Abstract
Whether there is one or multiple αβT cell antigen receptor (TCR) recognition modules in a given TCR/CD3 complex is a long-standing controversy in immunology. We show that T cells from transgenic mice that coexpress comparable amounts of two distinct TCRβ chains incorporate at least two αβTCRs in a single TCR/CD3 complex. Evidence for bispecific αβTCRs was obtained by immunoprecipitation and immunoblotting and confirmed on the surface of living cells both by fluorescence resonance energy transfer and comodulation assays by using antibodies specific for TCRβ-variable regions. Such (αβ)2TCR/CD3 or higher-order complexes were evident in T cells studied either ex vivo or after expansion in vitro. T cell activation is thought by many, but not all, to require TCR cross-linking by its antigen/major histocompatibility complex ligand. The implications of a multivalent (αβ)2TCR/CD3 complex stoichiometry for the ordered docking of specific antigen/major histocompatibility complex, CD4, or CD8 coreceptors and additional TCRs are discussed.
The T cell receptor complex (TCR/CD3) consists of disulfide-linked αβ heterodimers noncovalently associated with invariant proteins of the CD3 (CD3γ, δ, and ɛ) and the ζ (ζ, η, and FcɛRIγ) families (1–3). The TCRα and TCRβ components contain variable (V) and constant (C) domains homologous to those of antibodies (Ig), but they are not secreted and recognize antigen fragments embedded in molecules of the major histocompatibility complex (MHC) (4–6). The CD3/ζ subunits have both signaling and structural functions (7, 8). Whether one or multiple αβTCR heterodimers occur in a TCR/CD3 complex currently is unresolved. A reassessment of TCR stoichiometry began after the finding that two CD3ɛ chains occur per TCR/CD3 complex (9, 10), because early studies indicated a 1:1 ratio for TCR/CD3 chains using mAbs to TCR and CD3 for estimation of the number of binding sites (1). On the one hand, several groups have suggested that TCR, like Ig, is multivalent, [i.e., (αβ)2TCR and (H+L)2Ig]. It was argued that such an arrangement: i) maximizes the interactions between charged transmembrane residues in αβTCR (3+) and CD3/ζ (6−) chains, rendering a more stable complex, and ii) fits with the hydrodynamic measurements of TCR/CD3 complex size (11–13). On the other hand, recent biochemical analyses argue against a multivalent TCR model because immunoprecipitations with mAb to Vα or Vβ failed to reveal the predicted association of two TCRαs or two TCRβs in lysates of T cells bearing two distinct αβTCRs (14, 15).
Previous studies of the TCR/CD3 subunit stoichiometry (9, 10, 15) have relied on the ability to coprecipitate from surface-labeled cells two homologous forms of a given subunit that were distinguishable after gel electrophoresis and autoradiography. Although such an experimental design allowed to show that two CD3ɛ subunits, of human and mouse origin, occur per TCR/CD3 complex (9, 10), it may be not of universal application. We and others have noticed that the similarity between two distinct TCRα or TCRβ chains expressed by dual-receptor T cells impeded a neat discrimination using the surface labeling and gel electrophoresis methods alone (14–16). A sensitive and specific alternative method may be necessary to detect the small amounts of bispecific αβTCR/CD3 complexes predicted to occur on the surface of dual-receptor T cells bearing the putative multivalent TCR recognition modules, because the majority of TCR/CD3 complexes preferentially may be monospecific (i.e., bear multiple identical TCR recognition modules) (15). It was reasoned that TCRVβ regions may serve as a tag to directly identify by immunoblotting each component of a multivalent, bispecific αβTCR in immunoprecipitations from T cells that express two different TCRVβ chains done using mAbs specific for each TCRVβ in criss-cross fashion in side-by-side experiments. Herein the proposed approach was used to analyze T cells from Vβ2×Vβ8 double transgenic mice and obtain biochemical evidence that the TCR/CD3 complex contains more than one αβTCR recognition module. The close vicinity of two αβTCR recognition modules on the cell surface of intact cells was confirmed by using a fluorescence resonance energy transfer (FRET) method adapted to the study of single cells, as done before in studies of the CD3ɛ subunit stoichiometry (10).
MATERIALS AND METHODS
Animals.
Mice were bred at the Centro de Investigaciones Biológicas Animal Facility following institutional guidelines. Breeding pairs of TCRVβ2 (Tg2) and TCRVβ8.2 (Tg93) single transgenic mice were provided by A. Iglesias (17) and H. Bluethmann (18), respectively. Tg2 mice were crossed with Tg93 mice to raise the Vβ2×Vβ8 double transgenic mice (19). In our experiments, the double transgenic mice were screened and bred to favor that T cells expressed the dual TCR at homogeneous and similarly high levels (16). The phenotype of every individual mouse was checked at 4–6 weeks of age in peripheral blood samples by immunofluorescence with Vβ-specific mAbs and flow cytometry in an EPICS XL analyzer (Coulter) and analyzed again in the organ samples or cultured cells submitted to the biochemistry and FRET assays.
Antibodies.
