Abstract
The fusion (F) proteins of most paramyxoviruses are classical type I glycoproteins with a short hydrophobic leader sequence closely following the translation initiation codon. The predicted reading frame of the canine distemper virus (CDV) F protein is more complex, with a short hydrophobic sequence beginning 115 codons downstream of the first AUG. To verify if the sequence between the first AUG and the hydrophobic region is translated, we produced a specific antiserum that indeed detected a short-lived F protein precursor that we named PreF0. A peptide resulting from PreF0 cleavage was identified and named Pre, and its half-life was measured to be about 30 min. PreF0 cleavage was completed before proteolytic activation of F0 into its F1 and F2 subunits by furin. To test the hypothesis that the Pre peptide may influence protein activity, we compared the function of F proteins synthesized with that peptide to that of F proteins synthesized with a shorter amino-terminal signal sequence. F proteins synthesized with the Pre peptide were more stable and less active. Thus, the Pre peptide modulates the function of the CDV F protein. Interestingly, a distinct two-hit activation process has been recently described for human respiratory syncytial virus, another paramyxovirus.
Canine distemper virus (CDV) is a member of the genus Morbillivirus within the family Paramyxoviridae, order Mononegavirales. The genomes of morbilliviruses consist of single-stranded negative-sense RNA of about 16,000 nucleotides (nt) (9, 27). Six cistrons are transcribed consecutively, and from their mRNAs six structural and two nonstructural viral proteins are translated. Their coding regions are flanked by 5′ and 3′ untranslated regions (UTRs) that contain regulatory elements. In CDV, as in other morbilliviruses, the 5′ UTRs are 20 to 60 nt long. The exception is the longer region upstream of the predicted fusion (F) protein reading frame (5, 27).
The F proteins of paramyxoviruses, including most morbilliviruses, are classical type I glycoproteins, with a short hydrophobic leader sequence following the AUG translation initiation codon and a second hydrophobic sequence shortly preceding the stop codon. In the predicted F protein reading frame of CDV, however, a first AUG is situated 85 nt downstream of the 5′ end of the F mRNA, a second potential start codon follows at position 266, and the first codon of the hydrophobic leader-like sequence is at nt 428. Furthermore, there is an additional nonconserved in-frame AUG at position 461 of the large-plaque-forming variant of the vaccine strain Onderstepoort, which was initially discussed as the most probable site of translation initiation (7). However, it was recently shown that either one of the two conserved in-frame methionines (amino acid positions 1 and 61) is necessary for efficient F protein expression (6).
The comparative analysis of the sequence of different CDV strains has yielded interesting insights. In contrast to the high sequence conservation of the mature F protein, the nucleotide sequence between the first AUG and the predicted signal peptidase cleavage site at residue 137 varies by up to 21.4% (7, 15). Furthermore, a GC content of about 60% suggests extensive folding of the RNA (15). In addition to the two conserved in-frame start codons, two out-of-frame start codons are located at nt positions 228 and 408, of which the second is conserved among all strains (7, 15).
Processing of a classical type I glycoprotein involves recognition of the signal peptide by the signal peptide recognition particle, and subsequent translocation into the endoplasmic reticulum (ER), after which the signal peptide is cotranslationally cleaved. Protein maturation continues during transport through the ER and Golgi apparatus to the cell surface (reviewed in reference 17). Even though the sequences of different signal peptides share no common motifs, some structural requirements have been identified. The amino-terminal part ranges in length between 8 and more than 50 amino acids contains mostly positively charged residues. It is followed by the central hydrophobic region of 6 to 20 residues that will be inserted into the ER membrane, which is followed by another polar region. This domain contains helix breaking as well as small uncharged residues in positions −3 and −1, which determine the signal peptidase cleavage site (30). Interestingly, it has been shown for several viral glycoproteins that signal peptide cleavage can occur very late after translocation (12, 16). This posttranslational cleavage is inefficient and is thought to regulate the amount of functional protein and to optimize viral production (14, 16).
In this study, we examined the events that occur during synthesis of the CDV F protein in a transient-expression system as well as in the viral context. The functionality of F proteins with mutations and truncations in the amino-terminal sequence was assessed in a transient-expression assay. The mutations were then introduced into the CDV genome, recombinant viruses were recovered, and their phenotypes were examined.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATCC CCL-81) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 5% fetal calf serum (FCS). 293 cells (ATCC CRL-1573) were maintained in the same medium with 10% FCS. DH 82 cells (ATCC CRL-10389) were cultured in Eagle's minimal essential medium with nonessential amino acids and 15% FCS. All tissue culture media as well as supplements and FCS were purchased from Life Technologies. The small-plaque-forming variant of CDV Onderstepoort (CDVOS) and all recombinant viruses were propagated in Vero cells.
Construction of expression plasmids containing different mutants in the 5′ region of the F gene.
The plasmid pCG-FOS constituted the basis for all the mutants. The first and second in-frame start codons (residues 1 and 61) were changed to leucines (TTA) by site-directed mutagenesis (Quick-Change site-directed mutagenesis kit; Stratagene) either separately or in combination, resulting in pCG-FOSL1 (first ATG → TTA), pCG-FOSL61 (second ATG → TTA), and pCG-FOSml1/61 (first and second ATG → TTA).
