Abstract
DNA vaccines expressing the envelope (Env) of the human immunodeficiency virus type 1 (HIV-1) have been relatively ineffective at generating high-titer, long-lasting, neutralizing antibodies. In this study, DNA vaccines were constructed to express the gp120 subunit of Env from the isolate HIV-1R2 using both wild-type and codon-optimized gene sequences. Three copies of the murine C3d were added to the carboxyl terminus to enhance the immunogenicity of the expressed fusion protein. Mice (BALB/c) vaccinated with DNA plasmid expressing the gp120R2 using codon-optimized Env sequences elicited high-titer anti-Env antibodies regardless of conjugation to C3d. In contrast, only mice vaccinated with DNA using wild-type gp120R2 sequences fused to mC3d3, had detectable anti-Env antibodies. Interestingly, mice vaccinated with DNA expressing gp120R2 from codon-optimized sequences elicited antibodies that neutralized both homologous and heterologous HIV-1 isolates. To determine if the unique sequence found in the crown of the V3 loop of the EnvR2 was responsible for the elicitation of the cross-clade neutralizing antibodies, the codons encoding for the Pro-Met (amino acids 313–314) were introduced into the sequences encoding the gp120ADA (R5) or gp12089.6 (R5X4). Mice vaccinated with gp120ADA–mC3d3–DNA with the Pro–Met mutation had antibodies that neutralized HIV-1 infection, but not the gp12089.6–mC3d3–DNA. Therefore, the use of the unique sequences in the EnvR2 introduced into an R5 tropic envelope, in conjunction with C3d fusion, was effective at broadening the number of viruses that could be neutralized. However, the introduction of this same sequence into an R5X4-tropic envelope was ineffective in eliciting improved cross-clade neutralizing antibodies.
INTRODUCTION
At the end of 2003, approximately 42 million people were infected and living with the human immunodeficiency virus type 1 (HIV-1), the pathogen associated with the onset of acquired immunodeficiency syndrome (AIDS).1 Greater than 95% of new HIV infections occurred in developing countries (70% men, 30% women). AIDS is a state of immunodeficiency that weakens a patient's immune system resulting in the development of fatal opportunistic infections. Despite the effectiveness of highly active antiretroviral therapy (HAART), there are several drawbacks that prevent its worldwide use, particularly for individuals in developing nations.2–4 Therefore, one of the long-term goals of HIV/AIDS research has been the development of a safe and effective vaccine.
The induction of highly cross-reactive neutralizing antibodies is one of the goals of HIV vaccine development. A variety of vaccine strategies using envelope immunogens has failed to induce antibodies capable of neutralizing cross-clade, primary isolates.5–7 The elicitation of antibodies directed against Env appears to be a critical component of an AIDS vaccine.8 Cross-reactive antibodies that neutralize primary isolates have been infrequently described in sera from donors infected with HIV-1. However, reference serum prepared from a donor infected in the United States with a clade B strain of HIV-1 has neutralizing antibodies that cross-react extensively with primary HIV-1 isolates of various clades.9–11 The donor (HNS2) had a long-term nonprogressive HIV-1 infection for more than 10 years.12 Relatively cross-reactive antibodies that neutralize primary isolates have been described in sera from other donors with long-term nonprogressive HIV-1 infections.13
The EnvR2, expressed from one of the env genes cloned from patient HNS2, can be neutralized by sera from patients infected with HIV-1 from clades A, B, C, D, and F, as well as circulating recombinant forms (CRF).12 Virions pseudotyped with the EnvR2 can mediate CD4-independent infection. In addition, these viruses are sensitive to neutralization by a panel of monoclonal antibodies that recognizes conformation epitopes in envelope.14 A rare mutation found in the crown of the V3 loop, a proline (P) and methionine (M) (nucleotide position 313/314), appears responsible for the uncommon neutralization sensitivity phenotype and the capacity of this envelope to mediate CD4-independent infection.14 Recently, Dong et al.15 expressed the EnvR2 from a Venezuelan equine encephalitis (VEE) replicon and elicited high titer neutralizing antibodies in mice and rabbits following immunization. These properties are consistent with the possibility that the EnvR2 expresses one or more neutralization epitopes that are responsible for induction of cross-reactive neutralizing antibodies in the donor of HNS2.
DNA vaccination (genetic vaccination) induces protective immunity against a variety of pathogens.16–19 These genetic vaccines consist of eukaryotic expression plasmids that are inoculated into target cells in the skin, muscle, or mucosal surfaces of a host and are translated into proteins.7,20 DNA vaccination effectively induces both humoral and cellular immune responses to immunogens from diverse infectious agents.5,17–19,21–23 The use of a variety of HIV envelope immunogens has failed to induce antibodies capable of neutralizing more than a fraction of primary isolates.5,7 Unlike most immunogens, multiple DNA immunizations are required to elicit even modest titers of neutralizing antibody to the HIV envelope glycoprotein.17,18,22,24–30 However, the elicitation of neutralizing antibodies appears to be a critical component of any HIV vaccine.20,31
One approach advanced in our laboratory to enhance the immunogenicity to HIV-1 Env is the use of the complement protein, C3d, as a molecular adjuvant. Conjugation of multiple copies of C3d to an antigen enhances the immunogenicity of low or nonimmunogenic antigens.27,32 C3d, when fused to an antigen, but not when coinoculated, enhances the total IgG titer against the conjugated protein.27,32 Antibodies directed against (1) the hemagglutinin of influenza or measles virus,22,33,34 (2) the merozoite surface antigen, MSP1.19, of Plasmodium yoelii,35 (3) the capsular polysaccharide of serotype 14 Streptococcus pneumonia,36,37 (4) the hepatatis B surface antigen (L. Wang, personal communication) were increased between 1 and 3 logs following immunization with DNA-expressing C3d-conjugated antigens. The addition of three copies of murine or human C3d to soluble forms of HIV-1 envelope accelerated both the onset and the avidity maturation of antibody in vaccinated mice and enhanced neutralizing antibody titers compared to mice vaccinated with antigen alone.24,26,29,30 In addition, titers of neutralizing antibodies to influenza or measles viruses were increased using DNA plasmids expressing C3d-fused hemagglutinin.21,33,34 Interestingly, the precise mechanism of C3d enhancement is unclear, however, C3d appears to enhance antibody and cellular responses via both CR2-dependent and -independent mechanisms.38
Therefore, the goal of this study was to analyze the effectiveness of the unique V3 sequence from EnvR2 introduction into the env genes from two prototype R5 strains to determine if the Pro–Met mutation would confer increased neutralization capacity following DNA vaccination. These envelopes were expressed from either wild-type or synthetic codon-optimized sequences alone or in conjunction with mC3d3 and then analyzed for both total IgG and neutralization.