B. Malissen (Institut National de la Santé et de la Recherche Medicale–Centre National de la Recherche Scientifique, Marseilles, France), U. Staerz (University of Colorado Health Sciences Center, Denver), and H. von Boehmer (Institut National de la Santé et de la Recherche, Paris) kindly provided the mAbs specific for Vβ2 (B20.6.5) (20), Vβ8 (F23.1) (21), or Cα (H28–710) (22), respectively. These mAbs were purified from culture supernatants by affinity chromatography using protein A columns (Pharmacia) and either coupled to Sepharose beads or biotinated for the immunoprecipitation and Western blot experiments. Fluorochrome-labeled mAbs against CD3, CD4, CD8, CD11a, CD45, TCRVβ2, and TCRVβ8 were purchased from PharMingen. The specificity of anti-TCRVβ2 and anti-TCRVβ8 binding was confirmed in criss-cross experiments by staining T cell lines from either Vβ2 or Vβ8 single transgenic mice as well as in competition experiments by using the purified TCRVβ-specific mAb (16).
Cell Preparations, Immunofluorescence, and Flow Cytometry.
Blood was obtained by retroorbital bleeding, and mononuclear cells were purified by density gradient centrifugation over Ficoll/Paque (Pharmacia). Thymus and spleen were homogenized, and cells were recovered and washed twice by using ice-cold immunofluorescence buffer, PBS plus 2% BSA, plus 5 mM sodium azide. T cell lines were prepared as described previously (23). In the comodulation experiments (10), T cells were cultured at 37°C onto plastic wells precoated with either anti-TCRVβ2, -TCRVβ8, -CD3, or isotype and species-matched control mAbs, bound to promote TCR/CD3 complex down-regulation for the indicated times, before the immunofluorescence assay. ELISA plates (Costar 9018) were used to attain high-mAb-binding properties with a very low mAb release rate, as shown experimentally by cytometry-undetectable indirect immunofluorescence staining. All the fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-labeled mAbs and antisera were added at saturating titers and incubated with 5 × 105 cells for 20 min, before the cells were washed twice, in ice-cold immunofluorescence buffer using a refrigerated centrifuge. An EPICS XL analyzer was used immediately to quantitate the staining with FITC- and/or PE-conjugated mAbs at the single-cell level on the cell surface of intact cells, gated by their forward light-scatter and side-scatter signals. The viability of the gated cells was more than 99% as assessed independently by propidium iodide or 7-ADD staining. To allow for linearity and quantitation in the immunofluorescence analyses, a set of six calibration beads displaying predefined amounts of antibodies, QIFIKIT (DAKO), was used to set the optimal signal amplification and compensation levels (24).
Immunoprecipitation and Western Blot Analyses.
Transgenic cells were lysed in Brij96, or, alternatively, Nonidet P-40, digitonin, or 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, lysis buffer, and the lysate of 2 × 107 cells were immunoprecipitated with the indicated mAb, or species-matched irrelevant mAb, which was covalently coupled to protein A Sepharose beads using dimethylpimelimidate (Sigma). The precipitates were separated by SDS/12% PAGE under nonreducing conditions. In two-dimensional gels the second, vertical dimension was run under reducing conditions. After equilibration in transfer buffer, the proteins were transferred by wet blotting onto nitrocellulose membranes (Bio-Rad). Membranes were then blocked with nonfat 10% dry milk in PBS and incubated with the indicated mAb to TCRβ or TCRα domains in PBS containing 0.1% Tween-20. For detection of αβTCR proteins, biotin-conjugated mAbs were used as the first layer, followed by streptavidin-horse radish peroxidase (Southern Biotechnology). Immunoblots were revealed using enhanced chemiluminescence (Amersham) and x-ray film (Agfa) (25). Densitometry was done with a Computing Densitometer (Molecular Dynamics) on subsaturation-exposed film.
RESULTS AND DISCUSSION
T Cell Surface TCR/CD3 Complexes Accommodate Two or More αβTCR Recognition Modules.
Although TCR V region mAbs have been used widely as surrogate markers for αβT cell specificity in immunofluorescence, immunoprecipitation, and function analyses, there is scarce information on their reactivity in immunoblots (26). The specificity and sensitivity of TCR detection by Western blot analysis was assessed in preliminary criss-cross experiments (ref. 16; data not shown). Spleen T cells from mouse transgenic for TCRβ genes bearing either Vβ2 or Vβ8 regions (17, 18) were lysed under reducing conditions and subjected to Western blot analysis with mAbs to Vβ2 or Vβ8 TCRs (20, 21). The Vβ2- or Vβ8-bearing 45-kDa TCRβ proteins were revealed specifically by chemiluminescence. Instead, when samples were lysed and kept under nonreducing conditions during the assays, TCR Vβ-mAb recognized either ≈85-kDa disulfide-linked αβTCR complexes or free, nondisulfide linked, 45-kDa TCRβ chains. No crossreactivity for the other TCRβ chain was observed when an excess of more than 60-fold of the reciprocal TCRVβ was analyzed. The sensitivity of the assay was greater than 300,000 T cells [i.e., TCRs in the 10- to 100-fmol range assuming 30,000 TCRs per cell (16, 27)]. Transmembrane TCRβ chain is not exported to the T cell surface in the absence of TCRα or surrogate pre-TCRα chains (26). The 45-kDa band represents single TCRβ chains that, like other receptor subunits, are synthesized in excess and retained in the endoplasm reticule and degraded intracellularly if they do not assemble in “complete” TCR/CD3 complexes (2).