The mutants with increasing deletions in the 5′ region of the F gene (pCG-FOSΔ60, pCG-FOSΔ107, and pCG-FOSΔ114) were generated by PCR (Expand high-fidelity PCR system; Roche Biochemicals) with the forward primers 5′-TTTGGATCCGGCGCGCCATGAACAGGACCAGGTCCCGCAAGC-3′ (pCG-FOSΔ60), 5′-TTTGGATCCGGCGCGCCCCAATGGCAATCAACTCAGGCTCTC-3′ (pCG-FOS Δ107), and 5′-TTTGGATCCGGCGCGCCATGTGCACCTGGTTAGTCCTGTGGTGC-3′ (pCG-FOSΔ114), which introduce a BamHI and AscI site following the coding region (underlined), and the common reverse primer 5′-TGAAGTATTCTGGTCATATATCTCGCATGCATGTCCAAA-3′, which adds a SphI site (underlined) downstream of the open reading frame (ORF). In pCG-FOSΔ107 and pCG-FOSΔ114, an artificial ATG was introduced (bold letters). The correct sequences of all constructs were confirmed (ABI Prism 377 DNA sequencer; Perkin-Elmer Applied Biosystems).
Construction of CDV genomic full-length plasmids with alterations in the 5′ region of the F gene.
To facilitate the construction of full-length CDV plasmids with alterations in the 5′ region of the F gene, a unique restriction site was introduced in the 5′ UTR (AscI; nt 7046 to 7053) of the F ORF downstream of the first ATG by site-directed mutagenesis. The resulting plasmid, which also contains unique restriction sites upstream and downstream of the H ORF (31), was named pCDVII.
The mutants pCG-FOSL1, pCG-FOSL61, and pCG-FOSml1/61 were amplified from the pCG plasmids described above by using the forward primer 5′-TTTGGCGCGCCAGCCAGGGGCTGGAC-3′, which introduced an AscI site (underlined) upstream of the respective F coding region, and the reverse primer mentioned above. The PCR products were cloned into pCDVII by using the AscI site and an endogenous unique AflII site at positions 6669 to 6704. The resulting plasmids were named pCDV-FOSL1, pCDV-FOSL61, and pCDV-FOSL1/61, respectively. The constructs pCG-FOSΔ60, pCG-FOSΔ107, and pCG-FOSΔ114 already contained an AscI site upstream of the respective ATG in a way that respected the rule of six (21). Therefore, the inserts were generated by digesting the pCG plasmids with AscI and AflII and introducing fragments into pCDVII, yielding pCDV-FOSΔ60, pCDV-FOSΔ107, and pCDV-FOSΔ114. The sequences were confirmed.
Fusion assay.
A quantitative fusion assay based on the luciferase gene as the reporter gene and similar to that described previously (22) was established. To generate a suitable reporter gene plasmid, the luciferase gene (pGL2-Control vector; Promega) was subcloned into the pTM1 vector (20), in which an internal ribosomal entry site is located downstream of the T7 promoter to ensure efficient translation of the RNA transcribed by the T7 polymerase, yielding pTM1-luc. Vero cells were transfected with the different F expression plasmids together with pCG-HOS and pTM1-luc using a molar ratio of 1:1:0.7. Lipofectamine 2000 (Gibco BRL) was used as the transfection reagent, following the protocol of the supplier. Briefly, cells were seeded in 24-well plates so that they reached about 80% confluence for transfection. For each well to be transfected, 1.3 μg of DNA was diluted in 50 μl of OptiMEM (Gibco BRL). Another 50 μl of OptiMEM containing 2 μl of Lipofectamine 2000 was added, and the mixture was incubated at room temperature for 30 min. Before the solution was added to the cells, the culture medium was removed and replaced with 0.5 ml of DMEM without serum. For each well transfected, a second well of Vero cells was infected with modified vaccinia virus Ankara expressing the T7 polymerase (MVA-T7) (28) with a multiplicity of infection (MOI) of 1 at the time of transfection. Twelve hours after transfection or infection, the cells were washed twice with phosphate-buffered saline (PBS; Gibco BRL), and 50 μl of 0.25% trypsin-EDTA (Gibco BRL) was added to detach the cells. After incubation at 37°C for 5 min, each well of cells was resuspended in 1 ml of DMEM supplemented with 5% FCS, and the cells of one transfected and one infected well were mixed and centrifuged at 230 × g for 10 min. The pellet was resuspended in 2 ml of fresh DMEM with 5% FCS, transferred into two wells of a 24-well plate, and incubated for 36 h at 37°C. Following the visual grading of the fusion activity, the luciferase activity was determined with a luciferase assay system (Promega) and a 96-well plate-reading luminometer (Microlumat LB96P; EG & G Berthold). A fraction of each lysate was mixed with an equal amount of 2× Laemmli sample buffer (Bio-Rad) containing 0.5% β-mercaptoethanol and subjected to Western blot analysis.
Western blot analysis.
