Abstract
Astroviruses require the proteolytic cleavage of the capsid protein to infect the host cell. Here we describe the processing pathway of the primary translation product of the structural polyprotein (ORF2) encoded by a human astrovirus serotype 8 (strain Yuc8). The primary translation product of ORF2 is of approximately 90 kDa, which is subsequently cleaved to yield a 70-kDa protein (VP70) which is assembled into the viral particles. Limited trypsin treatment of purified particles containing VP70 results in the generation of polypeptides VP41 and VP28, which are then further processed to proteins of 38.5, 35, and 34 kDa and 27, 26, and 25 kDa, respectively. VP34, VP27 and VP25 are the predominant proteins in fully cleaved virions, which correlate with the highest level of infectivity. Processing of the VP41 protein to yield VP38.5 to VP34 polypeptides occurred at its carboxy terminus, as suggested by immunoblot analysis using hyperimmune sera to different regions of the ORF2, while processing of VP28 to generate VP27 and VP25 occurred at its carboxy and amino terminus, respectively, as determined by immunoblot, as well as by N-terminal sequencing of those products. Based on these data, the processing pathway for the 90-kDa primary product of astrovirus Yuc8 ORF2 is presented.
Human astroviruses (HAstV) have been found to be a frequent cause of gastroenteritis among young children worldwide (5, 7, 8, 15). The virions are formed by a nonenveloped protein capsid and a positive-stranded RNA genome of approximately 7 kb (9, 22). The RNA genome has three open reading frames (ORFs) (ORF1a, -1b, and -2), each encoding at least one polyprotein. The ORF1a contains viral serine protease and nuclear localization signal motifs, whereas the ORF1b has an RNA-dependent RNA polymerase motif (9, 22). The products of ORF1a and ORF1b are synthesized from the genomic RNA as two polyproteins, with the latter being produced as a polyprotein 1a-1b (approximately 160 kDa) through a frameshift translational mechanism (12-14). It is believed that the products of these ORFs, processed to smaller polypeptides by the viral protease, are involved in the viral RNA replication. ORF2, of approximately 780 amino acid residues, depending on the strain (21), codes for the structural virus polypeptides (18). The structural polyprotein is translated from a polyadenylated subgenomic RNA produced at high levels during infection, which is 3′-colinear with the genomic RNA (17). Based on the homology among strains belonging to different serotypes, at least two domains in the product of this ORF have been predicted (16, 21). The first domain includes amino acid residues 1 to 415, and it is highly conserved among all the human serotypes and some viruses from animal origin; the second domain (amino acid 416 to the end) is highly divergent among human serotypes (10, 16, 21, 23). Neutralizing epitopes have been mapped to the second domain (3, 20); therefore, it is likely that this hypervariable region is exposed on the viral particle. It is known that astrovirus infectivity is dependent on trypsin treatment (11); however, knowledge about the processing of the ORF2 structural polyprotein with trypsin is limited. Proteins of 20 to 90 kDa (2, 4, 18, 20) have been reported to be present in viral particles. Bass et al. reported that the initial product of ORF2 (a 90-kDa protein) is cleaved intracellularly at Arg70 to yield a protein of 79 kDa (2). In the absence of trypsin in the culture medium this protein was found in the astrovirus particles; however, in the presence of trypsin, the proteins found in the particles were 26, 29, and 34 kDa (2). The products of 29 and 26 kDa (VP29 and VP26), which have been consistently found in trypsin-treated viruses by several authors (2, 4, 18, 20), were shown to be generated by cleavage of the primary ORF2 HAstV-2 polyprotein at Arg361 and Arg395, respectively, sharing their carboxy terminus (20). Given this observation, the products of 34 and 29 kDa would not represent the whole protein of 79 kDa found in untreated particles, suggesting that additional polypeptides, which have not been identified, should result after trypsin treatment of the particles. In order to contribute to a better understanding of the processing of the ORF2 polyprotein of HAstV we have characterized the proteolytic pathway of the ORF2 polyprotein of a serotype 8 human astrovirus.
MATERIALS AND METHODS
Virus and cells.
The colon carcinoma cell line Caco-2, obtained from the American Type Culture Collection, was used throughout this work. The cells were cultured in a CO2 atmosphere at 37°C with minimum essential medium (Eagle's salts) (MEM), supplemented with glutamine, penicillin, streptomycin, and 15% fetal bovine serum. The astrovirus strain Yuc8 was isolated from a natural infection and adapted to grow in Caco-2 cells (16). During this work, virus passages 7 to 11 were used. To propagate the virus, cells were washed twice with MEM without fetal bovine serum and inoculated with Yuc8 previously treated with 10 μg of trypsin/ml for 30 min at 37°C (Trypsin 1:250; GIBCO), resulting in multiplicities of infection lower than 0.01. Adsorption of the virus to the cells was for 1 h at 37°C, the inoculum was removed, and MEM without serum containing 3.3 μg of trypsin/ml was added. Under our conditions, normal Caco-2 cells detached at higher trypsin concentrations for long incubation periods. The infected cells were incubated at 37°C and harvested 3 days postinfection, or when cytopathic effect was evident.