To determine whether each TCR/CD3 complex contains one or more αβTCR heterodimers, TCR/CD3 complexes that bear either Vβ2 or Vβ8 domains were immunoprecipitated from lysates of dual-receptor-bearing T cells from Vβ2×Vβ8 double transgenic mice (19). Association of two αβTCRs in single TCR/CD3 complexes was shown by the presence of the reciprocal TCRβ chain in the precipitates of each anti-Vβ-specific mAb, as revealed by immunoblotting (Fig. 1A). The coprecipitation of reciprocal TCR-Vβs was evident for both the 85-kDa and the 45-kDa bands from double transgenic T cells and occurred using four different detergents. No crossreactions were found in samples from single transgenic mice. The bispecific 85-kDa band, but not the 45-kDa band, incorporated TCRα as revealed by immunoblots with mAbs to Cα (Fig. 1B). The αβTCR heterodimers can be identified by two-dimensional (nonreducing/reducing) gel electrophoresis, in which the size diminution promoted by the break of the disulfide bond allows the TCRα and TCRβ subunits to migrate faster, below the diagonal where nondisulfide-bonded proteins run (26). Vβ2 and Vβ8 mAbs stained proteins off and on the diagonal that represent disulfide-linked and -unlinked TCRβ chains, respectively, in precipitates of lysates from double transgenic cells generated with the reciprocal Vβ-specific antibody (Fig. 1C Upper). Notably, TCRα staining was found only off the diagonal in two-dimensional gels (Fig. 1C Lower Left). It indicates that the 85-kDa band represents noncovalent associations of the “distinct” αβTCR disulfide-linked heterodimers, which cannot be resolved by size (14–16), whereas the 45-kDa band incorporates non-disulfide-linked associations of two TCRβs.
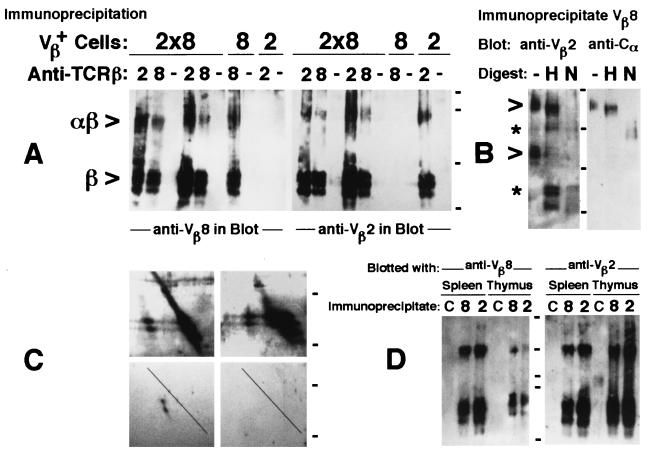
Figure 1.
Identification of bispecific αβTCRs on the surface of T cells from Vβ2×Vβ8 double transgenic mice. Immunoprecipitations of TCRβ from transgenic T cells were done with Vβ domain-specific mAbs or control reagents (−) and immunoblotted with mAbs to Vβ2, Vβ8, or Cα. Cell sources were either spleen T cells expanded in culture for 2–3 weeks with interleukin 2 (A–C) or freshly isolated single-cell suspensions from spleen and thymus (D) (10). Organs were either from Vβ2 or Vβ8 single transgenic mice or Vβ2×Vβ8 double transgenic F1 mice (17–19), as indicated (A). In additional control immunoprecipitations, lines labeled as C in D, Vβ2 and Vβ8 single transgenic cells were mixed in a 1:1 ratio before the immunoprecipitation with TCRVβ mAb, which had the reciprocal specificity to those used in immunoblotting. Double transgenic cells were used in the remaining experiments (B–D). Two-dimensional SDS/PAGE (C) of immunoprecipitates with mAb to Vβ2 (Left) were blotted with mAb to Vβ8 (Upper) and Cα (Lower), whereas immunoprecipitates with mAb to Vβ8 (Right) where blotted with anti-Vβ2 (Upper) and control (Lower) mAbs. Immunoprecipitates were digested (B) in the absence (−) or presence of endoglycosidase H (H) or N-glycosidase F (N) to assess the TCR glycosylation pattern (29). Sizes before and after the removal of N-linked sugars are indicated by open arrowheads and asterisks, respectively. Cells were lysed in 1% of either Nonidet P-40 (first three lines from the left in A) or Brij96 lysis buffer in the remaining experiments shown. Associations were also evident in the presence of two other detergents (0.25% saturated digitonin and 5 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, data not shown; ref. 16). Neat coprecipitations were attained in 30 independent experiments, each with different transgenic mice. Ticks indicate migration of molecular size markers (Bio-Rad), given in kDa: 101, 83, 50.6, and 35.5 kDa (A), 148, 60, 42, and 30 kDa (B), 60 and 42 kDa (C), and 145, 83, 60, 50, and 35 kDa (D).
To address the question of whether the bispecific αβTCRs were expressed on the cell surface, we took advantage of the wealth of information about the architectural editing of TCR/CD3 complexes that reach the plasma membrane (2, 3, 28, 29). The TCR glycoproteins in the bispecific αβTCRs/85-kDa band were biochemically distinct from those in the 45-kDa band in that their N-linked sugars were resistant to digestion with endoglycosidase H (Endo H, Fig. 1B). The endoglycosidase H-resistance pattern is a feature of αβTCRs expressed in the T cell surface (3, 29) and was demonstrated in the 85-kDa complexes, which retained their size after digestion (Fig. 1B, open arrowheads). The 45-kDa band instead was endoglycosidase H-sensitive and was reduced to the core polypeptide size of TCRβ (Fig. 1B, asterisks), as expected for the excess of individual receptor subunits in incomplete TCR/CD3 complexes retained in the endoplasm reticule (3, 29). N-glycosidase F digestions reduced both 85- and 45-kDa bands to their expected core polypeptide sizes, indicating the accessibility to digestion of the glycans bonded to the dual αβTCRs (29) (Fig. 1B). Taken together the results suggest that most 85-kDa dual-αβTCR-containing complexes are expressed on the T cell surface. TCRβ associations initiate intracellularly, and neither require interchain disulfide bond formation or TCRα integration into the assembling TCR/CD3 complexes.