MATERIALS AND METHODS
Plasmid vector DNA
pTR600, a eukaryotic expression vector, has been described previously.24,26,30,33,39 Briefly, the vector was constructed to contain the cytomegalovirus immediate-early promoter (CMVIE) plus intron A (IA) for initiating transcription of eukaryotic inserts and the bovine growth hormone polyadenylation signal [BGH poly(A)] for termination of transcription. The vector contains the ColE1 origin of replication for prokaryotic replication and the kanamycin resistance gene (Kanr) for selection in antibiotic media.
Construction of DNA vaccines
Previously, gp120ADA and gp12089.6 expression plasmids and gp120ADA–C3d3 and gp12089.6–C3d3 flusion constructs using wild-type Env gene sequences have been characterized.26,29,30 Briefly, the envelope sequences from the isolate, HIV-1R2, encoding almost the entire gp120 region were cloned into the pTR600 vaccine vector using unique restriction endonuclease sites (Fig. 1A). C3d sequences were cloned in frame at the 3′ end of the gp120 gene. The first 32 amino acids were deleted from the N-terminus of each sgp120 and replaced with a leader sequence from the trypsin plasminogen activator (tpA). The vectors expressing sgp120–mC3d3 fusion proteins were generated by cloning three tandem repeats of the mouse homologue of C3d in frame with the sgp120-expressing DNA. The construct design was based upon Dempsey et al.32 with glycine/serine linkers {(G4S)2} between gp120 and each C3d repeat.26,29,30 A second gp120R2 gene was synthesized to encode for the gp120 molecule (amino acids 1–526) using codons for enhanced expression in mammalian cells (GeneArt, Regensburg, Germany). In addition, the synthetic gp120R2 gene was cloned in frame with the mC3d3 gene.14,40 Lastly, oligonucleotides were constructed to introduce into the gp120ADA or gp12089.6 genes a proline/methionine (Pro–Met) at amino acids 313/314 in the V3 loop by polymerase chain reaction (PCR)-based mutagenesis (Fig. 1B). The same mutations were introduced into the gp120ADA–C3d3–DNA and gp12089.6–C3d3–DNA.
FIG. 1.
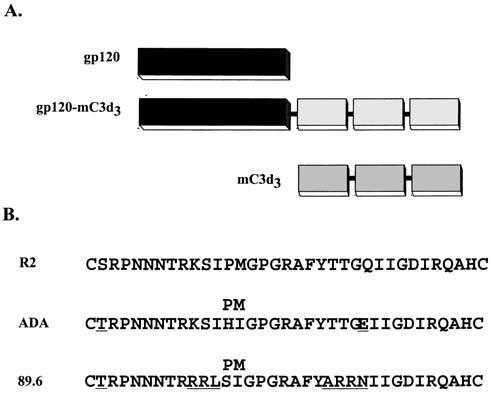
(A) The first schematic represents the secreted gp120 form of the envelope used as a vaccine insert. The second schematic represents the sgp120–C3d3 construct used as a vaccine insert. The third schematic represents the C3d3 construct used as a vaccine insert. Linkers composed of two repeats of four glycines and a serine {(G4S)2} were fused at the junctures between gp120 and C3d and between each C3d repeat. (B) V3 loop sequence representing R2, ADA, and 89.6. The superscript Pro–Met represents proline and methionine mutation introduced into gp120ADA and gp12089.6. Undelined amino acids indicate additional amino acids that differed with the R2 sequence.
The plasmids were amplified in Escherichia coli strain DH5α, purified using endotoxin-free, anion-exchange resin columns (Qiagen, Valencia, CA) and stored at –20°C in dH2O. Plasmids were verified by appropriate restriction enzyme digestion and gel electrophoresis. Purity of DNA preparations was determined by optical density reading at 260 and 280 nm and, therefore, each DNA vaccine inoculation contained >50 fg/μg of endotoxin per DNA inoculation.
Transfections and expression analysis
The human embryonic kidney cell line 293T (5 × 105 cells/transfection) was transfected with 1 μg of DNA using 12% lipofectamine according to the manufacturer's guidelines (Life Technologies, Grand Island, NY). Supernatants were collected and stored at –20°C. Cell lysates were collected in 300 μl of 1% Triton-X buffer and stored at –20°C. Quantitative antigen capture ELISAs were conducted as previously described.24,26,29 Alternatively, monoclonal antibodies (IgG1b12, F105, 2F5, 17b, 48d)25,41–43 were used to detect the fusion proteins in ELISA. All DNA expressing gp120 from wild-type sequences produced equal amounts of protein (∼1 μg/μl), which was three to four times higher than the amount of protein expressed from gp120–mC3d3–DNA. DNA expressing the same molecules from codon-optimized gene sequences elicited ∼10 times higher amounts of protein (data not shown). Similar results were observed for wild-type and codon-optimized Env gene sequences from isolates HIV-1ADA and HIV-189.6. The introduction of the Pro–Met mutation at amino acids 313/314 did not affect protein expression (data not shown).
Iodination and binding of sgp120 to cell surface receptors
The gp120 or gp120–mC3d3 molecules were labeled using Iodobead (Pierce, Rockford, IL) iodination of 10 μg of protein for 5 min in a 150 μl volume of phosphate-buffered saline (PBS), using 250 μCi of Na125I preincubated for 5 min with one Iodobead.44 125I incorporation rates of greater than approximately 50% resulted in oxidative destruction of protein that was no longer capable of binding any receptor. Radiolabeled proteins were purified from free Na125I by separation through a 0.3-ml Dowex column prepared in a 1-ml syringe and preequilibrated in a mixture containing 50 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM CaCl2, 1% bovine serum albumin (BSA), and 150 mM NaCl. Protein fractions were eluted in the void volume of the column, and the fractions containing peaks of labeled protein were combined. The integrity of gp120 after radiolabeling was verified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and autoradiography.
