to p27Gag (not shown).
FIG. 1.
(A). Thin section of Δp4Gag capsids released from E. coli cells. Magnification, ×105,000. (B) Δp4Gag capsids negatively stained with sodium silicotungstate (pH 8.2). Arrowheads point to peripheral rod-like projections. Magnification, ×225,000. (C) Round capsids released from aggregates by treatment with GNE-2 buffer. Note triangular shape in capsid labeled with arrowheads. Defects as seen on labeled capsid were not general. Negatively stained with sodium silicotungstate (pH 6.9). Magnification, ×200,000. (D) Tubular form of Δp4Gag capsid showing rows of fine black holes. Negatively stained with sodium silicotungstate (pH 6.9). Magnification, ×225,000. (E) Freeze-etched and shadowed Δp4Gag capsids. Note very regular pattern of holes on both spherical and tubular forms. Inset shows diffraction pattern of the upper tube. Shadows are dark. Magnification, ×175,000. (F) Tubular and spherical capsids of Δp4Gag prepared by freeze-etching and unidirectional shadowing. Large flat sheets (probably disrupted capsids) show the same surface pattern as the capsids. Note indication of triangular shape in the flat sheet (arrowheads). Magnification, ×175,000. Inset shows diffraction pattern of the tube. Bars, 100 nm.
A few tubular forms of the Δp4Gag capsids were also present, and negative staining of these aberrant structures revealed areas of regularly organized holes (Fig. 1D) with a center-to-center (CTC) spacing of 7.1 nm (SD = 0.9, n = 58). The ends of the tubular forms were occasionally hemispherical (Fig. 1E). Negatively stained spherical capsids measured approximately 86 nm in diameter, and tubular forms were approximately 80 nm in diameter.
Shadowed replicas of freeze-etched capsids (Fig. 1E and F) showed the three-dimensional nature of the capsids and a periodic organization of holes (appearing dark after photographic reversal) on the surface of spherical as well as tubular capsids. The CTC spacing of the holes obtained by direct measurement was 7.1 nm (SD = 0.7, n = 101). The cylindrical forms convincingly showed a hexagonal network which produced a typical diffraction pattern (Fig. 1E and F). The same periodicity and CTC spacing was revealed on flat sheets. In several instances, faults or breaks in the hexagonal arrays were observed (arrowheads in Fig. 1F).
The tubular structures were usually well preserved and provided a suitable area for image processing. Figure 2A shows one tubular form before and after quasi-optical filtration (see Materials and Methods). The filtered image revealed the presence of a hexagonal network on the whole surface of the tube. Figures 2B and C show images of the lattice after correction for distortion. The power spectrum (Fig. 2) clearly reveals the hexagonal nature of the lattice. Moreover, it confirms that negative staining produced a one-sided image of the tubular form. The image indicates a unit cell with a = 81.2 Å, b = 81.7 Å, and γ = 118.9°. Examination of the phase relationships by the program Allspace (56) showed that of 17 space groups tested, the lowest phase residuals were for the p3 and p6 plane groups, 16.3o and 15.7o, respectively. Thus, a threefold or sixfold symmetry was applied to the projection maps, but no visual difference was observed between the resulting images.
Figure 2D corresponds to the threefold symmetrized projection of Fig. 2C. The protein subunits appeared to form rings of six subunits contributed by three trimers, with one subunit common to neighboring rings. The center-to-center spacing of trimers in the processed tubular form was 8.2 nm, somewhat larger than the average spacing (7.1 nm) obtained by direct measurement of the ring distance of the spherical capsids. The central hole formed by the six subunits was approximately 4 nm in diameter.
Electron microscopy and image processing of Pro(−)CA.NC capsids.