Several groups have hypothesized that T cell receptors for antigen and their ligands may assemble upon ligand engagement in dimers of dimers termed superdimers (TCR-antigen-MHC)2, but in the absence of cognate antigen-MHC complexes, in resting T cells, TCRs would be monovalent (6, 30). It may be argued that the bispecific receptors used to reveal TCR multivalency were generated artificially or limited to a small fraction of T cells (i.e., because of detergent solubilization or activation of T cells). Several experiments were designed to address these possibilities. First, bispecific αβTCR 85-kDa bands were readily evident in freshly isolated, unstimulated cells from the thymus and spleen of Vβ2×Vβ8 double transgenic mice (Fig. 1D) and are not restricted to the in vitro activated T cells studied previously (Fig. 1 A–C). Second, no evidence for coprecipitation was attained when spleen and thymus cells from TCRVβ2 and TCRVβ8 single transgenic mice were mixed at a 1:1 ratio, centrifuged, and the cell pellet was lysed with detergent (Fig. 1D, lines C). Results indicate no bispecific receptor associations when immunoprecipitates from 4 × 107 single transgenic cell mixtures were subjected to immunoblotting with the reciprocal anti-Vβ mAb, whereas coprecipitation was always readily observed in the double transgenic T cell lysates from 2 × 107 cells run in parallel. Third, the vicinity of Vβ2 and Vβ8 TCRs was verified on the surface of single viable T cells kept at 4°C by using flow cytometric FRET as a spectroscopic ruler (10, 31). Efficient FRET causes quenching of the green FITC emission when the PE acceptor molecules are in close physical vicinity to the FITC donor dye (R0 = 10 nm; ref. 31). Quenching of the anti-Vβ2 and -Vβ8-FITC fluorescence was readily evident when the reciprocal PE-labeled TCRβ mAb was present (Fig. 2A and D, shaded histograms). FRET was also evident between TCRβs and the associated CD3 molecules, as reported before (10, 31). FRET is considered efficient when >2% energy transfer occurs (10, 31), and the observed specific shifts in the FITC fluorescence ranged up to 15–20%, similar to that observed for the TCR/CD3 couple (17–18%). Quenching was not detectable when the partners for Vβ2 or Vβ8 mAb in the two color staining were CD4-, CD8-, integrin CD11a-, or leukocyte common antigen CD45-specific mAb. It indicates specificity in the method because surface density of the latter acceptors is at least 2-fold higher than in both TCRs (1), which should favor FRET (10). Fourth, the relative amount of cell-surface TCR/CD3 complexes bearing two distinct TCRVβs was quantitated in comodulation experiments (1, 10). The down-regulation of either TCRVβ2 or TCRVβ8 was accompanied by comodulation of a sizable proportion (up to 20–40%) of the TCRs bearing the reciprocal TCRVβ (Fig. 3). That comodulation can affect up to half of unstimulated receptors in distinct dual-receptor T cells (i.e., also reaches a maximum of 40% vs. 80% stimulated TCRs; ref. 32) fits with a two αβTCR per TCR/CD3 complex stoichiometry. Finally, a computer model for the TCR-binding sites in tetrameric antigen/MHC–avidin complexes (33) was calculated by using the crystallographic coordinates available for the interacting molecules (4, 6, 34). Two important observations led to this latter experiment: i) tetrameric antigen/MHC complexes bind to T cells with remarkably higher avidity and slower dissociation rates than soluble monomeric antigen/MHC complexes, allowing for the immunological staining of T cells with different specificities (33), and ii) soluble divalent TCRs show markedly increased avidity for their MHC ligand when compared with monovalent TCRs (35). Our analysis of the tetrameric antigen/MHC–avidin model revealed that two neighbor antigen-binding sites in such an x-shaped TCR ligand would be closer than 8 nm, an order of magnitude nearer than expected for the random distribution of two TCR-binding sites in T cells bearing monovalent TCRs (16). If TCRs were monovalent, the latter distances would not allow for the experimentally observed cooperative binding, but two TCRs residing in the same TCR/CD3 complex may well be within the estimated 8-nm distance. Bispecific TCRs in double transgenic mice were used as a tool to reveal the association of two TCRs using three independent methods. The results show that, in our model, αβTCR/CD3 complexes occur constitutively as multivalent receptors on the surface of T cells. Because multimeric MHC ligands render saturating staining of normal T cells (33), we suggest that a large portion of, if not all, TCRs were also multivalent in normal αβT cells.
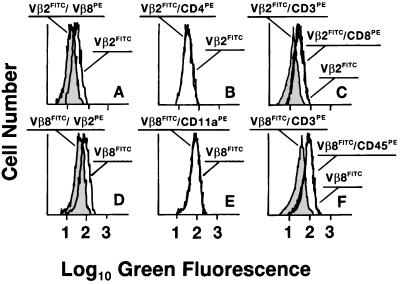
Figure 2.
