Abstract
The alpha/beta interferon (IFN-α/β)-induced STAT signal transduction pathway leading to activation of the ISGF3 transcription complex and subsequent antiviral responses is the target of viral pathogenesis strategies. Members of the Rubulavirus genus of the Paramyxovirus family of RNA viruses have acquired the ability to specifically target either STAT1 or STAT2 for proteolytic degradation as a countermeasure for evading IFN responses. While type II human parainfluenza virus induces STAT2 degradation, simian virus 5 induces STAT1 degradation. The components of the IFN signaling system that are required for STAT protein degradation by these paramyxoviruses have been investigated in a series of human somatic cell lines deficient in IFN signaling proteins. Results indicate that neither the IFN-α/β receptor, the tyrosine kinases Jak1 or Tyk2, nor the ISGF3 DNA-binding subunit, IFN regulatory factor 9 (IRF9), is required for STAT protein degradation induced by either virus. Nonetheless, both STAT1 and STAT2 are strictly required in the host cell to establish a degradation-permissive environment enabling both viruses to target their respective STAT protein. Complementation studies reveal that STAT protein-activating tyrosine phosphorylation and functional src homology 2 (SH2) domains are dispensable for creating a permissive STAT degradation environment in degradation-incompetent cells, but the N terminus of the missing STAT protein is essential. Protein-protein interaction analysis indicates that V and STAT proteins interact physically in vitro and in vivo. These results constitute genetic and biochemical evidence supporting a virus-induced, IFN-independent STAT protein degradation complex that contains at least STAT1 and STAT2.
The primary antiviral cytokines produced by higher eukaryotes are the alpha/beta interferons (IFN-α and IFN-β; referred to herein as IFN) that function directly on target cells by creating an antiviral state that blocks virus replication (24). The molecular basis for most antiviral effects induced by IFN requires IFN-induced mRNA and protein synthesis (46). IFN activates a transcriptional complex, ISGF3, composed of three proteins. Two subunits are members of the signal transducer and activator of transcription (STAT) family, STAT1 and STAT2, that heterodimerize and complex with a third protein, IRF9, a member of the interferon regulatory factor (IRF) family that provides DNA recognition.
The general mechanism leading to activation of ISGF3 has been well characterized (reviewed in references 20 and 46). IFN binding induces aggregation of a multichain receptor, causing the receptor-associated tyrosine kinases Jak1 and Tyk2 to phosphorylate the receptor cytoplasmic domain. The receptor phosphotyrosine provides a docking site for the src homology 2 (SH2) domain of the latent cytoplasmic STAT2 and/or STAT2-IRF9 complexes (28). STAT2 then becomes phosphorylated on tyrosine 690, providing a docking site for the latent STAT1. Following STAT1 phosphorylation on tyrosine 701, the two STATs heterodimerize via intermolecular SH2 domain-phosphotyrosine interaction (44) and, together with IRF9, form an active ISGF3 heterotrimer that can bind to IFN-stimulated gene (ISG) promoter IFN-stimulated response elements (ISRE). STAT proteins are long-lived, and their inactivation has been shown to involve dephosphorylation by a nuclear protein tyrosine phosphatase and recycling of the inactivated STATs (3, 17, 18, 33, 36).
It is not surprising to find that many, if not all, viruses have evolved strategies to impede host IFN responses (15). Evolution of enhanced IFN resistance can lead to highly infectious viruses and/or persistent infections (4, 11, 13, 14, 27, 47). Recently, the IFN antagonist strategies used by some negative-stranded RNA viruses have been determined to act directly on the ISGF3 STAT protein subunits. The paramyxovirus simian virus 5 (SV5) was found to evade IFN responses by specifically targeting the STAT1 protein for proteolytic degradation. This destruction of STAT1 was found to be mediated by expression of a single virus-encoded protein called V (11, 12, 54). Human parainfluenza virus 2 (HPIV2) blocks IFN signaling by preferentially inducing degradation of STAT2 and not STAT1 (40, 55). In common with SV5, the expression of the HPIV2 V protein from a cDNA clone is sufficient to abolish IFN-responsive transcription as a result of STAT2 destabilization (40). These two paramyxovirus V proteins have ∼50% amino acid sequence identity in their ∼220-amino-acid length, yet they specifically recognize and catalyze the destruction of only one of the two IFN-responsive STAT proteins.
The mechanistic basis for the selective STAT protein degradation mediated by paramyxovirus V proteins is not entirely understood, but the available evidence indicates that the V protein IFN antagonism involves the subjugation of cellular proteasome degradation systems (12, 40). As IFNs have been shown to regulate the expression and distribution of cellular proteases, proteasome subunits, and ubiquitin-like modifiers (16, 35, 56), one attractive mechanistic hypothesis for the observed virus-induced STAT degradation is that IFN signaling itself plays a role in establishing a cellular state permissive for degradation.
Human somatic cell lines that do not respond to IFN were tested for their ability to support specific STAT protein degradation in response to SV5 and HPIV2 infection. Evidence is provided that indicates that both STAT1 and STAT2 proteins are needed in the host cell to create a degradation-competent state, with one STAT acting as the degradation substrate while the other serves an essential accessory function. Despite this strict requirement for both STAT1 and STAT2 components of ISGF3, neither intact IFN signaling nor conventional STAT activation and dimerization is needed either for establishing degradation competence or for selective target recognition. Instead, complementation of defective degradation is achieved by expression of an N-terminal fragment of the missing STAT protein. The STAT proteins and V proteins interact in solution, suggestive of a multisubunit degradation complex. These findings indicate that while SV5 and HPIV2 have evolved to specifically target STAT1 or STAT2 for degradation, IFN antagonism is accomplished through the use of similar cellular mechanisms.
MATERIALS AND METHODS
Cells and viruses.
