Abstract
The stable propagation of a full-length transmissible gastroenteritis coronavirus (TGEV) cDNA in Escherichia coli cells as a bacterial artificial chromosome has been considerably improved by the insertion of an intron to disrupt a toxic region identified in the viral genome. The viral RNA was expressed in the cell nucleus under the control of the cytomegalovirus promoter and the intron was efficiently removed during translocation of this RNA to the cytoplasm. The insertion in two different positions allowed stable plasmid amplification for at least 200 generations. Infectious TGEV was efficiently recovered from cells transfected with the modified cDNAs.
Coronaviruses are enveloped, single-stranded, positive-sense RNA viruses that belong to the order Nidovirales (4). They have the largest RNA viral genome, about 30 kb. The recent construction of cDNAs encoding infectious RNAs for transmissible gastroenteritis coronavirus (TGEV) (1, 21), human coronavirus strain 229E (18), and infectious bronchitis virus (3) will facilitate the genetic manipulation of coronavirus genomes for the study of gene function and their use as expression vectors. The construction of these cDNAs had been hampered by the large size of the viral genome and the instability in Escherichia coli cells of some viral sequences located in open reading frame (ORF) 1a. Recently, the cloning and engineering of a full-length cDNA of TGEV in E. coli cells as a bacterial artificial chromosome (BAC) that reduced the number of plasmid copies per cell to a minimum were reported (1). Although this strategy led to the rescue of infectious TGEV from the cDNA, we observed that a residual toxicity remained in the cDNA involving sequences at the 3′ end of ORF 1a. This instability was observed by replicating the plasmid for more than 80 generations and led us to design a cloning strategy based on the manipulation of a virus cDNA without the toxic sequence and the insertion of this sequence before transfection into cells. To stabilize the BAC, we have defined a strategy that allows stabilization of the TGEV cDNA during its amplification in bacteria. The use of introns has proven very useful for the construction of stable infectious cDNAs expressing viral RNAs in vivo from a nuclear promoter (7, 10, 20). The insertion of an intron allows stable amplification of the plasmids in bacteria by disrupting the toxic sequence. This intron is removed by splicing during RNA translocation from the nucleus to the cytoplasm. In this report we describe the complete stabilization of the full-length TGEV cDNA by the insertion of a 133-nucleotide (nt) synthetic intron in two different positions and the rescue of infectious TGEV from the modified cDNAs.
Construction of a cDNA encoding the TGEV genome.
The assembly of a cDNA clone encoding an infectious full-length TGEV genome was reported previously (1). The construction of this cDNA started from a cDNA encoding the TGEV-derived defective minigenome DI-C, which had previously generated an RNA which was efficiently replicated by the wild-type virus (6, 11). This cDNA was assembled under the control of the cytomegalovirus (CMV) immediate-early promoter that expressed the viral RNA in the nucleus by the cellular RNA polymerase II.
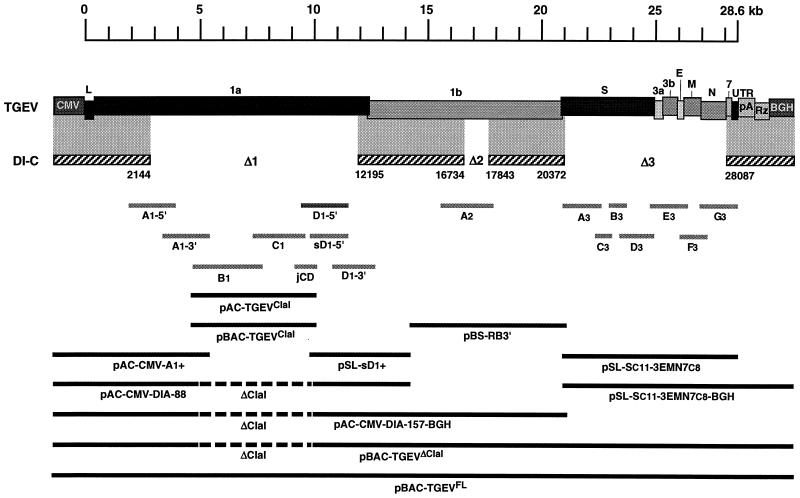
The minigenome DI-C presents three deletions in relation to the TGEV full-length genome. These deletions had to be filled in to generate a full-length infectious cDNA clone. The first deletion was located in ORF 1a (Δ1, including nt 2145 to 12194), the second one was in ORF 1b (Δ2, including nt 16735 to 17842), and the third one included almost all structural genes (Δ3, spanning nt 20373 to 28086) (Fig. 1). The initial strategy to assemble the infectious cDNA clone was based on the construction of the missing cDNA fragments by reverse transcriptase PCR (RT-PCR) amplification of viral RNA and the assembly of these fragments in the DI-C cDNA. To obtain an optimal replicase gene, the nucleotide changes previously described in DI-C genome were corrected to the sequence of TGEV PUR46-MAD strain genome (11, 13).
