Abstract
Ligand-dependent down-regulation that leads to rapid and extensive loss of protein is characteristic of several nuclear steroid receptors, including human progesterone receptors (PRs). In breast cancer cells, >95% of PRs are degraded 6 h after the start of progestin treatment. The mechanism for down-regulation is unknown. We examined the role of PR phosphorylation by mitogen-activated protein kinases (MAPKs) in this process. Lactacystin and calpain inhibitor I, specific inhibitors of the 26S proteasome, blocked progestin-induced down-regulation, and ubiquitinated conjugates of PR accumulated in cells. Ligand-dependent PR degradation was also blocked by specific inhibition of p42 and p44 MAPKs. To define the targets of phosphorylation by this kinase, two serine/proline MAPK consensus sites on PR were mutated. We demonstrate that mutation of PR serine-294 to alanine (S294A) specifically and completely prevents ligand-dependent receptor down-regulation. We also find that rapid, ligand-independent degradation of immature PR intermediates occurs by a proteasome-mediated pathway. These results demonstrate that PR destruction, by either of two alternate routes, is mediated by the 26S proteasome. Specifically, down-regulation of mature PRs occurs by a mechanism in which ligand binding activates PR phosphorylation by MAPKs at a unique serine residue, which then targets the receptors for degradation.
Advanced-stage breast cancers often lack steroid hormone receptors and/or are resistant to endocrine therapies. Progesterone receptors (PRs) are key markers of steroid hormone dependence and indicators of disease prognosis in breast cancer; their loss signals development of an aggressive tumor phenotype associated with acquisition of enhanced sensitivity to growth factors (1, 2). Among the factors that regulate PR levels are their ligands. Within 6 to 8 h after occupancy by progestins, the receptors are extensively down-regulated, by mechanisms that remain unknown.
Regulation of PR expression by ligands occurs at both the protein and mRNA levels. At the mRNA level, the effects of progestins on at least six PR messages have been defined in T47D human breast cancer cells (3, 4). PR mRNA levels decrease gradually between 4 and 20 h after progestin treatment and then return to pretreatment levels 24 to 48 h later. However, the relationship between PR mRNA fluctuation and levels of the two PR protein isoforms is unknown, because of the number and heterogeneity of PR transcripts, which can encode either one or both receptor isoforms (3). In addition to these fluctuations in PR mRNA levels, PR protein levels are also extensively and rapidly down-regulated in response to ligand binding. Endogenous PRs, labeled by biosynthetic incorporation of 2H, 13C, and 15N dense amino acids, turn over with a half-life of 21 h in control cells, compared with 6 h in progestin-treated cells (4). The rate and extent of PR protein decrease reflects the time course of receptor occupancy by ligand and the fractional saturation of receptors (4). Antiprogestins also induce PR down-regulation but with much slower kinetics than agonists, suggesting a relationship between transcriptional activity of PR induced by ligand and receptor down-regulation (C.A.L. and K.B.H., unpublished results).
The biological role of hormone-dependent down-regulation of PR and other steroid receptors is unknown. One reason that cells may expend the energy to clear activated receptors is to attenuate their own transcriptional responses. Alternatively, nuclear receptor turnover may provide a mechanism to “reset” the transcriptional apparatus after each stimulus, so that previously modified receptors can be replaced with newly synthesized, fully functional molecules. Thus, at steady state in tissues in which PRs are constantly exposed to changing levels of physiological progesterone, receptor down-regulation may allow for continual reactivation of transcription at PR-regulated genes.
To define the mechanisms for ligand-dependent PR down-regulation, we studied the role of phosphorylation and receptor degradation by the 26S proteasome. The timed destruction of regulatory proteins by the ubiquitin-proteasome pathway is emerging as an important mechanism for the tight control of diverse cellular processes, including signal transduction from cell surface receptors (5), gene transcription (6), angiogenesis (7), and cell cycle progression (ref. 8; reviewed in refs. 9 and 10). Aberrations in this pathway or its protein substrates are implicated in several disease states ranging from Alzheimer's disease (11) to cancer (reviewed in ref. 12), and inhibitors of ubiquitination are candidates for cancer clinical trials (13, 14).
We now demonstrate that PRs are targeted for down-regulation by phosphorylation. A highly specific inhibitor of the 26S proteasome blocks ligand-dependent PR protein loss. The same result is produced by inhibition of p42 and p44 MAPKs, as well as by mutation of a single MAPK consensus phosphorylation site on PR. These data demonstrate that liganded PRs are substrates for MAPK-induced phosphorylation, which targets the receptors for degradation by the 26S proteasome.
Materials and Methods
Cell Lines and Reagents.