Human HEC-1B cells (ATCC HTB-113), 2fTGH cells, and 2fTGH-derived cell lines U1A (Tyk2 deficient), U2A (IRF9 deficient), U3A (STAT1 deficient), U4A (Jak1 deficient), and U6A (STAT2 deficient) were grown in Dulbecco's modified Eagle's medium supplemented with 10% Cosmic Calf Serum (HyClone). U6A cells complemented with chimeric STAT2-STAT1 cDNA (originally called N2βT [30], but referred to herein as N2:C1) and an expression vector for the STAT1-STAT2 hybrid (originally called N1 [30], but referred to herein as N1:C2) were the generous gift of George Stark and Xiaoxia Li (Cleveland Clinic Research Foundation, Cleveland, Ohio). Expression plasmids for STAT2 Y690F and STAT2 R601K were the generous gift of James Darnell (Rockefeller University, New York). U6A cells were transfected to produce stably transfected cell lines in medium containing 500 μg of G418 per ml as described previously (23).
SV5 strain W3A (derived from a genetically defined recombinant virus system [19, 26]) and HPIV2 (Greer strain) were provided by Robert Lamb (Northwestern University) and Griffith Parks (Wake Forest University, N.C.) and were propagated and counted in simian CV1 cells. Plaque assays were performed on CV1 cells by using an overlay containing 0.5% agar with DMEM and 10 mM HEPES (pH 7.2). Cells were fixed at 4 to 6 days postinfection with 3.7% formaldehyde, and plaques were visualized after being stained with 0.1% crystal violet in 20% ethanol.
Infection, cell extraction, and immunoblotting.
For degradation assays, cells were infected with SV5 or HPIV2 at a multiplicity of infection (MOI) of 10 to 100 PFU/cell or mock infected and harvested for analysis at 16 h postinfection. Whole-cell extracts were prepared as described before (40). Total protein was quantitated, and equal amounts (15 to 30 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 7% gel and transferred to nitrocellulose filters. Immunoblotting was performed with commercial antiserum (Santa Cruz Biochemical) specific for STAT1α (C24) or STAT2 (C20) and processed for chemiluminescent detection. For detection of viral nucleocapsid proteins, antiserum raised against HPIV2-infected cells that cross-reacts with SV5 was used (Whittaker Biochemicals). Detection of the N2:C1 construct was accomplished by using antiserum specific for the Flag epitope tag (Zymed Biochemicals).
Reporter gene assays.
For luciferase assays, cells were transfected using Superfect reagent (Qiagen) by the manufacturer's method with a cytomegalovirus (CMV)-lacZ plasmid as a control for transfection efficiency, a reporter gene, and either empty vector or the cDNA expression plasmids indicated. For IFN-γ responses, the reporter gene contains four copies of the m67-SIE linked to a TATA box and the firefly luciferase open reading frame (ORF) (5). The IFN-α/β-responsive reporter gene contained five copies of the ISG54 ISRE element upstream of the TATA box and the firefly luciferase ORF. After 24 h, transfection medium was replaced with fresh medium or medium supplemented with IFN. Cells were harvested 6 h later in luciferase assay lysis buffer, and luciferase activity was measured according to the manufacturer's protocol (Promega). Values for luciferase activity were normalized to β-galactosidase activity. In all cases, average values of triplicate experiments are shown, normalized to IFN-treated controls. To express V proteins, STAT2, and STAT2 fragments, PCR products encompassing the indicated regions were generated and subcloned into the expression vector pEF-HA (gift of Netai Singha, Mt. Sinai Medical School), in frame with an N-terminal epitope tag. All constructs were verified by DNA sequencing.
Protein interaction analysis.
For glutathione S-transferase (GST) fusion proteins, SV5 and HPIV2 ORFs were excised from mammalian expression vectors and ligated to pGEX-5X (Pharmacia). Fusion proteins were induced with 0.5 mM IPTG (isopropylthiogalactopyranoside) and purified with glutathione-agarose by standard methods described elsewhere (1, 22). For identification of protein complexes, cellular protein whole-cell extracts (2 to 5 mg) were incubated with purified fusion protein-agarose beads in whole-cell extract buffer for 12 h and then washed five times with whole-cell extract buffer. Bound proteins were eluted by boiling in protein gel loading buffer and separated by SDS-PAGE for immunoblotting.
For protein immunoprecipitation, 60-mm dishes were transfected with cDNA encoding Flag epitope-tagged SV5 V, and whole-cell extracts were immunoprecipitated overnight with 5 μl of M2 affinity gel (Sigma). Pellets were washed five times with whole-cell extract buffer. Bound proteins were eluted by boiling in protein gel loading buffer and separated by SDS-PAGE for immunoblotting.
RESULTS
STAT1 and STAT2 but not IRF9 are necessary for STAT degradation.
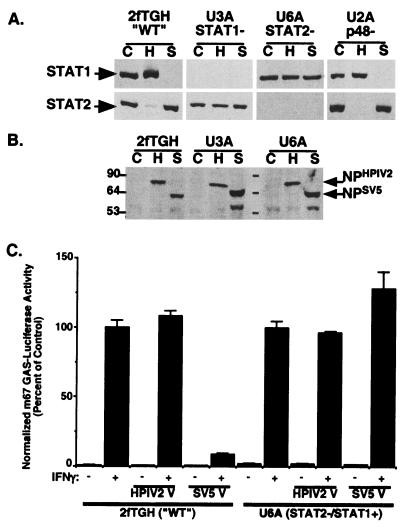
The ability of the paramyxoviruses SV5 and HPIV2 to induce a specific loss of cellular STAT1 (for SV5) and STAT2 (for HPIV2) is observed upon infection of human cell lines (Fig. 1A) (41). The 2fTGH cell line is the parent of IFN-unresponsive daughter cell lines (41) that contain single-gene defects in components of the IFN signaling pathway (Table 1) (reviewed in references 10 and 45). U3A cells are defective for STAT1 expression (34, 38), U6A cells are defective for STAT2 expression (29), and U2A cells are defective for IRF9 expression (25).