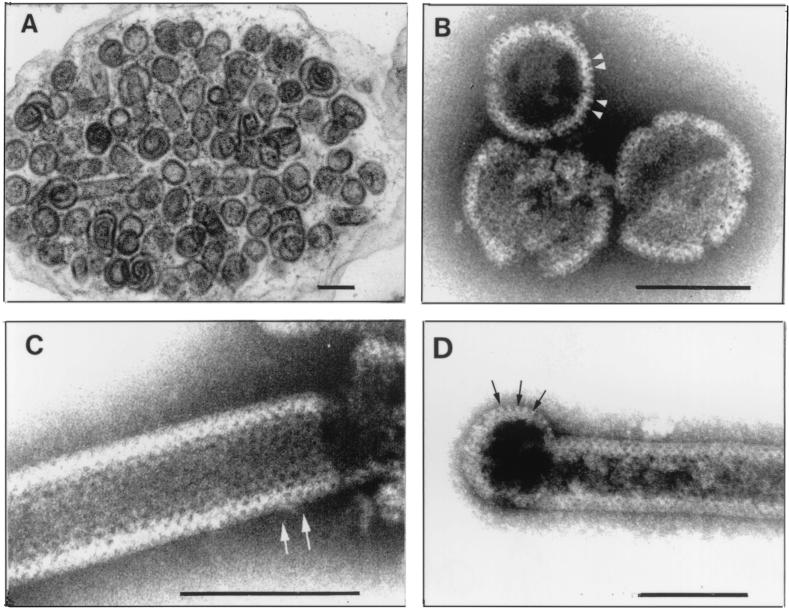
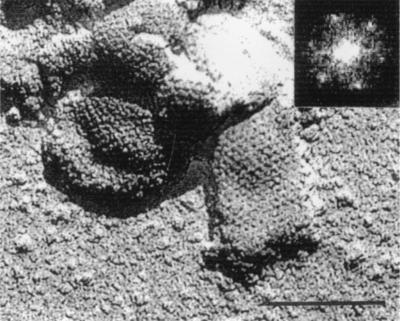
Thin sections of semipurified Pro(−)CA.NC capsids showed spherical shells about 80 to 100 nm in diameter (Fig. 3A). The shape and size of these capsids were not as uniform as those of Δp4Gag capsids. Occasional spiral shapes and tubular forms were also observed. Negative staining revealed mainly spherical shells (85 to 100 nm in diameter; Fig. 3B) but also a few tubular forms (60 to 80 nm in diameter; Fig. 3C), many with hemispherical ends (Fig. 3D). In both spherical and tubular capsids, regular arrays of holes were observed, and their average CTC spacing was 7.4 nm (SD = 0.7, n = 95). At the periphery of some capsids, regularly spaced rod-like projections were visible (Fig. 3B and D). These were approximately 11 nm long and 4 nm thick, with a CTC spacing of 6.6 nm (SD = 0.5, n = 43), leaving about 3 nm of space between rods. Surface replicas of fractured capsids revealed a hexagonal pattern of holes with CTC spacing of 7.3 nm (SD = 0.68, n = 97) obtained from direct measurement and from a few power spectra (Fig. 4). The unit cell of the periodic structure on the surface of the tubular form in this figure was a = 74.33 Å, b = 70.86 Å, and γ = 117°.
FIG. 3.
Capsids produced in E. coli by Pro(−)CA.NC construct. (A) Thin section of E. coli cell filled with spherical and a few tubular capsids. Magnification, ×66,000. (B) Spherical capsids negatively stained with sodium silicotungstate (pH 8.2) using Valentine's technique (55). Tiny black holes are visible. Arrowheads point to surface projections. (C) Cylindrical form displaying a regular pattern of holes (black dots). Arrows point to a hole with six neighbors. Magnification, ×360,000. (D) Example of tubular form with rounded end in which peripheral projections are visible (arrows). Sodium silicotungstate (pH 8.2). Magnification, ×240,000. Bars, 100 nm.
FIG.4.
Shadowed replica of freeze-etched Pro(−)CA.NC capsids. Shadows are dark. Magnification, ×200,000. Bar, 100 nm. Inset: Diffraction pattern of the tubular forms, tilted by +14o from the original image.
