Abstract
The effects of inserting reported nuclear localization signals (NLSs) into the Moloney murine leukemia virus (Mo-MuLV) integrase (IN) protein, within a replication-competent viral construct, were studied. In contrast to the virus harboring IN fused to the simian virus 40 (SV40) large T antigen NLS (SV40 NLS) (J. A. Seamon, M. Adams, S. Sengupta, and M. J. Roth, Virology 274:412-419, 2000), a codon-modified SV40 NLS was stably expressed during viral propagation. Incorporation of the codon-modified SV40 NLS into IN, however, altered the packaging of the Gag-Pol precursor in the virus; viral particles contained decreased levels of reverse transcriptase (RT) and IN. In addition, the virus showed delayed kinetics of viral DNA synthesis upon infection. A panel of infectious MuLVs containing alternative IN-NLS fusions was generated and assayed for cell cycle-independent infection. Viral infection with the NLS-tagged proteins, however, remained dependent on passage of the cells through mitosis. This finding has direct implications for engineering murine-based retroviral vectors for gene therapy.
Following entry into the cytoplasm of the host cell, reverse transcription of the retroviral RNA genome into double-stranded viral DNA occurs within large nucleoprotein complexes derived from the core of the infecting virion, termed preintegration complexes (PICs). These complexes must then gain access to host cell chromosomes within the nucleus to achieve stable integration. Human immunodeficiency virus type 1 (HIV-1) can infect certain types of nondividing cells (5, 22, 46, 74). Although the ability of lentiviruses to infect nondividing cells has been exploited in the development of vectors for human gene therapy, their application is limited by safety concerns. In contrast, nuclear accumulation of Moloney murine leukemia virus (Mo-MuLV) viral DNA increases only after infected cells pass through mitosis (61). This property limits the range of cells and target tissues that can use murine-based retroviral vectors for gene transfer or gene therapy.
Many studies put forth the model that HIV-1 PICs are actively imported into the nucleus. In support of this model, nuclear localization signals (NLSs) have been reported on a number of viral proteins that comprise the HIV-1 PIC. The role or roles that these sequences play in viral replication, however, remain unclear. In addition, a cis-acting viral DNA structure generated during lentivirus-specific reverse transcription, termed the central DNA flap, may also be involved in the nuclear import of the HIV-1 genome (77). To date, no NLSs have been identified in MuLV PIC components. This suggests that it is the disassembly of the nuclear envelope during mitosis that renders the chromosomes accessible to the MuLV PIC. Interestingly, the avian sarcoma virus (ASV), which has a novel NLS sequence (44, 45), presents an intermediate phenotype in which mitosis-independent replication can occur, but at a lower efficiency than HIV-1 (31).
Previous analysis indicated various molecular tags placed at the C terminus of the Mo-MuLV integrase (IN) protein could be incorporated into viral particles when expressed within the context of a replication-competent virus. IN proteins tagged with either six-His or hemagglutinin (HA) epitope tags were shown to be genetically stable during replication. In contrast, insertion of the SV40 large T-antigen NLS (SV40 NLS) was not stable, resulting in the selection and propagation of viruses in which the basic NLS sequence was disrupted (65). This study examines the stability of alternative NLS sequences within Mo-MuLV IN and the influence the NLS sequences have on the nuclear transport properties of Mo-MuLV. A similar approach to alter nuclear transport properties was reported to be successful for spleen necrosis virus (SNV), in which the efficiency of infecting nondividing cells was altered by incorporating an HIV-1 matrix (MA) protein NLS sequence into the SNV MA protein (56).
Expression of IN and IN-SV40 NLS proteins.