Purification of Yuc8.
Yuc8-infected Caco-2 cells were frozen and thawed three times, and the cell lysate was centrifuged at 25,000 rpm for 4 h (Beckman SW28 rotor). All the steps during viral purification were carried out at 4°C. The pellet was resuspended in TNE buffer (50 mM Tris [pH 7.4], 0.1 M NaCl, 10 mM EDTA) and extracted once with Genetron (trichlorotrifluoromethane), and the supernatant was collected after centrifugation at 6,000 rpm for 30 min (16F6-38 rotor; Brinkmann); this was further ultracentrifuged at 40,000 rpm for 2 h (SW50.1 rotor; Beckman), and the resulting pellet was resuspended in TNE buffer and loaded in a cesium chloride solution (initial density, 1.36). A density gradient was formed during ultracentrifugation for 16 to 18 h at 35,000 rpm (SW50.1 rotor; Beckman), and the opalescent band corresponding to viral particles was collected with a syringe and diluted in TNE buffer. This suspension was again ultracentrifuged at 40,000 rpm for 2.5 h (SW50.1 rotor; Beckman), and finally the pellet was resuspended in TNE buffer and stored at 4°C. Infectivity assays were performed to assure the identity of the purified particles. This preparation was used for further studies.
Astrovirus recombinant proteins and sera.
Clones expressing the recombinant astrovirus proteins were constructed in the pGEX4T vectors (Pharmacia), using standard methods (19). Proteins E1 (including amino acid residues 3 to 208 of ORF2), E2 (residues 209 to 341), and E3 (residues 386 to 594) were fused to the carboxy-terminus of glutathione-S-transferase (GST) and expressed in Escherichia coli. Since recombinant proteins were insoluble, they were purified by electroelution from sodium dodecyl sulfate (SDS)-polyacrylamide gels and quantified. Approximately 100 μg of each protein was mixed with Freund's complete adjuvant and subcutaneously inoculated into New Zealand rabbits. Three further inoculations of the protein into Freund's incomplete adjuvant were done at 15-day intervals. A peptide with the sequence KSNKQVTVEVSNGRNRC (named KSN), corresponding to residues 4 to 20 of HAstV-1 ORF2 (synthesized by Research Genetics Co.) was coupled with keyhole limpet hemocyanin (KLH) (Imject maleimide activated-KLH; Pierce) following the manufacturer's recommendations. The KLH-conjugated peptide was inoculated into BALB/c mice, following the same protocol used for the recombinant proteins. Sera were collected 2 weeks after the fourth inoculation.
Viral infectivity.
Caco-2 cells were grown on glass slides and infected as mentioned above. Sixteen to eighteen hours postinfection, cells were washed with phosphate-buffered saline (PBS) and fixed with 2% p-formaldehyde for 15 min at room temperature. Cells were permeabilized with 0.05% Triton X-100 in PBS for 15 min at room temperature and blocked for 1 h with 1% bovine serum albumin (fraction V; GIBCO). The serum raised against the E2 astrovirus protein described above was used as the primary antibody, and a secondary antibody conjugated to Alexa (goat anti-rabbit immunoglobulin G-Alexa488; Molecular Probes) was used for detection. Cells were washed four times with PBS-NH4Cl (50 mM) after every incubation step. The infectivity of the purified Yuc8 virus was determined by counting the infected cells and reported as fluorescent focus units per milliliter.
Trypsin digestion of purified virus and protein analysis.
Purified virus was treated with different trypsin concentrations (0 to 400 μg/ml) [l(tosylamido-2-phenyl)ethyl chloromethyl ketone (TPCK) treated, >180 U/mg; Worthington] for 1 h at room temperature. Each treated sample was split into fractions and used as follows: one fraction was employed to determine the viral infectivity immediately after trypsin treatment, and the other fractions were run in SDS-polyacrylamide gels. One gel was stained with silver, and the others were used for immunoblot analysis using the sera raised against the recombinant astrovirus proteins (each diluted 1:4,000 in PBS); a horseradish peroxidase-linked antibody to rabbit or mouse immunoglobulin G was then added for 1 h, and the antibody-antigen complex was detected with the Western blot chemiluminescence reagent (Renaissance; NEN Life Science Products). As a control, one well in each of the polyacrylamide gels was loaded with no virus but with the highest amount of trypsin used for virus digestion.
N-terminal protein sequencing.