Several negatively stained tubular forms exhibited a hexagonal diffraction pattern (Fig. 5B). This power spectrum clearly shows that both surfaces contributed to the projected image. After indexing (unit cell parameters: a = 97.6 Å, b = 100.6 Å, and γ = 122.2°), one side was processed as described above to obtain a projection map. The best phase residuals obtained were 18.2° for p3 and 19.1° for p6 symmetries. Figure 5 shows a processed image before (C) and after (D) threefold symmetrization of the upper (flattened) portion of a tubular form. The CTC spacing of the holes (9.7 nm) in this case was substantially larger than the above-mentioned average spacing, obviously due to air-dry negative staining The large central hole appeared more triangular and measured approximately 4.5 nm across. The pattern was in principle the same as that seen in processed Δp4Gag tubes (Fig. 2).
FIG. 5.
Results of image processing of a tubular form of Pro(−)CA.NC capsids. (A) Negatively stained tubular form with wider (flattened) upper part used for processing. Magnification, ×300,000. Bar, 100 nm. (B) Corresponding power spectrum. Processed image before (C) and after (D) threefold symmetrization reveals how three trimers form a ring made of six subunits. Protein is white.
DISCUSSION
An assessment of our observations requires discussion of several aspects of Gag protein organization in M-PMV and retroviruses in general. A conspicuous feature of retrovirus immature capsids is the presence of a cage-like network, as opposed to the closely packed submembrane shells of some other enveloped viruses such as the alphaviruses. We will discuss the implications of this finding for the architecture and assembly of M-PMV into spherical or cylindrical forms and the interpretation of the images obtained by the methods used. We will also compare the molecular organization of M-PMV capsids with the immature capsids of HIV and assess the contribution of our results to the question of icosahedral symmetry in retroviruses.
Multimethod approach and interpretation of the results obtained.
The tremendous progress in cryoelectron microscopy in virology (3, 11) has tended to push conventional preparative techniques so far into the background that they are often considered obsolete and not trustworthy (21). Despite this, we provide strong evidence from this study, using negative staining and freeze fracture/etching methods, of a continuing role for such approaches in studies of the molecular organization of retroviral Gag shells. Cryoelectron microscopy has revealed a hexagonal arrangement of HIV CA proteins in tubular or cone-like structures that have been assembled in vitro (24, 35) and has provided novel information on the organization of the submembrane Gag layer in immature forms of HIV and MuLV (19, 60). However, cryoelectron microscopy has failed to establish the structural organization of immature retroviral capsids or visualize the cage-like network described previously in HIV (41, 43) and here for M-PMV, in which we have employed the more conventional techniques of negative staining and freeze-etching.
While surface replicas of frozen-etched and shadowed material present only the surface features of an object, negative staining can also visualize some of the internal structures and at higher resolution than shadowing. The outcome of negative staining depends on the hydrophobic or hydrophilic properties of both the relevant protein and the supporting film (29, 38). In most cases, however, negative staining reveals those parts of the objects associated with the supporting film, and the height of the dried stain was estimated to be 10 to 20 Å (40), although this figure might be higher with larger and densely packed structures such as viruses. Air drying-caused flattening of larger structures has to be taken into account when size measurements are carried out.
In this study we have chosen to use sodium silicotungstate or methylamine tungstate at medium or high pH instead of very low pH uranyl acetate, which may obscure the holes due to shrinkage of the network and low penetration of the stain into intact capsids. A shrinkage effect of uranyl acetate has recently been reported (36). Thus, the data we have obtained from Δp4Gag and Pro(−)CA.NC capsids using negative staining and freeze-etching justify the application of conventional preparative techniques when cryoelectron microscopy meets serious obstacles.
Molecular organization of Δp4Gag and Pro(−)CA.NC capsids.
Previous work on HIV showed the presence of threefold symmetry in plasma membrane-associated Gag protein assemblies and in purified HIV Gag VLP (41, 43). In the present study, a similar fullerene-like organization of the M-PMV Gag polyprotein was observed in capsids produced in E. coli by Δp4Gag and by Pro(−)CA.NC constructs. In both cases, image processing revealed hexagonal rings that shared a single subunit. This pattern suggests that the six subunits may be contributed by three trimers, such as in HIV (Fig. 6B). This organization is supported by CTC spacing which is similar in both capsids as well as in the diagram and correlates well with the results of molecular modeling of the HIV MA trimeric network (16). It is also very close to the spacing observed in HIV VLP, in which an average from electron microscopy and X-ray crystallography of the HIV MA protein was 6.8 nm (43).