Vero cells were seeded into six-well plates, transfected with the different constructs or infected with virus at an MOI of 0.01, and incubated at 37°C for 48 h or until cytopathic effect (CPE) was observed. Cells were washed twice with PBS before the addition of 0.5 ml of lysis buffer (150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8.0]) with complete protease inhibitor (Roche Biochemicals) to each well. After incubation for 30 min at 4°C, the lysates were cleared by centrifugation at 5,000 × g for 15 min at 4°C and the supernatant was mixed with an equal amount of 2× Laemmli sample buffer (Bio-Rad) containing 0.5% β-mercaptoethanol. Samples were incubated for 10 min at 95°C, fractionated on SDS-10% polyacrylamide gels (Bio-Rad), and blotted on polyvinylidene difluoride membranes (Millipore). After blocking with 1% blocking reagent (Roche Biochemicals) overnight, the membranes were incubated with a rabbit antipeptide serum which recognizes the 14 carboxy-terminal residues of the CDV and measles virus F protein (Fcyt) (4). Following the incubation with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antiserum, the membranes were subjected to enhanced chemiluminescence detection (Amersham Pharmacia Biotech).
Recovery of recombinant viruses.
The recombinant viruses were recovered as described before (31) with an MVA-T7 based system (26). The first syncytia were observed 7 to 10 days after transfection. For each virus, three syncytia were picked and transferred onto fresh Vero cells in six-well plates. These infected cells were expanded into 75-cm2 flasks with 10 ml of DMEM supplemented with 2% FCS. When the CPE was pronounced, the cells were scraped into the medium and subjected once to freezing and thawing. The cleared supernatants were used for all further analysis.
Indirect immunofluorescence assay.
Subconfluent Vero cells were either transfected with pCG-FOS by using Lipofectamine 2000 as described above or infected with rCDVOS at an MOI of 0.01 and incubated for 48 h at 37°C. Then the cells were either shifted to 4°C and incubated unfixed with the primary antibody for 1 h or fixed with 2% paraformaldehyde, blocked with 0.5 M glycine, and permeabilized with 0.1% Triton X-100 before incubation with the primary antibody for 60 min at room temperature. Three primary antibodies were used for this experiment: the anti-Fcyt antiserum described above (1:200 dilution), a rabbit antiserum MC709 raised against the C-terminal residues (201 to 224) of the F2 subunit (1:100 dilution), and a rabbit antiserum (MC829) raised against residues 88 to 112 of the signal peptide of the FOS protein (1:100 dilution), which were generated by immunizing a rabbit with the respective keyhole limpet hemocyanin-coupled peptide. After incubation with the primary antibody, the cells were carefully washed twice and fixed as described above. The staining was performed with fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G (Amersham Pharmacia Biotech).
Radioimmunoprecipitation and pulse-chase analysis.
For each antiserum and time point, one well of a six-well plate seeded with Vero cells was transfected with the construct of interest using Lipofectamine 2000. Thirty-six hours after transfection, the cells were washed twice with PBS, and 2 ml of DMEM without glutamine, methionine, or cysteine was added. After incubation at 37°C for 1.5 h, the medium was exchanged for 1 ml of DMEM without glutamine, methionine, or cysteine, and 100 μCi of [35S]methionine (Amersham Pharmacia Biotech) was added to each well.
For steady-state analysis, the cells were labeled for 1.5 h at 37°C, washed three times with cold PBS, and lysed with 500 μl of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8.0]) with protease inhibitors (Complete; Roche Biochemicals) for 20 min at 4°C. The lysate was transferred into an Eppendorf tube and cleared at 5,000 × g for 15 min at 4°C, the supernatant was added to 50 μl protein A agarose beads (Bio-Rad), and the antibody was added at the appropriate concentration. After incubation at 4°C overnight, the beads were washed three times in RIPA buffer before 30 μl of 2× Laemmli sample buffer (Bio-Rad) containing 0.5% β-mercaptoethanol was added, and the samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) analysis, using a gel with a polyacrylamide concentration appropriate for the protein of interest. The gels were dried for 1 to 1.5 h at 70°C and exposed for 3 to 16 days using Biomax films (Kodak).
For pulse-chase experiments, cells were labeled for 30 min, washed three times with prewarmed PBS, and incubated with DMEM supplemented with 10% FCS. Samples were taken 0, 0.5, 1.5, 5, and 12 h after the pulse and treated as described for the samples for the steady-state analysis.
RESULTS
Translation initiation of the CDV F protein.
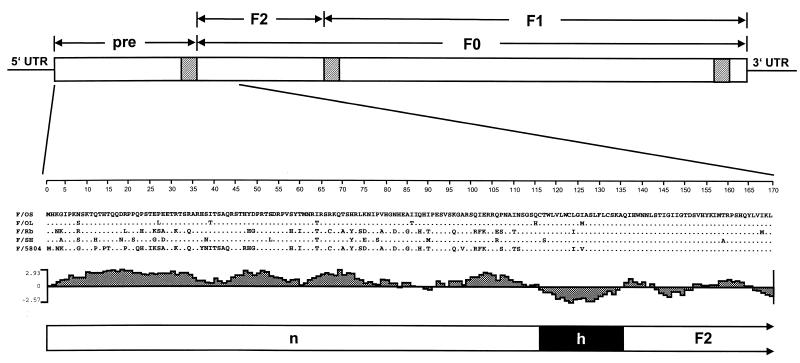
The predicted protein sequences of different CDV strains are shown in Fig. 1. The first 135 residues vary by up to 27.5% between strains, whereas the downstream amino acid sequence variation is 4% or lower (Fig. 1). The first 75 residues have a high hydrophilicity index, and they are followed by a stretch of 20 less hydrophilic residues (position 76 to 95) before another strong increase of hydrophilicity immediately upstream of a hydrophobic region (positions 96 to 115) (Fig. 1). The hydrophobic region is followed by a predicted signal peptide cleavage site, A↓QIHW (11). We refer to amino acids 1 to 135 as the Pre region, a potential long signal peptide.