Env binding assays were performed by resuspending HOS CCR5+ or HOS CXCR4+ cells in 75 μl of HEPES binding buffer [50 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM CaCl2, 5% BSA, 0.1% NaN3]. Labeled proteins were added to cells in 25 μl of binding buffer for a total volume of 100 μl. Cells were incubated at room temperature (RT) for 1 hr. Unbound radioactivity was removed by filtering cells through 25-mm filters (Whatman, Clifton, NJ) presoaked in 0.2% polyethyleneimine (Sigma, St. Louis, MO) and thoroughly washed. Filters were counted in a gamma counter and results are expressed as counts per minute. For competition experiments, radiolabeled gp120 and gp120–mC3d3 proteins were incubated with monoclonal antibody 17b (1 μg/μl) for 1 hr at 37°C. Cells were pelleted and washed with PBS and then analyzed in a gamma counter.
Animals and DNA immunizations
Six- to 8-week-old BALB/c mice (Harlan Sprague Dawley, Indianapolis, IN) were vaccinated intradermally using particle bombardment (gene gun) inoculations. Gene gun immunizations were performed on shaved abdominal skin of anethesized mice as described previously.26,34,45,46 Mice were immunized with two gene gun doses containing 1 μg of DNA per 0.5 mg of approximately 1 μm gold beads (Bio-Rad, Hercules, CA) at a helium pressure setting of 400 psi.
Immunological assays
An endpoint ELISA was performed to assess the titers of anti-Env IgG in immune serum28 using matched, recombinant HIV-1 gp120 protein to coat plates purified as previously described.24 Mouse sera from vaccinated mice were allowed to bind and subsequently were detected by antimouse IgG conjugated to horseradish peroxidase (Southern Biotechnology, Birmingham, AL). Endpoint titers were considered positive that were 2-fold higher than background. Similar ELISAs were performed to determine the isotype of IgG (IgG1 or IgG2a) or other immunoglobulin classes, IgM, IgE, and IgA, elicited by vaccination. All assays were performed in triplicate.
Avidity ELISAs were performed similarly to serum antibody determination ELISAs up to the addition of samples and standards.26,28–30,33,34,47–52 Samples were diluted to give similar concentrations of specific IgG by OD. Plates were washed three times with 0.05% PBS-Tween 20. Different concentrations of the chaotropic agent sodium thiocyanate (NaSCN) in PBS were then added (0, 1, 1.5, 2, 2.5, and 3 M NaSCN). Plates were allowed to stand at room temperature for 15 min and then washed six times with PBS-Tween 20. Subsequent steps were performed similarly to the serum antibody determination ELISA. Percent of initial IgG was calculated as a percent of the initial OD. All assays were done in triplicate.
Neutralization of virus infection
Pseudotyped viruses were prepared by transfection of 293T cells with pNL4-3.luc.E-R and envelope-expressing plasmids, as previously described.53,54 Infectivity and neutralization assays were carried out using HOS CD4+ CCR5+, HOS CD4+ CXCR4+, HOS CCR5+, or HOS CXCR4+ cells.12,40,53–55 Infectivity was determined on the basis of luminescence measured 3 days after infection. As previously described, neutralization assays were performed by preincubation of serial serum with pseudotyped viruses [ADA, 89.6, R2 (clade B), 92UG029 (clade A), or 92BR025 (clade C)] prior to cell infection.12,53,54 The average dilution of 50% of the mean luminescence from three independent experiments was determined for each serum sample for each pseudotyped virus. In addition, the nonspecific neutralizing titer from naive, age-matched mice at each dilution was subtracted from each value.
Statistics
For statistical analysis, a Student's t test was employed. The difference between gp120 fused to multiple copies of murine C3d and gp120 alone or fused to murine C3d was determined. Differences were considered statistically significant when p < 0.05.
RESULTS
Binding of monoclonal antibodies to envelope
The gp120R2 was more easily recognized by the monoclonal antibodies 17b and 48d,43,57–61 which recognize CD4-induced epitopes, compared to gp120ADA or gp12089.6 (Table 1). Similar results were observed with gp120ADA(Pro–Met) and gp12089.6(Pro–Met). The addition of mC3d3 did not inhibit the binding by any of the monoclonal antibodies (Table 1). All the gp120 molecules were recognized by the monoclonal antibodies F105 and 1b12 and the monoclonal antibody 2F5, which maps to a linear sequence in gp41, did not recognize any of the expressed gp120 envelopes. The addition of sCD4 to gp120ADA or gp12089.6 enhanced the binding of 17b and 48d (Table 1), however, sCD4 did not increase the binding of these antibodies to gp120R2. Therefore, CD4-induced epitopes are exposed on the gp120R2 molecule, as well as gp120, incorporating the Pro–Met mutation, which are recognized by antibodies that usually bind only following exposure to hCD4.
Table 1.
Binding of Monoclonal Antibodies to gp120 ± mC3d3
| Monoclonal antibody |
|||||
|---|---|---|---|---|---|
| Samples | F105 | 1b12 | 2F5 | 48d | 17b |
| R2 | |||||
| wt gp120 | 25,000a | 25,000 | <10 | 50,000 | 25,000 |
| wt gp120–mC3d3 | 25,000 | 25,000 | <10 | 12,500 | 12,500 |
| wt gp120 + sCD4 | 50,000 | 12,500 | <10 | 50,000 | 25,000 |
| wt gp120–mC3d3 + sCD4 | 25,000 | 50,000 | <10 | 25,000 | 50,000 |
| ADA | |||||
| wt gp120 | 25,000 | 25,000 | <10 | 1,600 | 800 |
| wt gp120–mC3d3 | 25,000 | 25,000 | <10 | 400 | 400 |
| wt gp120 + sCD4 | 50,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120–mC3d3 + sCD4 | 50,000 | 50,000 | <10 | 25,000 | 25,000 |
| ADA (PM) | |||||
| wt gp120 | 50,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120–mC3d3 | 25,000 | 25,000 | <10 | 12,500 | 25,000 |
| wt gp120 + sCD4 | 25,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120–mC3d3 + sCD4 | 50,000 | 25,000 | <10 | 25,000 | 25,000 |
| 89.6 | |||||
| wt gp120 | 25,000 | 25,000 | <10 | 3,200 | 400 |
| wt gp120–mC3d3 | 25,000 | 25,000 | <10 | 400 | 400 |
| wt gp120 + sCD4 | 50,000 | 25,000 | <10 | 25,000 | 50,000 |
| wt gp120–mC3d3 + sCD4 | 50,000 | 25,000 | <10 | 50,000 | 50,000 |
| 89.6 (PM) | |||||
| wt gp120 | 25,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120–mC3d3 | 25,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120 + sCD4 | 25,000 | 25,000 | <10 | 25,000 | 25,000 |
| wt gp120–mC3d3 + sCD4 | 25,000 | 25,000 | <10 | 50,000 | 50,000 |
Value represents the inverse of the endpoint dilution titer
Binding of gp120 to cell surface receptors
Radiolabeled gp120R2 directly bound to cells expressing hCCR5, but not to cells expressing hCXCR4 (Table 2). The addition of mC3d3 to the 3′ end of the fusion protein did not affect the binding of gp120R2 to the cell surface chemokine receptors. gp120ADA or gp12089.6 did not bind to coreceptor expressing cells, however, both molecules did bind to cells expressing hCD4 (data not shown). However, envelopes with the Pro–Met mutation efficiently bound to appropriate coreceptor expressing cells. The addition of anti-hCCR5 antibodies (Table 2) or nonradioactive gp120R2, gp120ADA(Pro–Met), or gp12089.6(Pro–Met) (data not shown) significantly (p > 0.05) competed with the radiolabeled proteins.
