Abstract
The packaging of a mature dimeric RNA genome is an essential step in human immunodeficiency virus type 1 (HIV-1) replication. We have previously shown that overexpression of a protease (PR)-inactive HIV-1 Gag-Pro-Pol precursor protein generates noninfectious virions that contain mainly monomeric RNA (M. Shehu-Xhilaga, S. M. Crowe, and J. Mak, J. Virol. 75:1834-1841, 2001). To further define the contribution of HIV-1 Gag and Gag-Pro-Pol to RNA maturation, we analyzed virion RNA dimers derived from Gag particles in the absence of Gag-Pro-Pol. Compared to wild-type (WT) dimeric RNAs, these RNA dimers have altered mobility and low stability under electrophoresis conditions, suggesting that the HIV-1 Gag precursor protein alone is not sufficient to stabilize the dimeric virion RNA structure. The inclusion of an active viral PR, without reverse transcriptase (RT) and integrase (IN), rescued the stability of the virion RNA dimers in the Gag particles but did not restore the mobility of the RNAs, suggesting that RT and IN are also required for virion RNA dimer maturation. Thin-section electron microscopy showed that viral particles deficient in RT and IN contain empty cone-shaped cores. The abnormal core structure indicates a requirement for Gag-Pro-Pol packaging during core maturation. Supplementing viral particles with either RT or IN via Vpr-RT or Vpr-IN alone did not correct the conformation of the dimer RNAs, whereas expression of both RT and IN in trans as a Vpr-RT-IN fusion restored RNA dimer conformation to that of the WT virus and also restored the electron-dense, cone-shaped virion core characteristic of WT virus. Our data suggest a role for RT-IN in RNA dimer conformation and the formation of the electron-dense viral core.
The gag and pol genes of human immunodeficiency virus type 1 (HIV-1) are initially expressed as the precursor polyproteins Gag and Gag-Pro-Pol (8, 41). During or immediately after budding these precursors are processed by the viral protease (PR) into their mature products (8). The 55-kDa Gag precursor generates matrix (MA), capsid (CA), spacer peptide p2, nucleocapsid (NC), spacer peptide p1, and p6gag. The 160-kDa Gag-Pro-Pol fusion protein generates MA, CA, p2, NC, p6pol, PR, reverse transcriptase (RT), and integrase (IN) (19). The Gag and Gag-Pro-Pol polyproteins are encoded by the same mRNA but are not synthesized at the same rate. An infrequent ribosomal frameshifting event generates a 20:1 ratio of Gag to Gag-Pro-Pol production. In HIV-1, the maintenance of this ratio is critical for viral infectivity and genomic RNA dimer stability (38).
Intracellular expression of Gag alone is sufficient to produce virion-like particles (VLPs) (for a review, see reference 43). Moreover, there is an important role for Gag and viral genomic RNA interactions in the assembly process, with the packaging and dimerization of the genomic RNA primarily occurring via RNA-Gag interactions (2, 10, 29). The NC domain of Gag binds to viral RNA and has been shown to facilitate both the RNA packaging and the dimerization processes (3, 4, 7, 9, 17). The initial interaction between genomic RNA and HIV-1 Gag is likely to occur via the NC sequences within the Gag precursor, as HIV-1 with defective viral PR still packages RNA (for a review see reference 12). Furthermore, analysis of wild-type (WT) and PR-defective (PR−) virions has revealed that dimerization of the genomic RNA in HIV-1 initiates prior to proteolytic processing (13), suggesting that Gag and Gag-Pro-Pol precursor proteins can support RNA dimerization independently of protein processing. However, to date, RNA dimer formation and the stability of genomic RNA in Gag-only particles and the contribution of Gag-Pro-Pol to dimer formation have not been well defined.
Morphological studies of HIV-1 particles have shown that infectious virions are characterized by a condensed, cone-shaped core structure surrounding the genomic RNA, which is thought to bind to NC, RT, and IN (8). Incorporation of Gag-Pro-Pol is critical for viral assembly, supplying the viral enzymes and mediating the selective packaging of the primer tRNA (22, 30, 32). In addition, DNA synthesis by an RT-defective virus cannot be rescued by exogenously supplied RT, suggesting that the Gag-Pro-Pol precursor is necessary to maintain RT in proper association with the ribonucleoprotein initiation complex (1). However, our understanding of the role of the precursor Gag-Pro-Pol in genomic RNA dimer formation and maturation is limited. The packaging of the retrovirus enzymes in the form of a precursor protein is in itself suggestive of a role for Gag-Pro-Pol in coordinating the placement of the viral enzymes within the mature virion and supporting the correct molecular arrangements for Gag-RNA interactions. Functional RT and IN proteins can be incorporated into viral particles when supplied in trans as Vpr fusion proteins to generate infectious virions (28, 45). Such an approach provides a powerful tool for investigating the role of IN and RT in viral genomic RNA dimerization outside the context of the Gag-Pro-Pol protein.
We have taken advantage of the supplementation approach described by Wu et al. (45) to investigate the role of Gag and Gag-Pro-Pol and its component enzymes in RNA dimerization. Isolation and analysis of viral genomic RNA from Gag-derived particles (GUAA) has facilitated studies of the conformation of genomic RNA dimers in the absence of the Pol-derived proteins. The inclusion of an active PR in the GUAA construct (PRUAA) provides a means to examine the role of Gag proteolysis in genomic RNA dimerization. Supplementation of RT and IN in trans, as a Vpr-RT, Vpr-IN, or Vpr-RT-IN fusion protein, enables an assessment of the role of these enzymes in viral assembly and RNA dimer maturation that is independent of the Gag-Pro-Pol precursor protein.
We show here that the HIV-1 Gag alone does not generate stable dimeric RNA in the correct conformation. While the stability of the genomic RNA isolated from these virus-like particles (VLPs) is reestablished by Gag proteolysis, the conformation of the RNA dimers is only partially restored. In addition, thin-section electron microscopy (TSEM) demonstrates that processing of Gag leads to the formation of cone-shaped, electron-lucent cores. Although independent packaging of Vpr-RT and Vpr-IN into processed Gag particles is not sufficient for RNA dimer maturation, virion packaging of the Vpr-RT-IN fusion protein restores the RNA dimer conformation and leads to the formation of an electron-dense, cone-shaped core similar to that observed in the WT particles.
MATERIALS AND METHODS
DNA plasmids.