Estrogen-resistant T47Dco breast cancer cells, their monoclonal PR-negative T47D-Y derivatives, and T47D-YB cells, which are T47D-Y cells engineered to stably express the PR-B isoform, were previously described (15, 16). Construction of HeLa:B cells—HeLa cervical carcinoma cells stably expressing PR-B—was described recently (17). Cells were routinely seeded at 1 × 106 cells per dish, cultured in 10-cm dishes, and incubated in 5% CO2 at 37°C in a humidified environment as described (16). Stably transfected cells were also maintained in 700 mg/ml neomycin analog G418 (Life Technologies, Gaithersburg, MD). For MAPK activation studies, 30 ng/ml epidermal growth factor (EGF), was added to cells cultured in serum-free medium for 16–18 h before growth factor addition. Phosphospecific and total p42/p44 MAPK antibodies, phosphospecific p38 MAPK antibodies, and the MEK1 inhibitor (PD98059) were purchased from New England Biolabs. The p38 MAPK inhibitors (SB202190 and SB203580) were purchased from Upstate Biotechnology (Lake Placid, NY). Horseradish peroxidase-conjugated secondary antibodies were obtained from Collaborative Biomedical Products (Bedford, MA). Hemagglutinin (HA) epitope tag antisera were purchased from Babco (Richmond, CA). R5020 was obtained from NEN and used at 20 nM. Lactacystin, calpain inhibitor I (LLnL, N-acetyl-Leu-Leu-Nle-CHO), calpain inhibitor II (ALLM, N-acetyl-Leu-Leu-Met-CHO), and geldanamycin (GA) were purchased from Calbiochem.
Immunoblotting.
For detection of PR-A- and -B isoforms or MAPKs in whole cells lysates, cells growing in 10-cm dishes were washed twice in 4 ml of PBS and lysed by scraping in extraction buffer [1% (vol/vol) Triton X-100/10 mM Tris⋅HCl, pH 7.4/5 mM EDTA/50 mM NaCl/50 mM NaF/20 μg/ml Aprotinin/1 mM PMSF/2 mM Na3VO4]. Lysates were clarified by centrifugation for 10 min at maximum speed in a refrigerated microfuge. Soluble proteins in clarified lysates were quantified by the method of Bradford (GIBCO/BRL), and equal amounts of protein were resolved by SDS/PAGE (10% acrylamide for MAPKs; 7.5% acrylamide for PRs) and detected by immunoblotting (18).
Coimmunoprecipitation.
HeLa cells were transiently transfected with 3.0 μg of the β-galactosidase expression plasmid pCH110 (Amersham Pharmacia) to monitor transfection efficiency, 1.5 μg of cDNA encoding flag-tagged PR-B (17), and 2.0 μg of cDNA encoding HA-tagged ubiquitin (6), by using calcium phosphate precipitation as described (19). Immunoprecipitation of PR:flag proteins from cell lysates (1.2 mg) and immunoblotting were performed as decribed by Richer et al. (17).
Results and Discussion
Ubiquitination and the 26S Proteasome.
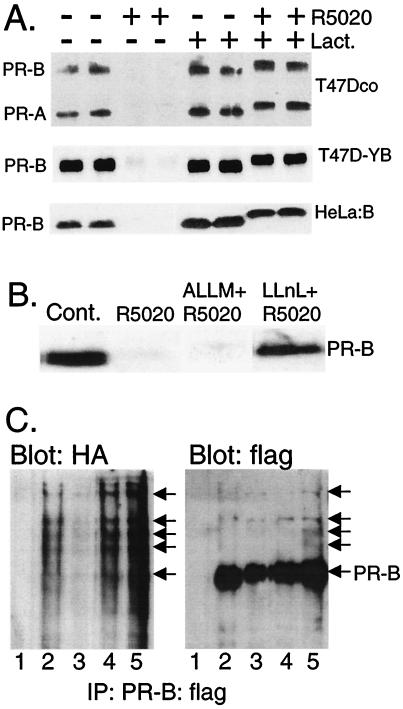
We examined the regulation of PR protein abundance by the ubiquitin-proteasome pathway after hormone treatment. T47Dco human breast cancer cells, which naturally express both the A and B isoforms of PR (20), were treated without or with the synthetic progestin R5020 to induce ligand-dependent receptor down-regulation (Fig. 1A). Disappearance of both PR isoforms was complete by 12 h of progestin treatment. However, R5020 had no effect on PR degradation in cells treated with lactacystin, a specific inhibitor of the 26S proteasome (ref. 21; reviewed in ref. 22). Similar results were obtained with progesterone (data not shown). Thus, human PRs appear to be substrates for regulated degradation by the 26S proteasome. Blockade of progestin-induced PR degradation by lactacystin also occurred in T47D-YB human breast cancer cells and in HeLa:B human cervical carcinoma cells, both of which stably express transfected recombinant human PR-B driven by the simian virus 40 (SV40) promoter (Fig. 1A). Because endogenous, naturally expressed receptors in T47Dco cells, as well as recombinant receptors whose stable expression is driven by heterologous promoters, are efficiently down-regulated, protein degradation rather than transcriptional regulation must account for this effect. The increased apparent molecular weight of lactacystin-stabilized, R5020-occupied PRs, as measured by their up-shift on SDS/PAGE gels (Fig. 1), reflects increased phosphorylation at multiple serine residues (23).
Figure 1.