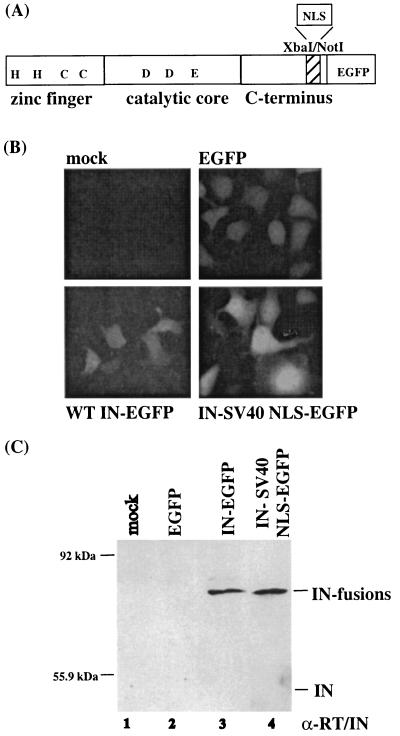
The effects of inserting the SV40 NLS into M-MuLV IN were first examined in an IN expression system in the absence of additional viral proteins. Deletion studies of the C terminus of MuLV IN have indicated that the C-terminal 28 amino acids are nonessential in vivo (62) and in vitro (41). The SV40 NLS sequence was inserted within this nonessential region, resulting in the truncation of the terminal 23 amino acids of IN (Fig. 1A) (65). The IN-SV40 NLS protein was generated by PCR amplification of the template plasmid pNCA-C-XN-SU8 harboring the NLS fusion (65). Full-length IN was PCR amplified with the template plasmid pNCA-C (17). Both products were inserted into a pcDNA3.1/Myc-His A (Invitrogen) expression vector as an EcoRI-XbaI fragment. The primers used for each construct are listed in Table 1. The C terminus of both wild-type IN and the IN-SV40 NLS proteins contained enhanced green fluorescent protein (EGFP) fusions, derived from replacement of BamHI-NotI fragments in pEGFP-1 (Clonetech). Both proteins were evaluated for subcellular localization in transfected canine osteosarcoma D17 cells (Fig. 1B). Localization was assayed via EGFP expression.
FIG. 1.
Localization of full-length IN and a truncated IN protein C-terminally fused to the SV40 NLS (IN-SV40 NLS). (A) Illustration of the three-domain structure of Mo-MuLV IN C-terminally fused to EGFP. The location of the NLS insertion, within a nonessential region of IN, is indicated. (B) Subcellular localization of IN and IN-SV40 NLS EGFP fusion proteins. Canine D17 cells (8 × 105/6-cm-diameter dish) were transfected by the Lipofectamine (Gibco BRL/Invitrogen) method according to the manufacturer's instructions and with 4 μg of plasmid DNA, 10 μl of Lipofectamine Plus reagent, 18 μl of Lipofectamine reagent, and 200 μl of serum-free medium for each complex. The complexes were added to 2.5 ml of serum-free medium and incubated on cells for 5 h. Subcellular localization of the vector control, the EGFP control, wild-type (WT) IN-EGFP, and IN-SV40 NLS-EGFP was analyzed by fluorescent microscopy 48 h after the addition of DNA. (C) Expression of IN-EGFP and IN-SV 40 NLS-EGFP fusion proteins. Transfected cells were lysed in 100 μl of 2× Laemmli buffer and spun for 3 min at 4°C. Lysates of D17 cells transfected with IN-EGFP (lane 3) and IN-SV 40 NLS-EGFP (lane 4) or the indicated controls (lanes 1 to 2) were analyzed by Western blotting with a polyclonal antibody (α-RT/IN) directed against Mo-MuLV RT and IN (rabbit 3, bleed 6) (72) and detected with the NEN ECL enhanced chemiluminescence system. The positions of wild-type IN and the IN fusion proteins are indicated to the right. The positions of migration of the prestained protein standards and their molecular masses are indicated to the side of each panel.
TABLE 1.