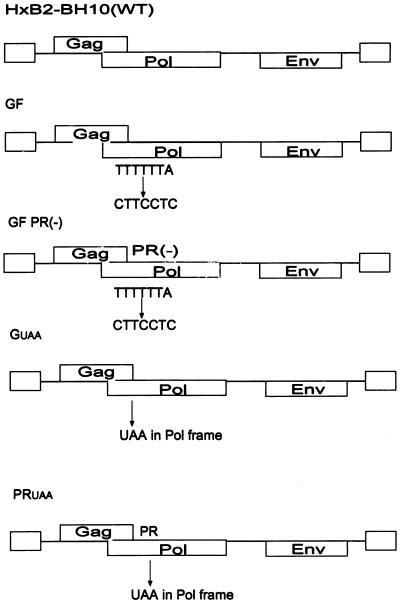
The WT HIV-1 plasmid used in this study, HXB2-BH10, has been previously described (42). PR-defective HIV-1 plasmid SVC21 PR(-), which has also been described previously (18, 30), was utilized as a control. The Gag frame (GF) mutant was constructed to reduce the level of Gag-Pro-Pol synthesis in the virus-producing cells. This mutation involves substitutions in the five-T stretch of nucleotides, which are responsible for the −1 ribosomal frameshifting, thus making it difficult for the ribosome to shift back and synthesize Gag-Pro-Pol. The tttttta sequence responsible for frameshifting was replaced with the cttcctt sequence by PCR stitch mutagenesis. Briefly, primers f1-sense (5′ggcaaagaagggcacacagcc 3′) and f1-antisense (5′cccgaggaagttagcctgtctctcagtac 3′) were used to amplify a 130-bp GF f1 fragment. Primers f2-sense (5′ggctaacttcctcgggaagatctggccttcc 3′) and f2-antisense (5′gttgacaggtgtaggtcctac 3′) were used to amplify a 400-bp GF f2 fragment. The GF f1 and GF f2 fragments were joined by PCR extension. The resulting PCR-amplified fragment was cloned into the HxB2-BH10 proviral DNA via restriction sites ApaI and BclI. To obtain the PR-defective form of GF [GF PR(-)], the aspartic acid at the active site of PR of the GF construct was replaced with asparagine via PCR stitch mutagenesis. The GUAA plasmid was constructed by PCR stitch mutagenesis to introduce a stop codon at amino acid position 13 of PR. As a result, this mutation does not allow the synthesis of full-length Gag-Pro-Pol or the expression of a functional PR. The PRUAA mutant was constructed to produce HIV-1 Gag particles that are cleaved by PR in the absence of RT and IN. Briefly, PCR stitch mutagenesis was used to introduce a stop codon within the Pol frame straight after the region encoding PR, allowing PR expression. The introduced mutations were confirmed by DNA sequencing. The Vpr-RT, Vpr-IN, and Vpr-RT-IN expression plasmids have been previously described (45).
Virus production.
The production of WT and mutant viral particles was achieved by the transfection of 10 μg of proviral DNA into 293T cells by a calcium phosphate method as previously described (24). For the supplementation studies 10 μg of Vpr-RT, Vpr-IN, or Vpr-RT-IN expression plasmid DNA was used in cotransfection with 10 μg of the PRUAA plasmid to generate viral particles that contain RT, IN, or RT-IN. An enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, Calif.) reporter plasmid (2 μg) was added to the DNA mixture to determine transfection efficiency.
Supernatants were collected 36 h posttransfection and centrifuged for 30 min at 2,000 × g and 4°C to remove cellular debris. The clarified supernatants were either frozen at −70°C or used immediately for further analysis. Cells were washed twice with either 1× phosphate-buffered saline (PBS) or 1× Tris-buffered saline (TBS buffer; 50 mM Tris [pH 7.4], 150 mM NaCl), followed by protein extraction using lysis buffer containing 1× TBS, 10 μl of Nonidet P-40/ml, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin. Cell lysates were collected and stored at −20°C for later use.
Intracellular viral protein analysis.
Cell lysates were rapidly frozen and thawed three times to weaken the cellular membrane. Cell debris was subsequently removed by centrifugation for 30 min at 4°C and 2,000 × g. The transfection efficiency of the samples was determined by measuring the level of EGFP from the reporter plasmid with a Bio Imaging Analyzer (Fuji Photo Film Co.). Cellular proteins from each sample, normalized for equivalent levels of EGFP, were mixed with 3 μl of sample buffer (100 mM Tris [pH 6.8], 3% sodium dodecyl sulfate [SDS], 33% glycerol, 0.03% bromophenol blue), heat-denatured for 10 min at 95°C, and resolved by 10% Tris-glycine SDS-polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred to a nitrocellulose membrane (Amersham). The membrane was blocked for 2 h in 3% casein dissolved in 2× TBS containing 0.3% Tween 20 (TBST) and probed overnight with pooled HIV-1-seropositive patient sera or an anti-p24 monoclonal antibody (NEN). After three washes with 1× TBST buffer the membrane was incubated with a horseradish peroxidase-conjugated antihuman secondary antibody (DAKO) for 2 h at room temperature. An enhanced chemiluminescence technique was used for visualization of HIV-1 proteins present in the cellular lysates (Amersham). Chemiluminescence was recorded by autoradiography.
Virion purification and protein analysis.
Supernatants from transfected cells were first clarified from cell debris by centrifugation for 30 min at 4°C and 2,000 × g. Viral particles were concentrated by ultracentrifugation through a 20% sucrose cushion using a Beckman ultracentrifuge (model L-90; SW 41 rotor) at 150,000 × g for 1 h at 4°C. Pellets were resuspended in 50 μl of TBS lysis buffer. Levels of virion proteins were first estimated via serial dilutions of virion proteins in Western dot blot analysis. HIV-1-positive patient serum was used as the primary antibody, followed by a horseradish peroxidase-conjugated antihuman secondary antibody. Enhanced chemiluminescence was then used for the quantification of total virion proteins. Equal amounts of virion proteins from each sample were mixed with 3 μl of sample buffer containing 5 mM β-mercaptoethanol, and the mixture was heated for 10 min at 95°C. Virion proteins were then resolved by 10% Tris-glycine SDS-PAGE as described above. The resolved virion protein samples were transferred onto nitrocellulose membranes by electrophoresis using a Bio-Rad transfer apparatus. Virion HIV-1 protein profiles of the samples were determined by Western analysis as described above.
Analysis of virion RNA packaging.