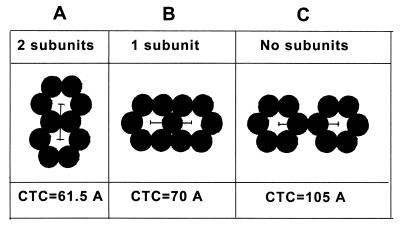
FIG. 6.
Diagram of rings with two (A), one (B), or no (C) subunits shared and the corresponding CTC spacing based on 35 Å diameter of the subunits. The results of image processing are in agreement with version B.
The other option when two subunits are shared (as found in fullerenes) (Fig. 6A) provides a substantially lower spacing than found in M-PMV capsids (∼7 nm), while in version C organization, the spacing is larger. This organization is only hexagonal, with no threefold symmetry. The presence of tubular forms in M-PMV capsids helped to increase the resolution of image processing and provide a clear picture of the fullerene-like network. In spite of the differences in size of M-PMV Gag domains, the molecular organization of the M-PMV Gag shell, including the spacing, is very similar to that in the HIV Gag shell. It appears that the fullerene-like pattern results from trimeric interaction of Gag trimers. This suggests that the basic building block is a trimer (not a hexamer) of the relevant protein [Δp4Gag and Pro(−)CA.NC].
The trimeric nature of HIV MA has been shown by X-ray crystallography (31) and received support from molecular modeling (16). Closer understanding of protein-protein interactions in M-PMV capsids is hampered by the absence of data from X-ray crystallography, though the solution structure of MA has been found compatible with the formation of trimers (12). Our observations certainly provide support for a trimeric interaction of certain Gag domains. The finding of basically the same pattern in Pro(−)CA.NC capsids as in Δp4Gag suggests that the CA.NC complex is critical for trimerization and assembly of the whole Gag polyprotein (see references 18 and 21 and references cited therein).
Trimer association versus dimer association of CA domains.
The pattern of the network produced by image processing indicates that the basic building block is a trimer of the whole Gag and more surprisingly also of the CA.NC domains. This finding is only partly supported by cross-linking experiments of retroviral Gag polyprotein or the CA domain. For example, Scarlata et al. (51) found that cross-linking HIV Gag protein with glutaraldehyde produced mainly dimers, though trimers, tetramers, and multimers were also observed using scanning transmission electron microscopy mass determination. Purified MA was found in a trimeric form only and CA was mainly dimers, tetramers, or multimers. Changes in fluorescence anisotropy revealed that Gag subunits associated in clusters of three on the surface of lipid membranes.
Cross-linking experiments on HIV and M-PMV CA and CA.NC with other reagents showed a prevalence of dimers, though small amounts of trimers and tetramers were also present (15, 28; M. Rumlova, unpublished results). The presence of a small amount of trimers reported by the above authors indicates that Gag domains can form trimers. Support for trimers also comes from a study by Morikawa et al. (37), who found that MA and MA.CA of HIV behaved as trimers on a glycerol gradient, while CA was a monomer under the same conditions. X-ray crystallography revealed that HIV (and simian immunodeficiency virus) MA formed trimers (31, 47), while HIV CA crystallized as dimers (7, 20, 59).
The threefold symmetry in CA.NC assemblies, though unexpected because of the presence of a dimerization subdomain in HIV CA found by X-ray crystallography (20), is in line with multiple interactions common to viruses with quasi-equivalent capsids in which identical protein subunits are found in different structural environments (33); for example, when icosahedral capsids are formed by a single protein which forms both hexagonal and pentagonal assemblies or when some viruses form capsids with T3 or T4 triangulation numbers, such as hepatitis B virus (14) and the yeast retrotransposon Ty-1 polypeptide A (1, 45). One study (1), in fact, is in agreement with our report on the presence of holes and trimers in HIV VLP (43). Rounded-up ends of tubular M-PMV capsids (Fig. 1E and 3D) demonstrate the ability of the proteins to form a hemispherical and possibly also a spherical structure with icosahedral symmetry.
Comparison of capsids assembled in E. coli with in vitro assemblies of other retroviral proteins.