The two distinct TCRβ chains are neighbors in the plasma membrane of live T cells bearing dual αβTCRs. Spleen T cell lines from Vβ2×Vβ8 double transgenic mice were stained (Upper) with mAb to Vβ2 conjugated with FITC alone or followed by PE-labeled mAb against either Vβ8, CD3, CD4, or CD8. Alternatively (Lower), cells were stained with FITC-conjugated anti-Vβ8 mAb alone or followed by PE-labeled mAb against either Vβ2, CD3, CD11a, or CD45. Flow cytometry analyses of average donor quenching (10, 31) measure the putative reduction in the green FITC fluorescence emission on the surface of viable T cells promoted by neighbor PE-acceptor molecules (10, 31). In cells stained with the two anti-TCRVβ mAbs (A and D), the FITC fluorescence distribution was reduced as indicated by the shift to the left in the presence of the quenching, reciprocal anti-TCRVβ mAb (shaded histograms). Both TCRβ chains were also in the vicinity of CD3 subunits (shaded histograms in C and F). Histograms for TCRVβ staining in the presence and absence of the PE-labeled mAb against CD4, CD8, CD11a, and CD45 instead are superimposed. The ratio of Vβ2 to Vβ8 mean fluorescence intensity ranged from 1:1 to 2:3 in samples from several mice. Similar results were observed for tetraplicate samples in three independent experiments as well as in analyses of freshly isolated spleen and thymus cells (data not shown).
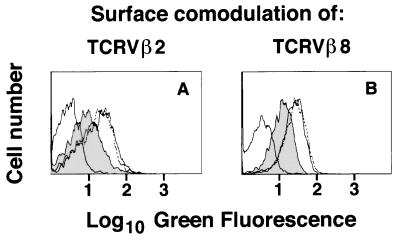
Figure 3.
Comodulation of unstimulated TCRs upon down-regulation of specific TCR/CD3 complexes in dual-receptor T cells. Immunofluorescence distribution profiles of spleen T cell lines from Vβ2×Vβ8 double transgenic mice submitted to different TCR down-regulation protocols and stained with FITC-labeled mAb against TCRVβ8 (A) or TCRVβ2 (B). Left histograms in both A and B represent the background staining with anti-Ig fluoresceinated control reagents that was superimposable to the autofluorescence (data not shown). Shaded histograms show the comodulation of the stained, unstimulated TCRs promoted after down-regulation of the reciprocal TCRβ, done by incubation at 37°C for 4 h in plates coated with 5 μg/ml of mAb specific for the reciprocal TCRVβ. Down-modulation is not promoted either in replicate plates kept in parallel at 4°C (dotted line) or by a control mAb [OKT3, which promotes TCR/CD3 comodulation in human CD3 transgenic T cells (10)] (continuous line), as shown in the right histograms. The mAbs do not promote modulation of the reciprocal TCRβ in single transgenic mice (data not shown). Results are representative of four experiments.
Punt et al. (15) advocated that each TCR/CD3 complex contains one αβTCR because their elegant biochemical approach did not reveal bispecific αβTCRs in tetratransgenic mice, which bear four fixed TCR chains (Vα3, Vα11, Vβ3, and Vβ8). It is difficult to preclude, as Punt et al. posed (15), that TCR complexes in the tetratransgenic mice contain only multiple, identical αβTCR pairs, but not the “detectable” bispecific TCRs, because of incompatibilities in the pairwise associations of two different αβTCRs. TCRα and TCRβ chains do not form surface-expressed TCR heterodimers in a random manner (36). Unlike tetratransgenic mice, T cells from Vβ2×Vβ8 double transgenic mice resemble the fraction of T cells from healthy individuals (37), which bear two TCRβ chains but a single nonfixed TCRα chain, which differs from clone to clone. The proportions of the two TCRs expressed on the cell surface show a continuous distribution in the different clones, reaching up to a 50-fold difference in their levels (36, 38). In our experiments, T cells express both TCRs at homogeneous and similarly high levels (Fig. 2), because the double transgenic mice were screened and bred to select that phenotype, which favors the detection of dual-TCR associations. The ratio of stimulated to unstimulated TCRs down-regulated in dual-receptor T cells varies markedly as a function of the stimulation conditions (13, 16, 32, 39), and it is evident only when the two TCRs were expressed in similar levels (16, 32). If there were just one TCR module per TCR/CD3 complex, the lack of down-modulation of unstimulated TCRs would be an essential feature of the serial-triggering theory of T cell activation (38, 39), and experiments showing comodulation of unstimulated TCRs may be viewed as a challenge to the theory (32). The down-regulation of unstimulated TCRs does not contradict the serial-triggering model when the αβTCR/CD3 complex is multivalent (16). Instead, it may provide another reason for the selectively inefficient down-regulation of bispecific TCRs observed in some systems (38, 39): bispecific (αβ)2 TCR/CD3 complexes are monovalent for each of two distinct ligands. Monovalent TCRs may have more stringent triggering requirements. Indeed, bispecific anti-CD3 mAbs do not down-regulate the TCR/CD3 complex unless the second binding site engages another receptor on the surface of T cells or antigen-presenting cells (APCs) (40, 41). Because bispecific CD3 mAbs bind at 1.5 times the level of divalent ones, such differences cannot be attributed to reduced receptor occupation (40). The latter result requires caution when using “conventional,” divalent mAbs in binding assays to estimate the TCR/CD3 ratio, for which either 1:1, 1:1.5, or 1:2 ratios were reported (1, 15, 27, 42).
Revision of the Monovalent Model of the TCR/CD3 Complex.