Sequences of the primers used in this study
| Construct | Primer sequence |
|---|---|
| IN EGFPa | 5′ CCGGAATTCCATGATAGAAAATTCATCACCCTACACC 3′ |
| 5′ CCAATTGGGCGCTCCGGGGGAACCTAGGCGC 3′ | |
| IN-SV40 NLS-EGFPa | 5′ CCGGAATTCCATGATAGAAAATTCATCACCCTACACC 3′ |
| 5′ CGCGGATCCAACTTTGCGGCCTTTCTCTTCTT 3′ | |
| Nucleoplasmin NLSb | 5′ GGCCGTAAAGAGCCCTGCGGCTACCAAAAAAGCAGGCCAGGCAAAGAAGAA 3′ |
| 3′ CATTTCTCGGGACGCCGATGGTTTTTTCGTCCGGTCCGTTTCTTCTTCCGG 5′ | |
| SV40 NLS (A-G)b | 5′ GGCCCCAAAGAAGAAGAGAAA 3′ |
| 3′ GGTTTCTTCTTCTCTTTCCGG 5′ | |
| HIV-1 MA NLSb | 5′ GGCCGGAAAGAAAAAATATAAATTAAAACATGG 3′ |
| 3′ CCTTTCTTTTTTATATTTAATTTTGTACCCCGG 5′ | |
| ASV IN NLSa | 5′ TGCGGCCGCAAAAACACCGATTCAAAAAC 3′ |
| 5′ TGCGGCCGCTTTTTCCCACTCCCCTGT 3′ | |
| hnRNP M9 NLSa | 5′ TGCGGCCGCAAATCAGTCTTCAAATTTTGG 3′ |
| 5′ TGCGGCCGCATAGCCACCTTGGTTTCG 3′ |
Primers contain NotI extensions and were used for PCR.
Oligonucleotides encode the NLS sequences and contain NotI extensions.
Expression of EGFP alone (∼27 kDa protein) resulted in a whole-cell pattern (Fig. 1B). This is consistent with its small size and passive diffusion within the cell. Fluorescent images of the IN-EGFP fusion protein (predicted molecular mass of ∼73 kDa) showed disperse localization throughout the cell, similar to that observed with the EGFP alone (Fig. 1B). Minimally, this localization suggests that the Mo-MuLV IN does not contain a strong NLS sequence. This result is consistent with previous observations that productive Mo-MuLV infection requires mitosis (61). In contrast, the IN-SV40 NLS-EGFP, a protein with a predicted molecular mass of ∼73 kDa, exhibited nuclear accumulation (Fig. 1B). Since the diffusion limit of nuclear pore complexes (NPCs) is ∼50 kDA for globular proteins (reviewed in reference 49), this result suggests that nuclear accumulation of the IN-SV40 NLS-EGFP resulted from a retention or active transport of the protein into the nucleus. Western blot analysis of intracellular lysates confirmed that the IN-SV40 NLS-EGFP fusion protein and wild-type IN-EGFP protein were expressed at similar levels and with limited breakdown of the fusion protein (Fig. 1C).
Generation of a panel of NLS-tagged IN proteins within replication-competent MuLV.
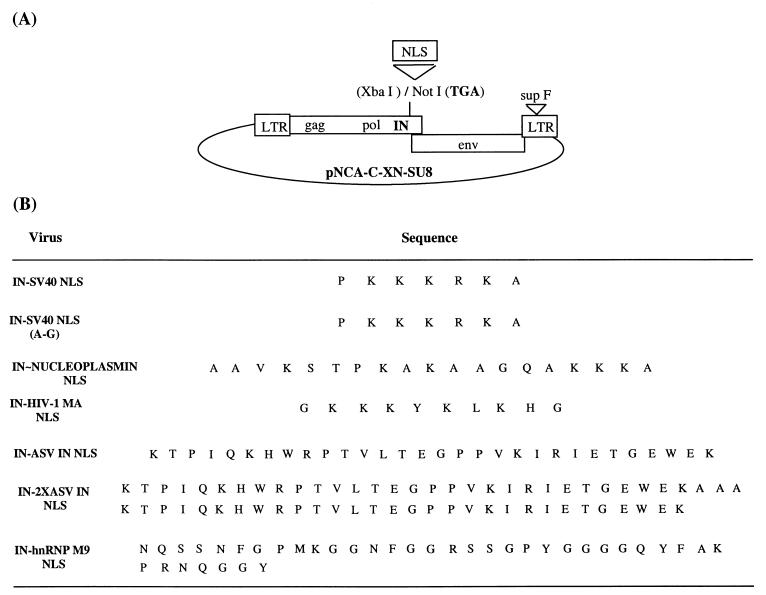
Since the independently expressed Mo-MuLV IN protein tagged with an NLS was actively transported to the nucleus, the ability of an NLS-tagged IN to affect the transport of Mo-MuLV PICs was investigated. The tight association of the Mo-MuLV IN protein with the viral DNA within PICs supports this approach. Ecotropic Mo-MuLV proviral clones were constructed with a panel of different NLS sequences fused to IN (Fig. 2). The NLS sequences were introduced into the Mo-MuLV IN protein within the region known to be nonessential in vivo (62, 65). This allowed for the expression of a truncated IN protein C-terminally fused to a particular NLS sequence without generating large deletions within the 5′ env leader sequence (65). To generate the proviral constructs, pNCA-C-XN-SU8 (65) was digested with NotI. Oligonucleotides that encode NLS sequences and contain NotI extensions were inserted. Alternatively, oligonucleotides with the NotI extensions were used for PCR amplification of the NLS sequences. The oligonucleotides used to generate each of the NLSs are shown in Table 1. The sequences of each of the inserted regions up to and including the stop codon were confirmed by sequence analysis as previously described (65).