FIG. 1.
STAT1- and STAT2-deficient cells are not permissive for STAT protein degradation. (A) 2fTGH, U3A, U6A, and IRF9/p48-deficient U2A cells were mock infected (lanes C) or infected with HPIV2 (H) or SV5 (S). Cell lysates were separated and transferred to membranes for immunoblotting with antiserum to STAT1 (top panels) or STAT2 (bottom panels). (B) Immunoblotting with antiserum for paramyxovirus nucleocapsid proteins. Sizes are shown in kilodaltons. (C) 2fTGH and U6A cells were transfected with an IFN-γ-dependent luciferase reporter gene in the presence or absence of coexpressed HPIV2 V protein or SV5 V protein as indicated. Data represent normalized luciferase values from triplicate samples, expressed as a percentage of that in IFN-γ-stimulated controls. WT, wild type.
TABLE 1.
Paramyxovirus degradation of STAT1 and STAT2a
| Cell line | STAT | Degradation competence
|
|
|---|---|---|---|
| HPIV2 | SV5 | ||
| 2fTGH (parent) | 1 | − | + |
| 2 | + | − | |
| U3A (STAT1 null) | 1 | Null | Null |
| 2 | − | − | |
| U3A + STAT1 (complemented) | 1 | − | + |
| 2 | + | − | |
| U3A + STAT1 Y701F | 1 | − | + |
| 2 | + | − | |
| U3A + STAT1 R602K | 1 | − | + |
| 2 | + | − | |
| U6A (STAT2 null) | 1 | − | − |
| 2 | Null | Null | |
| U6A + STAT2 (complemented) | 1 | + | + |
| 2 | + | − | |
| U6A + STAT2 Y690F | 1 | + | + |
| 2 | + | − | |
| U6A + STAT2 R601K | 1 | + | + |
| 2 | + | − | |
| U2A (IRF9 null) | 1 | − | + |
| 2 | + | − | |
| HEC-1B (IFNAR defect) | 1 | − | + |
| 2 | + | − | |
| U1A (Tyk2 null) | 1 | − | + |
| 2 | + | − | |
| U4A (Jak1 null) | 1 | − | + |
| 2 | + | − | |
| U6A + N1:C2 | 1 | − | − |
| N1:C2 | − | − | |
| U6A + N2:C1 | 1 | + | + |
| N2:C1 | + | − | |
| U3A + N1:C3 | N1:C3 | − | + |
| 2 | + | − | |
| U3A + N3:C1 | N3:C1 | − | − |
| 2 | − | − | |
2fTGH cells are intact for all IFN signaling components, U1A, U2A, U3A, U4A, and U6A are IFN-unresponsive daughter lines. HEC-1B lacks high-affinity IFN-α/β receptors. STAT1 and STAT2 were detected by immunoblotting with specific antisera, and the chimeras were detected with anti-Flag antiserum. The ability of HPIV2 or SV5 infection to induce loss of STAT protein is indicated by + (degrades) or − (does not degrade). Cells infected at an MOI of ≥10 were assayed at 16 h postinfection.
These STAT-deficient cell lines were subjected to virus infection and immunoblotting to detect STAT1 or STAT2. 2fTGH cell extracts stain positively for both STAT1 and STAT2, while U3A cells lack STAT1 expression and U6A cells lack STAT2 expression (Fig. 1A). Infection of 2fTGH cells with SV5 causes a disappearance of cellular STAT1 but not STAT2, and infection of 2fTGH cells with HPIV2 results in a disappearance of STAT2 but not STAT1 (Fig. 1A). In contrast, infection of U3A cells with HPIV2 resulted in no degradation of the endogenous STAT2 protein. Similarly, infection of U6A cells with SV5 resulted in no degradation of the endogenous STAT1 protein (Fig. 1A). Viral protein synthesis was observed in both U3A and U6A cells (Fig. 1B), indicating that they are susceptible to paramyxovirus infection. Therefore, the differential protein degradation profiles indicate that the cell lines deficient in STAT1 or STAT2 are inherently nonpermissive for paramyxovirus-induced STAT protein degradation. This result suggested that intact ISGF3 might be a prerequisite for degradation to occur.
To test this concept, STAT protein levels were assessed in virus-infected IRF9-deficient U2A cells. Infection of U2A cells with either SV5 or HPIV2 resulted in specific STAT protein degradation similar to that observed in parental 2fTGH cells. Together, these data indicate that while both STAT1 and STAT2 are required for paramyxovirus infection to induce specific STAT degradation, the trimeric ISGF3 factor itself is not the degradation target.
V protein-dependent antagonism of IFN-γ signaling is impaired in STAT2-deficient cells.
IFN-γ signaling activates a STAT1 homodimer, GAF (gamma-activated factor), that recognizes a distinct DNA response element, GAS (gamma-activated sequence) (9). U6A cells still express endogenous STAT1 that can be activated by IFN-γ to form the GAF transcription factor (29). To determine if the degradation incompetence of U6A cells can have a functional consequence for SV5-induced STAT1 antagonism, an IFN-γ-dependent GAS-luciferase reporter gene assay was carried out as a biologically meaningful endpoint.
Transfection of 2fTGH cells with an IFN-γ-responsive reporter gene produced a robust IFN-γ-dependent activation (Fig. 1C). Coexpression of the HPIV2 V protein had no effect on IFN-γ reporter gene activity, consistent with the fact that STAT2, the target of HPIV2 V, is not a participant in IFN-γ reporter gene transcription. In contrast, expression of the SV5 V protein dramatically reduced the IFN-γ response, reflecting the degradation of STAT1. Robust IFN-γ-dependent reporter gene activation was also observed in the U6A cells (Fig. 1C). As with the wild-type 2fTGH cells, reporter gene activity was normal upon expression of the HPIV2 V protein, but in the absence of STAT2, IFN-γ antagonism was lost upon expression of the SV5 V protein. This inability of the SV5 V protein to block IFN-γ signaling is consistent with the inability of SV5 infection to induce STAT1 degradation in U6A cells (Fig. 1A), reinforcing that both STAT1 and STAT2 are required for V protein-dependent STAT degradation. The absence of an accessory STAT protein creates a degradation-incompetent state.