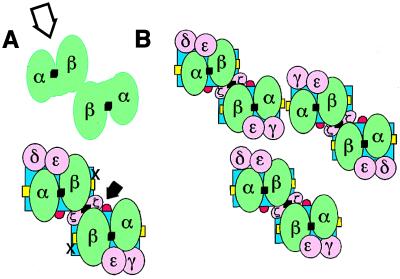
The finding that a TCR/CD3 complex can accommodate more than one αβTCR recognition module, together with the data of other authors, led us to review our minimal TCR/CD3 model (10), which is still widely accepted (15, 43). The proposal by Garboczi et al. of a general “interlocking” binding mode between αβTCR and MHC (5) allows us to envisage the footprint of a divalent αβTCR (Fig. 4A, green). The packing arises from a translation of a single TCR–antigen–MHC interaction (5) onto a template of two antiparallel MHC molecules (6). Vα domains would be positioned over the external halves of MHC (Fig. 4A, blue), and the N terminus of antigen (Fig. 4A, yellow) and Vβ domains would be positioned, neighbor, over the internal halves of MHC and the C terminus of antigen, but tilted 20–30° toward the diagonal (4, 5). The outer interface of the TCR constant domains shows a cavity (open arrow) flanked by the TCRβ elbow and the Cα transmembrane (4). Reinherz and coworkers have proposed that such a cave may accommodate the extracellular domain of a single CD3ɛ subunit (43). Because two CD3ɛ subunits occur per TCR/CD3 complex, and CD3ɛ subunits are incorporated into distinct dimers (i.e., CD3ɛγ and CD3ɛδ transduction modules, Fig. 4A, pink) (9–12), these authors argued that in a monovalent TCR only one of the two CD3 dimers can be accommodated in the single cave. It would leave a CD3 module exposed, apart from the docking site (43). However, a divalent TCR/CD3 complex provides a symmetric cave, which may harbor the second CD3 dimer. Thus, each CD3 module could be placed near a Cα transmembrane region, where they can interact cooperatively with either pair of Cα-Cβ domains (28). The elbow regions between V and C domains and the quite parallel Cα-Cβ intersection, which give the αβTCR its squat appearance, would place the Cβ and Cα transmembrane regions close to the symmetry axis and the indicated outer face of the αβTCR complexes, respectively (4). The third class of transduction module (i.e., disulfide-linked ζ-ζ chains; ref. 8) bears tiny extracellular domains. It could be placed in the symmetry axis, near the two Cβ transmembrane regions, bridging the two hemireceptors (44). The two positive-charged transmembrane residues either in each TCRα chain or in the TCRβ–TCRβ pair thus may be buried by the two negative charges present in each neighbor-parallel transduction module (11). Coreceptors are proposed to dock to the TCR/CD3 complex in an oriented fashion and only after the TCR engages the antigen/MHC ligand (6, 45). CD4 and CD8 coreceptors augment the TCR avidity for MHC class II and I molecules, respectively, after occupation of their binding sites in MHC molecules [i.e., red loop for β2 and cross for α1 sites in MHC class II (6), with a similar site in MHC class I (46)]. The interlocked mode of TCR–MHC interaction uncovers the docking location (Fig. 4A, solid arrow) for two coreceptors per antigen–receptor complex and may help to delineate how the coreceptor-associated p56lck tyrosine kinase targets the immune tyrosin receptor motifs in the transduction modules (45).
Figure 4.
Hypothetical model of (αβTCR)2/CD3 complex. (A) Packing of two αβTCR in a single TCR/CD3 complex. Disulfide bridges in each αβTCR heterodimer or in the ζ homodimer are depicted as black squares. The blue rectangles represent two antiparallel MHC class II molecules presenting the specific peptides (yellow strands). The surface of the αβTCR interface opposite to the CD3 pocket is flat; for simplicity, ovals have been used in most cases. Different isoforms of TCR have been reported with regard to the CD3/ζ module usage, which is reviewed in ref. 54. For simplicity, only the three most common transduction modules are included. (B) The hexameric antigen/MHC packing arises after homotypical aggregation of TCRs and coreceptors, as observed from the T cell nuclei looking toward the APC. Oligomerization occurs in the two dimensions of the T cell and APC membrane after the TCR/CD3 complexes have engaged specific antigen/MHC complexes. The specific TCR ligands are monomers that are rare and occur naturally dispersed on the APC surface among a majority population of irrelevant antigen/MHC complexes but may be concentrated to the T cell/APC interface by the constitutively divalent TCRs.
The possibility for intra-TCR/CD3 complex cross-linking by antigen does not preclude higher degrees of TCR oligomerization (Fig. 4B). Davis and coworkers have shown ligand-specific oligomerization of αβTCRs, using solubilized monomeric molecules, that are titratable clearly beyond (TCR–antigen–MHC)2 arrangements (47). A chain-docking of TCRs implies multiple “oligomerization sites” per αβTCR/MHC “superdimer.” Whereas the proposed TCRβ/TCRβ and ζ-ζ interactions may interlock constitutive αβTCR dimers, TCRα may nucleate serial oligomerization of divalent αβTCR/CD3 complexes plus coreceptors, triggered by ligand engagement. Candidate domains for Vα dimerization, placed by Fields et al. in the vicinity of the switched C" strand (30), are located suggestively facing outward of the left and right wings of the superdimer. Linear arrays of TCRs could form patches of complexes by oligomerization in the second dimension of the membrane, perhaps aided by the dimeric association of coreceptors (48, 49), shown at the high concentration proposed for the T cell/APC interface (50). TCR occupation may successfully enrich the latter interface for the small portion of MHC complexes presenting agonist antigen (50), which then may dimerize (47) (irrelevant antigen–MHC complexes are not depicted for simplicity because they are in excess of 500- to 1,000-fold). If divalent TCRs then were internalized serially, a small, “preformed” patch of agonist/MHC ligands (i.e., a hexamer) could sustain an “efficient” trailing engagement of more than 1,000 TCRs per cluster (39) and account for the internalization of TCRs as dimers (51). The scheme agrees with the idea that antigen/MHC oligomers were the epitopes that efficiently trigger T cell activation, being trimers or higher-order complexes the best immunogens (refs. 52 and 53; and M. M. Davis, personal communication). Divalent T cell receptors may provide a “built-in” kinetic proofreading mechanism for paired agonist/antagonist ligand-titratable discrimination. The relevance of “interlocked” and “titratable discrimination” models of TCR–antigen/MHC recognition for specific selection and sensitive T cell activation has been discussed elegantly by other authors (5, 47). Our model combines features of both proposals and adds that the TCR is divalent, built as a three-dimensional nucleation unit for the essential ligands and signal transduction elements needed for “successful” T cell triggering.