FIG. 1.
The F gene ORF and the amino-terminal 170 residues of the CDV F protein. (Top) Schematic drawing of the F gene. The predicted reading frame beginning with the first in-frame AUG is boxed. Hydrophobic regions are hatched. (Center) Comparison of the amino acid sequence of the CDV strains Onderstepoort (OS and OL, small- and large-plaque-forming variants), Rockborn (Rb), Snyder Hill (SH), and 5804Han89 (5804) starting from the first in-frame translation initiation codon in the F ORF. (Bottom) Predicted hydrophilicity and structural organization of the first 170 residues of CDVOS. n and h, N-terminal and hydrophobic subdomains. The cleavage site between position 135 and 136 (A↓QIHW) was predicted by the method of Ladunga et al. (11).
To verify if the Pre region is translated, a set of mutants was generated based on the plasmid pCG-FOS (F protein of the small-plaque-forming variant of the vaccine strain Onderstepoort). Initially, the two in-frame methionines (residues 1 and 61) were mutated to leucines either individually or simultaneously. The mutant with a leucine codon (TTA) in place of the first methionine codon was called L1, that with a leucine at position 61 was called L61, and the double mutant was called L1/61. In addition, mutants with deletions of increasing portions of the N terminus were generated and named Δ60, Δ107, and Δ114.
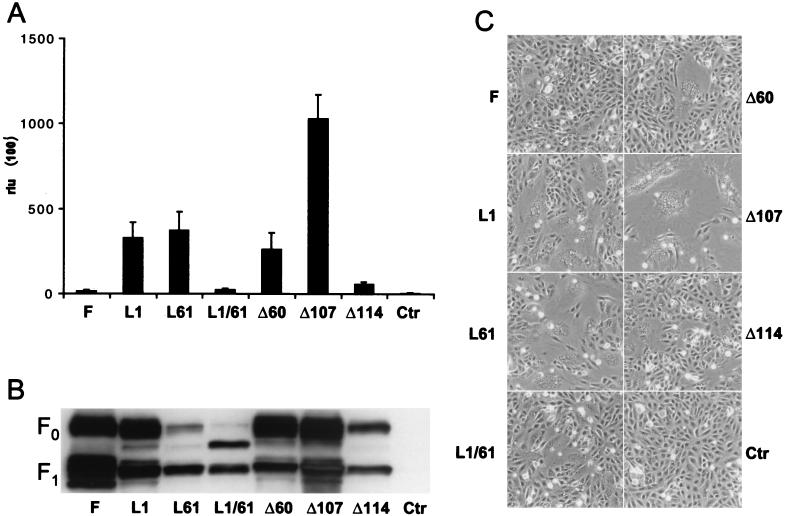
These mutants were coexpressed with the CDVOS H protein, and their activity was determined in a cell fusion assay (22). Significant differences in fusion efficiencies were observed. Forty-eight hours after transfection, the fusion activity of the unaltered F protein barely exceeded the background value despite strong expression (Fig. 2). The mutation of either in-frame methionine to leucine led to an ∼20-fold increase of fusion activity, even though the F protein levels produced were higher for the parental than for the altered proteins (Fig. 2). The construct in which both methionines are mutated displays a fusion activity similar to that of the unaltered construct (Fig. 2A and C), but the amount of protein expressed was extremely reduced (Fig. 2B). The strong band that is detected above the F1 signal is thought to be the product of translation initiation at the first in-frame methionine of this construct at residue 158, which would not be translocated into the ER due to the lack of a signal sequence and therefore would not be further processed. To a lower degree, this band can also be observed in the other constructs.
FIG. 2.
Characteristics of the different F proteins. (A) Quantitative fusion assays. Vero cell monolayers were either infected with MVA-T7 (MOI of 1) or transfected with the different F constructs, the plasmid coding for the HOS protein (pCG-HOS), and the plasmid containing the luciferase gene under the control of the T7 promoter (pTM1-luc). At 12 h after transfection, the cell populations were mixed and seeded into fresh plates. After 36 h at 37°C, fusion was quantified by measuring luciferase activity. The mean values of four independent experiments in duplicate are shown. (B) Protein analysis. Proteins were extracted from cells used for fusion assays, separated by reducing SDS-PAGE (10%), and blotted onto polyvinylidene difluoride membranes. The membranes were incubated with the anti-Fcyt rabbit antipeptide serum. (C) Phase-contrast image of the Vero cells from the fusion experiment described above, 48 h after cotransfection.