IFN signaling is not required for STAT protein degradation.
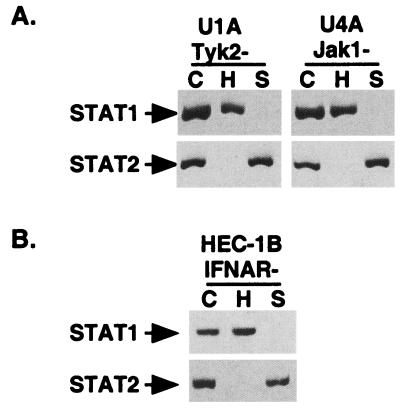
The observed lack of STAT protein degradation in STAT-deficient cell lines could be readily explained if an IFN-regulated gene product or signaling event were required to facilitate degradation. To examine the involvement of tyrosine kinases that are required for IFN signaling, two additional 2fTGH-derived cell lines were used. U1A cells lack expression of the Tyk2 tyrosine kinase, and U4A cells lack expression of the Jak1 tyrosine kinase (34, 37, 41). Upon infection with SV5 or HPIV2, both U1A and U4A were found to support appropriate STAT protein degradation (Fig. 2A). Therefore, JAK kinase-mediated phosphorylation events are not required for STAT degradation.
FIG. 2.
IFN signaling is not required for degradation competence. (A) JAK kinases are not required for STAT degradation. U1A and U4A cells were subjected to infection with paramyxoviruses and analyzed as in Fig. 1. (B) IFN receptor signaling is not required for STAT degradation. Infection and analysis of IFN-unresponsive (IFNAR−) HEC-1B cells were carried out as in Fig. 1.
As nonphosphotyrosine signals emanating from the IFN receptor might equally contribute to a permissive STAT degradation environment, HEC-1B, a human cell line that does not express high-affinity IFN receptors and is refractory to all measured aspects of IFN signaling (6, 48, 51, 53), was used to determine if any aspect of IFN receptor signaling is required for the destruction of STATs. HEC-1B cells contain ample endogenous STAT1 and STAT2 and are permissive for virus-induced STAT degradation (Fig. 2B). These results demonstrate that components of IFN signaling upstream of STAT1 and STAT2 are not required for paramyxoviruses to degrade latent STAT proteins.
STAT1, STAT2, and V proteins can form a complex.
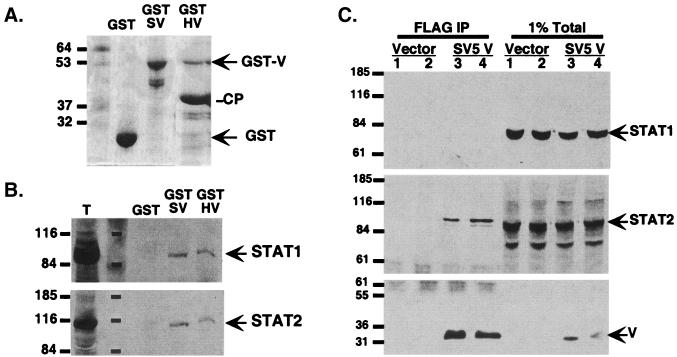
The somatic cell mutants provide genetic evidence suggestive of a degradation factor that is composed minimally of STAT1, STAT2, and the V protein. To test this possibility biochemically, affinity precipitation by bacterially expressed GST fusion proteins was performed. Both GST-SV5 V and GST-HPIV2 V gave rise to full-length proteins of ∼53 kDa that were bound to the beads, but GST-HPIV2 V was found to be proteolyzed intracellularly, resulting in an abundant cleavage product (Fig. 3A). GST control, GST-SV5 V, and GST-HPIV2 V beads were analyzed for the ability to bind to cellular STAT proteins from a 2fTGH cell extract. In support of the genetic data, the GST-SV5 V and the GST-HPIV2 V were able to retain both STAT1 and STAT2 (Fig. 3B). Importantly, the GST carrier alone did not bind to either STAT protein under these conditions. This finding indicates that the paramyxovirus V proteins are capable of associating with both STAT1 and STAT2.
FIG. 3.
V proteins bind both STAT1 and STAT2. (A) GST carrier and GST-V fusion proteins were purified from Escherichia coli with glutathione-agarose and separated by SDS-PAGE, and the gel was stained with Coomassie blue. SV, SV5 V protein; HV, HPIV2 V protein; CP, cleavage product of GST-HV. Sizes are shown in kilodaltons. (B) Binding of STATs from a cell extract. GST proteins were incubated with 2fTGH whole-cell extracts, washed extensively, and then evaluated for STAT1 or STAT2 binding by immunoblot. Lane T, 1% of total extract. Sizes are shown in kilodaltons. (C) Coimmunoprecipitation of STAT2 with SV5 V. Lysates from cells transfected with Flag-SV5 V were immunoprecipitated (IP) with Flag M2 affinity gel and probed for copurified STAT1 and STAT2. V protein was detected by Flag Western blot. Sizes are shown in kilodaltons.
A verification of this protein complex was obtained by coimmunoprecipitation assays (Fig. 3C). Flag epitope-tagged SV5 V protein was immunoprecipitated from transfected cells and processed for Western blotting. In the V-expressing cells, STAT1 is degraded and therefore not detected in the postdegradation complex. STAT2 remains tightly associated with the precipitated V protein. Together, these biochemical assays support the genetic evidence in favor of a functional V-STAT1-STAT2 protein complex.
Restoration of STAT degradation competence.