FIG. 1.
Assembly of the full-length TGEV cDNA. The assembly of a TGEV infectious cDNA (top) was started from the DI-C cDNA (discontinuous sequence indicated by the striped boxes). DI-C RNA has three deletions: Δ1 (10 kb), Δ2 (1.1 kb), and Δ3 (7.7 kb), which were filled in with newly synthesized RT-PCR products covering the rest of the viral genome (light grey bars). The sequence for the Δ1 deletion was amplified in six independent fragments: A1-5′, A1-3′, B1, C1, D1-5′, and D1-3′. The fragment D1-5′ (nt 9349 to 11173) could not be stably cloned. Nevertheless, the D1-5′ sequence was split into two overlapping clones (jCD [nt 8978 to 9758] and sD1-5′ [nt 9581 to 11173]) that were fully stable. Thus, the full-length cDNA was assembled in two large fragments that were finally joined by a ClaI9615 restriction site that split the toxic region. For that purpose, the ClaI4417 site was used as the other end of these fragments, while a third ClaI site, at position 18997, was removed by directed mutagenesis. A plasmid encoding the smallest fragment, including the viral sequence from nt 4310 to 9758 (pAC-TGEVClaI), was assembled by joining clones B1, C1, and jCD. Clones A1-5′ and A1-3′ were joined to the CMV immediate-early promoter (CMV) plus the 5′ end of the TGEV genome to make plasmid pAC-CMV-A1+. On the other hand, clones sD1-5′ and D1-3′ and the 5′ end of ORF 1b were joined to build plasmid pSL-sD1+, which was put together with pAC-CMV-A1+ to obtain a plasmid containing the 5′ third of the TGEV cDNA except for the ClaI deletion, called pAC-CMV-DIA-88. The Δ2 deletion was filled in with clone A2, which was joined to other ORF 1b sequences in plasmid pBS-RB3′. This was assembled with plasmid pAC-CMV-DIA-88 to obtain pAC-CMV-DIA-157-BGH. The Δ3 deletion was filled in with clones A3 to G3, from which plasmid pSL-Sc11-3EMN7c8 was assembled. The S gene was derived from the PUR46-C11 TGEV strain (Sc11) and genes 3, E, M, N, and 7 were from the PUR46-C8 strain (3EMN7c8), as well as the rest of the genome. The sequences necessary to make an accurate 3′ end, i.e., a synthetic poly(A) tail of 24 A residues (pA), the hepatitis delta virus ribozyme (Rz), and the bovine growth hormone termination and polyadenylation sequences (BGH) were added, yielding plasmid pSL-Sc11-3EMN7c8-BGH. Then a cDNA containing the whole TGEV genome except the ClaI4417-ClaI9615 fragment was assembled in the low-copy-number plasmid pACNR1180 (not shown). To improve the stability of the TGEV-derived cDNA sequences during their amplification in bacteria, the cDNA was transferred to pBeloBAC11, making plasmid pBAC-TGEVΔClaI. The ClaI4417-ClaI9615 restriction fragment was also cloned into pBeloBAC11, leading to plasmid pBAC-TGEVClaI. Insertion of the ClaI4417-ClaI9615 restriction fragment from pBAC-TGEVClaI into ClaI-linearized pBAC-TGEVΔClaI led to the full-length TGEV cDNA (pBAC-TGEVFL). Letters above the TGEV cDNA indicate viral genes. L, leader; UTR, untranslated sequences. Numbers below DI-C cDNA indicate positions of deletions with respect to the TGEV genome. ΔClaI, gap between ClaI4417 and ClaI9615 restriction sites.