Table 2.
Binding of 125I Radiolabeled HIV-1 Envelope to HOS Cells Expressing the Coreceptors hCCR5 or hCXCR4
| Coreceptor expressing cellsa |
|||
|---|---|---|---|
| Samples | hCCR5 | hCXCR4 | Anti-hCCR5b |
| R2 | |||
| wt gp120 | 22,501 ± 3,510c | 355 ± 127 | 174 ± 57 |
| wt gp120–mC3d3 | 22,013 ± 1,568 | 241 ± 205 | 256 ± 32 |
| co gp120 | 17,129 ± 3,629 | 267 ± 130 | 50 ± 29 |
| co gp120–mC3d3 | 18,219 ± 578 | 97 ± 75 | 117 ± 92 |
| ADA | |||
| wt gp120 | 323 ± 510 | 200 ± 110 | 152 ± 71 |
| wt gp120–mC3d3 | 287 ± 168 | 41 ± 23 | 129 ± 86 |
| co gp120 | 297 ± 62 | 67 ± 130 | 96 ± 92 |
| co gp120–mC3d3 | 253 ± 57 | 97 ± 75 | 117 ± 41 |
| ADA (PM) | |||
| wt gp120 | 19,445 ± 2,520 | 65 ± 12 | 74 ± 17 |
| wt gp120–mC3d3 | 15,553 ± 1,678 | 79 ± 135 | 67 ± 44 |
| co gp120 | 23,894 ± 2,004 | 234 ± 108 | 95 ± 20 |
| co gp120–mC3d3 | 16,283 ± 1,741 | 197 ± 107 | 150 ± 37 |
| 89.6 | |||
| wt gp120 | 384 ± 139 | 105 ± 62 | 113 ± 30 |
| wt gp120–mC3d3 | 446 ± 43 | 333 ± 143 | 129 ± 43 |
| 89.6 (PM) | |||
| wt gp120 | 21,490 ± 3,102 | 17,265 ± 1,009 | 372 ± 170 |
| wt gp120–mC3d3 | 29,835 ± 4,688 | 9,312 ± 4,510 | 174 ± 74 |
HOS cells expressed either hCCR5 or hCXCR4, but not hCD4
10 mg/ml of 17b mixed with protein prior to cell incubation
Value represents the inverse of the endpoint dilution titer. All values represent the average counts per minute of three independent experiments ± SEM
Antibody response to gp120-mC3d3 DNA immunizations
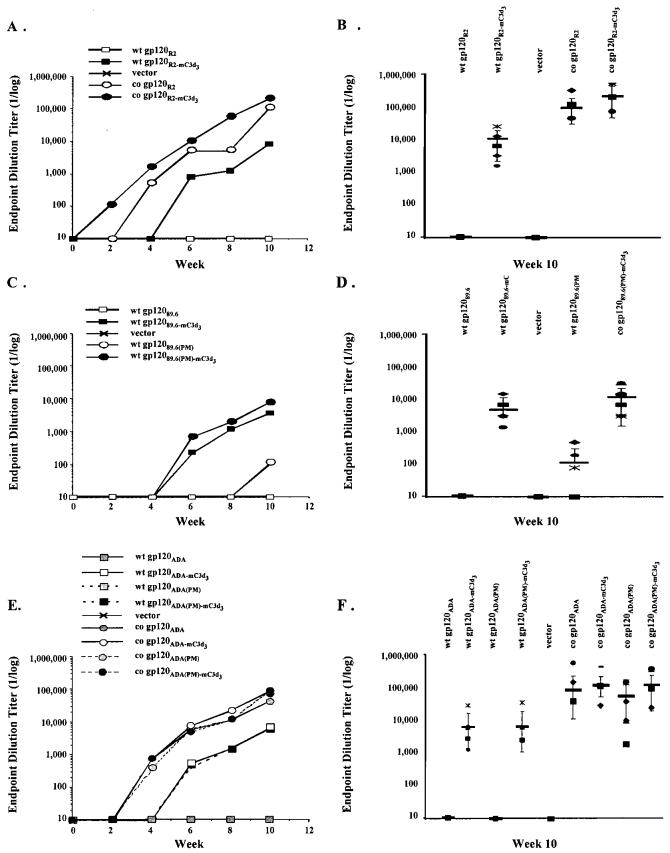
As previously reported,24,26,29,30,33 mice vaccinated with DNA plasmid (2 μg) expressing the gp120R2–mC3d3 from wild-type sequences (day 1 and then boosted at weeks 4 and 8) raised detectable titers of anti-Env antibody (1:6200 at week 8) (Fig. 2A). Few, if any, anti-EnvR2 antibodies were detected in mice vaccinated with gp120R2–DNA following three inoculations. In contrast, mice vaccinated with DNA expressing gp120R2 or gp120R2–mC3d3 from codon-optimized sequences (2 μg) had detectable titers (both IgG1 and IgG2a) after the first immunization, which were boosted following additional inoculations (Fig. 2A). Mice vaccinated with gp120R2–mC3d3–DNA using codon-optimized sequences had an average peak titer that was greater than 1 log higher (>1:000,000 at week 8) than mice vaccinated with gp120R2–mC3d3–DNA using wild-type sequences (<1:10,000 at week 8). In addition, mice vaccinated with gp120R2–DNA using codon-optimized sequences elicited similar, or in some cases higher, titers than C3d-conjugated forms (Fig. 2A). Similar results were observed in collected sera from mice vaccinated with vaccines using the gp120ADA, gp120ADA(Pro–Met), gp12089.6, or gp12089.6(Pro–Met) (Fig. 2B and C). The introduction of the Pro–Met mutation into the gp120ADA or gp12089.6 did not appear to affect the titers of IgG elicited in vaccinated mice, regardless of whether the proteins were expressed from wild-type or codon-optimized sequences. Therefore, at this dose of inoculum, the C3d enhancement effect was masked when the fused protein was expressed from DNA using codon-optimized gene sequences.