The deletion of the first 60 amino acids (Δ60) led to an ∼15-fold increase of fusion activity (Fig. 2A and C), with high levels of protein expression (Fig. 2B). This recapitulated the phenotype of L1. The further reduction of the precursor sequence to eight residues, which corresponds to the length of a classical signal peptide (Δ107), led to a 70-fold increase of fusion activity and high levels in protein expression (Fig. 2). Minimal fusion activity and protein expression were observed when the first 114 residues were deleted (Fig. 2). In summary, the standard F protein showed the highest level of expression but the lowest activity. Mutant proteins with no standard start codons or a very short sequence upstream of the hydrophobic region were poorly expressed (L1/61 and Δ114). Mutants of intermediate length or with a leucine at position 61 were expressed at intermediate levels and had high activity (L1, L61, and Δ60).
An intriguing observation was that the L61 mutant, differing only in one amino acid from the standard protein, also gained function. To verify the significance of this observation, we produced mutants with an alanine or isoleucine at position 61. These mutants were functionally similar to the parental protein (data not shown), implying a peculiar effect of the L61 amino acid change. These results were consistent with F protein translation starting at the first AUG, resulting in the production of a long signal peptide. They also suggested a negative effect of this sequence on F protein function because the mutant with a short signal peptide-like amino-terminal sequence was the most active (Δ107).
Recovery and characterization of recombinant viruses with alterations in the F protein amino-terminal region.
To characterize the effect of these alterations within the viral background, recombinant viruses were generated. The parental F gene was exchanged for the mutated F genes in pCDVII as AscI-AflII fragments. In that way, pCDVII-FOSL1, pCDVII-FOSL61, pCDVII-FOSL1/61, pCDVII-FOSΔ60, pCDVII-FOSΔ107, and pCDVII-FOSΔ114 were constructed. Subsequently, recovery of the recombinant viruses was attempted. Within 2 to 5 days after the transfer of the transfected 293 cells onto Vero cells, multiple syncytia were detected in all dishes but those transfected with pCDVII-FOSL1/61. Despite several attempts, a virus based on pCDVII-FOSL1/61 could not be recovered. The identity of the recombinant viruses was confirmed by reverse transcription-PCR and sequence analysis of the F genes, which showed that no point mutations had occurred compared to the transfected plasmid.
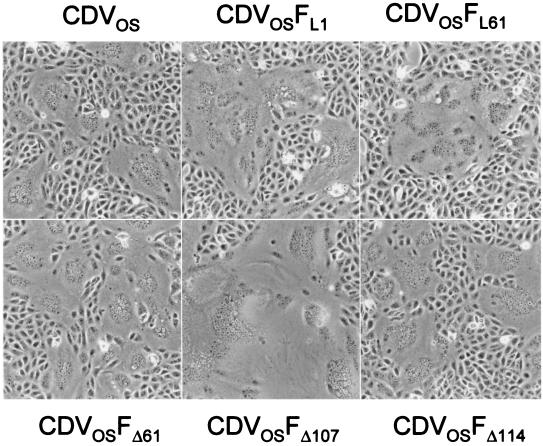
To characterize the recombinant viruses, growth curves in Vero and DH 82 cells were performed using a MOI of 0.01. All viruses reached titers within 1 logarithm of that of the parental virus (data not shown). The fusion activity of the five new recombinant viruses was compared to that of the parental virus (Fig. 3). The fusion activity of the parental virus (Fig. 3) was comparatively stronger than that produced by transfection of plasmids encoding the parental F and H proteins (Fig. 2), indicating that the in vitro assay only partially reflects the fusion activity in the context of a viral infection. Nonetheless, the growth phenotype of the recombinant viruses (Fig. 3) mirrored the fusion activity observed after coexpression of the respective F protein with H (Fig. 2C). It is thus apparent that the amino-terminal region of the F protein has a negative effect on the fusion efficiency also in the context of a viral infection.
FIG. 3.
CPE of the recombinant viruses with alterations in the amino-terminal region of the F protein. Vero cells were infected with the parental and recombinant viruses and photographed 48 h after infection at an MOI of 0.01.
Detection of F precursor proteins.
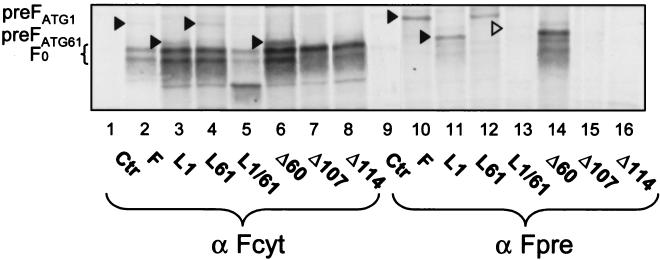
To investigate if the postulated F0 protein precursor can be detected, a peptide corresponding to residues 88 to 112 was synthesized, and a rabbit antiserum against this peptide was produced and named Fpre. The radioimmunoprecipitation shown in Fig. 4 indicated that the antiserum recognizes bands at approximately 78 kDa (Fig. 4, lane 10) and at about 68 kDa (lane 11) in the lysate of cells transfected with the different expression plasmids. Plasmids with an ATG codon at position 1 produce the longer protein (Fig. 4, lanes 10 and 12); plasmids with a TTA codon at position 1 or a deletion of 60 residues but an ATG codon at position 61 produce the shorter protein (Fig. 4, lanes 11 and 14). As expected, Fpre antiserum does not recognize any of these proteins when extracts of cells transfected with plasmids that have a long deletion or mutations in both start codons are examined (Fig. 4, lanes 13, 15, and 16). Nevertheless, a weak band of intermediate size is detected in the FL1/61 extracts (Fig. 4, lane 13), suggesting inefficient translation initiation on a non-AUG codon. The two F precursors can also be detected with the Fcyt antiserum, which recognizes the cytoplasmic tail (Fig. 4, lanes 2, 3, 4, and 6), suggesting that cleavage of the F protein amino-terminal extensions may be posttranslational. We named the long F protein produced by the unmodified F plasmid (apparent molecular mass, 78 kDa) PreF0.