All of the cDNA fragments required to fill in the Δ2 and Δ3 deletions were obtained and stably cloned, as well as the cDNAs for the Δ1 deletion, except for the clone containing cDNA fragment D1-5′ (nt 9349 to 11173), which grew in very small colonies or did not grow at all, in both high- and low-copy-number plasmids using several E. coli strains (XL1-Blue, DH5α, DH10B, etc). In the few cases in which some plasmid DNA was obtained, the introduction of changes disrupting the ORF, like stop codons or small insertions or deletions, located between nt 9711 and 10009 was observed. Interestingly, this fragment sequence was easily amplified in two independent overlapping clones spanning nt 8978 to 9758 and nt 9581 to 11173 (jCD and sD1-5′, respectively), with the correct sequence. These results indicated that none of the individual fragments but instead their combined sequences were the source of the toxicity. Thus, the assembly strategy was redesigned to delay the joining of both sequences as much as possible. In the last step, just before cDNA transfection into bacteria, the ClaI4417-ClaI9615 fragment was inserted into the plasmid encoding the rest of the TGEV genome, previously cloned in the low-copy-number (10 to 12 copies/cell) vector pACNR1180 (14) to increase stability. However, E. coli cells did not tolerate the full-length cDNA, implying that toxicity originating from the insertion of the sequences around the ClaI9615 site was still significant despite the reduction in the number of plasmid copies per cell (data not shown).
To increase the stability of TGEV sequences in E. coli cells, the last assembly step was performed in the very-low-copy-number BAC vector pBeloBAC11 (19). This vector replicates up to a maximum of two copies per cell and has been proved to stably maintain large eukaryotic DNA fragments (up to 300 kb) (12, 16). This strategy allowed the construction of a full-length TGEV cDNA (pBAC-TGEVFL) and its correct amplification in E. coli DH10B cells for 80 generations.
Stability of the full-length cDNA in E. coli cells.
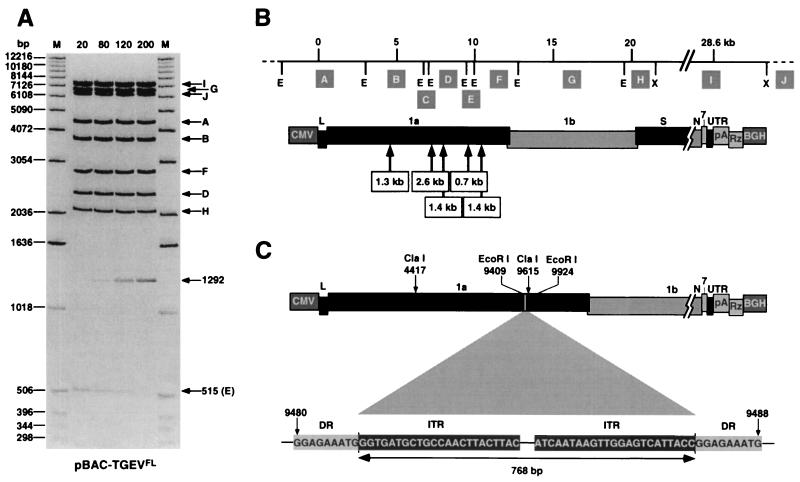
To analyze the stability of the full-length TGEV cDNA through its amplification in E. coli cells, this plasmid (pBAC-TGEVFL) and the plasmids from which it was derived (pBAC-TGEVΔClaI and pBAC-TGEVClaI) were grown for 200 generations in DH10B cells, as described previously (1). Plasmid DNA was extracted and analyzed by studying its restriction endonuclease pattern. Plasmids pBAC-TGEVΔClaI and pBAC-TGEVClaI were fully stable for more than 200 generations (data not shown), while the plasmid encoding the full-length TGEV cDNA remained stable for about 80 generations (Fig. 2A). In fact, at this point a new band of 1.2 kb was observed, accompanied by the disappearance of the 0.5-kb band, indicating an insertion of 0.7 kb. However, this stability was sufficient to rescue infectious TGEV on a regular basis (1).