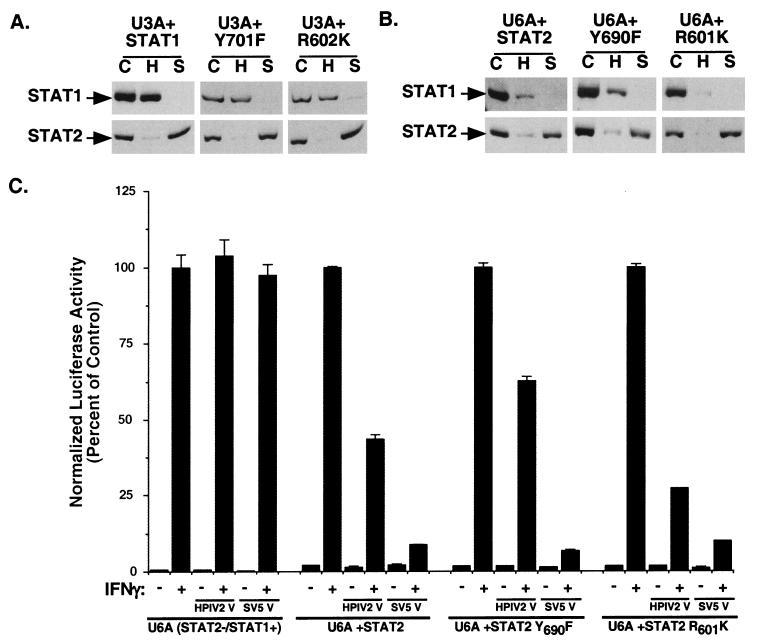
To define the STAT protein features required to reconstitute a permissive degradation environment and to test the susceptibility of the defective STATs as degradation substrates, STAT-deficient cell lines complemented with wild-type and mutated STATs were subjected to virus infection and degradation assays. Two defining features of STAT proteins are C-terminal tyrosine phosphorylation and conserved SH2 domains. Complementation of U3A cells with wild-type full-length STAT1α cDNA completely restores ISGF3 signaling as well as antiviral IFN responses, but expression of U3A cells with a STAT1α mutant that lacks either the activating tyrosine residue (Y701F) or a functional SH2 domain (R602K) fails to restore IFN signaling (21, 38).
Infection of wild-type STAT1α-complemented U3A cells with SV5 and HPIV2 fully restores STAT1 or STAT2 degradation similar to parental 2fTGH cells (Fig. 4A). Expression of either Y701F or R602K STAT1 protein also resulted in reconstitution of degradation competence, as indicated by specific protein degradation by both viruses. These observations indicate that restoration of a permissive paramyxovirus degradation environment in U3A cells requires only the latent STAT1 protein and not activated or dimeric STAT1.
FIG. 4.
STAT protein activation and dimerization are unnecessary for complementation of degradation competence. (A) U3A cells expressing wild-type STAT1 or either the Y701F or R602K mutant were infected and analyzed for STAT1 and STAT2. C, mock infection; H, HPIV2 infection; S, SV5 infection. (B) U6A cells expressing wild-type STAT2 or either the Y690F or R601K mutant were infected and assayed for degradation of STATs. All reconstituted cell lines restored degradation competence. (C) HPIV2 antagonizes IFN-γ signaling in reconstituted U6A cells. U6A cells and U6A cells expressing STAT2 or the Y690F or R601K mutant were subjected to an IFN-γ-dependent luciferase reporter gene assay in the presence or absence of coexpressed HPIV2 V protein or SV5 V protein.
Similarly, complementation of U6A cells with wild-type STAT2 can restore IFN signaling and ISGF3-dependent transcription, but expression of a STAT2 activating tyrosine mutant (Y690F) or SH2 domain mutant (R601K) fails to restore IFN responses (29, 42). Infection of wild-type STAT2-complemented U6A cells with SV5 and HPIV2 restores degradation competence to the nonpermissive U6A cells. However, the HPIV2-induced degradation response exhibited a loss of fidelity in the complemented cells, as HPIV2 infection induced a partial loss of endogenous STAT1 protein as well as STAT2 in the U6A cells (Fig. 4B). The ability of SV5 to induce specific STAT1 degradation was maintained in the U6A cells, as observed for parental 2fTGH cells and STAT1-complemented U3A cells. Expression in U6A cells of either Y690F or R601K mutated STAT2 proteins also resulted in complementation of degradation competence. As with STAT2-complemented U6A cells, HPIV2 exhibited a loss of accuracy, but SV5 remained specific for STAT1. These complementation results indicate that degradation incompetence is a single-gene defect.
The functional significance of HPIV2-induced STAT1 degradation in all three STAT2-reconstituted U6A cell lines was tested with an IFN-γ-dependent reporter gene assay. U6A cells are insensitive to expression of either HPIV2 or SV5 V protein, but the IFN-γ response in STAT2-complemented U6A cells is blunted by HPIV2 and inhibited by SV5 V regardless of STAT2 mutations (Fig. 4C). This transient assay confirms that the loss of fidelity is due to a difference in the cellular condition and not to more mundane reasons relating to virus contamination or V protein mutations that may have occurred during virus propagation. While STAT2 is absolutely required for IFN antagonism, specificity of targeting might be influenced by other loci. In all complemented U3A and U6A cells, degradation of the mutated STAT proteins was observed, indicating that STAT protein tyrosine phosphorylation and SH2 domain functions are nonessential for substrate target recognition and degradation.
STAT N terminus is required for degradation.