Pelleted virions were prepared by ultracentrifugation as described above and were resuspended in 500 μl of Trizol (Gibco) for genomic RNA extraction. Samples were subsequently incubated for 30 min in ice to allow complete lysis to occur, and 200 μl of chloroform was added to remove the organic solvent and virion proteins. Samples were mixed and centrifuged for 20 min at 20,000 × g at 4°C. The aqueous layer containing the RNA was transferred to a fresh Eppendorf tube and precipitated with 1/10 volume of 3 M sodium acetate-1 ml of ice-cold 100% ethanol at −20°C overnight. Precipitated RNA samples were centrifuged for 30 min at 20,000 × g and 4°C. The RNA pellets were washed with 200 μl of 70% ethanol and then air dried for 1 h at room temperature. Each pellet was resuspended in 10 μl of RNase-free water and stored at −20°C until use.
Genomic packaging of virion RNA was assessed by RNA dot blotting using a Bio-Rad dot blot apparatus. Serial 10-fold dilutions of virion RNA samples, which were normalized by total virion protein, were used to construct a standard curve. Each sample was mixed with 29 μl of RNA denaturation buffer (37% formaldehyde, 60% deionized formamide, 3% 20× SSC [3 M NaCl, 0.3 M sodium citrate, pH 7.0]) and incubated at 65°C for 10 min for RNA denaturation. After incubation, samples were immediately chilled on ice. To each tube, 78 μl of 20× SSC buffer was added, and samples were loaded onto a Hybond membrane (Amersham) via a dot blot apparatus (Bio-Rad). The wells were washed twice with 10× SSC, and the membrane was air dried overnight at room temperature. The membrane was exposed to UV light for 90 s to cross-link the RNA onto the membrane and then blocked for 1 h at 42°C with 10 ml of hybridization buffer (40 ml of the hybridization buffer contains 8 ml of 5× SSPE [750 mM NaCl, 63 mM NaH2PO4, 0.5 mM EDTA], 20 ml of deionized formamide, 4 g of dextran sulfate, 400 μl of salmon sperm DNA [10 mg/ml], 3 ml of 20% SDS, and 5 ml of double-distilled H2O).
The amount of genomic RNA in the viral RNA samples was quantified by using radioactive antisense oligonucleotide probe 790H (5′CTGACGCTCTCGCACCC3′), which has been previously described (31). Briefly, the 790H oligomer binds specifically to the HIV-1 genomic RNA sequence between positions 338 and 354 (DNA positions 791 to 807). The 790H oligomer was 5′ end labeled with T4 polynucleotide kinase and γ-32P-labeled ATP (3,000 Ci/mmol; Amersham). The membrane containing the RNA samples was incubated overnight at 42°C with the radioactively labeled probe. The membrane was washed once for 30 min with 1× SSC-0.1% SDS and twice with 0.2× SSC-0.1× SDS for 30 min. The results were visualized by autoradiography.
Analysis of virion RNA dimerization.
Virion pellets were resuspended in 500 μl of dimeric RNA lysis buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1% SDS, 50 mM NaCl, 10 U of proteinase K), extracted with phenol-chloroform, and isolated for melting curve analysis as previously described (13, 14).
Similar amounts of genomic RNA were used to analyze the stability of the virion RNA dimer in each preparation. Samples were heated at various temperatures (indicated at the bottom of each gel) for a period of 10 min, followed by a quick chill on ice. Heat-denatured dimeric and monomeric RNAs were separated by electrophoresis in a 1% native agarose gel in 0.5× Tris-borate-EDTA buffer. Samples were transferred overnight onto a nitrocellulose (Hybond N) membrane (Amersham). The membrane containing the RNA samples was air dried for 2 h at room temperature and exposed to UV light for 90 s for cross-linking. The membrane was blocked for 1 h at 42°C with 10 ml of hybridization buffer and incubated overnight with a radioactive riboprobe (pGEM7zHIV-1), which is complementary to the 5′ end of the HIV-1 genomic RNA sequences, as previously described (38). The radioactive riboprobe was synthesized by linearizing pGEM7z HIV-1 with BamHI, followed by T7 RNA polymerase-directed in vitro transcription (Promega) in the presence of [α-32P]CTP (NEN). After being probed, the membrane was washed once for 30 min with 1× SSC-0.1% SDS buffer and twice for 30 min with 0.2× SSC-0.1% SDS buffer. The results were visualized by autoradiography.
Metabolic labeling of virion proteins.
293T cells were transfected with 10 μg of each plasmid DNA by the calcium phosphate method described above. Twelve hours posttransfection cells were washed twice with PBS and incubated for an additional 12 h in fresh media. Subsequently, cells were washed with PBS and starved for 1 h in methionine- and cysteine-free Dulbecco’s modified Eagle medium containing 10% dialyzed fetal calf serum (FCS) (ICN Biochemicals). Cells were washed twice with PBS and cultured in methionine- and cysteine-free medium containing [35S]methionine-cysteine (70 mCi/ml; specific activity, 1,175 Ci/mmol; ICN Biochemicals). After a 1-h incubation at 37°C, 10% FCS was replenished and virus production was monitored every 6 h by measuring RT levels. To determine the RT levels for each virus, 10 μl of supernatant from each sample was mixed with 10 μl of NP-40. Samples were incubated for 30 min at room temperature for inactivation of the virus, and the RT activities of the viruses in the supernatant were measured by a micro-RT assay as previously described (15).
Labeled virus, isolated 36 h posttransfection, was first clarified from cell debris at 3,000 rpm for 30 min at 4°C in a Beckman centrifuge and pelleted through a 20% sucrose cushion with a Beckman model L-90 ultracentrifuge (SW 41 rotor). Standardized amounts of HIV proteins and genomic RNA extracted from labeled, pelleted WT and mutant viruses were loaded onto a nitrocellulose (Hybond N) membrane (Amersham) for the detection of 35S-labeled proteins. The specific activity of the [35S]methionine-cysteine labeling was 124,800 cpm/μg of labeled HIV-1 proteins. The presence of the 35S-labeled proteins was monitored by phosphorimaging (Fuji Photo Film Co.), which has a detection limit of the equivalent of 1 ng of our labeled protein.
TSEM.
Transfected 293T cells were harvested 36 h posttransfection and processed for TSEM essentially as described previously (27). Briefly, cells were washed in cacodylate (CAC) buffer (pH 7.3) and pelleted by centrifugation at 500 × g for 5 min. The cell pellet was then fixed in 2% (wt/vol) glutaraldehyde-CAC buffer for 1 h at 4°C. After several washes in CAC buffer, the pellet underwent secondary fixation in 1% (wt/vol) osmium tetroxide-CAC buffer for 1 h at 4°C. Pellets were rinsed in distilled water, dehydrated in graded ethanols, cleared in propylene oxide, and embedded in Spurr resin (Ladd Research, Inc., Williston, Vt.). Ultrathin sections with silver and gold interference colors were mounted on uncoated 200-mesh copper grids and stained with uranyl acetate and lead citrate. The sections were then examined with a CM12 electron microscope (Philips).