FIG. 2.
Stability of the TGEV full-length cDNA in E. coli cells. (A) EcoRI-XhoI restriction pattern of plasmid pBAC-TGEVFL extracted from E. coli DH10B cells that were grown for the number of generations shown at the top. The DNA bands were observed after ethidium bromide staining. The lower arrow (E) indicates the disappearance of the 515-bp band. The arrow corresponding to a band of 1292 bp shows the appearance of a new band. M, molecular mass markers. Letters on the right (A to J) indicate the restriction endonuclease fragments (see panel B) except fragment C (254 bp), which is not shown in the gel. (B) Sizes and relative positions of fragments inserted in the TGEV full-length cDNA during different repetitions of the stability experiments. Simultaneous insertion of two or more fragments was never observed. The positions of EcoRI (E) and XhoI (X) restriction sites are indicated below the top bar. Letters in shaded boxes (A to J) represent restriction fragments with the following sizes: A, 4,458 bp; B, 3,759 bp; C, 254 bp; D, 2,372 bp; E, 515 bp; F, 2,834 bp; G, 6,671 bp; H, 2,069 bp; I, 7,421 bp; J, 6,357 bp. Dashed lines at the ends of the top bar correspond to pBeloBAC11 vector sequences. (C) Structure of the 768-bp insertion sequence type 1 located between positions 9480 and 9488 of TGEV genome cDNA. The sequences of the direct repeats (DR) of 9 bp originated by the insertion and the inverted terminal repeats (ITR) of 23 bp are shown. Abbreviations are as in Fig. 1.
To study full-length-cDNA stability, aliquots of cell cultures carrying the full-length cDNA at a different number of generations were grown on agar plates. A mixture of small and large colonies was observed for all generations tested. Small colonies corresponded to plasmids with the restriction endonuclease pattern expected for the correct cDNA sequence and were still toxic for E. coli cells, while large colonies contained rearranged plasmids that were apparently no longer harmful to the bacteria. A larger number of generations correlated with a greater ratio of large to small colonies: the percentages of large colonies at 80, 120, and 200 generations were around 20, 90, and 100%, respectively. Plasmid DNA from a number of large colonies was analyzed by EcoRI-XhoI digestion to identify the nature of rearrangements. All the clones contained different insertions types, from 0.7 to 2.6 kb in length, always located in ORF 1a (Fig. 2B). The smallest one, a ∼0.7 kb insertion located between the restriction endonuclease sites EcoRI9409 and EcoRI9924, was sequenced. This insertion corresponded to E. coli insertion sequence 1 (IS 1), a transposon of 768 bp that includes a 23-bp inverted terminal repeat on either side (9). This sequence is known to be randomly inserted, generating 9-bp direct repeats in the target DNA (Fig. 2C).
Insertion of an intron to stabilize the TGEV full-length cDNA.