PR down-regulation is mediated by the 26S proteasome. (A) T47Dco breast cancer cells, which express the natural B and A isoforms of PRs constitutively (15), and T47D-YB (16) and HeLa:B cells (17), which stably express the recombinant B isoform of PRs, were treated without or with the progestin R5020 (10 nM) for 12 h in the absence (DMSO solvent) or presence of lactacystin [Lact. (10 μM)], and PR protein (100 μg of total protein per lane) was detected by immunoblotting with PR-specific monoclonal antibodies. (B) Inhibition of PR down-regulation by calpain inhibitor I, but not calpain inhibitor II. T47D-YB cells were treated as in A, except with calpain inhibitor II [ALLM (25 μM)] or calpain inhibitor I [LLnL (25 μM)]. Each compound alone had no effect on PR abundance. (C) PR-ubiquitin conjugates in cells transiently overexpressing ubiquitin and PR-B. HeLa cells were transiently cotransfected with expression vectors encoding HA-tagged ubiquitin and epitope-flag-tagged wild-type PR-B (PR-B:flag) and treated for 4 h without (lane 2) or with R5020 (lane 3; 10 nM) in the absence or presence of lactacystin (lane 4; 10 μM) or LLnL (lane 5; 25 μM). PRs were immunoprecipitated by using anti-flag M2 affinity gel and visualized by immunoblotting with either HA- or flag-specific antibodies. Lane 1, nonspecific antibody and similar affinity gel. High-molecular-weight ubiquitinated forms of PR-B:flag are indicated (arrows).
PR degradation in the presence of R5020 appears to be specifically mediated by the 26S proteasome, because ALLM, which inhibits the activity of several degradative enzymes but not that of the 26S proteasome, had no effect on PR degradation (Fig. 1B). In contrast, LLnL, a related compound that inhibits several neutral cysteine proteases, including the cysteine protease activity of the 26S proteasome, effectively blocked PR degradation (Fig. 1B).
Although ubiquitin-independent degradation of substrates by the 26S proteasome is known (24), most proteins are targeted for destruction through this pathway by ligation to 76-amino acid ubiquitin molecules, assembled as polyubiquitin chains (reviewed in ref. 25). Polyubiquitinated proteins are often difficult to visualize. Only ≈3% of cyclin proteins exist as ubiquitin conjugates during early mitosis; these species exhibit a 90-s half-life (8). The proportion of PRs that exist as ubiquitin conjugates at a given time point may be even lower, because the receptors have a much longer half-life of several hours (4). To demonstrate ubiquitin-conjugated PR, flag-tagged PR-B and HA-tagged ubiquitin were transiently overexpressed in HeLa cells by cotransfection of the respective expression vectors (17). Cells were treated with R5020 for 4 h in the absence or presence of 26S proteasome inhibitors, to allow accumulation of PR-ubiquitin conjugates. flag-tagged PR-Bs were immunoprecipitated with anti-flag antibodies and visualized by immunoblotting with either HA- or flag-specific antibodies (Fig. 1C). Note that transiently overexpressed PRs are often incompletely down-regulated in response to R5020 and remain visible in immunoprecipitates; a 4-h time course was used to trap PR-ubiquitin conjugates. Additionally, PR:flag species of large molecular size accumulated in lysates from cells treated with R5020 in the presence of lactacystin or LLnL (Fig. 1C). These PR-B species were also recognized by HA-specific antibodies, indicating that they represent polyubiquitinated forms of B-receptors (Fig. 1C). Similar, large-molecular-sized chick PR complexes were recently identified in ubiquitin immunoprecipitates from progesterone-treated oviducts (26). Fig. 1C shows that ubiquitin conjugates were also present in immunoprecipitates containing unliganded PR:flag, suggesting that some ligand-independent ubiquitination occurs in this overexpression system (see below).
MAPK and Phosphorylation.
The yeast E2 ubiquitin-conjugating enzyme Cdc34 targets phosphorylated substrates for proteolytic degradation (reviewed in refs. 9 and 27), and phosphorylation is a positive signal for the ubiquitination and targeting of several proteins to the 26S proteasome, including cyclin E (28, 29), cyclin D1 (30), and IκBα (31). PRs are basally phosphorylated at multiple serine residues, and their phosphorylation is enhanced in response to hormone binding (ref. 32; reviewed in refs. 33 and 34). Specific functions for PR phosphorylation remain unassigned (35). At least two phosphorylation sites common to the A and B isoforms of PR, serine-294 and serine-345, are predominantly and latently phosphorylated after treatment of cells with progestins (36). Both of these sites are serine–proline consensus sites, PXX-S/T-P, for proline-directed kinases of the MAPK superfamily (37), but whether MAPK phosphorylates them is unknown. Furthermore, the ability of progestins, as well as estrogens, to activate MAPK in T47D breast cancer cells has been well-documented (38–40). We find that treatment of serum-starved T47D-YB cells with R5020 for 5 min modestly activates p42 and p44 MAPK, but not p38 MAPK, as measured by the presence of phosphorylated active kinase in immunoblots (data not shown).