FIG. 2.
Anti-Env IgG raised by DNAs expressing gp120 in gene gun vaccinated mice. Mice were primed with DNA at day 0 and boosted at weeks 4 and 8. Sera were obtained from mice every 2 weeks. Serum collected at the indicated times from each mouse was assayed for specific IgG levels by ELISA. Plates (96-well) were coated with recombinant gp120R2 protein. Data are represented as the average of 10 mice. Preimmune serum from mice had no detectable specific IgG. Endpoint dilution titers were conducted by diluting the sera until OD values reached background. (A) Time course of DNA expressing R2 immunogens. (B) Range of endpoint dilution titer from collected serum (week 10) from mice vaccinated with DNA expressing R2 immunogens. (C) Time course of DNA expressing 89.6 immunogens. (D) Range of endpoint dilution titer from collected serum (week 10) from mice vaccinated with DNA expressing 89.6 immunogens. (E) Time course of DNA expressing DNA immunogens. (F) Range of endpoint dilution titer from collected serum (week 10) from mice vaccinated with DNA-expressing ADA immunogens.
Interestingly, mice vaccinated with DNA expressing non-C3d versions of each gp120 molecule had detectable titers of IgG1, but little or no titers of IgG2a at week 10 (data not shown). However, mice vaccinated with DNA expressing C3d-conjugated gp120 molecules elicited similar titers of IgG1 and IgG2a. The use of codon-optimized gene sequences did not alter the ratio between the two immunoglobulin classes, even though the overall titer was approximately 1 log higher.
The avidity of the antibody generated with DNA expressing gp120–mC3d3 was consistently higher than antisera from gp120–DNA vaccinated mice. Antisera from mice vaccinated with gp120–DNA had an ED50 of ∼0.5 M. However, mice vaccinated with DNA expressing gp120–mC3d3 had an ED50 of ∼1.5 M (week 10). Similar results were observed from mice vaccinated with DNA expressing gp120ADA(Pro–Met) or gp12089.6(Pro–Met). These results indicate that the Pro–Met mutation did not affect the maturation of the anti-Env antibody.
Envelopes with the Pro–Met mutation elicit cross-reactive neutralizing antibodies
Mice vaccinated with DNA expressing gp120 using wild-type gene sequences were unable to neutralize viral infection (week 10) due to the lack of anti-Env antibodies (Fig. 2), whereas, the addition of C3d raised antibodies capable of neutralizing homologous infection (1:16 dilution) (Table 3). However, DNA plasmid expressing neither gp120ADA–mC3d3, nor gp12089.6-mC3d3 was able to elicit antibodies that significantly prevented heterologous viral infection.
Table 3.
Elicitation of Neutralizing Antibodies by DNA Vaccines
| Viral isolatea |
|||||
|---|---|---|---|---|---|
| Samples | R2 | ADA | 89.6 | A | C |
| R2 | |||||
| wt gp120 | <10b | <10 | <10 | <10 | <10 |
| wt gp120–mC3d3 | 16 | <10 | <10 | <10 | <10 |
| co gp120 | 80 | <10 | <10 | <10 | <10 |
| co gp120–mC3d3 | 256 | 32 | 56 | 24 | 48 |
| ADA | |||||
| wt gp120 | <10 | <10 | <10 | <10 | <10 |
| wt gp120–mC3d3 | <10 | 32 | <10 | <10 | <10 |
| co gp120 | <10 | <10 | <10 | <10 | <10 |
| co gp120–mC3d3 | 20 | 72 | <10 | <10 | <10 |
| ADA (PM) | |||||
| wt gp120 | <10 | <10 | <10 | <10 | <10 |
| wt gp120–mC3d3 | <10 | 24 | <10 | <10 | <10 |
| co gp120 | <10 | <10 | <10 | <10 | <10 |
| co gp120–mC3d3 | 40 | 96 | 20 | 24 | 20 |
| 89.6 | |||||
| wt gp120 | <10 | <10 | <10 | <10 | <10 |
| wt gp120–mC3d3 | <10 | <10 | 40 | <10 | <10 |
| 89.6 (PM) | |||||
| wt gp120 | <10 | <10 | <10 | <10 | <10 |
| wt gp120–mC3d3 | <10 | <10 | 20 | <10 | <10 |
Clade B, R2, ADA, 89.6; clade A, 92UG029; clade C, 92BR025
Value represents the inverse of the endpoint dilution titer, n = 10. Clade 50% neutralization of viral infection. The average of three independent experiments
Mice vaccinated with DNA expressing gp120R2 from codon-optimized gene sequences elicited high titers of neutralizing antibodies against HIV-1R2 (1:80 dilution). The addition of C3d to gp120 raised the neutralizing antibodies (1:256 dilution) that neutralized homologous virus infection. Furthermore, conjugation of C3d to gp120R2 broadens the neutralizing capacity of the antisera, preventing virus infection by the heterologous viruses (Table 3). Mice vaccinated with DNA expressing gp120ADA ± mC3d3 from codon-optimized gene sequences did not broaden the number of viruses neutralized compared to non-C3d-conjugated gp120ADA. In addition, mice vaccinated with gp120ADA(Pro–Met)–mC3d3–DNA using codon-optimized genes neutralized both homologous and heterologous viruses (Table 3). Overall, codon optimization or C3d fusion enhanced the titer of neutralizing antibody. However, the induction of the highest titers of cross-reactive, neutralizing antibodies was elicited by C3d coupled to gp120R2 or to a gp120 incorporating the unique V3 loop mutations found in EnvR2.
DISCUSSION
In this study, DNA plasmids expressing the envelope from the HIV-1R2 (isolated from donor HNS2) were used to elicit high titer neutralizing antibodies in vaccinated mice (Table 3). Serum collected from donor HNS2, a long-term nonprogressor, had antibodies that neutralized primary isolates from a broad range of clades.15 Interestingly, when the EnvR2 is pseudotyped onto a virion, the virus is easily neutralized by anti-HIV sera.14 Therefore, we hypothesized that the gp120 subunit of EnvR2 would elicit cross-reactive neutralizing antibodies expressed from a DNA plasmid following vaccination. To enhance titers of anti-Env antibodies by DNA plasmids expressing gp120R2, two strategies were employed: (1) codon optimization of the gp120R2 gene sequences62,63 and (2) the use of C3d conjugated to gp120R2.24,26,29,30,64 Each strategy raised high titer anti-Env antibodies that neutralized homologous HIV-1R2 viral infection, however, the combination of both codon optimization and C3d to gp120R2 elicited the broadest neutralizing antibody responses (Table 3).