Purified virus particles were treated with 10 and 200 μg/ml of trypsin, as described above. Samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (PVDF-Plus 0.1 μm; Osmonics) for 1 h at 50 V in CAPS [3-(cyclohexylamino)-1-propane sulfonic acid] buffer (10 mM CAPS [pH 11], 10% methanol). All reagents used were freshly prepared and filtered through a 0.22-μm-pore-size membrane. Gels were allowed to polymerize overnight at room temperature. Transferred proteins were visualized by staining with Ponceau-S, and the bands were excised, washed thoroughly with MilliQ water, and dried in a dust-free environment. PVDF membranes were sent for sequencing to the Harvard Microchemistry Facility (Harvard University). VP70 was obtained from samples with no trypsin treatment; VP28 was obtained from digestion of purified particles with 10 μg of trypsin/ml; and VP34, VP27, and VP25 were obtained from samples treated with 200 μg of trypsin/ml (see Fig. 2).
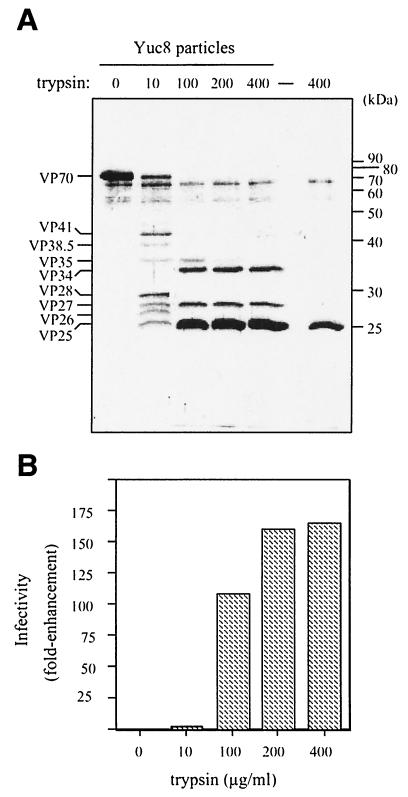
FIG. 2.
Trypsin treatment of purified Yuc8 particles cleaves VP70 into several polypeptides and enhances the virus infectivity. Purified Yuc8 astrovirus was treated with the indicated concentrations of TPCK-treated trypsin for 1 h at room temperature. Each sample was then divided in two portions which were either run in an SDS-12.5% polyacrylamide gel and stained with silver (A) or used to determine viral infectivity (B). (A) Trypsin alone (400 μg/ml) was used as a control, migrating as a 25-kDa protein (rightmost lane). The molecular mass standards (in kilodaltons) (protein ladder; GIBCO) and the positions of the viral proteins are marked. The assignment of the molecular mass for the viral proteins was made based on their average migration in several experiments and different standards, as reference. The infectivity was determined by quantification of the infected cells detected by immunofluorescence and expressed as n-fold enhancement.
RESULTS
The intracellular processing of the ORF2 primary product of Yuc8 is limited.
To study the cleavage pathway of the primary product encoded by a HAstV serotype 8 (strain Yuc8) ORF2, three regions of this ORF containing either amino acid residues 3 to 208 (polypeptide E1), 209 to 341 (E2), or 386 to 594 (E3) were synthesized in bacteria as C-terminal fusion products with GST, and the fusion proteins were used to generate region-specific hyperimmune rabbit sera (see Fig. 5A). Also, antibodies to a synthetic peptide (named KSN) comprising amino acids 4 to 20 of the HAstV-1 ORF2 were generated in mice.
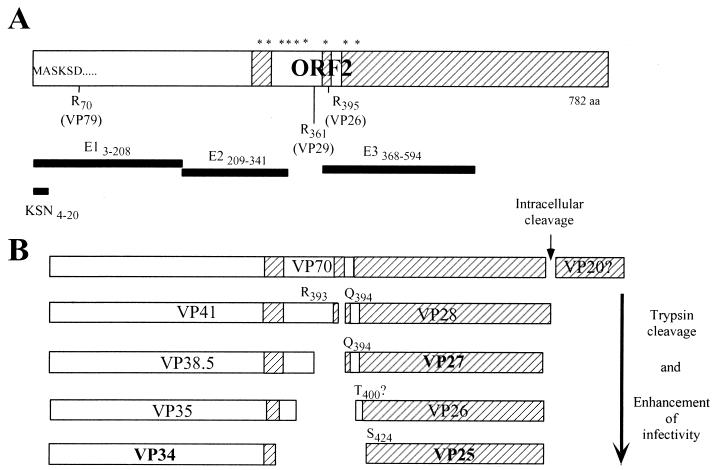
FIG. 5.
(A) Diagram of the ORF2 of HAstV and recombinant astrovirus proteins. ORF2 is represented as a box, and the recombinant proteins and the KSN peptide are represented as thick black lines. The diagram is to scale, and the relative positions of the astrovirus Yuc8 recombinant proteins E1, E2, and E3 and the peptide KSN is shown (subscripts indicate the amino acid residues included in each protein). The hypervariable regions found among human astrovirus serotypes (16) are shown as striped boxes. The arginine (R) residues identified by Bass (2) and Sanchez-Fauquier (20) as cleavage sites in the capsid polyprotein of HAstV-1 and HAstV-2 and the protein products proposed (in parentheses) to be generated by these cleavages are indicated. Asterisks represent susceptible trypsin sites conserved among astroviruses belonging to different serotypes, which could be cleaved to yield the VP41- and VP28-derived polypeptides. (B) Proposed trypsin processing pathway for the ORF2 polyprotein of astrovirus Yuc8. Boxes represent the products observed during virus activation; the final products, present in fully activated particles, are indicated in boldface type. The N-terminal amino acid residue of the VP28-derived products is shown. See details in the text.