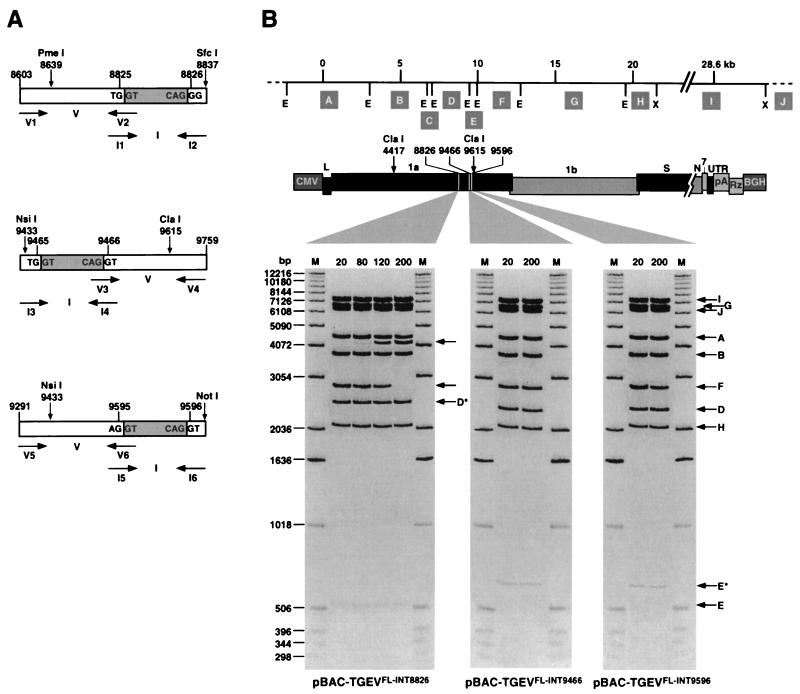
The spontaneous insertion of heterologous sequences into ORF 1a, mainly between nt 7500 and 11000, allowed the stable growth of plasmid pBAC-TGEVFL in large colonies in DH10B cells. The same results were obtained in previous experiments when TGEV cDNA fragments from nt 7978 to 9758 and from nt 9581 to 13960 were stably cloned when split by a 271-bp spacer during the subcloning process (data not shown). Thus, in order to stabilize the TGEV full-length cDNA we decided to introduce a known spacer sequence within the area of ORF 1a that could not be cloned in one piece in low-copy-number plasmids. One possibility was the insertion of an intron. In vivo transcription of the intron-containing full-length clones from the CMV promoter would most likely splice the intron in the nucleus before RNA translocation to cytoplasm in the transfected cells, thus restoring the ORF and the infectivity of the plasmid-derived RNA. For that purpose, a 133-nt synthetic intron obtained from the vector pRL-CMV (Promega) was used. The sites of insertion of the intron into the virus sequence were designed to generate 5′ and 3′ intron splice sites matching the consensus sequences of mammalian introns (15). The theoretical probability of splicing for every insertion site was calculated with the help of a program for the prediction of splice sites in human DNA sequences (HSPL) (17). The three sites with the highest splicing probabilities, at positions 8826, 9466, and 9596, were selected. The intron was placed in the appropriate position of the viral sequence by overlapping PCR (Fig. 3A), using the oligonucleotides shown in Table 1. The final PCR products were cut with the indicated restriction enzymes and inserted into the plasmid pBAC-TGEVClaI. Then the ClaI4417-ClaI9615 fragment was introduced into the plasmid pBAC-TGEVΔClaI. The plasmids containing the full-length cDNAs with the intron were called pBAC-TGEVFL-INT8826, pBAC-TGEVFL-INT9466, and pBAC-TGEVFL-INT9596.
FIG. 3.
Insertion of an intron to stabilize the TGEV full-length cDNA. (A) Strategy for the insertion of the 133-nt intron between positions 8825 and 8826, 9465 and 9466, and 9595 and 9596 of the TGEV sequence. For each position, the upper bar represents the viral cDNA fragment with the intron inserted (grey box). These cDNA fragments are the product of overlapping PCR from the partial PCR products V and I. Fragments V were amplified from TGEV cDNA and fragments I were amplified from the pRL-CMV plasmid, by using the oligonucleotides (horizontal arrows) indicated in Table 1. The restriction sites used to insert the final PCR products into the plasmid pBAC-TGEVClaI are shown (vertical arrows). The letters inside the boxes correspond to the nucleotides located in the intron-TGEV sequence junctions. (B) Stability of the three intron-containing full-length TGEV cDNAs in E. coli cells. The EcoRI-XhoI restriction patterns of plasmids pBAC-TGEVFL-INT8826 (left), pBAC-TGEVFL-INT9466 (middle), and pBAC-TGEVFL-INT9596 (right), extracted from E. coli DH10B cells grown for the indicated number of generations are shown. The positions of EcoRI (E) and XhoI (X) restriction sites are indicated below the top bar. Letters in shaded boxes (A to I) represent restriction fragments identical to those indicated in Fig. 2. Dashed lines at the ends of the top bar correspond to pBeloBAC11 vector sequences. Gels were stained with ethidium bromide. In the pBAC-TGEVFL-INT8826 restriction pattern the middle arrow indicates the disappearance of the ∼2.8-kb band, while the upper arrow shows the appearance of a new band of ∼4.1 kb. M, molecular mass markers. Asterisks indicate fragments with a size increase of 133 nt corresponding to intron insertion.