RESULTS
Reduction in synthesis of the Gag-Pro-Pol protein is associated with a decrease in p24-CA.
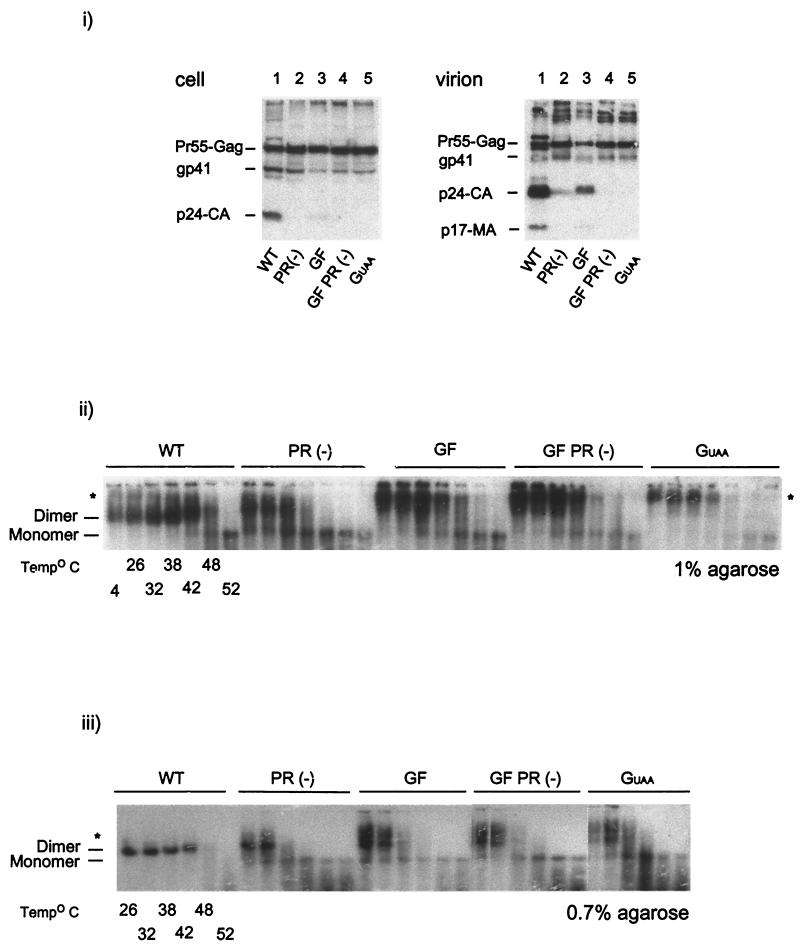
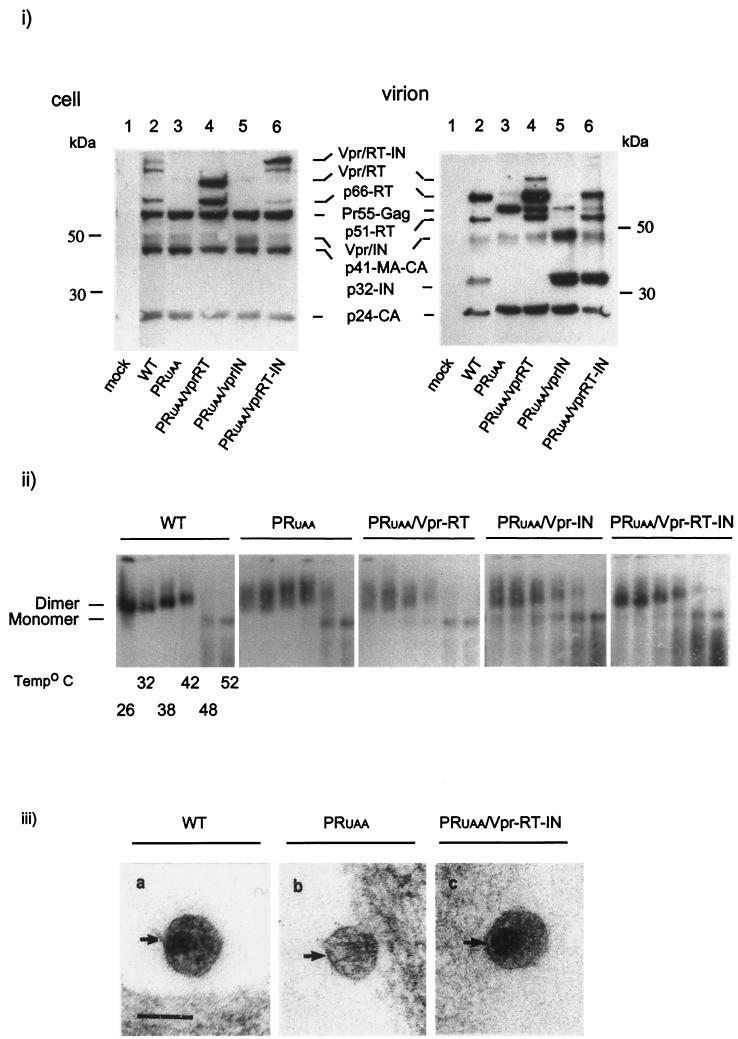
293T cells were transfected with the WT control, HXB2-BH10, or mutants derived from this infectious clone, namely GF, GF PR(-), and GUAA, and a PR− control (Fig. 1). The PR− control showed no proteolytic processing of Gag and Gag-Pro-Pol polyproteins due to inactivation of viral PR (Fig. 2i, virion, lane 2). Mutants GF, GF PR(-), and GUAA each restrict Pol protein synthesis. Assessment by Western blot analysis showed intracellular protein profiles that were consistent with the introduced mutations (Fig. 2i, cell). Restricting the synthesis of Pol proteins resulted in a corresponding decrease of p24-CA in VLPs (Fig. 2i, virion). The GF construct, which contains a mutation that alters the frameshifting site within the HIV-1 genome, generated a 2.5-fold decrease in p24-CA in VLPs compared to the level in WT virions as determined by phosphorimaging (data not shown), demonstrating that this mutation hinders but does not abolish Gag-Pro-Pol synthesis (Fig. 2i, virion, lane 3). The GF PR(-) construct produced no p24-CA due to the mutation of the PR active site within the GF. As the GUAA construct did not support Gag-Pro-Pol synthesis, both cellular and virion protein profiles showed only Gag (Fig. 2i, cell and virion, lane 5, respectively).