The EnvR2 has a rare mutation in the proximal limb of variable region 3 (V3), amino acids 313–314, Pro–Met.12,54 The neutralization sensitivity of this envelope has been mapped to this unique region of V3.14 Recently, Zhang et al.14 demonstrated that the EnvR2 was highly sensitive to neutralization by the monoclonal antibody 19b, which is directed against conformation-sensitive epitopes at the crown of the V3 loop. The binding of specific monoclonal antibodies that recognize CD4-induced epitopes is also dependent on this region (Table 1). A unique double turn is located at the apex of the V3 loop (GPGRAF) that is highly conserved across clade B isolates.65 A proline residue immediately 5′ to the V3 loop apex could significantly alter the structure of the V3 loop, resulting in exposure of the coreceptor binding site.66 Unlike most HIV-1 envelopes, the gp120R2 specifically bound to cells expressing hCCR5 (Table 2), indicating that the conformation of this envelope allows for coreceptor binding without the necessity of prior hCD4 interaction. The exposure of the coreceptor binding site in EnvR2 could explain the enhanced sensitivity of this envelope to neutralization.
The EnvR2, or the unique sequences located in EnvR2, could be an attractive immunogen for inducing neutralizing antibodies in humans following vaccination.67,68 Recently, Dong et al.15 demonstrated that neutralizing antibodies can be induced using a VEE replicon system expressing the EnvR2. Therefore, in this study, the unique sequence at the crown of the V3 loop in EnvR2 was introduced into two envelopes that do not normally elicit high titer neutralizing antibodies. Even though there are differences in the V3 loop sequences of EnvR2 and EnvADA (four amino acids) or Env89.6 (eight amino acids), the apex sequence is conserved in all three envelopes (Fig. 1). Mice vaccinated with each of these mutated envelopes did not dramatically enhance the neutralizing titers to homologous virus challenge regardless of whether the genes were expressed from wild-type or codon-optimized sequences (Table 3). However, the addition of C3d to these envelopes not only enhanced the anti-Env titer, but also broadened the number of viruses neutralized by collected sera (Table 3).
This study confirms that C3d can act as a molecular adjuvant to enhance antibody responses to the conjugated antigen. However, the mechanism(s) of action of C3d are still unclear. Recently, we demonstrated that unexpectedly, C3d can function as an effective adjuvant in the absence of CD21/35 expression.38 C3d has been shown to bind to CD21 (CR2) resulting in the coligation of CD19 to CD21, which, in turn, stimulates a signaling cascade that leads to the proliferation of B lymphocytes.32 However, the only direct evidence supporting this conclusion was that pretreatment of mice with an antibody against CD21 suppressed the effect elicited by C3d. Although anti-CD21 monoclonal treatment is known to inhibit humoral immune responses to a variety of antigens,69–71 anti-CD21 monoclonal antibody treatment may have effects on B cell function beyond blocking C3d binding.72 Therefore, C3d may enhance immune responses, including cellular responses,24,64 by multiple mechanisms. C3d may possibly enhance the uptake and processing of conjugated antigens by B cells, which results in enhanced antigen presentation. Also, the process by which C3d enhances neutralizing antibodies remains unclear, however, this enhancement is dependent on the neutralization potential of the conjugated immunogen. The gp120 proteins with a modified V3 loop used in this study and trimeric forms of Env (gp140)24 both elicited neutralizing antibodies that were enhanced by conjugation to C3d.
Despite the delayed appearance of neutralizing antibody in infected patients47 (6–8 months to achieve affinity maturation), this component of the immune response appears important in controlling infection. Immunogens that elicit high titer anti-envelope antibodies are one component of a vaccine needed for a multifaceted immune response against HIV-1. Even though DNA-expressing gp120 molecules conjugated to C3d or expressed using codon-optimized gene sequences elicited higher titer anti-Env antibodies, only mice vaccinated with EnvR2 had cross-reactive neutralizing antibodies (Table 3). Interestingly, the introduction of the unique Pro–Met mutation into a prototypic R5 envelope (ADA) or a prototypic dual tropic envelope (89.6) enhanced the neutralizing capacity of these proteins. Mice vaccinated with DNA using codon-optimized gp120ADA(Pro–Met) genes fused to mC3d3 elicited 4- to 8-fold high titers of neutralizing antibodies than nonmutated envelopes (Table 3). Therefore, the Pro–Met mutation in EnvR2 is most likely responsible for the constitutively exposed coreceptor binding site on the molecule. In addition, envelope immunogens incorporating the EnvR2 sequence induced broadly cross-reactive neutralizing antibodies following immunization.
ACKNOWLEDGMENTS
This research was supported by NIH grant Awards AI44325 to T.M.R. The authors thank Douglas Fearon for supplying the murine C3d constructs. In addition, the authors thank Gerry Quinnan for providing the wild-type EnvR2-expressing plasmid, as well as critical comments and discussions of the manuscript. Ms. Yvonne Brooks was a participant in the High School Honors Medicine Program from Junius H. Rose High School. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: HIV-Ig from NABI and NHLBI, the 92UG029 and 92BR025 virus stocks were provided by The UNAIDS Network for HIV, and the HIV-1 gp120 monoclonal antibodies from James E. Robinson (17b and 48d), from Dennis Burton and Paul Parren (IgG1b12), from Marshall Posner and Lisa Cavacini. (F105), and from Hermann Katinger (2F5).