Acknowledgments
This paper is dedicated to the memory of the late P. Fernández-Barrado. We thank C. Martínez-A. and K. Karjalainen for critical comments; A. Valencia and R. Giraldo for help with molecular modeling; E. Calvo-Alcocer for manuscript editing; B. Malissen, H. von Boehmer, and U. Staerz for reagents; and the staff at the Centro de Investigaciones Biológicas animal facility for help. This research was supported by Comision Interministerial de Ciencia y Tecnologia and Comunidad Autonoma de Madrid fellowships (G.F.M. and E.S.) and Comision Interministerial de Ciencia y Tecnologia Grants SAF96-201 (A.H.), MEC HA-161 (A.H. and A.I.), PM95-5 (B.A.) and Comunidad Autonoma de Madrid Grants 98/96 and 47/96 (A.H. and B.A.).
ABBREVIATIONS
- APC
antigen presenting cell
- FITC
fluorescein isothiocyanate
- FRET
fluorescence resonance energy transfer
- MHC
major histocompatibility complex
- PE
phycoerythrin
- TCR
T cell receptor
References
- 1.Meuer S C, Acuto O, Hussey R E, Hodgdon J C, Fitzgerald K A, Schlossman S F, Reinherz E L. Nature (London) 1983;303:808–810. doi: 10.1038/303808a0. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H, Alarcon B, Wileman T, Terhorst C. Annu Rev Immunol. 1988;6:629–662. doi: 10.1146/annurev.iy.06.040188.003213. [DOI] [PubMed] [Google Scholar]
- 3.Klausner R D, Lippcott-Schwartz J, Bonifacino J S. Annu Rev Immunol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 4.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 5.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 6.Brown J H, Jardetzsky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 7.Chan A C, Irving B A, Weiss A. Curr Opin Immunol. 1992;4:246–251. doi: 10.1016/0952-7915(92)90072-m. [DOI] [PubMed] [Google Scholar]
- 8.Wegener A M, Letourneur F, Hoeveler A, Brocker T, Luton F, Malissen B. Cell. 1992;68:83–95. doi: 10.1016/0092-8674(92)90208-t. [DOI] [PubMed] [Google Scholar]
- 9.Blumberg R S, Ley S, Sancho J, Lonberg N, Lacy E, McDermott F, Schad V, Greenstein J L, Terhorst C. Proc Natl Acad Sci USA. 1990;87:7220–7224. doi: 10.1073/pnas.87.18.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Hera A, Müller U, Olsson C, Isaaz S, Tunnacliffe A. J Exp Med. 1991;173:7–17. doi: 10.1084/jem.173.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green N M. Nature (London) 1991;351:349–350. doi: 10.1038/351349a0. [DOI] [PubMed] [Google Scholar]
- 12.Terhorst C. In: T Cell Receptors. Bell J I, Owen M J, Simpson E, editors. New York: Oxford Univ. Press; 1995. pp. 369–402. [Google Scholar]
- 13.Exley M, Wileman T, Mueller B, Terhorst C. Mol Immunol. 1995;32:829–839. doi: 10.1016/0161-5890(95)00046-h. [DOI] [PubMed] [Google Scholar]
- 14.Hou X, Dietrich J, Kulhmann J, Wegener A M, Geisler C. Eur J Immunol. 1994;24:1228–1233. doi: 10.1002/eji.1830240534. [DOI] [PubMed] [Google Scholar]
- 15.Punt J A, Roberts J L, Kearse K P, Singer A. J Exp Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Miguel G. Ph.D. Thesis. Madrid: Alcalá University; 1998. [Google Scholar]
- 17.Iglesias A, Hansen-Hagge T, Bonin A V, Weltzien H U. Eur J Immunol. 1992;22:335–341. doi: 10.1002/eji.1830220208. [DOI] [PubMed] [Google Scholar]
- 18.Uetmatsu Y, Ryser S, Dembic Z, Borgulya P, Krimperfort P, von Boehmer H, Steinmetz M. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 19.van Meerwijk J P, Romagnoli P, Iglesias A, Bluethmann H, Steinmetz M. J Exp Med. 1991;174:815–819. doi: 10.1084/jem.174.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grégoire C, Rebaï N, Schweisguth F, Necker A, Mazza G, Auphan N, Millward A, Schmitt-Verhulst A M, Malissen B. Proc Natl Acad Sci USA. 1991;88:8077–8081. doi: 10.1073/pnas.88.18.8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staerz U, Rammensee H G, Benedetto J D, Bevan M J. J Immunol. 1985;134:3994–4000. [PubMed] [Google Scholar]
- 22.Becker M L, Near R, Mudgett-Hunter M, Margolies M N, Kubo R T, Kaye J, Hedrick S M. Cell. 1989;58:911–921. doi: 10.1016/0092-8674(89)90943-4. [DOI] [PubMed] [Google Scholar]
- 23.Tunnacliffe A, Olsson C, de la Hera A. Int Immunol. 1989;1:546–550. doi: 10.1093/intimm/1.5.546. [DOI] [PubMed] [Google Scholar]
- 24.Roman L I, Manzano L, de la Hera A, Abreu L, Rossi I, Alvarez-Mon M. Gastroenterology. 1996;110:1008–1019. doi: 10.1053/gast.1996.v110.pm8612987. [DOI] [PubMed] [Google Scholar]