TABLE 1.
Oligonucleotides used to introduce the intron into three different positions of the TGEV full-length cDNA
| Oligonucleotide | Sequencea | |
|---|---|---|
| Long name | Short name | |
| TGEV-8603-vs | V1 | 5′-CCTAGTTTTCCTCTATGACTCACTCCC-3′ |
| pRLi8826-5′-rs | V2 | 5′-CCTTGATACTTACCATTGAACCTGTGTAGTAC-3′ |
| pRLi8826-5′-vs | I1 | 5′-CAGGTTCAATGGTAAGTATCAAGGTTACAAG-3′ |
| pRLi8826-3′-rs | I2 | 5′-CCATTCTGTAGTCAGCTTCTCCCTGTGGAGAGAAAG-3′ |
| pRLi9466-3′-vs | V3 | 5′-CCTTTCTCTCCACAGGTTCCAATTTTGAAGGAGAAATG-3′ |
| TGEV-9759-rs | V4 | 5′-CCTTGTTGAGTCTTACGATGC-3′ |
| pRLi9466-5′-vs | I3 | 5′-GTATACATGCATCACTTAGAACTTGGAAATGGCTCGCATGTTGGTAAGTATCAAGGTTACAAG-3′ |
| pRLi9466-3′-rs | I4 | 5′-CCTTCAAAATTGGAACCTGTGGAGAGAAAGGCAAAG-3′ |
| TGEV-9291-vs | V5 | 5′-GCTTGTTATGAAGGATGTC-3′ |
| PRLi9596-5′-rs | V6 | 5′-TGATACTTACCTTTCACCATTGATAAGTG-3′ |
| PRLi9596-5′-vs | I5 | 5′-AATGGTGAAAGGTAAGTATCAAGGTTACAAG-3′ |
| PRLi9596-3′-rs | I6 | 5′-TGACATCGATGTGTTTGTAACAAACCACCTGTGGAGAGAAAGGCAAAG-3′ |
Bold letters indicate the sequence corresponding to the pRL-CMV intron.
The stability of the intron-containing TGEV full-length cDNAs was studied by analyzing the restriction endonuclease pattern of the cDNAs at different generation numbers (Fig. 3B). The plasmid with the intron introduced at position 8826 was apparently stable for about 80 generations, and then an insertion sequence type 10 right arm (IS10R) of 1,329 bp was inserted, as confirmed by sequencing. In contrast, the plasmids containing the intron at either position 9466 or position 9596 showed a stable restriction pattern after 200 generations, strongly suggesting that they were very stable. To determine whether these plasmids were stabilized by point mutations or by small insertions or deletions, which were not detectable by restriction analysis, the region from nt 8500 to 10500 was sequenced in both plasmids. No changes were observed in the sequence, confirming that stabilization of the TGEV full-length cDNA and its safe propagation in E. coli cells had probably been achieved.
Recovery of infectious virus from the intron-containing full-length cDNAs.
Since pBAC-TGEVFL-INT8826 was not completely stable, rescue experiments were not attempted with this BAC. Instead, rescue of infectious TGEV was studied with pBAC-TGEVFL-INT9466 and pBAC-TGEVFL-INT9596, as well as pBAC-TGEVFL as a positive control. These plasmids were transfected separately into BHK-pAPN cells as described previously (1). To increase the transfection efficiency, BHK-pAPN cells (2), which are BHK cells constitutively expressing the TGEV receptor (porcine aminopeptidase-N), were chosen because they are transfected with an efficiency about 10-fold higher than that of the ST cells previously used (unpublished results). At 6 h posttransfection, cells were removed with trypsin and were plated over a confluent monolayer of ST cells in order to increase TGEV replication titers and because the cytopathic effect induced by TGEV is more easily detected in ST cells. At 72 h posttransfection, supernatants from passage 0 were used to infect ST cells. This procedure was repeated four times every 24 h. Cytopathic effect was clearly seen in passages 1 to 4, indicating that infectious TGEV had been recovered from the full-length cDNAs originally containing the intron. A silent mutation (C9584T) that was introduced by PCR in the plasmid pBAC-TGEVFL-INT9596 was present in the recovered virus, indicating that it came from the cDNA and was not a contamination product (data not shown). The region where the intron had been inserted was sequenced in all recovered viruses. All of them showed the wild-type sequence, indicating that the intron was precisely removed, restoring the sequence of the parental virus in the virus progeny.