FIG. 4.
Radioimmunoprecipitation of the different F proteins. Transfected Vero cells were labeled for 1.5 h, lysed, and immunoprecipitated with anti-Fcyt or anti-Fpre rabbit antipeptide serum. Equivalent aliquots of the protein A eluates were separated by reducing SDS-PAGE (7.5%). The open triangle indicates the possible precursor of the mutant without in-frame methionines (L1/61).
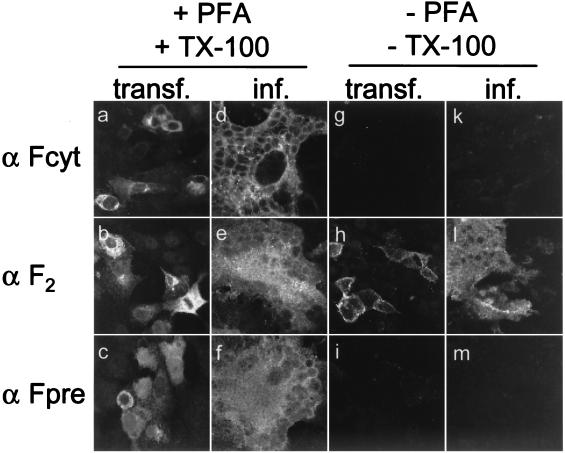
The PreF0 protein does not reach the cell surface. We took advantage of antisera directed against the amino-terminal region of F, the F2 subunit, or the intracellular tail of the F1 subunit to characterize the cellular localization of PreF0 (Fig. 5a to f). As expected, anti-Fcyt did not detect its epitope in nonpermeabilized cells at 4°C (Fig. 5g and k), whereas anti-F2 did (Fig. 5h and l). The Fpre antiserum did not detect any protein in nonpermeabilized cells (Fig. 5i and m), indicating either that PreF0 is not transported to the cell surface or that the amino-terminal region is not translocated.
FIG. 5.
Immunofluorescence staining of Vero cells transfected with pCG-FOS or infected with CDVOS by using antibodies against different parts of the F protein. Cells were either fixed with paraformaldehyde (PFA) and permeabilized with Triton X-100 (TX-100) 48 h after transfection or infection with an MOI of 0.01 (a to f) or incubated unfixed with the primary antibody at 4°C before treatment with paraformaldehyde (g to m). Anti-Fcyt, anti-F2, or anti-Fpre rabbit antipeptide serum was used as the primary antibody.
An immunoprecipitation experiment with surface-biotinylated cells using the three antisera mentioned above revealed that in contrast to the Fcyt and F2 antisera, which precipitated F0 as well as their respective subunit, the Fpre antiserum did not precipitate any specific band (data not shown). In combination with the results from the immunofluorescence, this suggests that the Pre sequence is cleaved before the mature protein is transported to the cell surface.
Characterization of PreF0 and of two small cleavage products.
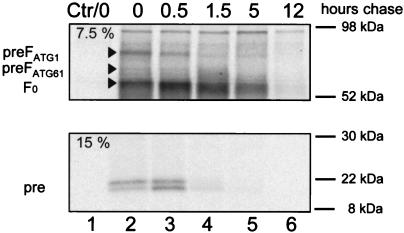
F protein maturation was characterized by pulse-chase analysis. Immediately after labeling, two large proteins (approximately 78 and 68 kDa) were detected by the Fpre antiserum (Fig. 6, upper panel, lane 2). The upper band was strong and had an apparent molecular weight suggesting translation initiation on AUG 1, and it was named accordingly. The lower band was much weaker and had an apparent weight suggesting initiation on AUG 61. The intensity of the PreFATG1 precursor band was reduced to about 70% after a 30-min chase (Fig. 6, upper panel, lane 3) and to about 50% after a 1.5-h chase (Fig. 6, upper panel, lane 4).
FIG. 6.
Pulse-chase analysis of cells transfected with pCG-FOS. Transfected Vero cells were pulse-labeled for 30 min and then chased for the indicated periods with fresh growth medium containing methionine and cysteine. Equivalent cell lysate samples were immunoprecipitated with anti-Fpre rabbit antipeptide serum. Equivalent aliquots of the protein A eluates were separated by reducing SDS-PAGE (top, 7.5%; bottom, 15%). The positions of F0, the precursors PreFATG1 and PreFATG61, and the possible signal peptide cleavage products Pre are indicated on the left.