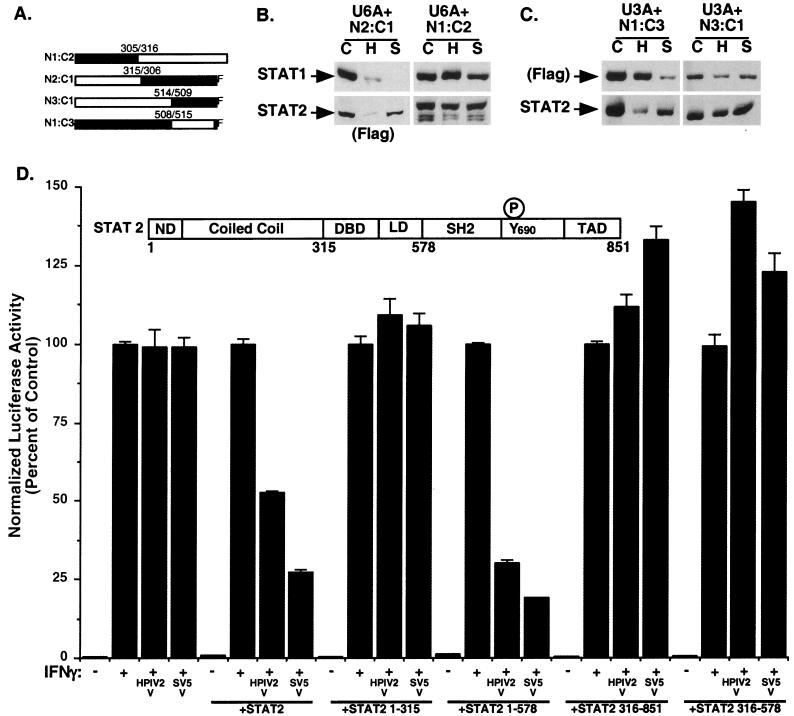
STAT protein N-terminal regions can mediate several protein-protein interactions via a lengthy coiled coil, while the C-terminal domains participate in signal transduction, dimerization, DNA binding, and transcriptional activation (7, 50; reviewed in reference 20). To localize the regions of STAT2 that confer a permissive degradation environment, U6A cells expressing chimeric STAT2-STAT1 proteins (Fig. 5A) were subjected to infection with SV5 and HPIV2. Expression of N2:C1, a fusion consisting of the N terminus of STAT2 (amino acids 1 to 315) fused to the C-terminal domain of STAT1β (amino acids 306 to 712 fused to a Flag epitope tag) (30), complemented the degradation defect of U6A cells (Fig. 5B) comparably to wild-type STAT2. HPIV2 degradation exhibited characteristic lost accuracy, but SV5 targeted STAT1 and not STAT2. In contrast, the opposite hybrid STAT protein, N1:C2 (amino acids 1 to 305 of STAT1 fused to amino acids 316 to 851 of STAT2 [30]) did not reconstitute a permissive degradation environment for either virus. These results indicate that the amino-terminal 315 amino acids of STAT2 are needed to complement the U6A degradation impairment.
FIG. 5.
Restoration of degradation competence maps to the STAT protein N terminus. (A) Diagrammatic representation of STAT1-STAT2 hybrids used to restore U6A cell lines (adapted from reference 30) and STAT1-STAT3 hybrids used to restore U3A cell lines (adapted from reference 23). Numbers indicate the amino acids of each STAT at the fusion junction. F refers to Flag epitope tag. (B) Hybrid STAT protein-complemented U6A cells were subjected to infection with HPIV2 and SV5 and analyzed as in Fig. 1 except that the N2:C1 hybrid was detected with antiserum specific for its C-terminal Flag epitope tag. C, mock infection; H, HPIV2 infection; S, SV5 infection. (C) Hybrid STAT1-STAT3 protein-complemented U3A cells were subjected to virus infection and analyzed for STAT degradation. The N1:C3 hybrid was detected with antiserum specific for its C-terminal Flag epitope tag. (D) Complementation of IFN antagonism with STAT2 fragments. Inset depicts STAT2 domain structure. ND, N domain; DBD, DNA-binding domain; LD, linker domain; TAD, transcription activation domain. U6A cells were subjected to IFN-γ-dependent luciferase reporter gene assays in the presence or absence of coexpressed HPIV2 V protein or SV5 V protein in the presence or absence of coexpressed STAT2 fragments as indicated.
In a corresponding experiment, U3A cells stably expressing STAT1-STAT3 chimeras were subjected to paramyxovirus infection and degradation assays (Fig. 5A) (23). When the N1:C3 chimera (amino acids 1 to 508 of STAT1 fused to amino acids 515 to 770 of STAT3) was expressed in U3A cells, the endogenous STAT2 protein was degraded following infection with HPIV2, and the chimeric STAT1-STAT3 fusion protein was degraded in SV5-infected cells (Fig. 5C). The complementary N3:C1 chimera (amino acids 1 to 514 of STAT3 joined to amino acids 509 to 750 of STAT1) did not reconstitute STAT degradation. Together, these findings support the conclusion that the STAT N-terminal domains are required for complementing defective degradation due to STAT protein deficiency. The observation that the hybrid proteins were successfully degraded in infected cells also suggests that the N-terminal region of STAT2 might harbor a substrate recognition region for the degradation system.
STAT2 fragments complement defective degradation.
To further map the complementing region of STAT2, full-length STAT2 or individual STAT2 fragments encoding amino acids 1 to 315, 1 to 578, 316 to 578, and 579 to 851 were expressed in U6A cells and analyzed for their ability to permit V protein antagonism of IFN-γ-responsive transcription. Coexpression of the viral V proteins with the full-length STAT2 suppressed the IFN-γ-responsive transcription, as in the stably complemented U6A cell lines (Fig. 5D). The N-terminal fragment of STAT2 (amino acids 1 to 315) did not permit coexpressed V proteins to antagonize the reporter gene. Expression of a longer STAT2 fragment that contains the STAT N-terminal domains as well as the STAT DNA-binding and linker domain regions (amino acids 1 to 578) was sufficient to complement IFN-γ antagonism. An overlapping C-terminal STAT2 fragment (amino acids 316 to 851) failed to complement IFN antagonism in U6A cells, as did the isolated DNA-binding and linker domain (amino acids 316 to 578). Together with the results from the STAT chimeras, the data indicate that the STAT2 protein N-terminal regions are necessary and sufficient to establish degradation competence in U6A cells.