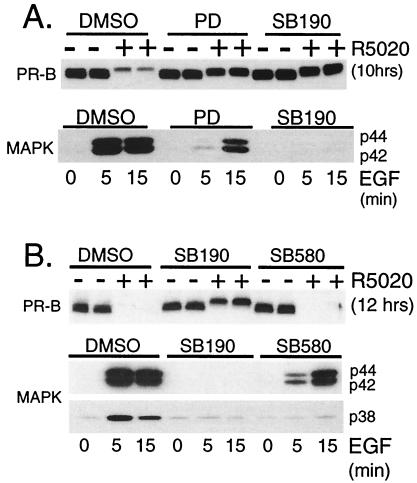
Because progestins activate MAPK (39) and phosphorylation of PRs occurs at two serine–proline MAPK consensus sites in response to progestins (36), we tested whether progestin-induced PR down-regulation depended on MAPK activation. R5020 was added to T47D-YB cells pretreated with either the MEK1 inhibitor, PD (41), or a putative specific inhibitor of p38 MAPK, SB202190 (SB190; ref. 42). The effects of these inhibitors on ligand-dependent PR down-regulation were assayed by immunoblotting (Fig. 2A). PD (100 μM) blocked PR protein loss by ≈70% in T47D-YB cells treated with R5020 for 10 h, and SB190 (40 μM) blocked down-regulation completely. To ensure the effectiveness and specificity of each inhibitor at the concentrations used, we examined p42 and p44 MAPK activity in whole-cell lysates from EGF-treated T47D-YB cells (Fig. 2A). PD (100 μM) effectively inhibited the robust activation of p42 and p44 MAPKs by EGF at 5 min, compared with vehicle-treated controls. Surprisingly, however, this effect was transient, and MAPK activity was only weakly inhibited by PD in cells treated with EGF for 15 min. In contrast, treatment of cells with EGF in the presence of SB190 (40 μM) resulted in complete inhibition of p42 and p44 MAPKs at all time points tested, indicating that this compound is not a selective inhibitor of p38 MAPK alone. Total MAPK levels remained constant and unchanged under these conditions, as measured by immunoblotting (data not shown). These results suggest that activation of either p42 and p44 MAPKs or p38 MAPK or activation of both contribute to ligand-dependent PR down-regulation.
Figure 2.
Activation of MAPK triggers ligand-dependent PR down-regulation. (A) MEK inhibitors block ligand-induced PR down-regulation. T47D-YB cells were pretreated with vehicle (DMSO) or with PD98059 [PD (100 μM)] or SB202190 [SB190 (40 μM)] for 30 min before challenge with R5020 [10 nM (Upper)] or EGF [30 ng/ml (Lower)] for the indicated times. PR-B protein levels (Upper) and MAPK activity (Lower) were measured by immunoblotting with PR- or MAPK-phosphospecific antibodies, respectively. Total MAPK levels remained unchanged (data not shown). (B) Ligand-induced PR down-regulation requires activation of p42 and p44 MAPKs, but not p38 MAPK. T47D-YB cells were pretreated with vehicle (DMSO), SB202190 [SB190 (40 μM)], or SB203580 [SB580 (40 μM)] for 30 min before challenge with R5020 [10 nM (Upper)] or EGF [30 ng/ml (Lower)] for the indicated times. PR-B protein levels (Upper) were measured with specific antibodies, and p42/p44 and p38 MAPK activities (Lower) were measured with phosphospecific antisera for each kinase; 100 μg of protein was loaded per lane.
To distinguish between p42 and p44 MAPKs vs. p38 MAPK, we tested SB203580 (SB580; ref. 42), an additional inhibitor of p38 MAPK (Fig. 2B). The activities of p42 and p44 MAPKs and p38 MAPK were concomitantly assayed in EGF-treated cells from the same experimental set. Again, SB190 blocked PR down-regulation. This compound inhibited both EGF-stimulated p42 and p44 MAPKs and p38 MAPK activities after 5 and 15 min of EGF treatment. In contrast, SB580 did not block PR degradation. This compound weakly inhibited activation of p42 and p44 MAPKs in cells treated for 5 min with EGF, but was unable to inhibit MAPK activity in cells treated with EGF for 15 min. Under the same conditions, SB580 fully inhibited p38 MAPK activity. Thus, protection of PRs from ligand-induced down-regulation correlated with the ability of each inhibitor to block p42 and p44 MAPK activation. Several experimental conditions and inhibitor concentrations led to similar conclusions (data not shown). PD was a more effective inhibitor of p42 and p44 MAPKs than SB580 at 5 min, also the peak of MAPK activation by progestins (39). Thus, early activation of p42 and p44 MAPKs, but not p38 MAPK, may be required for progestin-induced PR down-regulation. Because PR phosphorylation is also a rapid event (23), neither kinase activation nor PR phosphorylation appears to be rate limiting for PR degradation.
Phosphorylation of Serine-294.
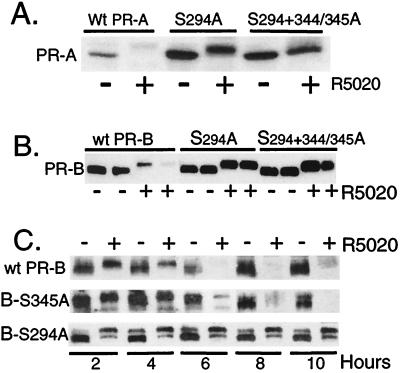
To assess whether the requirement for MAPK results from a direct effect on PR phosphorylation, serine-294 alone or together with serine-344 or -345 was replaced with alanine in both the A and B isoforms of PR. Ligand-dependent receptor down-regulation of the mutants was then measured in transient expression systems (Fig. 3 A and B). Mutant A and B receptors were fully functional as measured by transcriptional assays (data not shown; see ref. 35). HeLa cells transiently expressing either wild-type or mutant PR-A (Fig. 3A) or PR-B (Fig. 3B) were treated without or with R5020, and the amount of PR protein was measured in whole-cell lysates by immunoblotting. PR-A and PR-B, containing mutations at serine-294 alone or at both phosphorylation sites, were completely resistant to R5020-induced down-regulation. Thus, mutation of the single amino acid serine-294 to alanine was sufficient to stabilize both PR isoforms in the presence of R5020. Mutant PRs still underwent the characteristic ligand-induced upward mobility shifts on SDS/PAGE gels, which was caused by phosphorylation at multiple sites (23, 35). Clearly, phosphorylation at serine-294 is not required for this up-shift (ref. 36; Fig. 3). It is interesting that the basal expression levels of mutant PR-A were consistently stabilized, even in the absence of ligand (Fig. 3A), suggesting that PR-As undergo a low level of ligand-independent turnover (see Figs. 5 and 6).