Li et al. (35) studied in vitro-assembled cylindrical and conical forms of HIV CA using cryoelectron microscopy followed by image processing. This approach revealed a different organization from that described in this study. The N-terminal regions of CA formed hexagonally organized rings with a small central hole (about 25 Å as opposed to 40 Å holes in the M-PMV capsids). In the CA tubes described by Li et al. (35), the hexagonal rings do not share any subunits; individual rings are connected with neighboring rings via the C-terminal dimerization domains of each CA molecule (see Fig. 6C for a simplified sketch of this organization). In this case, the CTC spacing of the central holes was about 10.7 nm, substantially more than the ∼7-nm spacing in M-PMV capsids.
It is obvious that this organization is different from the organization in Δp4Gag and Pro(−)CA.NC capsids, and it is possible that the reason for this difference lies in the structure of the CA domains that were assembled in each case. Li et al. used the mature form of CA, in which the N terminus is in the form of a β-hairpin due to interaction of the N-terminal proline residue with an aspartate at position 51, while in the M-PMV Pro(−)CA.NC capsids, the N-terminal proline was deleted (48). The six coordinated rings found in HIV CA by Li et al. may represent a situation similar to that in virus cores, while the M-PMV Δp4Gag and Pro(−)CA.NC constructs assemble into spherical structures and are thus more likely to reflect the interactions that exist in the immature capsid. Interestingly, in a cryoelectron microscopic analysis of MuLV immature and mature viruses, Yeager et al. (60) found evidence for a hexagonal paracrystalline lattice with a unit cell spacing of 80 Å for the immature particles and 115 Å for the cores in mature particles, values that are similar to the ∼70 Å center-to-center spacing we describe here for p4Gag immature shells and the 107 Å spacing described by Li et al. (37) for mature CA tubes.
It might be argued that the subunits which make up the hexagonal organization observed in our analysis of M-PMV capsids each represent a parallel dimer of Δp4Gag or Pro(−)CA.NC subunits and that the hexamers might therefore represent a condensed form of the arrangement observed by Li et al. (35). This arrangement would be incompatible with MA trimers in the case of Δp4Gag and would be at variance with the observed pattern (rings sharing one subunit). In addition, the number of Gag monomers involved in each assembled capsid would double. Recent mass calculations from scanning transmission electron microscopic analyses of M-PMV immature capsids have shown that the median number of Gag precursors or immature M-PMV capsid is approximately 2,000 (46). If one calculates the number of Gag molecules that would be required to occupy the surface area of a spherical immature capsid 85 nm in diameter, based on the CTC spacing (∼7 nm) of the trimers observed by negative stain and freeze-etching approaches described here, the number would be approximately 1,750. This figure is close to the number of Gag molecules in Rous sarcoma virus (57). In contrast, if each component of the hexamer were a dimer of Gag, the number of Gag precursors per immature capsid would total 3,500.
Barklis et al. (4, 5) studied assembly of MuLV His.CA and HIV His.CA domains at lipid membranes, mimicking the Gag CA-membrane interactions. Their experiments showed a cage-like pattern with hexagonal as well as trigonal holes in the protein two-dimensional crystals. The rings were formed by six CA subunits, with two subunits shared between rings (Fig. 6A). Interestingly a similar pattern (rings with two subunits shared) was recorded in tubular structures assembled in vitro from MuLV His.CA.NC plus RNA and visualized by negative staining (62). This is at variance with our results, which showed that M-PMV CA.NC formed spherical shells in E. coli when Pro1 was deleted or when a few residues were attached to the CA N terminus (48).
Implications of cage-like network for virus assembly (incorporation of viral glycoproteins and cellular proteins).
The finding of a cage-like hexagonal network in M-PMV and HIV logically argues against a dense packing of the Gag polyprotein in the immature capsid. Support for this organization comes also from the calculation of the number of Gag monomers (∼1,750) that would complete a spherical capsid 85 nm in diameter, as described above. This figure is also similar to the number of MA copies (1,680) in a mature form of HIV Gag shell (84.2 nm in diameter) deduced from molecular modeling of an MA trimeric network (16).