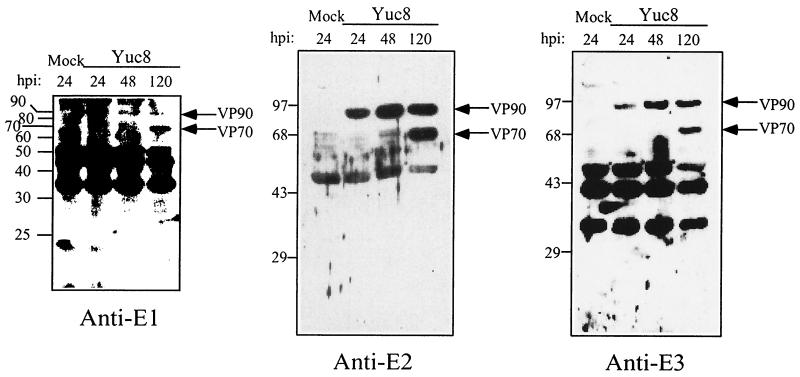
Total proteins of Yuc8-infected cells, harvested at different times postinfection, were analyzed by immunoblotting with the sera mentioned above. The anti-E1, anti-E2, and anti-E3 sera recognized proteins of 90 and 70 kDa (hereafter named with the prefix VP) in infected, but not in mock-infected, cells (Fig. 1). VP90 most probably represents the primary translation product of ORF2 (whose calculated molecular mass based in its amino acid sequence is 87 kDa), while VP70 appears to be derived from VP90. The cleavage of VP90 to yield VP70 seems to be cell-associated since the protein samples used in this analysis were harvested after removing the culture medium; however, it does not represent a cleavage equivalent to that previously reported by Bass (2), based on the VP70 recognition by antibodies to the residues 4 to 20 of ORF2 (anti-KSN; see next section and Fig. 3A). The 20-kDa polypeptide that is presumably cleaved off from VP90 was not detected with any of the available sera. The antibodies to KSN detected the same viral proteins as the antibodies to E1, E2, and E3 (not shown).
FIG. 1.
Sera to recombinant Yuc8 proteins recognize 90- and 70-kDa proteins in Yuc8-infected cells. A monolayer of Yuc8-infected Caco-2 cells was washed with PBS twice at 24, 48, and 120 h postinfection (hpi) and lysed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.5% SDS, plus phenylmethylsulfonyl fluoride and leupeptin as protease inhibitors. A lysate of mock-infected cells was included as a control. Cell lysates were homogenized and centrifuged at 14,000 × g to discard the pellet. Supernatant was collected and loaded in an SDS-15% polyacrylamide gel for immunoblot analysis with the indicated sera. The migration of the molecular mass markers (in kilodaltons) (indicated by a protein ladder [GIBCO] for anti-E1 and prestained protein standards [GIBCO] for anti-E2 and anti-E3) and of the viral proteins is indicated.
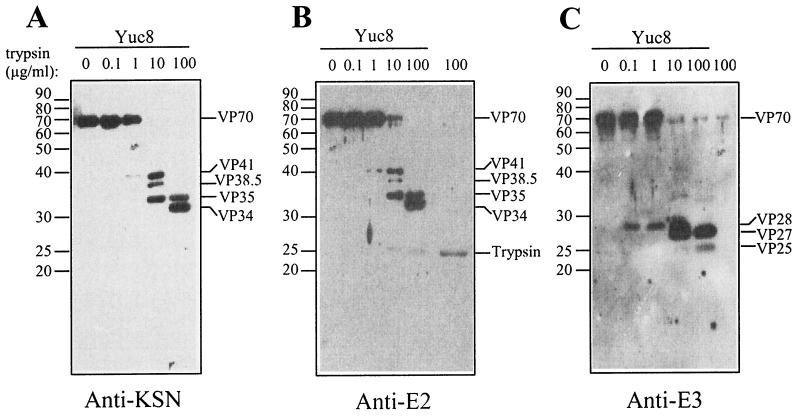
FIG. 3.