REFERENCES
- 1.UNAIDS www.unaids.org
- 2.Katzenstein TL. Molecular biological assessment methods and understanding the course of the HIV infection. APMIS Suppl. 2003;114:1–37. [PubMed] [Google Scholar]
- 3.Pozniak A. Optimizing efficacy and tolerability in today's HAART. AIDS Read. 2003;13:S4–8. [PubMed] [Google Scholar]
- 4.Rutherford G, Sangani P, Kennedy G. Three- or four-versus two-drug antiretroviral maintenance regimens for HIV infection. Cochrane Database Syst Rev. 2003;4:CD002037. doi: 10.1002/14651858.CD002037. [DOI] [PubMed] [Google Scholar]
- 5.Graham BS. NIH Vaccine Research Center Clinical Studies. http://www.vrc.nih.gov/VRC/clinstudies.htm 2001
- 6.Mascola JR. Defining the protective antibody response for HIV-1. Curr Mol Med. 2003;3:209–216. doi: 10.2174/1566524033479799. [DOI] [PubMed] [Google Scholar]
- 7.Spearman P. HIV vaccine development: Lessons from the past and promise for the future. Curr HIV Res. 2003;1:101–120. doi: 10.2174/1570162033352093. [DOI] [PubMed] [Google Scholar]
- 8.Amara RR, Smith JM, Staprans SI, et al. Critical role for Env as well as Gag-Pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J Virol. 2002;76:6138–6146. doi: 10.1128/JVI.76.12.6138-6146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Souza M, Durda P, Hanson C, et al. Evaluation of monoclonal antibodies to HIV-1 by neutralization and serological assays: An international collaboration. AIDS. 1991;5:1061–1070. doi: 10.1097/00002030-199109000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Vujcic L, Katzenstein D, Martin M, et al. International collaborative study to compare assays for antibodies that neutralize human immunodeficiency virus. AIDS Res Hum Retroviruses. 1990;6:847–853. doi: 10.1089/aid.1990.6.847. [DOI] [PubMed] [Google Scholar]
- 11.Vujcic LK, Quinnan GV., Jr Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retroviruses. 1995;11:783–787. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- 12.Quinnan GV, Jr, Zhang PF, Fu DW, et al. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res Hum Retroviruses. 1999;15:561–570. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- 13.Connor RI, Korber BT, Graham BS, et al. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Bouma P, Park E, et al. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independence infectivity of human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol. 2002;76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong M, Zhang PF, Grieder F, et al. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J. Virol. 2003;77:3119–3130. doi: 10.1128/JVI.77.5.3119-3130.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly JJ, Ulmer JB, Liu MA. DNA vaccines. Dev Biol Strand. 1998;95:43–53. [PubMed] [Google Scholar]
- 17.Liu MA, Fu TM, Donnelly JJ, et al. DNA vaccines. Mechanisms for generation of immune responses. Adv Exp Med Biol. 1998;42:187–191. [PubMed] [Google Scholar]
- 18.Robinson HL. DNA vaccines for immunodeficency viruses. AIDS. 1997;11(Suppl A):S109–119. [PubMed] [Google Scholar]
- 19.Robinson HL. DNA vaccines: Basic mechanism and immune responses (Review) Int J Mol Med. 1999;4:549–555. doi: 10.3892/ijmm.4.5.549. [DOI] [PubMed] [Google Scholar]
- 20.Graham B. Clinical trials of HIV vaccines. Annu Rev Med. 2002;53:207–221. doi: 10.1146/annurev.med.53.082901.104035. [DOI] [PubMed] [Google Scholar]
- 21.Green TD, Newton BR, Rota PA, et al. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine. 2001;20:242–248. doi: 10.1016/s0264-410x(01)00266-3. [DOI] [PubMed] [Google Scholar]
- 22.Lu S, Arthos J, Montefiori DC, et al. Simian immunodeficiency virus DNA vaccine trial in macaques. J Virol. 1996;70:3978–3991. doi: 10.1128/jvi.70.6.3978-3991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson HL, Pertmer TM. DNA vaccines for viral infections: Basic studies and applications. Adv Virus Res. 2000;55:1–74. doi: 10.1016/s0065-3527(00)55001-5. [DOI] [PubMed] [Google Scholar]
- 24.Bower JF, Yang X, Sodroski J, et al. Elicitation of improved neutralizing antibodies using DNA vaccines expressing soluble stabilized HIV-1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78:4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchacher A, Predl R, Strutzenberger K, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 26.Green TD, Montefiori DC, Ross TM. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J Virol. 2003;77:2046–2055. doi: 10.1128/JVI.77.3.2046-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu S, Santoro J, Fuller D, et al. Use of DNAs expressing HIV-1 env and noninfectious HIV-1 particles to raise antibody responses in mice. Virology. 1995;209:147–154. doi: 10.1006/viro.1995.1238. [DOI] [PubMed] [Google Scholar]
- 28.Richmond JFL, Lu S, Santoro C, et al. Screening of HIV-1 Env glycoproteins for the ability to raise neutralizing antibody using DNA immunization and recombinant vaccinia virus boosting. Virology. 1997;230:265–270. doi: 10.1006/viro.1997.8478. [DOI] [PubMed] [Google Scholar]
- 29.Ross TM, Xu Y, Green TD, et al. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with gp120-C3d fusion proteins. AIDS Res Hum Retroviruses. 2001;17:829–835. doi: 10.1089/088922201750252025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toapanta FR, Ross TM. Mouse strain-dependent differences in enhancement of immune responses by C3d. Vaccine. 2004;22:1773–1781. doi: 10.1016/j.vaccine.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 31.Letvin NL. Strategies for an HIV vaccine. J Clin Invest. 2002;110:15–20. doi: 10.1172/JCI15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dempsey PW, Allison MED, Akkaraju S, et al. C3d of complement as a molecular adjuvant: Bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell JA, Green TD, Bright RA, et al. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine. 2003;21:902–914. doi: 10.1016/s0264-410x(02)00539-x. [DOI] [PubMed] [Google Scholar]
- 34.Ross TM, Xu Y, Bright RA, et al. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1:127–131. doi: 10.1038/77802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steward M, Cox V, Oldroyd R, et al. Towards a human C3d-based vaccine for malaria. Mol Immunol. 2001;38:118. [Google Scholar]
- 36.Haas KM, Hasegawa M, Steeber DA, et al. Complement receptors CD21/35 link innate and protective immunity during Streptococcus pneumoniae infection by regulating IgG3 antibody responses. Immunity. 2002;17:713–723. doi: 10.1016/s1074-7613(02)00483-1. [DOI] [PubMed] [Google Scholar]
- 37.Test ST, Mitsuyoshi J, Connolly CC, et al. Increased immunogenicity and induction of class switching by conjugation of complement C3d to pneumococcal serotype 14 capsular polysaccharide. Infect Immun. 2001;69:3031–3040. doi: 10.1128/IAI.69.5.3031-3040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haas KM, Toapanta FR, Oliver JA, et al. C3d functions as a molecular adjuvant in the absence of CD21/35 expression. J Immunol. 2004;172:5833–5837. doi: 10.4049/jimmunol.172.10.5833. [DOI] [PubMed] [Google Scholar]