Virus titers were determined at different passages (Fig. 4A). At passage 0, the virus coming from plasmid pBAC-TGEVFL had grown to levels 1 and 2 logarithmic units higher than those of the viruses originated from pBAC-TGEVFL-INT9596 and pBAC-TGEVFL-INT9466, respectively. Nevertheless, viral titers were almost identical at passage 2 and reached a plateau in all cases.
FIG. 4.
Functional analysis of the intron-containing full-length cDNAs. (A) Growth kinetics of viruses originating either from the full-length cDNA without the intron (NO INT) or from the full-length cDNAs with the intron (INT9596 and INT9466). BHK-pAPN cells were transfected with the indicated plasmids or mock transfected (MOCK) and reseeded over ST cells. Passage 0 supernatant was collected 72 h posttransfection and passaged on fresh ST cells. Passages 1 to 4 were collected 24 h postinfection. (B) Efficiency of intron removal in viral RNAs transcribed in vivo from the intron-containing full-length cDNAs. BHK-pAPN cells were transfected with the full-length cDNA with the intron at position 9466 (INT9466) or 9596 (INT9596), and cytoplasmic and nuclear RNAs were extracted 20 and 40 h posttransfection (hpt). The region that includes the intron was amplified by RT-PCR, and bands of 601 and 468 bp were obtained in the cases of nonspliced and spliced RNAs, respectively. Gels were stained with ethidium bromide. C, cytoplasmic RNA; N, nuclear RNA; R, TGEV RNA; D, pBAC-TGEVFL-INT9596 DNA; M, molecular mass markers.
BHK-pAPN cells transfected with full-length cDNAs, either with or without the intron, were analyzed by immunofluorescence assay to test the expression of viral proteins. When monoclonal antibodies specific for the three major TGEV structural proteins (S, M, and N) were used, cells transfected with the two cDNAs containing the intron at positions 9466 and 9596 showed the same pattern as cells transfected with the cDNA without the intron (results not shown).
To study whether the difference in the titers of supernatants from cells transfected with plasmids pBAC-TGEVFL-INT9466 and pBAC-TGEVFL-INT9596 at passage 0 correlated with the extent of splicing of the genomic RNA, the proportion of viral RNA molecules with the native (spliced) sequence accumulated in the cytoplasm of BHK-pAPN cells transfected with these plasmids was analyzed. To fractionate the nuclei and cytoplasms, cells were lysed with a 0.5% NP-40-containing buffer at 20 and 40 h posttransfection. After centrifugation, the supernatants were treated with a buffer containing 8 M urea-1.5% sodium dodecyl sulfate, and then cytoplasmic RNA was extracted with phenol-chloroform, while the pellets were resuspended in 0.25 M sucrose-1.5% (wt/vol) sodium citrate and spun through a 0.88 M sucrose-1.5% (wt/vol) sodium citrate layer to purify nuclei (8). This procedure yields nuclei with a purity of over 95% as determined by optical microscopy. The nuclear RNA was obtained after phenol-chloroform extraction and ethanol precipitation. Both cytoplasmic and nuclear RNA were treated with RNase-free DNase I to remove contaminating plasmid DNA (5). A viral RNA fragment that included the two positions of insertion of the intron (from nt 9291 to 9758) was amplified by RT-PCR. PCR products were analyzed on a 2% agarose gel, and the intensities of the bands corresponding to amplification of spliced and nonspliced RNAs for each situation were estimated by using a gel documentation system (model 2000; Bio-Rad). These bands represent the amount of each RNA accumulated in both cell compartments. It should be noted that the amount of RNA accumulated in the cytoplasm corresponds to the viral RNA spliced in the nucleus and translocated to the cytoplasm plus the newly synthesized RNA produced in the cytoplasm by the viral replicase. About 10 to 15% of total genomic RNA molecules accumulated in the cells corresponded to the spliced form, when the intron had been inserted at position 9466 or 9596 (Fig. 4B). The length of the RNA molecules with the intron removed was similar at 20 h posttransfection independent of the position where the intron was inserted. At 40 h posttransfection most of the RT-PCR product from cytoplasmic RNA presented the “spliced” size, probably due to the amplification of TGEV RNA by the viral replicase and not to the greater extent of splicing.