The presence of hexagonal rings formed by three HIV MA trimers correlates well with a specific interaction of gp41 trimers with the underlying MA domain (54). Several papers have reported deletions or point mutations in the hole-facing area of MA, which prevented incorporation of viral glycoproteins into virus particles (see references 5, 10, and 18 and references cited therein). This Env-accommodating function of the cage-like network was also postulated for assemblies of MuLV CA and CA.NC proteins by Barklis et al. (4, 5) and Zuber and Barklis (61).
In M-PMV the cytoplasmic tail of gp20 is 38 amino acids long, and recent mutational analyses of the M-PMV MA domain point to a similar Env incorporation function for this region of the MA protein (D. Song and E. Hunter, unpublished data). It is difficult to reconcile the closely packed Gag shell with the need to provide space for Env proteins and cellular proteins, such as members of the major histocompatibility complex (2) and other cellular proteins (27). Wilk et al. (58) attributed an increase in electron density in membranes of frozen-hydrated wild-type HIV to the presence of Env glycoproteins. This, however, is inconsistent with the observation that Gp120/41 projections are 21 nm apart (22), a distance sufficient for the incorporation of cellular proteins (16).
Conclusion.
We provide strong evidence in this study that Gag protein molecules are not closely packed in M-PMV capsids, but form a cage-like network similar to the fullerene-like model in HIV (41, 43). In this model, three Gag trimers form hexagonal rings that share a single subunit. Our model is in good agreement with data from X-ray crystallography of HIV MA (31, 47) and M-PMV nuclear magnetic resonance structure (12). Molecular modeling (16) showed the plausibility that the HIV MA trimers may form the triangular facets of an icosahedron.
Our previous work (48) argues strongly that it is the structure of the CA domain, constrained from forming its mature form by the N-terminal domains of the Gag precursor, which allows Gag to associate in these trimeric interactions and form the immature capsid. Nevertheless, it is likely that other domains of Gag, such as MA, NC, and spacer peptides, contribute to the efficient and stable formation of the Gag trimeric complex. The similarity of the molecular organization of the Gag protein shell in HIV and M-PMV suggests that the fullerene-like model might be common to all retroviruses.
Acknowledgments
We thank Eric Barklis and Wes Sundquist for communicating recent work to us prior to publication and Mark Forster and Barbara Mulloy for comments on the manuscript. We also acknowledge skillful technical assistance by Morag Jackson.
This work was supported by grants 203/00/1005 and 203/98/P151 from the Czech Republic Grant Agency, by Fogarty International Award TW00050, and partly by the Medical Research Council in London and the Wellcome Trust International Grant.
REFERENCES
- 1.Al-Khayat, H. A., D. Bhella, J. M. Kenney, J.-F. Roth, A. J. Kingsman, E. Martin-Rendon, and H. R. Saibil. 1999. Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J. Mol. Biol. 292:65-73. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, L. O., J. W. Bess, R. C. Sowder II, R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryoelectron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barklis, E., J. McDermott, S. Wilkens, et al. 1997. Structural analysis of membrane-bound capsid proteins. EMBO J. 16:1199-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barklis, E., J. McDermott, S. Wilkens, S. D. Fuller, and D. Thomson. 1998. Organization of HIV-1 capsid proteins on a lipid monolayer. J. Biol. Chem. 273:7177-7180. [DOI] [PubMed] [Google Scholar]
- 6.Bernhard, W. 1958. Electron microscopy of tumor cells and tumor viruses. A review. Cancer Res. 18:491-509. [PubMed] [Google Scholar]
- 7.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celma, C. C. P., J. M. Manrique, J. L. Affranchino, E. Hunter, and S. A. Gonzales. 2001. Domains in the simian immunodeficiency virus gp41 cytoplasmic tail required for envelope incorporation into particles. Virology 283:253-261. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, W., R. M. Burnett, and R. L. Garcea. 1997. Structural biology of viruses. Oxford University Press, Oxford, England.