- 39.Young KR, Smith JM, Ross TM. Characterization of DNA vaccines expressing a non-infectious human immunodeficiency virus-like particle. Virology. 2004;327:262–272. doi: 10.1016/j.virol.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Zhang PF, Chen X, Fu DW, et al. Primary virus envelope cross-reactivity of the broadening neutralizing antibody response during early chronic human immunodeficiency virus type 1 infection. J Virol. 1999;73:5225–5230. doi: 10.1128/jvi.73.6.5225-5230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posner MR. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquired Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- 42.Thali M. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. Virology. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thali M, Furman C, Helseth E, et al. Lack of correlation between soluble CD4-induced shedding of the human immunodeficiency virus type 1 exterior envelope glycoprotein and subsequent membrane fusion events. J Virol. 1992;66:5516–5524. doi: 10.1128/jvi.66.9.5516-5524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doranz BJ, Baik SS, Doms RW. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J Virol. 1999;73:10346–10358. doi: 10.1128/jvi.73.12.10346-10358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haynes JR, Fuller DH, Eisenbraun MD, et al. Accell particle-mediated DNA immunization elicits humoral, cytotoxic, and protective immune responses. AIDS Res Hum Retroviruses. 1994;10:S43–45. [PubMed] [Google Scholar]
- 46.Pertmer TM, Eisenbraun MD, McCabe D, et al. Gene gun based nucleic acid immunization eliciting of humoral and cytotoxic T lymphocyte response following epidermal delivery of nanogram quantitation of DNA. Vaccine. 1995;13:1427–1430. doi: 10.1016/0264-410x(95)00069-d. [DOI] [PubMed] [Google Scholar]
- 47.Cole K, Rowles J, Jagerski B, et al. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall JT, Heckel C. Thiocyanate elution estimation of relative antibody avidity. J Immunol Methods. 1988;115:153–154. doi: 10.1016/0022-1759(88)90324-9. [DOI] [PubMed] [Google Scholar]
- 49.Ito KY, Takeuchi Y, Ito K, et al. S train-dependent antibody response induced by DNA immunization. Immunol Lett. 2000;74:245–250. doi: 10.1016/s0165-2478(00)00266-2. [DOI] [PubMed] [Google Scholar]
- 50.Leung L, Srivastava IK, Kan E, et al. Immunogenicity of HIV-1 Env and Gag baboons using a DNA prime/protein boost regimen. AIDS. 2004;18:991–1001. doi: 10.1097/00002030-200404300-00006. [DOI] [PubMed] [Google Scholar]
- 51.MacDonald RA, Hosking CS, Jones CL. The measurement of relative antibody affinity by ELISA using thiocyanate elution. J Immunol Methods. 1988;106:191–194. doi: 10.1016/0022-1759(88)90196-2. [DOI] [PubMed] [Google Scholar]
- 52.Shiver JW, Davies ME, Yasutomi Y, et al. Anti-HIV Env immunities elicited by nucleic acid vaccines. Vaccine. 1997;15:884–887. doi: 10.1016/s0264-410x(96)00251-4. [DOI] [PubMed] [Google Scholar]
- 53.Park EJ, Vujcic LK, Anand R, et al. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J Virol. 1998;72:7099–7107. doi: 10.1128/jvi.72.9.7099-7107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinnan GV, Jr, Zhang PF, Fu DW, et al. Evolution of neutralizing antibody response against HIV type 1 virions and pseudovirions in multicenter AIDS cohort study participants. AIDS Res Hum Retroviruses. 1998;14:939–949. doi: 10.1089/aid.1998.14.939. [DOI] [PubMed] [Google Scholar]
- 55.Park EJ, Gorny MK, Zolla-Pazner S, et al. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J Virol. 2000;74:4183–4191. doi: 10.1128/jvi.74.9.4183-4191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park EJ, Quinnan GV., Jr Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J Virol. 1999;73:5707–5713. doi: 10.1128/jvi.73.7.5707-5713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clapham P, McKnight A, Weiss R. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: Induction and enhancement by soluble CD4. J. Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan N, Sun Y, Satterntau Q, et al. CD4-induced conformational changes in the HIV-1 gp120 glycoprotein: Consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins BA, Buge S, Aldrich K, et al. Resistance of HIV-1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding or multiple sites. J Virol. 1996;70:8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyatt R, Kwong P, Desjardins E, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 61.Wyatt R, Sullivan N, Thali M, et al. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andre S, Seed B, Eberle J, et al. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haas J, Park EC, Seed B. Coden usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 64.Liu F, Mboudjeka I, Shen S, et al. Independent but not synergistic enhancement to the immunogenicity of DNA vaccine expressing HIV-1 gp120 glycoprotein by codon optimization and C3d fusion. Vaccine. 2004;22:1764–1772. doi: 10.1016/j.vaccine.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 65.Ghiara JB, Ferguson DC, Satterthwait AC, et al. Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site. J Mol Biol. 1997;266:31–39. doi: 10.1006/jmbi.1996.0768. [DOI] [PubMed] [Google Scholar]
- 66.Lu M, Kim PS. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J Biomol Struct Dyn. 1997;15:46–71. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 67.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puffer BA, Pohlmann S, Edinger AL, et al. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol. 2002;76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heyman B, Wiersma EJ, Kinoshita T. In vivo inhibition of the antibody response to a complement receptor-specific monoclonal antibody. J Exp Med. 1990;172:665–668. doi: 10.1084/jem.172.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thyphronitis G, Kinoshita T, Inoue K, et al. Modulation of mouse complement receptors 1 and 2 suppress antibody responses in vivo. J Immunol. 1991;147:224–230. [PubMed] [Google Scholar]
- 71.Wiersma E, Kinoshita T, Heyman B. Inhibition of immunological memory and T-independent humoral responses by monoclonal antibodies specific for murine complement receptors. Eur J Immunol. 1991;21:2501–2506. doi: 10.1002/eji.1830211029. [DOI] [PubMed] [Google Scholar]
- 72.Chakravarty L, Zabel MD, Weis JJ, et al. Depletion of Lyn kinase from the BCR complex and inhibition of B cell activation by excess CD21 ligation. Int Immunol. 2002;14:139–146. doi: 10.1093/intimm/14.2.139. [DOI] [PubMed] [Google Scholar]