Processing of the VP70 protein by trypsin is ordered. Purified Yuc8 astrovirus particles were treated with the indicated trypsin concentrations, as mentioned in Fig. 2, and each sample was divided in equal parts to run three independent SDS-12.5% polyacrylamide gels and transferred to nitrocellulose. Each blot was incubated with either anti-KSN (A), anti-E2 (B), or anti-E3 (C) antibodies. As a control, 100 μg of trypsin/ml was included (rightmost lane in panels B and C). Note that anti-E2 (B), but not anti-E3 (C), partially cross-reacts with trypsin (25 kDa). An additional ∼69-kDa band detected by anti-E3 (C) is not of viral origin, since it appears in the trypsin-alone lane. The position of the viral proteins and of trypsin are indicated as well as of the molecular mass markers (protein ladder; GIBCO). The infectivities of these samples were also determined, confirming that the enhancement of infectivity increased slightly with trypsin at 10 μg/ml and was higher at 100 μg/ml, as shown in Fig. 2.
Trypsin treatment of Yuc8 virus results in an ordered processing of VP70 and enhancement of the viral infectivity.
Cesium chloride-purified untreated Yuc8 virus particles were found to contain exclusively VP70 (Fig. 2); however, when the virions were treated with trypsin (10 μg/ml), seven polypeptides in the range of 25 to 41 kDa were observed (Fig. 2A). At the highest trypsin concentration used, three predominant products of 34, 27, and 25 kDa accumulated. No products below 25 kDa were detected in gels with a higher concentration of polyacrylamide (not shown). When the effect of trypsin treatment on viral infectivity was determined, we found that the virus containing only VP70 showed a low basal infectivity, which varied around 104 fluorescent focus units/ml in the various experiments. The virus infectivity increased only slightly when the virions were treated with trypsin at 10 μg/ml, but it was enhanced more than 100-fold after digestion with trypsin concentrations higher than 100 μg/ml (Fig. 2B). Thus, the highest viral infectivity reached correlated with the presence of proteins VP34, VP27, and VP25, which seem to be the final trypsin processing products of the VP90 polyprotein precursor.
To determine the processing pathway of VP70, purified virus particles were treated with 10-fold-increasing trypsin concentrations (starting at 0.1 μg/ml) and the processing protein intermediates were identified by immunoblot using the sera to the GST-fusion proteins described above. Treatment of Yuc8 purified particles with various trypsin amounts revealed that VP70 was processed in an orderly fashion (Fig. 3). The first two detected cleavage products (at 1 μg/ml) were VP41 and VP28, which represent the amino and carboxy termini of VP70, respectively, since VP41 was recognized by the antibodies to KSN (Fig. 3A), E1 (not shown), and E2 (Fig. 3B), while VP28 was detected by the anti-E3 serum (Fig. 3C). VP41 was further processed into the polypeptides VP38.5 and VP35 (Fig. 3A and B, lane 10 μg/ml). With trypsin at 100 μg/ml, VP41 and VP38.5 disappeared to yield polypeptides VP35 and VP34 (Fig. 3A and B), while at the highest trypsin concentration employed (400 μg/ml [Fig. 2A]), VP34 was the only product derived from VP41 that was detected. The reactivity of all these polypeptides with the KSN antiserum, raised to residues 4 to 20 of ORF2, indicates that they are produced by processing of the carboxy terminus of VP41 (Fig. 3A). Antibodies to E2 recognized a 25-kDa band with trypsin at 100 μg/ml; however, that band seems to correspond to trypsin, based on the signal observed in the well loaded with the enzyme alone (Fig. 3B).
Further cleavage of VP28 by trypsin resulted in the generation of polypeptides VP27 and VP25 (Fig. 3C, 10 and 100 μg of trypsin/ml). These proteins were recognized by the anti-E3 serum and not by antibodies to E2 or KSN. In Fig. 3C, the lower band recognized by anti-E3 seems to correspond to VP25 and not to VP26, since the latter protein was in very low amounts and transiently produced (detected only at 10 μg of trypsin/ml). The VP25 protein observed by silver staining in Fig. 2A comigrates with trypsin, which raised the possibility that this protein was not of viral origin. However, as observed in Fig. 3C, that band is recognized by antibodies to E3 when virus was present (lane Yuc8, 100 μg/ml), but not when trypsin alone was loaded into the gel (Fig. 3C, the rightmost lane).
N-terminal sequence of the viral proteins.