When other aliquots of the same protein extracts were separated on a more concentrated protein gel (Fig. 6, lower panel), a double band at approximately 15 to 17 kDa was detected. Of the two small cleavage products, the lowest gained in intensity after a 30-min chase and remained detectable after 1.5 h (Fig. 6, lane 4). The size of these peptides is compatible with cleavage at or around the predicted signal peptidase recognition sequence.
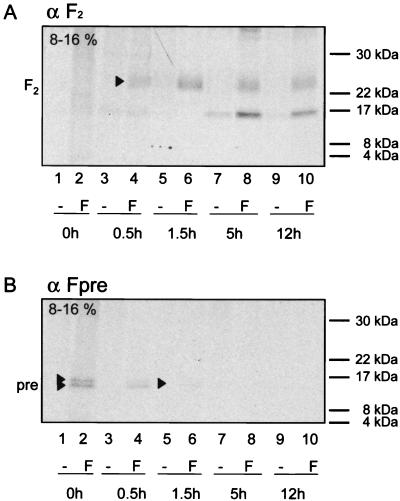
A similar pulse-chase experiment was performed to assess the temporal relation between Pre region cleavage and furin cleavage of F0 into F1 and F2. The F2 subunit was detected with the anti-F2 antiserum after a 30-min chase as a broad band at approximately 22 to 25 kDa (Fig. 7A, lane 4). The intensity of this band increased after a 1.5-h chase and decreased only slightly after 5 and 12 h (Fig. 7A, lanes 6, 8, and 10), indicating that the processing of the F protein into its mature form is rather slow and results in a relatively stable protein.
FIG. 7.
Pulse-chase analysis of CDV FOS maturation. Vero cells transfected with pCG-FOS (F) or mock transfected (−) were pulse-labeled for 30 min and then chased for the indicated periods with fresh growth medium containing methionine and cysteine. Equal cell lysate samples were immunoprecipitated with anti-F2 or anti-Fpre rabbit antipeptide serum. Equivalent aliquots of the protein A eluates were separated by reducing SDS-PAGE (8-to-16% gradient). The positions of F2 and the signal peptide cleavage products Pre are indicated on the left. The origin of the band of about 16 kDa detected 5 and 12 h postpulse in both transfected and control cells is unknown.
The two small cleavage products of PreF0 were detected at the highest level immediately after the pulse, and their intensity decreased by about one-half after a 30-min chase (Fig. 7B, lanes 2 and 4). After a 1.5-h chase, only a very faint signal was observed, which disappeared after 5 h. These findings, and the fact that a band corresponding to the F2 subunit with the Pre region still attached was never detected with any of the antisera used, suggest that the long precursor peptide cleavage occurs before furin cleavage.
DISCUSSION
The unique features of the region between the M and F proteins of morbilliviruses have raised questions about its function in viral replication and virulence (2, 6, 7, 10, 15, 25). In this study, we present evidence that the amino-terminal extension (residues 1 to 135) of the CDV F protein is more than a classical signal sequence. We show that deletions in this >100-residue-long sequence lead to an increase in the activity of the mature protein, indicating that this region has regulatory function.
The F protein amino-terminal extension modulates fusion function.
The classical signal peptide of a type I glycoprotein is usually cleaved off cotranslationally and immediately degraded (1). Thus, it is not expected to have a significant influence on the biological activity of the protein. However, more complex roles for certain cellular signal peptides have been characterized (17), and it has been shown for other viral glycoproteins that their signal peptides are cleaved not cotranslationally but rather posttranslationally (14, 16, 29). Similar to the situation in CDV, those signal peptides are unusually long and positively charged, which may delay protein processing (13, 18). Consequently, reduction of the charge of these signal peptides, or replacement with short signal peptides from efficiently transported glycoproteins, leads to an increase in cleavage efficiency and subsequently to an increase of mature and active protein (13).
The CDV F protein long amino-terminal extension has similar characteristics. The remarkable new property of this sequence is that it modulates function: mature F proteins derived from a PreF0 precursor are less fusogenic than those derived from shorter precursors. It is as yet unclear if this is due to a slightly different primary sequence of these proteins or to the effect of the extension on folding of the rest of the glycoprotein. The recent observation that the measles virus glycoproteins hetero-oligomerize in the ER (24) suggests the possibility that the glycoproteins of all morbilliviruses including CDV may do the same. In that case, a mechanism limiting fusion of intracellular membranes may be required. There are clear indications that the interactions between the glycoprotein cytoplasmic tails and the M protein limit fusion (3, 4), but in addition the conformation and the interactions of the glycoprotein ectodomains may influence fusion activity (R. K. Plemper, A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo, unpublished results).
It is also of interest that human respiratory syncytial virus, classified within the genus Pneumovirus of the family Paramyxoviridae, has developed another noncanonical mechanism to control membrane fusion. In this virus, proteolytic cleavage of the F0 precursor at two closely spaced furin recognition sites has recently been detected (8, 32), and it has been proposed that the two-hit cleavage process may delay fusion activation and thus allow release of active particles.
A role in pathogenicity for the F protein Pre peptide?