These data can explain the initial delay in raising virus titer in supernatants from cells transfected with pBAC-TGEVFL-INT9466 or pBAC-TGEVFL-INT9596 in comparison with plasmid pBAC-TGEVFL, because of the initially lower proportion of infectious viral RNA molecules accumulated after splicing. But, despite the initial delay with respect to virus from pBAC-TGEVFL, titers of viruses originated from the cDNAs with the intron were similar to those from RNA genomes without introns in just two passages. These experiments confirm that the introduction of an intron is a very useful strategy to overcome the toxicity caused by TGEV cDNA sequences in E. coli cells. Insertion of the intron enhances the stability of the cDNA encoding a full-length TGEV RNA for more than 200 generations, which permits amplification of the cDNA to gram quantities, sufficient to perform almost any reverse genetics engineering of the infectious cDNA clone. This strategy will probably be of general use for the cloning and expression of other large-size functional RNA molecules.
Nucleotide sequence accession number.
The nucleotide positions mentioned in this article correspond to the TGEV PUR46-MAD strain sequence deposited in the EMBL database with accession no. AJ271965.
Acknowledgments
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874). J.M.G. received a fellowship from the Spanish Department of Education and Culture. Z.P., F.A., and E.C. received fellowships from the Biotechnology and Fishery and Agro-Industrial Research Programs of the European Community.
REFERENCES
- 1.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benbacer, L., E. Kut, L. Besnardeau, H. Laude, and B. Delmas. 1997. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 71:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, New York, N.Y.
- 5.Huang, Z., M. J. Fasco, and L. S. Kaminsky. 1996. Optimization of DNase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. BioTechniques 20:1012-1020. [DOI] [PubMed] [Google Scholar]
- 6.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Méndez, J. Plana-Durán, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansen, I. E. 1996. Intron insertion facilitates amplification of cloned virus cDNA in Escherichia coli while biological activity is reestablished after transcription in vivo. Proc. Natl. Acad. Sci. USA 93:12400-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones, P., J. Qiu, and D. Rickwood. 1994. RNA isolation and analysis, p. 15-46. BIOS Scientific Publishers Ltd, Oxford, United Kingdom.
- 9.Lewin, B. 1994. Genes V, p. 999-1032. Oxford University Press, Oxford, United Kingdom.
- 10.López-Moya, J. J., and J. A. García. 2000. Construction of a stable and highly infectious intron-containing cDNA clone of a plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 68:99-107. [DOI] [PubMed] [Google Scholar]
- 11.Méndez, A., C. Smerdou, A. Izeta, F. Gebauer, and L. Enjuanes. 1996. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology 217:495-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruggli, N., J. D. Tratschin, C. Mittelholzer, and M. A. Hofmann. 1996. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J. Virol. 70:3479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senapathy, P., M. B. Shapiro, and N. L. Harris. 1990. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183:252-278. [DOI] [PubMed] [Google Scholar]
- 16.Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak, Y. Tachiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solovyev, V. V., A. A. Salamov, and C. B. Lawrence. 1994. Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res. 22:5156-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiel, V., J. Herold, B. Schelle, and S. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 19.Wang, K., C. Boysen, H. Shizuya, M. I. Simon, and L. Hood. 1997. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques 23:992-994. [DOI] [PubMed] [Google Scholar]
- 20.Yamshchikov, V., V. Mishin, and F. Cominelli. 2001. A new strategy in design of (+)RNA virus infectious clones enabling their stable propagation in E. coli. Virology 281:272-280. [DOI] [PubMed] [Google Scholar]
- 21.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]