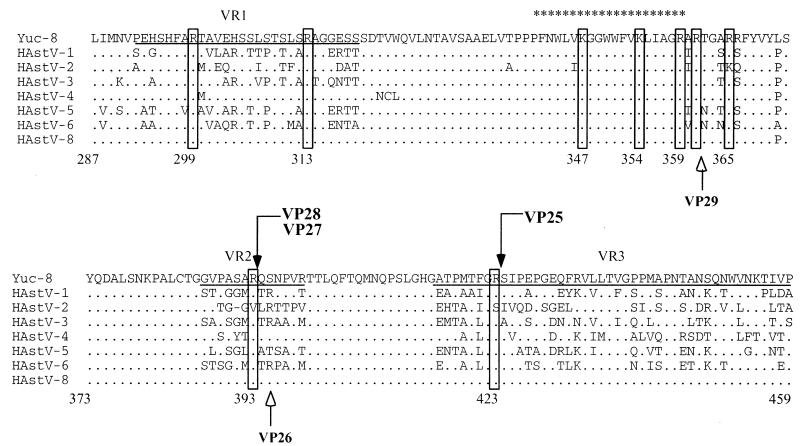
To determine the exact trypsin cleavage sites in VP70, proteins VP70, VP34, VP28, VP27, and VP25 were separated by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes for N-terminal sequencing. VP70 and VP34 were found to have a blocked amino terminus. VP28 and VP27 were found to have the same amino-terminal sequence (QSNPVRTTLQFT…), indicating that the cleavage that generates them occurs at Arg393 and that the difference between them is at their carboxy-end. The N-terminal sequence of VP25 (SIPEPGEQFRVL…) revealed that this protein is in fact derived from VP28 as a result of a cleavage at Arg423. Although VP25 and trypsin comigrate, sequencing of the 25-kDa band by Edman degradation revealed exclusively the astrovirus protein, probably because the N terminus of trypsin may be blocked. Figure 4 shows the N-terminal sequence of VP28, VP27, and VP25 of Yuc8 in the context of the aligned sequences of eight human astrovirus serotypes.
FIG. 4.
N-terminal sequence of VP28-derived proteins. Purified Yuc8 particles were trypsin treated and electroblotted to PVDF membranes. VP28, VP27, and VP25 were purified, and the N termini were sequenced. The sequence alignment of eight HAstV serotypes between residues 287 and 459 of ORF2 is shown. The cleavage sites for the proteins VP28, VP27, and VP25 (boxes marked by black arrows) and the amino acid residues which could potentially represent the carboxy end of the VP41-derived proteins (boxes at the amino acid residues 299, 313, 347, 354, 359, 361, and 365) are indicated. The cleavage sites for the proteins VP29 and VP26 of HAstV-2, previously described (20), are also indicated (white arrows). The dots along the sequence denote identity, and only amino acid changes are marked. The previously described variable regions VR1 and VR2 and part of VR3 (16) are underlined. The common astrovirus epitope predicted (21) is marked with asterisks. The numbers below the sequences indicate the amino acid position based on the Yuc8 sequence. Sequence alignment was made by Clustalw analysis (http://www.ebi.ac.uk/clustalw/) using sequences with accession numbers L23513 (HAstV-1), A45695 (HAstV-2), AF141381(HAstV-3), Z33883 (HAstV-4), U15136 (HAstV-5), Z46658 (HAstV-6), Z66541 (HAstV-8), and AF260508 (Yuc8).
DISCUSSION
In this study we describe the processing pathway of a serotype 8 human astrovirus ORF2 polyprotein. The initial processing of the primary product, VP90, seems to be cell associated and yields VP70, which is assembled to form the virion. In contrast to what was described by Bass and Qiu for a serotype 1 strain (2), the cell-associated processing of Yuc8 VP90 does not occur at Arg70 but rather occurs at its carboxy terminus, since VP90 and VP70 are both recognized by the KSN antiserum, which is directed to the residues 4 to 20 of the ORF2. Processing at Arg70 in HAstV-1 (2) would eliminate an arginine-rich fragment from the mature capsid protein, which is highly conserved among astroviruses. Arginine residues have a high affinity for RNA (24), and similar basic regions have been suggested to be important for genome encapsidation in some RNA viruses, like alfalfa mosaic (1) and Sindbis (6) viruses. Thus, it makes sense that this fragment is preserved in VP70, where it may play an important structural role in the mature particle, by interacting with the RNA genome. The 20-kDa protein, which would be cleaved off from the carboxy-terminus of VP90 in Yuc8, to yield VP70, was not detected with the available sera, and further studies are required to determine the fate of this polypeptide. It also remains to be identified which protease is responsible for the VP90 cleavage. Although intracellular processing of VP90 to VP70 seems to be slow, given that VP70 is barely detected at 48 h with anti-E2 but clearly detected at 120 h with all sera (Fig. 1), it is possible that VP70, once produced, is assembled and released to the supernatant. Accordingly, Sanchez-Fauquier (20) reported that the protein assembled in the virion is mainly present in the culture supernatant.
Based on the present work, we propose a processing pathway for VP90 (Fig. 5). VP90 yields VP70, which is present in the virions. Trypsin treatment of purified Yuc8 virions results in the ordered processing of VP70 with a concomitant enhancement of the viral infectivity. Based on the reactivity of the trypsin cleavage products with the sera to peptide KSN and to the recombinant proteins E1, E2, and E3, and on the N-terminal sequence of VP28, the first cleavage seems to occur at Arg393, to yield proteins VP41 and VP28, which represent the amino and the carboxy termini of VP70, respectively. VP41 is further cleaved to yield VP38.5, VP35, and VP34. Protein VP38.5 seems to be processed quickly, since it was barely detected by silver staining (Fig. 2) and it was a minor band by immunoblotting with anti-KSN and anti-E2 sera (Fig. 3). Since VP34, VP35, VP38.5, VP41, and VP70 are all recognized by anti-KSN antibodies, we propose that the processing of VP90, VP70, and VP41 occurs at their carboxy termini. There are several trypsin susceptible sites upstream to Arg393 (including amino acid residues 299, 313, 347, 354, 361, and 366 [Fig. 4]), where cleavages could yield proteins with sizes similar to those of the observed products. In fact, it is known that Arg361 is used as a cleavage site in HastV-2 (20). It is not known if the small peptides cleaved off from the carboxy termini of these proteins remain associated to the virion or are released from it.