Figure 3.
Mutation of serine-294 to alanine at a MAPK consensus site stabilizes PR in the presence of progestin. (A) Mutant S294A and S294+S344/345A PR-As are resistant to R5020-induced receptor down-regulation. HeLa cells were transiently transfected with cDNA vectors (1 μg) encoding either wild-type PR-A or the S294A or S294+344/345A mutants of PR-A and then treated without or with R5020 (10 nM) for 8 h. PR protein was measured in whole-cell lysates (100 μg) by immunoblotting. (B) Mutant S294A and S294+344/345A PR-Bs are resistant to R5020-induced receptor down-regulation. Duplicate cultures of HeLa cells were transfected with wild-type PR-B or each phosphomutant of PR-B and treated without or with R5020 for 18 h. Protein (100 μg per lane) was loaded, and PR levels were measured by using PR-specific monoclonal antibodies. (C) The S294A PR-Bs stably expressed in T47D-Y cells are resistant to ligand-induced down-regulation. PR-negative breast cell lines stably expressing either an S344/345A (B-S345A) mutant or the S294A (B-S294A) mutant PR-B receptors were produced by transfection of receptor expression vectors containing the neomycin-resistance gene into T47D-Y cells and selected for growth in G418. Neoresistant clonal cell lines were screened for PR expression, and S344/345A or S294A PR-containing cells or wild-type PR-B-containing T47D-YB cells (wt-PR-B) were treated without or with R5020 (10 nM) for 2–10 h; protein levels were measured with PR-specific antibodies. 150–200 μg of protein was loaded per lane.
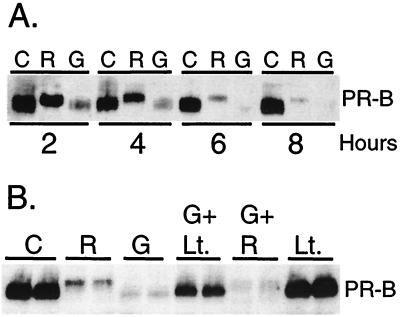
Figure 5.
Ligand-independent degradation of immature PRs. (A) Immature PRs are rapidly degraded in the absence of ligand. T47D-YB cells were treated without [C (EtOH vehicle)] or with R5020 alone [R (10 nM)] or with the hsp 90-binding agent GA (G) alone (10 μg/ml) for 2, 4, 6, and 8 h, and PR-B protein in whole-cell lysates was measured by immunoblot analysis. (B) Ligand-independent degradation is mediated by the 26S proteasome. Duplicate cultures of T47D-YB cells were treated as indicated, without [C (EtOH vehicle)] or with R5020 [R (10 nM)] alone, GA [G (10 μg/ml)] alone, or with GA plus lactacystin [Lt. (10 nM; 30-min pretreatment)], GA plus R5020, or lactacystin alone for 6 h. PR-B protein in whole-cell lysates (100 μg of protein per lane) was measured by immunoblot analysis with PR-specific antisera.
Figure 6.
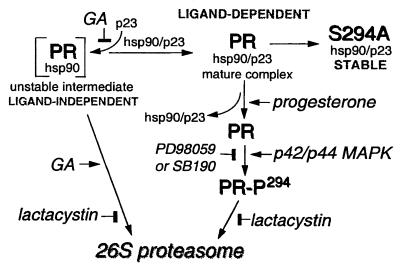
Model for regulated loss of PR protein by the 26S proteasome and for ligand-independent and ligand-dependent PR down-regulation. Mature PR complexes bind progesterone, leading to ligand-dependent phosphorylation of serine 294 by p42/p44 MAPKs, which serves as a signal for PR degradation by the 26S proteasome. Inhibition of p42/p44 MAPKs (PD98059 or SB190) or the 26S proteasome (lactacystin) blocks loss of wild-type PR in the presence of progestins. S294A-mutant PRs are resistant to ligand-dependent down-regulation. In the presence of GA, rapid ligand-independent degradation of immature PR complexes also occurs by a 26S proteasome-dependent pathway.