- 25.Sanz E, de la Hera A. J Exp Med. 1996;183:2693–2698. doi: 10.1084/jem.183.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling H J, von Boehmer H. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 27.Saizawa K, Rojo J, Janeway C A., Jr Nature (London) 1987;328:260–263. doi: 10.1038/328260a0. [DOI] [PubMed] [Google Scholar]
- 28.Manolios N, Letourneur F, Bonifacino J S, Klausner R D. EMBO J. 1991;10:1643–1651. doi: 10.1002/j.1460-2075.1991.tb07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcón B, Berkhout B, Breitmeyer J, Terhorst C. J Biol Chem. 1988;263:2953–2961. [PubMed] [Google Scholar]
- 30.Fields B A, Ober B, Malchioldi E L, Lebedeva M, Braden B C, Ysern X, Kin J, Shao X, Ward E, Mariuzza R A. Science. 1995;270:1821–1824. doi: 10.1126/science.270.5243.1821. [DOI] [PubMed] [Google Scholar]
- 31.Szabò G, Jr, Pine P S, Weaver J L, Karasi M, Aszalos A. Biophys J. 1992;61:661–670. doi: 10.1016/S0006-3495(92)81871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niedergang F, Dautry-Varsat A, Alcover A. J Immunol. 1997;159:1703–1710. [PubMed] [Google Scholar]
- 33.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 34.Pugliese L, Coda A, Malcovati M, Bolognesi M. J Mol Biol. 1993;231:698–710. doi: 10.1006/jmbi.1993.1321. [DOI] [PubMed] [Google Scholar]
- 35.O’Herrin S M, Lebowitz M S, Bieler J G, al-Ramadi B K, Utz U, Bothwell A L M, Schneck J P. J Exp Med. 1997;186:1333–1345. doi: 10.1084/jem.186.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Sussman J L, Ashwell J D, Germain R N. J Immunol. 1989;143:3379–3384. [PubMed] [Google Scholar]
- 37.Padovan E, Giachino C, Cella M, Valititti S, Acuto O, Lanzavecchia A. J Exp Med. 1995;181:1587–1591. doi: 10.1084/jem.181.4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viola A, Lanzavecchia A. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 39.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Nature (London) 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 40.Roosnek E, Lanzavecchia A. J Exp Med. 1989;170:297–302. doi: 10.1084/jem.170.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roosnek E, Tunnacliffe A, Lanzavecchia A. Eur J Immunol. 1990;20:1393–1396. doi: 10.1002/eji.1830200627. [DOI] [PubMed] [Google Scholar]
- 42.Thibault G, Bardos P. J Immunol. 1995;154:3814–3820. [PubMed] [Google Scholar]
- 43.Wang J, Lim K, Smolyar A, Teng M, Liu J, Tse A, Liu J, Hussey R E, Chrishti Y, Thomson C, et al. EMBO J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.San José E, Sahuquillo A G, Bragado R, Alarcón B. Eur J Immunol. 1998;28:12–21. doi: 10.1002/(SICI)1521-4141(199801)28:01<12::AID-IMMU12>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 45.Dianzani U, Shaw A, al-Ramadi B K, Kubo R T, Janeway C A., Jr J Immunol. 1992;148:678–688. [PubMed] [Google Scholar]
- 46.Jones E, Tormo J, Reid S, Stuart D I. Immunol Rev. 1998;163:121–128. doi: 10.1111/j.1600-065x.1998.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 47.Reich Z, Boniface J J, Lyons D S, Borochov N, Wachtel E J, Davis M M. Nature (London) 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 48.Sakihama T, Smolyar A, Reinherz E L. Proc Natl Acad Sci USA. 1995;92:6444–6448. doi: 10.1073/pnas.92.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H, Kwong P D, Hendrickson W A. Nature (London) 1997;387:527–530. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 50.Davis M M. Nature (London) 1995;375:104. doi: 10.1038/375104a0. [DOI] [PubMed] [Google Scholar]
- 51.Batchmann M F, Salzmann M, Oxenius A, Ohashi P S. Eur J Immunol. 1998;28:2571–2579. doi: 10.1002/(SICI)1521-4141(199808)28:08<2571::AID-IMMU2571>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 52.Abastado J P, Lone Y C, Casrouge A, Boulot G, Kourilsky P. J Exp Med. 1995;182:439–447. doi: 10.1084/jem.182.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rötzschke O, Falk K, Strominger J L. Proc Natl Acad Sci USA. 1997;94:14642–14647. doi: 10.1073/pnas.94.26.14642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dave V P, Cao Z, Browne C, Alarcón B, Fernández-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes D J. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]