The residue Arg393 is conserved in all human astrovirus serotype strains but in serotype 2, in which cleavage at Arg395 occurs to generate a 26-kDa protein (20); in this virus, an additional cleavage to generate a 29-kDa polypeptide was shown to occur at Arg361. In Yuc8, we found two proteins (VP28 and VP27) of similar size to those reported by Sanchez-Fauquier (20), which have the same amino-terminal sequence, indicating that they differ in the carboxy-end, and not in the amino terminus, as reported for VP29 and VP26 (20). Thus, VP27 seems to be a product of processing at the carboxy region of VP28. Given that the nature of the protease that cleaves VP90 to yield VP70 is not known, the exact carboxy-end of VP70, and therefore of VP28, cannot be predicted; however, considering the size of these two proteins, that cleavage could occur around amino acid residue 635. In Yuc8, susceptible trypsin sites which could yield carboxy-truncated VP28 protein are located at positions 586 and 618.
VP25 is produced by cleavage at Arg423 and is one of the predominant viral products when the purified viral particles show the highest infectivity. A protein equivalent to VP25 (one cleaved at Arg423) has not been described before as a component of the activated virus. Based on the viral products observed after complete activation of HAstV-2 and Yuc8 with trypsin, VP29 and VP26 (20) could be functionally equivalent to VP27 and VP25 (this work), respectively; however, differences in the N termini between VP26 and VP25 suggest that processing, and possibly conformation, is partially different between these strains. The product of 26 kDa observed in Fig. 2, when the virus was treated with trypsin at 10 μg/ml, was not analyzed by sequence since it was transiently produced and barely detected; however, it could be an intermediate product of cleavage at Arg400 and therefore not equivalent to VP26 of HAstV-2 (20).
Bass and Qiu (2) reported that cleavage of the HAstV-1 capsid precursor to yield a protein of 29 kDa occurs at Arg395 and not at residue 361, as reported by Sanchez-Fauquier (20) for a protein of a similar size. Thus, the cleavage that yielded the 29-kDa protein of HAstV-1 does seem to be equivalent to the cleavage that generates VP28 and VP27 in Yuc8. Unfortunately, the amino terminus of the 26-kDa protein of HAstV-1 was not determined, so that it is not known if in the case of HAstV-1 both proteins have the same amino (as described in this study for Yuc8) or the same carboxy (as described for HAstV-2 [20]) terminus.
In contrast to previous observations with other astrovirus strains (2, 18, 20), purification of Yuc8 particles cultivated in the presence of trypsin resulted in a virus containing a single polypeptide (VP70), and not in virus containing the smaller VP34, VP27, and VP25 proteins. These proteins were only observed after treatment of the purified Yuc8 particles with high trypsin concentrations (more than 200 μg/ml), via protein intermediates. The fact that VP70 or the protein intermediates leading to the smaller protein products were not detected in other reports (2, 4, 20) might be due to differences in the susceptibility of the astrovirus strains analyzed to trypsin or to the specific activity of the trypsin preparation used.
The study of the processing pathway of the astrovirus Yuc8 by trypsin has revealed a complex and ordered process, which may be partially different in different astrovirus strains. This is supported by the fact that the smallest cleavage products of two astrovirus strains after complete trypsin digestion of the capsid, VP25 of Yuc8 and VP26 of HAstV-2 (20), have a different N terminus. The different cleavage products could be the result of the protein assembled in the capsid (i.e., VP70 in Yuc8 versus VP79 in HAstV-1) or the consequence of alternative trypsin cleavage sites present in two equivalent proteins having different amino acid sequence.
Although the mechanism of the enhancement of astrovirus infectivity by trypsin remains to be determined, we know now that the largest enhancement of Yuc8 infectivity by trypsin was observed only after several cleavages occurred, when VP34, VP27, and VP25 accumulated. The cleavages responsible for the enhancement of Yuc8 infectivity seem to be dependent on previous cleavages of VP70, which probably expose new trypsin-susceptible sites, suggesting that conformational changes occur during virus activation. In agreement with this idea, Bass and Qiu (2) observed that trypsin-treated and -untreated HAstV-1 particles show different reactivity to monoclonal antibodies directed to different epitopes of the structural protein. It is possible that trypsin-treated particles gradually change their conformation to favor a better attachment, entry, or uncoating of the virions during infection.