FIG. 2.
Schematic diagram illustrating insertion of various NLS sequences into the C terminus of Mo-MuLV IN protein within the context of replication-competent proviruses. (A) Illustration of the Mo-MuLV proviral constructs. Insertions within IN were performed in the vector pNCA-C-XN-SU8, encoding the complete Mo-MuLV proviral DNA genome, including the long terminal repeats (LTR) and the gag, pol, and env genes. The plasmid contains an SuIII tyrosine suppressor tRNA, which was inserted into a nonessential region of the 3′ LTR (in31SuIII) (48). The IN protein is encoded at the 3′ terminus of the pol gene of Mo-MuLV. The env gene products overlap the IN protein and are expressed from an alternate reading frame. The vertical line indicates position 6215 (69), where the XbaI site was destroyed by insertion of a NotI linker (62). Each of the NLSs was inserted individually at this position, immediately upstream of a TGA stop codon. (B) The amino acid sequence of each of the NLSs is listed.
The first set of NLS sequences used in this study include those that encode strong basic or dibasic regions, exemplified by the SV40 NLS and nucleoplasmin NLS sequences, respectively. Nuclear import of molecules containing these signals is conferred through the importin β proteins (1, 27, 36, 58). Previous analysis of the insertion of the SV40 NLS sequence into the C terminus of Mo-MuLV IN indicated that the virus inactivated the NLS within a stretch of six consecutive adenine (A) residues (65). To suppress this pathway of inactivation, the codon usage within the SV40 NLS sequence of Lys was changed from AAA to AAG [IN-SV40 NLS (A-G)]. An additional construct was generated that encoded a basic NLS sequence reported to function within the HIV-1 MA protein (IN-HIV-1 MA NLS) (6).
Alternative NLS sequences that are not basic in nature were investigated. The IN protein of the avian retrovirus ASV encodes a novel proline-rich sequence that functions as an NLS (44, 45). This signal alone on the IN protein appears to be sufficient for moderate levels of transduction into nondividing cells (43). Two constructs containing either one (IN-ASV IN NLS) or two (IN-2X ASV IN NLS) copies of the ASV IN NLS sequence inserted in the C terminus of the MuLV IN protein were generated. These NLS sequences were created by PCR amplification of the RCASBP(A) vector (35) and subcloned into a pTZ vector (U.S. Biochemical Corp.) before excision with NotI.
Finally, the heterogeneous nuclear ribonucleoprotein (hnRNP) A1 M9 sequence was appended to the C terminus of MuLV IN. This sequence was generated by PCR amplification from the GSTM9 vector (a gift from Gideon Dreyfuss) (8). The M9 sequence of hnRNP is different from more common NLS motifs in that it is much larger (38 amino acids) and is rich in glycine and aromatic amino acids, while essentially devoid of clusters of basic residues (70). The M9 sequence was first identified as an NLS and was found to bind to a nuclear receptor termed transportin. This region was later reported as having a nuclear export signal (NES) (52) and has now been defined within an emerging class of transport signals known as nucleocytoplasmic shuttling (NS) signals that direct both nuclear import and export (51, 52).
Effects on the viral life cycle of the viral clones harboring the IN-NLS sequences.