We noted that progestin-induced PR down-regulation is slowed in transient expression systems (Fig. 3 A and B), probably because excessive amounts of receptor proteins are produced. The ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3) required for polyubiquitination of protein substrates may be limiting in such overexpression systems (8, 9, 43). To analyze PR down-regulation under more physiological conditions, either S294A mutant or S344A/S345A mutant PR-B receptors were stably introduced into PR-negative T47D-Y cells (16) and compared with wild-type PR-B, also stably expressed in T47D-Y cells (Fig. 3C). Cells were treated with R5020 for 2–10 h, and the abundance of receptor protein was monitored by immunoblotting. The S294A PR-B mutant was completely resistant to progestin-induced down-regulation over the 10-h time course, whereas both S344A/S345A and wild-type receptors were largely degraded after 6–8 h (Fig. 3C). Mutant S294A PR-B receptors were resistant to degradation for as long as 42 h of R5020 treatment (data not shown). Other independently derived clones of T47D-Y cells stably expressing PR mutants gave similar results.
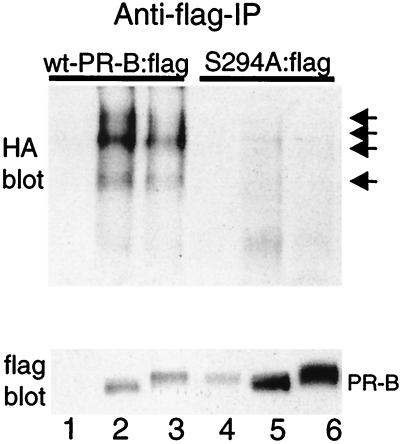
To test whether S294A mutant PR were ubiquitinated in response to R5020 treatment, flag-tagged wild-type or S294A mutant PR-B and HA-tagged ubiquitin were transiently overexpressed in HeLa cells (ref. 17; as in Fig. 1). Cells were treated with R5020 for 4 h, and flag-tagged wild-type or mutant PR-Bs were immunoprecipitated with anti-flag antibodies and visualized by immunoblotting with either HA- or flag-specific antibodies (Fig. 4). Transiently overexpressed wild-type PRs were incompletely down-regulated in response to R5020, as previously observed (Fig. 1 and 3). Immunoblotting with HA-specific antibodies revealed PR:flag species of large molecular size in lysates from cells transiently overexpressing wild-type PR-Bs, but not S294A mutant PR-Bs, indicating that S294A mutant PR-B fails to undergo efficient ubiquitination (Fig. 4). Immunoblotting with flag-specific antibodies demonstrated much higher levels of unconjugated S294A mutant PR-B protein compared with wild-type PR-B, reflecting the greater stability of S294A mutant PR-B (Fig. 3). Antiflag antibodies also recognized ubiquitin-conjugated wild-type PR-B species (data not shown).
Figure 4.
Mutant S294A PR-Bs fail to undergo ubiquitination. HeLa cells were transiently cotransfected with expression vectors encoding HA-tagged-ubiquitin and either epitope-flag-tagged wild-type PR-B [wt-PR-B:flag (lanes 1–3)] or epitope-flag-tagged S294A mutant PR-B [S294A:flag (lanes 4–6)] and treated for 4 h without (lanes 2 and 5) or with R5020 (lanes 3 and 6). PRs were immunoprecipitated by using an anti-flag M2 affinity gel and were visualized by immunoblotting with either HA- or flag-specific antibodies. Lanes 1 and 3, nonspecific antibody and similar affinity gel. High-molecular-weight ubiquitinated forms in immunoprecipitates from lysates of cells containing wild-type but not S294A mutant PR-Bs are indicated by arrows.
Thus, specific phosphorylation of PR on serine-294 by MAPK appears to be required for progestin-induced PR ubiquitination and degradation by the 26S proteasome. Ligand binding may alter PR conformation such that it becomes a better substrate for p42 and p44 MAPKs. Whether PRs are uniquely phosphorylated in response to progestin-activated MAPKs, or in response to basally activated or preactivated MAPKs, is unknown. Previous studies to define a function for PR phosphorylation have relied heavily on the use of transient transfection assays (35, 44), but the behavior of such overexpressed receptors may differ significantly from that of natural or stably expressed proteins. This study is the first demonstration of a clear and specific role for any phosphorylated site on human PRs, in which at least nine sites have been documented (33, 34).
Ligand-Independent Degradation.
Because expression of S294A mutant PR-As appeared to be stabilized in a ligand-independent manner (Fig. 3A), we determined whether PR protein degradation via the 26S proteasome can also proceed in the absence of ligand. We took advantage of the fact that unliganded mature PRs exist in heteromeric complexes with several heat-shock protein (hsp) chaperones, including p23, hsp 70, and hsp 90. GA is a benzoquinone that blocks addition of the chaperone-assembly factor, p23, to the PR/hsp90 complex and thereby blocks PR maturation to a ligand-binding form (45). In the presence of GA, unliganded immature PR-Bs are degraded at an even faster rate than that induced by ligand (Fig. 5A). However, PRs in GA-treated cells fail to undergo the characteristic upshifts associated with ligand-induced receptor phosphorylation, indicating that they are not similarly phosphorylated (Fig. 5A; 2–4 h). To determine whether GA-induced loss of immature PRs is also mediated by the 26S proteasome, cells were treated with GA in the absence or presence of lactacystin (Fig. 5B). Lactacystin blocked GA-induced PR protein loss compared with controls containing either GA or lactacystin alone, indicating a role for the 26S proteasome in ligand-independent PR degradation. PR levels were not significantly raised by lactacystin, indicating minimal accumulation of newly synthesized PRs over this time course. It is unclear how immature PR heterocomplexes are targeted to the 26S proteasome in the absence of ligand and whether phosphorylation plays a role in this process. Indeed, we found that S294A mutant PR-Bs were also rapidly degraded in the presence of GA (data not shown), suggesting that the targeting mechanism for unliganded PRs differs from that of liganded PRs. Rapid degradation of other hsp 90-chaperoned proteins, including c-erbB2 (5), Raf-1 (46), and glucocorticoid receptors (47), in the presence of GA also occurs by an ubiquitin-dependent proteasomal mechanism (reviewed in ref. 48).