The processing of the Yuc8 ORF2 polyprotein by trypsin to yield infectious particles seems to be a complex event regulated in a cascade. Understanding of this event and the mechanism by which trypsin enhances astrovirus infectivity should help to elucidate the initial interactions of the virus with the host cell.
Acknowledgments
We thank Maria P. E. Salas for technical assistance and Elizabeth Mata for assistance with animal care. We also thank Pavel Isa for critical reading of the manuscript.
This work was partially supported by grants MENSE31739 from the National Council for Science and Technology—Mexico, grant IN200999 from DGAPA-UNAM, and grants 75197-527106 and 55000613 from Howard Hughes Medical Institute.
REFERENCES
- 1.Baer, M., F. Houser, L. Loesch-Fries, and L. Gehrke. 1994. Specific RNA binding by amino-terminal peptides of alfalfa mosaic virus coat protein. EMBO J. 13:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass, D. M., and S. Qiu. 2000. Proteolytic processing of the astrovirus capsid. J. Virol. 74:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, D. M., and U. Upadhyayula. 1997. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J. Virol. 71:8666-8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Capsid protein composition of reference strains and wild isolates of human astroviruses. Virus Res. 49:49-57. [DOI] [PubMed] [Google Scholar]
- 5.Dennehy, P. H., S. M. Nelson, S. Spangenberger, J. S. Noel, S. S. Monroe, and R. I. Glass. 2001. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 184:10-15. [DOI] [PubMed] [Google Scholar]
- 6.Geigenmuller-Gnirke, U., H. Nitschko, and S. Schlesinger. 1993. Deletion analysis of the capsid protein of Sindbis virus: identification of the RNA binding region. J. Virol. 67:1620-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero, M. L., J. S. Noel, D. K. Mitchell, J. J. Calva, A. L. Morrow, J. Martinez, G. Rosales, F. R. Velazquez, S. S. Monroe, R. I. Glass, L. K. Pickering, and G. M. Ruiz-Palacios. 1998. A prospective study of astrovirus diarrhea of infancy in Mexico City. Pediatr. Infect. Dis. J. 17:723-727. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann, J. E., D. N. Taylor, P. Echeverria, and N. R. Blacklow. 1991. Astroviruses as a cause of gastroenteritis in children. N. Engl. J. Med. 324:1757-1760. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Stine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 90:10539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonassen, C. M., T. O. Jonassen, Y. M. Saif, D. R. Snodgrass, H. Ushijima, M. Shimizu, and B. Grinde. 2001. Comparison of capsid sequences from human and animal astroviruses. J. Gen. Virol. 82:1061-1067. [DOI] [PubMed] [Google Scholar]
- 11.Lee, T. W., and J. B. Kurtz. 1981. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J. Gen. Virol. 57:421-424. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, T. L., and S. M. Matsui. 1996. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J. Virol. 70:2869-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis, T. L., and S. M. Matsui. 1997. Studies of the astrovirus signal that induces (−1) ribosomal frameshifting. Adv. Exp. Med. Biol. 412:323-330. [DOI] [PubMed] [Google Scholar]
- 14.Marczinke, B., A. J. Bloys, T. D. Brown, M. M. Willcocks, M. J. Carter, and I. Brierley. 1994. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J. Virol. 68:5588-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver, C. J., G. Hansman, P. White, J. C. Doultree, M. Catton, and W. D. Rawlinson. 2001. Diagnosis of enteric pathogens in children with gastroenteritis. Pathology 33:353-358. [PubMed] [Google Scholar]
- 16.Mendez-Toss, M., P. Romero-Guido, M. E. Munguia, E. Mendez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 17.Monroe, S. S., B. Jiang, S. E. Stine, M. Koopmans, and R. I. Glass. 1993. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J. Virol. 67:3611-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monroe, S. S., S. E. Stine, L. Gorelkin, J. E. Herrmann, N. R. Blacklow, and R. I. Glass. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J. Virol. 65:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual., 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sanchez-Fauquier, A., A. L. Carrascosa, J. L. Carrascosa, A. Otero, R. I. Glass, J. A. Lopez, C. San Martin, and J. A. Melero. 1994. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology 201:312-320. [DOI] [PubMed] [Google Scholar]
- 21.Wang, Q. H., J. Kakizawa, L. Y. Wen, M. Shimizu, O. Nishio, Z. Y. Fang, and H. Ushijima. 2001. Genetic analysis of the capsid region of astroviruses. J. Med. Virol. 64:245-255. [DOI] [PubMed] [Google Scholar]
- 22.Willcocks, M. M., T. D. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 75:1785-1788. [DOI] [PubMed] [Google Scholar]
- 23.Willcocks, M. M., J. B. Kurtz, T. W. Lee, and M. J. Carter. 1995. Prevalence of human astrovirus serotype 4: capsid protein sequence and comparison with other strains. Epidemiol. Infect. 114:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarus, M. 1993. An RNA-amino acid affinity, p. 205-217. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world, vol. 24. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.