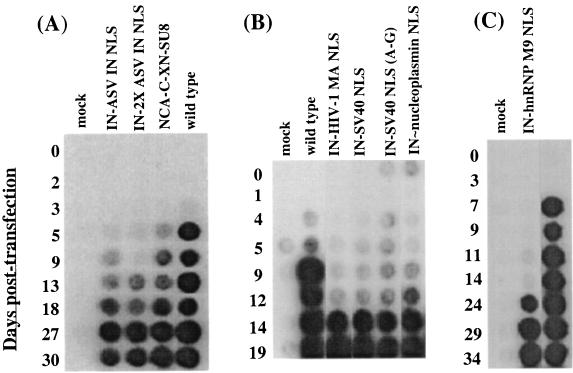
The effects of the various NLS sequences on the Mo-MuLV life cycle were examined. Plasmids containing proviral DNA encoding the IN-NLS proteins were individually introduced into D17/pJET cells (2, 55) in the presence of DEAE-dextran (50). Transient expression of the plasmid DNA allows for assembly and release of viral particles and subsequent reinfection. Viable virus can then spread throughout the culture. Viral propagation was monitored by the release of reverse transcriptase (RT) activity into the media (26). The proviral constructs were transfected and tested in triplicate. In the wild-type Mo-MuLV (pNCA-C)-transfected cultures, RT activity was detected by approximately day 5 (Fig. 3A). The parental viral clone that contains the truncated IN, pNCA-C-XN-SU8, showed no delay in the detection of RT activity compared to that of the wild type (Fig. 3A). The Mo-MuLV viral clones IN-HIV-1 MA NLS, IN-SV40 NLS, IN-SV40 NLS (A-G), IN∼nucleoplasmin NLS, IN-ASV IN NLS, and IN-2X ASV IN NLS showed minor delays in viral spreading compared with the wild type, with detection of RT activity between days 9 and 14 (Fig. 3A and B). Levels equivalent to those of wild-type Mo-MuLV were reached after approximately 2 weeks of passage for each of the IN-NLS viruses tested. This suggested that the fusion of these NLS sequences did not grossly impede function in vivo. In contrast, the IN-hnRNP M9 NLS fusion produced a much longer delay in the appearance of virus in the culture supernatants. RT activity was not detected until day 24 (Fig. 3C).
FIG. 3.
Time course of generation of the viruses harboring IN-NLS fusions following transfection. (A, B, and C) Plasmid DNA containing the complete proviral Mo-MuLV bearing the specific IN-NLS sequences was transiently introduced into D17/pJET cells. At various days posttransfection, supernatant from cultured medium was collected and assayed for RT activity (26) to monitor viral production. The specific tagged viruses are indicated above the lanes. The number of days after the DNA was introduced into the cells is indicated to the left.
Sequence analysis of unintegrated viral DNA.
Minor delays in viral spreading may reflect slow propagation of the viruses containing the NLSs, or it may reflect the outgrowth of a mutant in the culture. To address this issue, the stability of the panel of NLSs tested in our replication-competent system was examined. Unintegrated, extrachromosomal viral DNA from cells freshly infected with the various NLS viruses was isolated, and the region encoding IN was PCR amplified. DNA from the total population was sequenced to examine the genotype of the viruses spreading during viral propagation. Sequence analysis of the unintegrated viral DNA from virus encoding IN tagged with the wild-type SV40 NLS codon usage, in this independent round of transfection, revealed an insertion of an A residue within a stretch of six A residues of the SV40 NLS sequence. The shift in the reading frame introduced negatively charged amino acids within the basic NLS and restored the original reading frame of the C terminus of IN (65). Thus, consistent with previous analysis, the selection for this mutation was confirmed. In contrast, the IN-SV40 NLS (A-G) virus, generated from the construct engineered to suppress this insertion by altered codon usage, was stable during viral propagation (Fig. 4).
FIG. 4.