DISCUSSION
The speed and inaccuracy of virus replication enable pathogens to evolve rapidly to thwart host defense mechanisms. The STAT proteins typically possess long half-lives, but the paramyxoviruses HPIV2 and SV5 have evolved to induce destruction of the IFN-α/β-responsive STATs to eliminate the selection pressure of innate antiviral immunity. SV5 efficiently targets cellular STAT1 protein, while HPIV2 targets STAT2. The data presented here demonstrate that both STAT1 and STAT2 are required to be present in the cell to render it degradation competent for either virus to target an individual STAT protein. In the absence of STAT1, HPIV2 could not induce degradation of the endogenous STAT2, and in the absence of STAT2, SV5 did not induce degradation of the endogenous STAT1. Unexpected was the finding that while both STAT1 and STAT2 are required for selective paramyxovirus-induced degradation to occur, IFN signal transduction and ISGF3 themselves are entirely dispensable.
As a consequence of this degradation impairment, STAT-deficient cells are insensitive to V protein-induced IFN antagonism, as illustrated by the inability of the SV5 V protein to suppress STAT1-dependent IFN-γ-responsive transcription in the absence of STAT2. This result indicates that the STAT protein-targeting property of the V protein is its sole IFN antagonistic action and also reinforces that in the absence of the accessory STAT protein, the cells can no longer support V-mediated STAT interference.
Despite the importance of the two STAT proteins to complement STAT degradation, in the absence of upstream cellular IFN signaling proteins, the cellular environment is permissive for either virus to induce specific STAT protein degradation (summarized in Table 1). Thus, neither JAK tyrosine kinase nor IFN receptor deficiencies disrupt the capacity for STAT protein degradation, and furthermore, the ISGF3 trimer is not the endpoint complex targeted by the viruses. Nonetheless, the results provide evidence suggesting that a protein complex containing at least STAT1 and STAT2 is recognized by the virus-induced V protein-dependent degradation system.
Somatic cell lines deficient in individual IFN signaling components were essential to these studies, as they enabled complementation analysis to be performed. Complementation of the STAT protein deficiencies with cDNA expression vectors revealed the lost capacity for virus-induced degradation to be a single-gene defect and demonstrated that both activating tyrosine phosphorylation- and SH2 domain-mediated receptor recognition and dimerization are dispensable for degradation to occur. These functions of the STAT protein are not needed either for complementation of degradation competence or for targeting and destruction of STAT proteins. Unexpectedly, loss of targeting specificity was consistently observed upon HPIV2 infection of STAT2-complemented U6A cells. As this lost fidelity was characteristic of both stable cell lines and transient-transfection assays, it appears to be a specific property of the U6A cell line. It remains possible that very precise levels of STAT2 expression or STAT2-STAT1 ratios are required in the cell to maintain HPIV2 targeting specificity. Notably, specificity of SV5 targeting was not altered in U6A-derived cells, and HPIV2 exhibited high fidelity in STAT1-complemented U3A cell lines.
Complementation of STAT2-deficient cell lines with hybrid STAT1-STAT2 fusion proteins revealed that only cells containing the amino-terminal 315 amino acids of STAT2 fused to STAT1 amino acids 316 to 712 reverted to permissive STAT protein degradation. However, these 315 amino acids expressed alone, outside the context of a full-length STAT molecule, were insufficient to complement the defective degradation. A longer fragment (amino acids 1 to 578) was able to complement the degradation defect. The STAT2 DNA-binding and linker domains encompassed by amino acids 315 to 578 are insufficient to complement defective degradation by themselves, but these regions are highly homologous between STAT1, STAT2, and STAT3. It is possible that the conserved residues contribute to the complementation by N2:C1.
Similar STAT1-STAT3 hybrids revealed that the STAT1 N terminus could complement defective degradation in STAT1-deficient U3A cells. However, in some of these complemented cell lines, the chimeric STAT did not precisely recapitulate the accuracy and efficiency of degradation observed with complementation by native STAT1. This discrepancy might be the result of different protein abundances between cell lines or reflect unique structures in the hybrid proteins that do not faithfully imitate the missing STAT.
The STAT N terminus contains two functional domains that have been implicated in protein-protein interactions. The N-domain (STAT1 amino acids 1 to 123) has been implicated in several protein-protein interactions affecting transcription, enabling dimerized STATs to bind tandem DNA elements cooperatively (49, 52). The second domain is a coiled coil that projects outward from the DNA-binding and dimerization domains of the C-terminal STAT core (2, 7). The coiled-coil domain presents a large surface that interacts with several cellular proteins (7, 22, 28, 57). Both of these STAT domains have the potential for interacting specifically with the viral V protein to form a cross-link between the two STATs, possibly forming a nucleation site that presents a novel surface to attract cellular proteolytic recognition factors.
At present, the molecular partners mediating V protein effects and their interactions with STAT targets remain to be elucidated, but the genetic and biochemical evidence presented here suggests a molecular complex that includes at least the viral V proteins, the cellular STAT1 and STAT2 proteins, and undefined components of the cellular degradation machinery. SV5 and HPIV2 V proteins were found to be capable of interacting with the DDB1 subunit of a damaged-DNA-binding factor, DDB, that is defective in some group E xeroderma pigmentosum patients (32). Expression of DDB1 can partially restore changes in the cell cycle induced by chronic SV5 V protein expression (31). It may prove to be relevant to STAT degradation that a second subunit of DDB (p48/DDB2) has a high affinity for and is targeted by cullin 4A, a member of a family of cellular proteins that possess ubiquitin ligase activity and participate in regulated proteolysis as members of macromolecular targeting complexes (8, 39, 43).