FIG. 1.
Schematic representation of proviral DNA constructs used in the study. The WT plasmid is HxB2-BH10 and has been previously described (42). A PCR-amplified fragment was cloned into the HxB2-BH10 proviral DNA via restriction sites ApaI and BclI to alter −1 ribosomal frameshifting (GF). To obtain the GF PR(-) mutant, the aspartic acid at the active site of PR of the GF construct was replaced with asparagine via PCR mutagenesis. A stop codon introduced within the frame of the Pol protein terminated Gag-Pro-Pol synthesis in amino acid 13 of PR (GUAA) and abolished PR activity.
FIG. 2.
The impact of mutations within the Gag and Gag-Pro-Pol regions of HIV-1 on the protein compositions of purified virions and genomic RNA dimer maturation. (i) Viral proteins were semiquantified by Western blot analysis. Purified virions were resolved by 10% Tris-glycine SDS-PAGE. Resolved proteins were probed with pooled sera from HIV-1-infected individuals as described in Materials and Methods. (ii) The impact of mutations within the Gag-Pro-Pol region in RNA dimer formation and stability was determined by using a melting curve combined with electrophoretic analysis of WT and mutant dimers. Virion RNA was resuspended in RNA dimerization buffer and heat denatured for 10 min at the indicated temperatures. Dimers and monomers were electrophoresed in a 1% native agarose gel and probed with an HIV-1 riboprobe, as described in Materials and Methods. (iii) Electrophoresis using a lower percentage of agarose gel (0.7%) further resolved the slow-migrating genomic RNAs into doublets. Stars indicate the location of the slow-migrating dimers.
Genomic RNAs isolated from Gag-derived particles display altered mobility in comparison to mature WT genomic RNA dimers and immature PR− RNA dimers.
The WT genomic virion RNA migrated as sharp dimeric RNA and started dissociating into monomeric RNA at 48°C (Fig. 2ii) (13). In contrast, the immature RNA dimers in PR− virions migrated with slightly less mobility a more diffused pattern and dissociated into monomer RNA at approximately 5 to 10°C below the temperature at which WT RNA dissociated. However, genomic RNA isolated from mutant virion particles that have reduced levels of Gag-Pro-Pol [such as GF and GF PR(-)] or that lack Gag-Pro-Pol (i.e., GUAA) migrated in a 1% agarose gel with mobilities that are distinct from those for both the WT mature and PR− immature RNA dimers (Fig. 2ii). These mutant RNA dimers also exhibited stability lower than that of WT genomic RNA but comparable to that of PR− genomic RNA. Interestingly, it was noted that the slower-running RNA bands, observed in the genomic RNA, derived from GF, GF PR(-), and GUAA virions were also detected in small amounts within the WT genomic RNA samples. Electrophoresis using a gel with a lower percentage of agarose (0.7%) further resolved the slow-migrating genomic RNAs into doublets (Fig. 2ii). Differences in the migration mobilities observed for these RNAs by using 1 and 0.7% agarose may, in part, be due to variations in resistance of agarose gels. The higher-migrating RNA band within the doublet appeared to be present in small amounts in the unheated PR− sample (Fig. 2ii). The proportion of this slow-migrating genomic RNA was, however, inversely correlated with the presence of Gag-Pro-Pol in these mutant virion particles.
The slow RNA dimer migration of Gag-derived virions is not due to cellular or viral protein contamination.
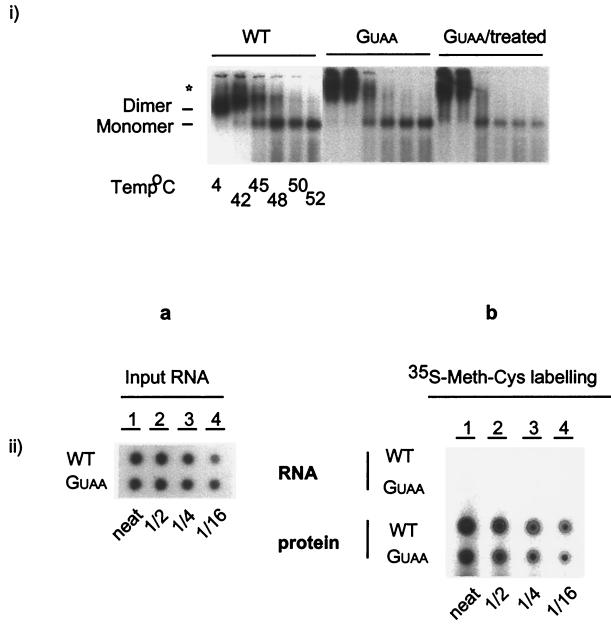
No detectable levels of viral proteins were present in the RNA samples as determined by Western blot analysis (data not shown). Additional proteinase K treatment of the GUAA-derived genomic RNA was used to rule out the presence of a protein-RNA aggregation within the GUAA RNA preparations as the cause of their low electrophoretic mobility (Fig. 3i). RNA dimerization analysis of samples before and after extended proteinase K treatment showed no differences in migration or stability among the Gag-derived genomic RNAs. In addition, the more sensitive procedure of [35S]Met-Cys metabolic labeling was employed. The genomic virion RNA was prepared from an equivalent of approximately 13 μg of labeled HIV-1 proteins, as measured by the Lowry assay. While 35S labeling routinely yielded approximately 124,800 cpm/μg of protein, no radioactive proteins were detected in the genomic RNA samples (Fig. 3ii). The phosphorimager used for the monitoring the 35S radioactivity present in HIV-1 proteins has a detection limit of as low as 1 ng of labeled protein. Thus, protein contamination is not responsible for the slow migration of the Gag-derived genomic RNA (GUAA).
FIG. 3.
Metabolic labeling of virion proteins and electrophoretic analysis of proteinase K (PK)-treated genomic RNAs. (i) WT, GUAA, and PK-treated GUAA RNA, isolated as described in Materials and Methods, was treated at the indicated temperatures for RNA dimerization analysis. The star indicates the location of the slow-migrating dimers. (ii) Twofold dilutions of equal amounts of 35S-labeled virion proteins and genomic RNA were loaded in a nitrocellulose (Hybond N) membrane. Results were visualized by autoradiography.
Proteolytic processing of Gag alone is sufficient for genomic RNA dimer stability.