Summary.
Substrates for the ubiquitin-proteasome pathway often contain specific amino acid sequence motifs required for their degradation (8). Examination of the amino acid sequences immediately surrounding serine-294 of PRs revealed the presence of such a motif; a 9-amino acid consensus “destruction box” originally described in cyclin molecules and required for their destruction by the ubiquitin-proteasome pathway (8, 49). It is interesting that, in PRs, this sequence is nested within the serine-294 MAPK consensus site. Another consensus destruction box motif is located further downstream at amino acid positions 399–407 of human PRs, encompassing basal phosphorylation site serine-400, which is an in vitro target of cyclin-dependent kinase 2 (Cdk2) (50). In addition to other basal phosphorylation sites in human PR, this site also undergoes a modest increase in phosphorylation in response to ligand (36). Although the functional significance of these two sequences is unknown, their presence at different positions in the PR N terminus may explain why destruction of immature receptor intermediates and ligand-induced phosphorylation of mature PRs, followed by their down-regulation, ultimately proceed via targeting to a common degradation pathway. Phosphorylation of PRs at serine-294 by MAPK may expose one destruction box motif(s) for targeted destruction by the 26S proteasome. Phosphorylation of the second site may lead to degradation of immature or unliganded receptors. Indeed, structural analysis of the human PR-A amino terminus indicates that both phosphorylation sites are exposed on the surface of the protein molecule (D. L. Bain and K.B.H., unpublished results). Similarly, phosphorylation may also target mouse glucocorticoid receptors for degradation (51). Simultaneous mutation of a minimum of three phosphorylation sites resulted in more stable glucocorticoid receptors in the presence of ligand; the kinases involved have not been defined. Estrogen receptors recently have been shown to be degraded by the ubiquitin-proteasome pathway in vitro (44), but the role of phosphorylation, if any, is unknown.
In summary (Fig. 6), these results demonstrate that rapid, ligand-independent destruction of immature PR intermediates is mediated by the 26S proteasome. Additionally, we show that ligand-dependent down-regulation of mature PRs occurs by a mechanism involving phosphorylation of PRs by p42/p44 MAPKs at serine-294, thus targeting PRs for ubiquitination and destruction by the 26S proteasome. This is a demonstration of a specific effect of the MAPK pathway on a hormone-dependent nuclear receptor phosphorylation site and a definition of the function of this phosphorylation. Cross-talk between MAPK- and PR-signaling pathways suggests mechanisms by which steroid hormone resistance and acquisition of growth factor responsiveness are integrated in advanced breast cancer (17, 18).
Acknowledgments
We are grateful to Bruce E. Clurman, Fred Hutchinson Cancer Research Center, Seattle, WA, for the gift of the HA-tagged ubiquitin construct; Glenn S. Takimoto, University of Colorado Health Sciences Center, for the gift of the S344/345A PR-A mutant receptor construct; and Jennifer K. Richer, University of Colorado Health Sciences Center, for the gift of the HeLa:B cells stably expressing epitope flag-tagged human PR-B and for the PR:flag expression vector construct. This work was supported by The National Institutes of Health Grants DK53825 (C.A.L.) and DK48238 and CA26869 (K.B.H.).