An approximately 16-kDa cleavage product was detected, and its half-life was determined to be about 30 min. The fact that this peptide is not immediately degraded suggests the possibility that it may have another function. In human immunodeficiency virus (HIV)-infected cells, several calmodulin-dependent processes involved in immune defense are disrupted (19), which is thought to be associated with the interaction of the signal peptide of the HIV glycoprotein with calmodulin (18). Calmodulin recognizes positively charged, amphiphilic α-helical stretches of 16 to 35 residues (baa helix) (23). The signal sequence of the HIV-1 glycoprotein has an extended amino-terminal region that can potentially form such a baa helix (residues 1 to 35), and it has been shown that a synthetic peptide corresponding to the amino-terminal 23 residues of the HIV glycoprotein has high affinity for calmodulin and efficiently inhibits Ca2+-calmodulin-dependent phosphodiesterase in vitro (18). Since a potential baa helix can be identified in the amino-terminal region of the signal peptide of the CDV F protein, a similar interaction with calmodulin is conceivable, which may influence CDV pathogenesis.
Acknowledgments
We thank Sompong Vongpunsawad for excellent technical support and Erick Poeschla for constructive discussion of the manuscript.
This work was supported by grants from the Mayo and Siebens Foundations and by a Emmy Noether award from the German Research Foundation (DFG) to V.V.M.
REFERENCES
- 1.Blobel, G., P. Walter, C. N. Chang, B. M. Goldman, A. H. Erickson, and V. R. Lingappa. 1979. Translocation of proteins across membranes: the signal hypothesis and beyond. Symp. Soc. Exp. Biol. 33:9-36. [PubMed] [Google Scholar]
- 2.Cathomen, T., C. J. Buchholz, P. Spielhofer, and R. Cattaneo. 1995. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214:628-632. [DOI] [PubMed] [Google Scholar]
- 3.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherpillod, P., K. Beck, A. Zurbriggen, and R. Wittek. 1999. Sequence analysis and expression of the attachment and fusion proteins of canine distemper virus wild-type strain A75/17. J. Virol. 73:2263-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, S. A., G. J. Belsham, and T. Barrett. 1990. The role of the 5′ nontranslated regions of the fusion protein mRNAs of canine distemper virus and rinderpest virus. Virology 177:317-323. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe et al. (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 10.Heider, A., S. Santibanez, A. Tischer, E. Gerike, N. Tikhonova, G. Ignatyev, M. Mrazova, G. Enders, and E. Schreier. 1997. Comparative investigation of the long non-coding M-F genome region of wild-type and vaccine measles viruses. Arch. Virol. 142:2521-2528. [DOI] [PubMed] [Google Scholar]
- 11.Ladunga, I., F. Czako, I. Csabai, and T. Geszti. 1991. Improving signal peptide prediction accuracy by simulated neural network. Comput. Appl. Biosci. 7:485-487. [DOI] [PubMed] [Google Scholar]
- 12.Li, Y., J. J. Bergeron, L. Luo, W. J. Ou, D. Y. Thomas, and C. Y. Kang. 1996. Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport. Proc. Natl. Acad. Sci. USA 93:9606-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Y., L. Luo, D. Y. Thomas, and C. Y. Kang. 1994. Control of expression, glycosylation, and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology 204:266-278. [DOI] [PubMed] [Google Scholar]
- 14.Li, Y., L. Luo, D. Y. Thomas, and C. Y. Kang. 2000. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology 272:417-428. [DOI] [PubMed] [Google Scholar]
- 15.Liermann, H., T. C. Harder, M. Lochelt, V. von Messling, W. Baumgartner, V. Moennig, and L. Haas. 1998. Genetic analysis of the central untranslated genome region and the proximal coding part of the F gene of wild-type and vaccine canine distemper morbilliviruses. Virus Genes 17:259-270. [DOI] [PubMed] [Google Scholar]
- 16.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 18.Martoglio, B., R. Graf, and B. Dobberstein. 1997. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 16:6636-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, M. A., T. A. Mietzner, M. W. Cloyd, W. G. Robey, and R. C. Montelaro. 1993. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retrovir. 9:1057-1066. [DOI] [PubMed] [Google Scholar]
- 20.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 21.Murphy, S. K., and G. D. Parks. 1997. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus simian virus 5. Virology 232:145-157. [DOI] [PubMed] [Google Scholar]
- 22.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Neil, K. T., and W. F. DeGrado. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem. Sci. 15:59-64. [DOI] [PubMed] [Google Scholar]
- 24.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 25.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider, H., P. Spielhofer, K. Kaelin, C. Dotsch, F. Radecke, G. Sutter, and M. A. Billeter. 1997. Rescue of measles virus using a replication-deficient vaccinia-T7 vector. J. Virol. Methods 64:57-64. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu, M. S., W. Husar, S. D. Cook, P. C. Dowling, and S. A. Udem. 1993. Canine distemper terminal and intergenic non-protein coding nucleotide sequences: completion of the entire CDV genome sequence. Virology 193:66-72. [DOI] [PubMed] [Google Scholar]
- 28.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 29.Verschoor, E. J., E. G. Hulskotte, J. Ederveen, M. J. Koolen, M. C. Horzinek, and P. J. Rottier. 1993. Post-translational processing of the feline immunodeficiency virus envelope precursor protein. Virology 193:433-438. [DOI] [PubMed] [Google Scholar]
- 30.von Heijne, G. 1984. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. EMBO J. 3:2315-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]