Acknowledgments
We gratefully acknowledge George Stark (Lerner Research Institute) for providing the 2fTGH and IFN-insensitive derivative cell lines used in these studies and Mairead Commaine and Xiaoxia Li (Lerner Research Institute) for hybrid STAT1-STAT2 expression plasmids and cell lines. Thanks also to Bob Lamb (Northwestern University) and Griffith Parks (Wake Forest University) for providing viruses and for advice on their propagation as well as comments on the manuscript and Daniel Besser, James E. Darnell, Jr. (The Rockefeller University), Stuart Aaronson, Julie Talon, and Tom Moran (Mount Sinai) for providing reagents and expertise.
This work was supported in part by the New York City Council Speaker's Fund for Biomedical Research and a Mt. Sinai Research Enhancement Award to C.M.H.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Becker, S., B. Groner, and C. W. Muller. 1998. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394:145-151. [DOI] [PubMed] [Google Scholar]
- 3.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besser, D., J. F. Bromberg, J. E. Darnell, Jr., and H. Hanafusa. 1999. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol. Cell. Biol. 19:1401-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. Y., T. Sato, A. Fuse, T. Kuwata, and J. Content. 1981. Resistance to interferon of a human adenocarcinoma cell line, HEC-1, and its sensitivity to natural killer cell action. J. Gen. Virol. 52:177-181. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X., U. Vinkemeier, Y. Zhao, D. Jeruzalmi, J. E. Darnell, Jr., and J. Kuriyan. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827-839. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA binding proteins are targets of Cul4A-mediated ubiquitination and degradation. J. Biol. Chem. 22:22. [DOI] [PubMed] [Google Scholar]
- 9.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 10.Darnell, J. E., Jr., I. M. Kerr, and G. M. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, T. A., D. Nandi, M. Cruz, H. J. Fehling, L. V. Kaer, J. J. Monaco, and R. A. Colbert. 1998. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-γ)-inducible subunits. J. Exp. Med. 187:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haspel, R. L., and J. E. Darnell, Jr. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 96:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haspel, R. L., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends on a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 20.Horvath, C. M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25:496-502. [DOI] [PubMed] [Google Scholar]
- 21.Horvath, C. M., and J. E. Darnell, Jr. 1996. The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 protein. J. Virol. 70:647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horvath, C. M., Z. Wen, and J. E. Darnell, Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984-994. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs, A., and J. Lindemann. 1957. Virus interference. I. The interferons. Proc. R. Soc. Lond. B 147:258-267.13465720 [Google Scholar]
- 25.John, J., R. McKendry, S. Pellegrini, D. Flavell, I. M. Kerr, and G. R. Stark. 1991. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol. Cell. Biol. 11:4189-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller, M. A., S. K. Murphy, and G. D. Parks. 2001. RNA replication from the simian virus 5 antigenomic promoter requires three sequence-dependent elements separated by sequence-independent spacer regions. J. Virol. 75:3993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45:195-200. [DOI] [PubMed] [Google Scholar]
- 28.Lau, J. F., J.-P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 15:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X., S. Leung, I. M. Kerr, and G. R. Stark. 1997. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol. Cell. Biol. 17:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 33.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 88:11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monaco, J. J., and D. Nandi. 1995. The genetics of proteasomes and antigen processing. Annu. Rev. Genet. 29:729-754. [DOI] [PubMed] [Google Scholar]
- 36.Mowen, K., and M. David. 2000. Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol. 20:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller, M., J. Briscoe, C. Laxton, D. Guschin, A. Ziemiecki, O. Silvennoinen, A. G. Harpur, G. Barbieri, B. A. Witthuhn, C. Schindler, S. Pellegrini, A. F. Wilks, J. N. Ihle, G. R. Stark, and I. M. Kerr. 1993. The protein tyrosine kinase JAK1 complements defects in interferon-α/β and -γ signal transduction. Nature 366:129-135. [DOI] [PubMed] [Google Scholar]
- 38.Muller, M., C. Laxton, J. Briscoe, C. Schindler, T. Improta, J. E. Darnell, Jr., G. R. Stark, and I. M. Kerr. 1993. Complementation of a mutant cell line: central role of the 91-kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathway. EMBO J. 12:4221-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qureshi, S. A., S. Leung, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1996. Function of Stat2 protein in transcriptional activation by IFN-α. Mol. Cell. Biol. 16:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 44.Shuai, K., C. M. Horvath, L. H. Tsai-Huang, S. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 45.Stark, G. R. 1997. Genetic analysis of interferon and other mammalian signaling pathways. Harvey Lect. 93:1-16. [PubMed] [Google Scholar]
- 46.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 47.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhaegen, M., M. Divizia, P. Vandenbussche, T. Kuwata, and J. Content. 1980. Abnormal behavior of interferon-induced enzymatic activities in an interferon-resistant cell line. Proc. Natl. Acad. Sci. USA 77:4479-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vinkemeier, U., S. L. Cohen, I. Moarefi, B. T. Chait, J. Kuriyan, and J. E. Darnell, Jr. 1996. DNA binding of in vitro activated Stat1α, Stat1β, and truncated Stat1: interaction between NH2 terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 15:5616-5626. [PMC free article] [PubMed] [Google Scholar]
- 50.Vinkemeier, U., I. Moarefi, J. E. Darnell, Jr., and J. Kuriyan. 1998. Structure of the amino-terminal protein interaction domain of STAT-4. Science 279:1048-1052. [DOI] [PubMed] [Google Scholar]
- 51.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, X. A., Y. L. Sun, and T. Hoey. 1996. Cooperative DNA binding and sequence selective recognition conferred by the Stat amino terminal domain. Science 273:794-797. [DOI] [PubMed] [Google Scholar]
- 53.Yonehara, S., M. Yonehara-Takahashi, and A. Ishii. 1983. Binding of human interferon alpha to cells of different sensitivities: studies with internally radiolabeled interferon retaining full biological activity. J. Virol. 45:1168-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, X., M. H. Wrzeszczynska, C. M. Horvath, and J. E. Darnell, Jr. 1999. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]