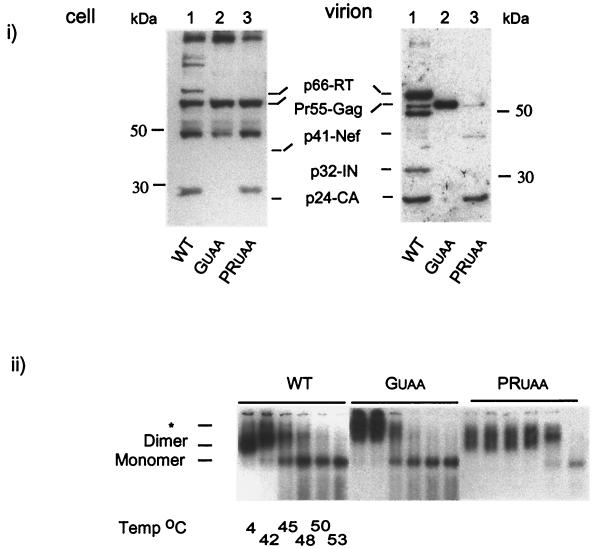
As RNA rearrangement in a dimeric conformation with optimal stability occurs spontaneously with or immediately after Gag processing by PR, we attempted to determine the role of the Gag proteolysis by PR in RNA dimer maturation, in the absence of Gag-Pro-Pol. We introduced an active PR into the GUAA construct to generate PRUAA, which produces VLPs with mature Gag proteins without RT and IN (Fig. 4i). The genomic RNA extracted from the PRUAA viral particles showed stability similar to (and sometime better than) that of the WT genomic RNA (Fig. 4i). However, the migration pattern of these RNAs in 1% agarose gel was not completely restored to that of the WT, with a high-molecular-weight band that often appeared as a doublet (Fig. 4ii). The phenotype of these PRUAA RNA dimers resembled one representative of an intermediate state between the GUAA and WT RNA genome. These results suggest that the proteolytic processing of Gag alone in VLPs is sufficient for genomic RNA dimer stability but not for restoring WT RNA dimer conformation.
FIG. 4.
Efficient Gag processing by PR of Gag and its effect on RNA dimer formation and stability. (i) Intracellular viral protein levels and total virion protein levels were standardized by EGFP and dot blot analysis, respectively (data not shown). Transfected cell lysates and purified virions (see Materials and Methods) were resolved by 10% Tris-glycine SDS-PAGE. Resolved proteins were probed with pooled sera from HIV-1-infected individuals as described in Materials and Methods. Lanes 1, WT cellular viral protein (cell) and virion protein (virion) profiles; lanes 2, cellular viral protein and virion protein profiles of GUAA; lanes 3, cellular viral proteins and virion protein profiles of PRUAA. (ii) To examine RNA dimer formation and stability, genomic RNA isolated from the WT and GUAA and PRUAA mutants was heated to the indicated temperatures and electrophoresed in a 1% agarose gel, transferred, and probed with a HIV-1 riboprobe. Results were visualized by autoradiography. The star indicates the location of the slow-migrating dimers.
Proteolytic processing of HIV-1 Gag alone is not sufficient for the maturation of the virion core.
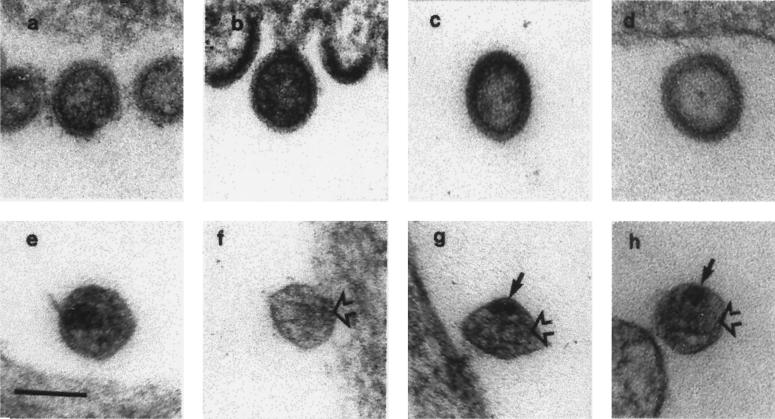
Consistent with previous reports (21), both immature and mature virions were detected in the WT preparations by electron microscopy (EM) analysis (Fig. 5d and e). However, only immature virions were detected within the PR− and GUAA preparations (Fig. 5a and b). Immature particles were round or ovoid in shape and bound by two outer concentric layers (Fig. 5a to d). These particles measured about 90 to 110 nm in diameter. The distance between the two outer layers was about 10 nm. PRUAA preparations contained particles with cone-shaped aberrant cores (Fig. 5f to h) and immature virions resembling the PR-defective particles (Fig. 5c). In longitudinal sections of the WT viral particles, the central core structure extended along the particle and frequently contained dark-staining cone-shaped core material (Fig. 5e). Although similar to WT virions, the PRUAA virions had a cone-shaped core structure (Fig. 5f to h); the electron-dense dark-staining bodies were absent from the inside of the cores but could occasionally be found adjacent to the empty core (Fig. 5g and h). Particles in both WT and PRUAA preparations were round or oval and measured about 90 to 95 nm in diameter.
FIG. 5.
EM analysis of the mutant viral particles. Thin-section electron micrographs of the main HIV-1 particle types detected in the three mutant preparations and the control preparation. (a) Immature-particle PR− preparation; (b) immature-particle GUAA preparation; (c) immature-particle PRUAA preparation; (d) immature-particle WT control preparation; (e) mature-particle WT control preparation; (f to h) mature-particle PRUAA preparations. The core typically appeared empty (open arrows) although a dark-staining body was sometimes seen adjacent to the core (solid arrows). Bar = 100 nm (same scale for all panels).
Supplementation of the RT-IN complex in trans restores the conformation of HIV-1 RNA dimers and the virion core.