Sequence analysis of unintegrated viral DNA from cells freshly infected with viruses bearing IN-NLS fusions that encode A6 stretches. Sequence analysis of NLS viruses. A portion of the expanded DNA sequence from the input parental proviral constructs is shown for the INSV40 NLS, IN-SV40 NLS (A-G), IN∼nucleoplasmin NLS, and IN-HIV-1 MA NLS viruses with the amino acid sequence under each codon. The in-frame stop codon is marked in boldface and boxed. The stretch of six adenine (A) residues within each of the NLSs is in boldface. For each virus, unintegrated viral DNA was isolated from freshly infected cells by the Hirt method of extraction (33) and sequenced as previously described (65). Sequence analysis of the IN-SV40 NLS and the IN∼nucleoplasmin NLS revealed that a frameshift mutation had occurred. The insertion site for the mutations is marked in boldface, and the resulting amino acid insertions are placed in italics, with the out-of-frame TGA stop codon boxed. The IN-SV40 NLS (A-G) and the IN-HIV-1 MA NLS viruses remained stable during viral propagation.
The results described above suggest that there is a strong selection for mutations within the A6 region, since the selection of such mutants was highly reproducible. To determine whether a similar selection would occur in other NLS sequences with A6 stretches, the NLS based on the bipartite nucleoplasmin and the putative NLS from the HIV-1 MA protein (amino acid sequence positions 25 to 33) were further examined. Sequence analysis of the unintegrated viral DNA from the IN∼nucleoplasmin NLS virus revealed that a similar frameshift mutation had occurred, resulting in the insertion of an A residue within the A6 stretch encoding the dibasic NLS sequence. The shift in reading frame introduced negatively charged amino acids downstream of the insertion site, disrupting the second of the two basic regions. The insertion also restored the original reading frame of the C terminus of IN (Fig. 4). Moreover, identical insertion events were observed for the IN∼nucleoplasmin NLS virus in three independent experiments. In contrast, sequence analysis of the unintegrated viral DNA from the IN-HIV-1 MA NLS virus showed that the HIV-1 MA NLS was stably maintained during propagation. Thus, in contrast to the well-characterized basic and bipartite sequences of the SV40 large T antigen and the nucleoplasmin NLSs, respectively, Mo-MuLV could tolerate the KKKYK sequence of the putative HIV-1 MA NLS (amino acid sequence positions 25 to 33) within the C terminus of IN. It should be noted that there are conflicting reports regarding the ability of this putative HIV-1 MA NLS to function, in and of itself, in postentry events (6, 23, 24, 59, 73). Although an NES that directs the cytoplasmic localization of the MA protein was identified just upstream of this NLS sequence (6), the NES in its entirety was not present in the sequence appended to the IN.
The observation that a putative NLS of HIV-1 MA was tolerated raised the possibility that NLSs from other retroviral proteins might also be compatible with infectivity. To address this, the stability of the IN-ASV IN NLS and IN-2X ASV IN NLS was examined. Following viral propagation, sequence analysis indicated that both IN-ASV IN NLS and IN-2X ASV IN NLS were stably maintained. It is interesting to note that the sequence coding for the ASV IN NLS contains two regions of A5 stretches.
The virus bearing the nucleocytoplasmic shuttling sequence-IN fusion, IN-hnRNP M9 NLS, was also analyzed. Sequence analysis of the unintegrated viral DNA of this virus showed no alteration in the sequence encoding for the NLS. Interestingly, however, appending hnRNP M9 NLS sequences onto the truncated C terminus of IN produced a much longer delay in the appearance of virus in the culture supernatants compared to the wild type (Fig. 3C). Taken together, our results show that the stability of the viruses encoding the IN-NLSs varied, depending on the nature of the individual NLS sequence inserted. The results suggest that variation in the strength of the NLS, the specific nuclear pathway used, and/or their shuttling properties have direct effects on the compatibility of the IN-NLS in the viral life cycle.
Analysis of the IN-SV40 NLS (A-G) viral proteins.