Abbreviations
- PR
progesterone receptor
- MAPK
mitogen-activated protein kinase
- EGF
epidermal growth factor
- HA
hemagglutinin
- LLnL
N-acetyl-Leu-Leu-Nle-CHO
- ALLM
N-acetyl-Leu-Leu-Met-CHO
- GA
geldanamycin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Horwitz K B, McGuire W L. Steroids. 1975;25:497–505. doi: 10.1016/0039-128x(75)90027-6. [DOI] [PubMed] [Google Scholar]
- 2.Elledge R M, McGuire W L, Osborne C K. Semin Oncol. 1992;19:244–253. [PubMed] [Google Scholar]
- 3.Wei L L, Krett N L, Francis M D, Gordon D F, Wood W M, O'Malley B W, Horwitz K B. Mol Endocrinol. 1988;2:62–72. doi: 10.1210/mend-2-1-62. [DOI] [PubMed] [Google Scholar]
- 4.Nardulli A M, Katzenellenbogen B S. Endocrinology. 1988;122:1532–1540. doi: 10.1210/endo-122-4-1532. [DOI] [PubMed] [Google Scholar]
- 5.Mimnaugh E G, Chavany C, Neckers L. J Biol Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 6.Treier M, Staszewski L M, Bohmann D. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 7.Oikawa T, Sasaki T, Nakamura M, Shimamura M, Tanahashi N, Omura S, Tanaka K. Biochem Biophys Res Commun. 1998;246:243–248. doi: 10.1006/bbrc.1998.8604. [DOI] [PubMed] [Google Scholar]
- 8.Glotzer M, Murray A W, Kirschner M W. Nature (London) 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 9.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 10.Pagano M. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 11.Kim T W, Pettingell W H, Hallmark O G, Moir R D, Wasco W, Tanzi R E. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 12.Spataro V, Norbury C, Harris A L. Br J Cancer. 1998;77:448–455. doi: 10.1038/bjc.1998.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg D. The Scientist. 1998;12:10–13. [Google Scholar]
- 14.Mikulski S M, Viera A, Deptala A, Darzynkiewicz Z. Int J Oncol. 1998;13:633–644. doi: 10.3892/ijo.13.4.633. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz K B, Mockus M B, Lessey B A. Cell. 1982;28:633–642. doi: 10.1016/0092-8674(82)90218-5. [DOI] [PubMed] [Google Scholar]
- 16.Sartorius C A, Groshong S D, Miller L A, Powell R L, Tung L, Takimoto G S, Horwitz K B. Cancer Res. 1994;54:3868–3877. [PubMed] [Google Scholar]
- 17.Richer J K, Lange C A, Manning N G, Owen G, Powell R, Horwitz K B. J Biol Chem. 1998;273:31317–31326. doi: 10.1074/jbc.273.47.31317. [DOI] [PubMed] [Google Scholar]
- 18.Lange C A, Richer J K, Shen T, Horwitz K B. J Biol Chem. 1998;273:31308–31316. doi: 10.1074/jbc.273.47.31308. [DOI] [PubMed] [Google Scholar]
- 19.Sartorius C A, Tung L, Takimoto G S, Horwitz K B. J Biol Chem. 1993;268:9262–9266. [PubMed] [Google Scholar]
- 20.Horwitz K B, Alexander P S. Endocrinology. 1983;113:2195–2201. doi: 10.1210/endo-113-6-2195. [DOI] [PubMed] [Google Scholar]
- 21.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 22.Fenteany G, Schreiber S L. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- 23.Sheridan P L, Evans R M, Horwitz K B. J Biol Chem. 1989;264:6520–6528. [PubMed] [Google Scholar]
- 24.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Nature (London) 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 25.Pickart C M. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 26.Syvala H, Vienonen A, Zhuang Y H, Kivineva M, Ylikomi T, Tuohimaa P. Life Sci. 1998;63:1505–1512. doi: 10.1016/s0024-3205(98)00417-2. [DOI] [PubMed] [Google Scholar]
- 27.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Won K A, Reed S I. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 29.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 30.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 32.Rao K V, Peralta W D, Greene G L, Fox C F. Biochem Biophys Res Commun. 1987;146:1357–1365. doi: 10.1016/0006-291x(87)90799-6. [DOI] [PubMed] [Google Scholar]
- 33.Takimoto G, Horwitz K. Trends Endocrinol Metab. 1993;4:1–7. doi: 10.1016/1043-2760(93)90056-k. [DOI] [PubMed] [Google Scholar]
- 34.Weigel N L. Biochem J. 1996;319:657–667. doi: 10.1042/bj3190657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takimoto G S, Hovland A R, Tasset D M, Melville M Y, Tung L, Horwitz K B. J Biol Chem. 1996;271:13308–13316. doi: 10.1074/jbc.271.23.13308. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Beck C A, Poletti A, Edwards D P, Weigel N L. Mol Endocrinol. 1995;9:1029–1040. doi: 10.1210/mend.9.8.7476977. [DOI] [PubMed] [Google Scholar]
- 37.Davis R J. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 38.Migliaccio A, Domenico M D, Cstoria G, Falco A D, Bontempo P, Nola E, Auricchio F. EMBO J. 1996;15:1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 39.Migliaccio A, Piccolo D, Castoria G, Di Domenico M, Bilancio A, Lombardi M, Gong W, Beato M, Auricchio F. EMBO J. 1998;17:2008–2018. doi: 10.1093/emboj/17.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Improta-Brears T, Whorton A R, Codazzi F, York J D, Meyer T, McDonnell D P. Proc Natl Acad Sci USA. 1999;96:4686–4691. doi: 10.1073/pnas.96.8.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, et al. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 43.Haas A L, Siepmann T J. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 44.Nawaz Z, Lonard D M, Dennis A P, Smith C L, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith D F, Whitesell L, Nair S C, Chen S, Prapapanich V, Rimerman R A. Mol Cell Biol. 1995;15:6804–6812. doi: 10.1128/mcb.15.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulte T W, Blagosklonny M V, Ingui C, Neckers L. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 47.Whitesell L, Cook P. Mol Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 48.Pratt W B. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 49.King R W, Glotzer M, Kirschner M W. Mol Biol Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Beck C A, Poletti A, Clement J P, Prendergast P, Yip T T, Hutchens T W, Edwards D P, Weigel N L. Mol Endocrinol. 1997;11:823–832. doi: 10.1210/mend.11.6.0006. [DOI] [PubMed] [Google Scholar]
- 51.Webster J C, Jewell C M, Bodwell J E, Munck A, Sar M, Cidlowski J A. J Biol Chem. 1997;272:9287–9293. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]