We cotransfected the PRUAA plasmid with Vpr-RT, Vpr-IN, or Vpr-RT-IN constructs to express RT and IN individually or simultaneously as a fusion protein (RT-IN). Lysates of cells cotransfected with PRUAA and the Vpr-RT expression vectors showed a band with a molecular mass of approximately 81 kDa, consistent with the molecular mass of Vpr-RT, as well as a band with a molecular mass 66 kDa (RT) at the same position as WT RT (Fig. 6i, cell, lane 3). Cotransfection with PRUAA and Vpr-IN resulted in a band of approximately 47 kDa, which is consistent with the molecular mass of the Vpr-IN fusion (Fig. 6i, cell, lane 4). Lysates of cells cotransfected with the PRUAA and Vpr-RT-IN expression vectors showed overall intracellular protein patterns similar to that of the WT and an additional band (Vpr-RT-IN) of approximately 120 kDa (Fig. 6i, cell, lane 5). The immunoblotting of the purified virions showed efficient processing and virion packaging of RT, IN, and RT-IN (Fig. 6i, virion, lanes 4 to 6, respectively). A small amount of unprocessed Vpr-RT complex previously observed in the cellular lysates was also detected in the virions packaging RT in trans (Fig. 6i, virion, lane 4). These results are consistent with the initial report that showed successful packaging and proteolytic processing of RT, IN, and RT-IN in the virions when provided in trans as part of a Vpr fusion (45).
FIG. 6.
Supplementation with RT, IN, and RT-IN in trans: cellular expression and virion incorporation and processing. Provision of RT-IN in trans restores virion RNA conformation and core condensation. (i) Similar amounts of intracellular and virion proteins were resolved by 10% Tris-glycine SDS-PAGE and gel electrophoresis. Transferred proteins were probed with HIV-1 sera to assess the presence of Vpr-RT, Vpr-IN,and Vpr-RT-IN in transfected cells and the levels of RT and IN packaged within the virions. (ii) The effect of supplementation with RT, IN, and RT-IN within PRUAA on conformational changes of genomic RNA was examined by melting curve and electrophoretic analyses of WT and mutant RNAs. Samples were electrophoresed in 1% native agarose gel and probed with an HIV-1 riboprobe as described in Materials and Methods. (iii) Virions packaging RT-IN provided in trans as a Vpr fusion protein show a morphology similar to that of the WT (condensed, electron-dense cores) and distinct features, unlike virions deficient in RT and IN (empty cone-shaped cores).
Genomic RNA isolated from virions showing incorporation of Vpr-RT, Vpr-IN, and Vpr-RT-IN fusion proteins (Fig. 6ii) were assayed for RNA dimer formation and stability. Provision of RT and IN separately as Vpr fusion proteins did not rescue the dimeric conformation of the genomic RNA in these mutant particles. However, the genomic RNA isolated from PRUAA/Vpr-RT-IN particles displayed dimer stability and electrophoretic mobility similar to that of the dimeric RNA in WT HIV-1 (Fig. 6ii). Furthermore, the empty cone-shaped core in PRUAA was also restored to WT upon the supplementation of Vpr-RT-IN fusion protein (Fig. 6iii), demonstrating the importance of the HIV-1 Pol protein in the complete maturation of the virion RNA dimers and the formation of the mature virion core.
DISCUSSION
We have identified a high-molecular-weight, unstable form of viral genomic RNA, which may be an intermediate conformation of the RNA dimers that occurs prior to the full maturation of the dimeric RNA genome. We further show here that Gag proteolysis stabilizes RNA dimers but fails to facilitate the complete rearrangement of genomic RNA into the dimeric form found in the WT. Our results also substantiate current evidence that RNA packaging is distinct from RNA dimerization and highlight the importance of Gag-Pro-Pol in RNA dimer maturation during HIV-1 assembly. The provision of RT and IN in trans as a Vpr-RT-IN fusion protein restores the conformation of the genomic RNA dimers to that found in WT virions and also restores the electron-dense cone-shaped core. Data presented here reconfirm the strong correlation between RNA dimer maturation and core condensation that we have reported recently (39).
The process of RNA packaging and dimerization is directly linked to Gag, whether in its precursor form as the Gag polyprotein or following processing by viral PR to its constituent proteins (2, 29). Extensive studies have shown that specific and nonspecific binding of the basic residues of Gag to the HIV-1 RNA genome ensures proper packaging of the viral genomic RNA within the virion. The N terminus of NC, including some specific basic residues and its zinc finger(s), has a distinctive role in RNA dimerization and packaging in Moloney murine leukemia virus, Rous sarcoma virus, and HIV-1 (3, 4, 7, 9, 16, 17, 39). The finding that PR-defective viral particles encapsidate dimeric RNA (13) demonstrates that Gag is sufficient for the initiation of the RNA dimerization process. However, the in vivo role of the Gag precursor alone in dimer maturation has not been described. While Gag-derived particles packaged normal levels of viral genomic RNA, the dimeric RNA is less stable than that of the WT and migrates more slowly in a native gel. The lower stability of these RNA dimers is consistent with the low stability of RNA dimers in PR-defective immature HIV-1 described by Fu et al. (13). The altered mobility of virion genomic RNAs in GUAA is clearly distinct from that of virion RNA dimers found in WT mature and PR-defective immature particles. To our knowledge, the genomic RNA complexes found in GUAA have not previously been described.
Our analysis ruled out viral protein contamination as the cause of this altered mobility. However, several groups have shown that the in vitro assembly of HIV-1 and other retroviruses can be facilitated by nonviral RNAs (5, 6, 33). Therefore, it is possible that RNAs of cellular origin could potentially form aggregates with the viral genomic RNA, thus altering the conformation of the packaged dimeric RNA within these particles. Muriaux et al. have recently demonstrated that virion packaging of cellular RNA can occur in viral RNA packaging-deficient Moloney murine leukemia virus, thus compensating for the lack of nascent virion RNA (35). Yet, we found no major differences in viral genomic RNA packaging between GUAA and WT particles; consequently, it appears that the altered mobility of viral RNA species found in the Gag-derived particles is due to genomic RNA that has a conformation different from that of WT RNA. Further analysis of these RNA dimers showed the presence of some dimers resembling the WT RNA dimers and others that migrate more slowly than WT dimers on a lower-percentage nondenaturing gel. Laughrea and Jette have previously shown that two forms of RNA dimers (tightly and loosely associated) can be generated via in vitro RNA dimerization (26). While it is difficult to compare the two in vitro forms of RNA dimers described by Laughrea and Jette with the distinct forms of RNA dimers shown in this study, it is possible that our respective studies refer to RNA dimers with similar conformations.
Introduction of an active PR to the Gag-derived particles (PRUAA) restored dimer stability. This is consistent with previous reports (10, 11, 34, 38, 39) which show that genomic RNA dimer stability is under the control of mature NC. However, in our study, while immature dimers were formed in the complete absence of the Pol protein and the active PR restored dimeric RNA stability to WT levels, Gag proteolysis did not correct the RNA conformation. This finding suggests that regions in the Pol protein other than PR are also important for coordinating the arrangement of viral genomic RNA.