- 12.Conte, M. R., M. Klikova, E. Hunter, T. Ruml, and S. Matthews. 1997. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 16:5819-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 14.Crowther, R. A., N. A. Kiselev, B. Bottcher, J. A. Berriman, G. P. Borisova, V. V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 15.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster, M., B. Mulloy, and M. V. Nermut. 2000. Molecular modelling study of HIV p17 gag (MA) protein utilising data from electron microscopy and X-ray crystallogaphy. J. Mol. Biol. 298:841-857. [DOI] [PubMed] [Google Scholar]
- 17.Frank, J., B. Shimick, and H. Dowse. 1981. SPIDER-A modular software for electron image processing. Ultramicroscopy 6:343-358. [Google Scholar]
- 18.Freed, E. O. 1998. HIV Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 19.Fuller, S. D., T. Wilk, B. E. Gowen, H.-G. Krausslich, and V. M. Vogt. 1997. Cryoelectron microscopy reveals ordered domains within the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 20.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1997. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 21.Garnier, L., J. B. Bowzard, and J. W. Wills. 1998. Recent advances and remaining problems in HIV assembly. AIDS 12(Suppl. A):S5-S16. [PubMed] [Google Scholar]
- 22.Gelderblom, H. R., E. H. Hausman, M. Ozel, G. Pauli, and M. A. Koch. 1987. Fine structure of human immunodeficiency virus and immunolocalization of structural proteins. Virology 156:171-176. [DOI] [PubMed] [Google Scholar]
- 23.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. de Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 24.Grättinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Muller, and H.-G. Kräusslich. 1999. In vitro assembly properties of wild-type and cyclophilin binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 25.Gross, I., H. Hohenberg, C. Huckhagel, and H.-G. Kräusslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross, I., H. Hohenberg, T. Wilk, C. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H.-G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammarstedt, M., K. Wallengren, K. W. Pedersen, N. Roos, and H. Garoff. 2000. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc. Natl. Acad. Sci. USA 97:7527-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen, M. S. T., and E. Barklis. 1995. Structural interactions between retroviral Gag proteins examined by cysteine cross-linking. J. Virol. 69:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris, J. R. 1997. Negative staining and cryoelectron microscopy: the thin film techniques. Bios Scientific Publishers Ltd., Oxford, England.
- 30.Henderson, R., J. M. Baldwin, K. H. Downing, J. Lepault, and F. Zemlin. 1986. Structure of purple membrane from Halobacterium halobium: recording, measurement and evaluation at 3.5 Å resolution. Ultramicroscopy 19:147-178. [Google Scholar]
- 31.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hockley, D. J., R. D. Wood, J. P. Jacobs, and A. J. Garrett. 1988. Electron microscopy of human immunodeficiency virus. J. Gen. Virol. 69:2455-2469. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, J. E. 1996. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. USA 93:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klikova, M., S. S. Rhee, E. Hunter, and T. Ruml. 1995. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, S., C. P. Hill, W. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 21:410-413. [DOI] [PubMed] [Google Scholar]
- 36.McDermott, J., K. Mayo, and E. Barklis. 2000. Three-dimensional organization of retroviral capsid proteins on a lipid monolayer. J. Mol. Biol. 309:121-133. [DOI] [PubMed] [Google Scholar]
- 37.Morikawa, Y., W.-H. Zhang, D. J. Hockley, M. V. Nermut, and I. M. Jones. 1998. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein p17. J. Virol. 72:7659-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nermut, M. V. 1982. Advanced methods in electron microscopy of viruses, p. 1-58. In C. R. Howard (ed.), New developments in practical virology. Alan R. Liss, Inc., New York, N.Y. [PubMed]
- 39.Nermut, M. V., and L. D. Williams. 1977. Freeze-fracturing of monolayers capillary layers of cells, membranes and viruses: some technical considerations. J. Microsc. 110:121-132. [DOI] [PubMed] [Google Scholar]