Several studies have collectively demonstrated that Gag-Pro-Pol synthesis and its incorporation within the viral particle are critical for HIV-1 function or morphogenesis (23, 25, 37, 38). The 20:1 ratio of Gag to Gag-Pro-Pol production supports the greater requirement for structural proteins over enzymatic proteins during viral assembly, and maintenance of the ratio is critical for viral function (23, 38). It has been shown that Gag-Pro-Pol interacts with Gag during virus particle formation (36, 40). Specific viral functions linked to Gag-Pro-Pol synthesis and incorporation include proteolytic processing, reverse transcription, integration, and tRNA3Lys packaging (30-32). In a recent study we reported that efficient incorporation of Gag-Pro-Pol in trans supports the enzymatic function of HIV-1 (20). Here, to assess the role of Gag-Pro-Pol-derived enzymes RT and IN in RNA dimer maturation, independently of the Gag-Pro-Pol precursor, we provided these proteins in trans during cotransfection with PRUAA either individually as Vpr-RT and Vpr-IN or as fusion protein Vpr-RT-IN (45). While the packaging of either RT or IN did not fully restore RNA dimer conformation, the packaging of the RT-IN fusion protein generated RNA dimers that are similar to those of WT HIV-1. By showing that the genomic RNA structure of the RT-IN-supplemented virions closely resembles that of the WT virus, our data suggest a specific role for RT-IN in RNA rearrangement during viral assembly and viral particle maturation.
It has been proposed that RT interacts with NC in order to form the RNP complex and that a direct RT-NC interaction is required for reverse transcription in vivo (44). It is possible that RT and NC interact at different stages of viral assembly, contributing not only to the formation of the RNP complex but also to the maturation of the virion core. A role for RT and IN in virion core formation is supported in our study by the results of EM analysis of the Gag-derived particles. While Gag-derived virions appeared as empty immature particles, the processed Gag particles (PRUAA) were detected as aberrant virions with empty but cone-shaped cores. As expected, provision of RT-IN as part of Vpr fusion resulted in the formation of electron-dense cone-shaped cores. Our data underline the importance of Gag-Pro-Pol in virion morphogenesis by showing that Gag-Pro-Pol and in particular RT-IN are critical for the formation of the infectious virion core.
Acknowledgments
We thank John Mills for the critical review of the manuscript and Jean-Luc Darlix, Nicolas Deacon, and Damian Purcell for helpful discussions and suggestions. We thank Katherine Kedzierska for her help with the protein quantification analysis.
Miranda Shehu-Xhilaga is a recipient of a NHMRC Dora Lush Ph.D. scholarship. Melissa Hill is a recipient of a Burnet Centenary postdoctoral fellowship. Suzanne M. Crowe is supported by a grant from the Australian National Council of HIV/AIDS and Related Diseases, the Australian National Centre in HIV Virology Research, and the MBC Research Fund. Johnson Mak is a recipient of an NHMRC Peter Doherty postdoctoral fellowship.
REFERENCES
- 1.Ansari-Lari, M. A., and R. A. Gibbs. 1996. Expression of human immunodeficiency virus type 1 reverse transcriptase in trans during virion release and after infection. J. Virol. 70:3870-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz, R. D., J. Luban, and S. P. Goff. 1993. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 67:7190-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burniston, M. T., A. Cimarelli, J. Colgan, S. R. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding site for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 9.Darlix, J.-L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 10.Feng, Y.-X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, Y.-X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 10:1-15. [DOI] [PubMed] [Google Scholar]
- 13.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorelick, R. J., J. S. M. Nigida, J. J. W. Bess, L. O. Arthur, L. E. Henderson, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, L. E., M. A. Bowers, R. Sowder, S. A. Serabyn, D. G. Johnson, J. J. Bess, L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, M. K., C. W. Hooker, D. Harrich, S. M. Crowe, and J. Mak. 2001. Gag-Pol supplied in trans is efficiently packaged and supports viral function in human immunodeficiency virus type 1. J. Virol. 75:6835-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hockley, D. J., R. D. Wood, J. P. Jacobs, and A. J. Garrett. 1988. Electron microscopy of human immunodeficiency virus. J. Gen. Virol. 69:2455-2469. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., A. Khorchid, J. Wang, M. A. Parniak, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1997. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into human immunodeficiency virus type 1. J. Virol. 71:4378-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, M., P. Patel, S. Davis, and S. R. Green. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 Gag-Pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 26.Laughrea, M., and L. Jette. 1997. HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5′ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry 36:9501-9508. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. Y., J. A. Marshall, and D. S. Bowden. 1992. Replication complexes associated with the morphogenesis of Rubella virus. Arch. Virol. 122:95-106. [DOI] [PubMed] [Google Scholar]
- 28.Liu, H., X. Wu, H. Xiao, J. A. Conway, and J. C. Kappes. 1997. Incorporation of functional human immunodeficiency virus type 1 integrase into virions independent of the Gag-Pol precursor protein. J. Virol. 71:7704-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mak, J., A. Khorchid, Q. Cao, Y. Huang, I. Lowy, V. R. Prasad, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 1997. Effects of mutations in Pr160gag-pol upon tRNA3Lys and Pr160gag-pol incorporation into HIV-1. J. Mol. Biol. 265:419-431. [DOI] [PubMed] [Google Scholar]
- 32.Marquet, R., C. Isel, C. Ehresmann, and B. Ehresmann. 1995. tRNAs as primer of reverse transcriptase. Biochimie 77:113-124. [DOI] [PubMed] [Google Scholar]
- 33.Morikawa, Y., T. Goto, and K. Sano. 1999. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J. Biol. Chem. 274:27997-28002. [DOI] [PubMed] [Google Scholar]
- 34.Muriaux, D., H. De Rocquigny, B. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 35.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retroviral particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, J., and C. D. Morrow. 1992. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J. Virol. 66:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, J., and C. D. Morrow. 1991. Overexpression of the Gag-Pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehu-Xhilaga, M., H. G. Kraeusslich, R. Swanstrom, S. Pettit, J.-Y. Lee, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the P2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, A. J., N. Srivivasakumar, M.-L. Hammarskjöld, and D. Rekosh. 1993. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J. Virol. 67:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanstrom, R., and J. W. Wills. 1997. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Terwilliger, E. F., E. A. Cohen, Y. C. Lu, J. G. Sodroski, and W. A. Haseltine. 1989. Functional role of human immunodeficiency virus type 1 Vpu. Proc. Natl. Acad. Sci. USA 86:5163-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral Gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 44.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]