- 40.Nermut, M. V., and J. Perkins. 1979. Consideration of the three-dimensional structure of the adenovirus hexon from electron microscopy and computer modelling. Micron 10:247-266. [Google Scholar]
- 41.Nermut, M. V., D. J. Hockley, J. B. M. Jowett, I. M. Jones, M. Garreau, and D. Thomas. 1994. Fullerene-like organization of HIV gag protein shell in virus-like particles produced by recombinant baculovirus. Virology 198:288-296. [DOI] [PubMed] [Google Scholar]
- 42.Nermut, M. V., and D. J. Hockley. 1996. Comparative morphology and structural classification of retroviruses. Curr. Top. Microbiol. Immunol. 214:1-24. [DOI] [PubMed] [Google Scholar]
- 43.Nermut, M. V., D. J. Hockley, P. Bron, D. Thomas, W.-H. Zhang, and I. M. Jones. 1998. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J. Struct. Biol. 123:143-149. [DOI] [PubMed] [Google Scholar]
- 44.Nermut, M. V., K. Wallengren, and J. Pager. 1999. Localization of actin in Moloney murine leukaemia virus by immunoelectron microscopy. Virology 160:23-34. [DOI] [PubMed] [Google Scholar]
- 45.Palmer, K. J., W. Tichelaar, N. Myers, N. R. Burns, S. J. Butcher, A. J. Kingsman, S. D. Fuller, and H. R. Saibil. 1997. Cryoelectron microscopy structure of yeast Ty retrotransposon virus-like particles. J. Virol. 71:6863-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker, S. D., J. S. Wall, and E. Hunter. 2001. An analysis of Mason-Pfizer monkey virus Gag particles by scanning transmission electron microscopy. J. Virol. 75:9543-9548. [DOI] [PMC free article] [PubMed]
- 47.Rao, Z., A. S. Belyaev, E. Fry, P. Roy, I. M. Jones, and D. I. Stuart. 1995. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature 378:743-747. [DOI] [PubMed] [Google Scholar]
- 48.Rumlova-Klikova, M., E. Hunter, M. V. Nermut, I. Pichova, and T. Ruml. 2000. Analysis of Mason-Pfizer monkey virus Gag domains required for capsid assembly in bacteria. J. Virol. 74:8452-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakalian, M., S. D. Parker, R. A. Weldon, Jr., and E. Hunter. 1996. Synthesis and assembly of retrovirus Gag precursors in immature capsids in vitro. J. Virol. 70:3706-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakalian, M., and E. Hunter. 1999. Separate assembly and transport domains within the Gag precursor of Mason-Pfizer monkey virus. J. Virol. 73:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarlata, S., L. S. Ehrlich, and C. A. Carter. 1998. Membrane-induced alterations in HIV-1 gag and matrix protein-protein interactions. J. Mol. Biol. 277:161-169. [DOI] [PubMed] [Google Scholar]
- 52.Sonigo, B., C. Barker, E. Hunter, and S. Wain-Hobson. 1986. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive retrovirus. Cell 45:375-385. [DOI] [PubMed] [Google Scholar]
- 53.Steward, M. 1988. Introduction to the computer image processing of electron micrographs of two-dimensionally ordered biological structures. J. Electron Microsc. Tech. 9:301-324. [DOI] [PubMed] [Google Scholar]
- 54.Turner, B. G., and M. F. Summers. 1999. Structural biology of HIV. J. Mol. Biol. 285:1-32. [DOI] [PubMed] [Google Scholar]
- 55.Valentine, R. C., B. M. Shapiro, and E. R. Stadtman. 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7:2143-2152. [DOI] [PubMed] [Google Scholar]
- 56.Valpuesta, J. M., J. L. Carrascosa, and R. Henderson. 1994. Analysis of electron microscopy images and electron diffraction patterns of thin crystals of Φ29 connectors in ice. J. Mol. Biol. 240:281-287. [DOI] [PubMed] [Google Scholar]
- 57.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H.-G. Kräusslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D 55:85-92. [DOI] [PubMed] [Google Scholar]
- 60.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber, G., and E. Barklis. 2000. Atomic force microscopy and electron microscopy analysis of retrovirus Gag proteins assembled in vitro on lipid bilayers. Biophys. J. 78:373-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuber, G., J. McDermott, S. Karanjia, W. Zhao, M. F. Schmid, and E. Barklis. 2000. Assembly of retrovirus capsid-nucleocapsid proteins in the presence of membranes or RNA. J. Virol. 74:7431-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]