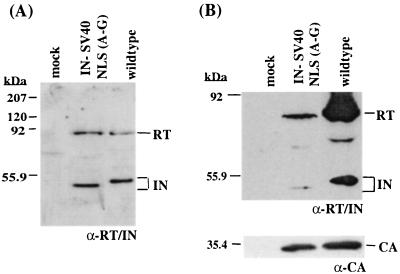
The stable production of the IN-SV40 NLS (A-G) virus presented a unique opportunity to investigate the selection against the basic tract within the parental IN-SV40 NLS virus (65). In addition to mediating integration itself, the integrity of IN is also important for virus assembly and morphogenesis. In the viral assembly pathway, IN is expressed as part of the Gag-Pol precursor protein, which is subsequently processed in the virion by the viral protease releasing the mature products. Mutations within IN that alter the efficiency of the proteolytic processing have previously been identified (63, 68). To determine whether the insertion of the SV40 NLS (A-G) into Mo-MuLV IN affected the proteolytic processing of the IN protein within the virions, virus was collected from producer cells after transfection and passage in D17/pJET cells. Samples of wild-type and IN-SV40 NLS (A-G) virus were normalized by the level of RT activity in the media and were analyzed by Western blotting with polyclonal antisera directed against MuLV RT and IN (Fig. 5A). With both viruses, the 80-kDa RT protein was readily detected, with proportional levels of IN. As predicted, the IN-SV40 NLS (A-G) protein ran at a faster mobility than the wild-type IN due to the net result of the truncation of the 23 amino acids at the C terminus of IN plus the 11 amino acids encoding the SV40 NLS insertion (Fig. 5A). The truncated IN-SV40 NLS (A-G) protein was stable and observed at levels equivalent to those of the wild-type IN. No accumulation of precursor Gag-Pol or Pol intermediates was detected. These results indicate that the insertion of the SV40 NLS (A-G) into IN did not affect the proteolytic processing of the precursor protein or the stability of the IN protein.
FIG. 5.
Impaired viral assembly of the IN-SV40 NLS (A-G) virus. (A) Analysis of viral proteins. One milliliter of concentrated viral particles was collected from an overnight incubation of 5 ml of medium on a 10-cm-diameter plate of producer cells and pelleted through 200 μl of a 25% sucrose cushion at 25,000 × g. Isolated viral particles were normalized with respect to RT activity (26). Viral proteins from mock (lane 1), IN-SV40 NLS (A-G) (lane 2), and wild-type (lane 3) viruses were analyzed by Western blotting with an Mo-MuLV RT and IN polyclonal antibody (α-RT/IN) (rabbit 3, bleed 6) (72). Normalization required eightfold more IN-SV40 NLS (A-G) supernatant than wild-type IN supernatant. (B) Virion incorporation. Virions were prepared from the supernatant of mock, IN-SV40 NLS (A-G), or wild-type viral producer cells and normalized to the amount of CA protein by using anti-CA antibodies (α-CA) (no. 75S-287; Quality Biotech). Following normalization, Western blot analyses were performed with the Mo-MuLV RT and IN polyclonal antibody (rabbit 3, bleed 6 [top panel]) or the CA-specific polyclonal antibody (bottom panel). The positions of RT and IN are indicated to the right. The positions of protein standards and their molecular masses are shown to the left of each panel.
Interestingly, Western blot analysis of virion proteins normalized to viral capsid (CA) protein, rather than RT activity, identified a major difference between the wild-type Mo-MuLV and the virus bearing the IN-SV40 NLS (A-G). Figure 5B shows the Western blot analyses of the concentrated virions detected by an RT/IN antibody. Quantitative analysis of the relative amount of RT in both wild-type and IN-SV40 NLS (A-G) viruses, normalized to amounts of CA, revealed a 10-fold decrease in RT packaged within the IN-SV40 NLS (A-G) virus. This result indicates that the presence of the SV40 NLS sequence at the C terminus of IN alters the level of Gag-Pol precursor that is incorporated into the viral particle. The Gag-Pol protein that is incorporated, however, is processed correctly. Consistent with this finding, immunocytochemistry examination of cells assembling and producing IN-SV40 NLS (A-G) and wild-type viruses revealed an overall retention of Gag-Pol precursor protein within the IN-SV40 NLS (A-G) producer cells. The Pol protein was predominantly localized around the nucleus, with a low level found within the nucleus (data not shown). This suggests that the SV40 NLS sequesters the Pol precursor proteins away from the plasma membrane, where assembly takes place.
Analysis of viral DNA synthesis in vivo of IN-SV40 NLS virus and IN-SV40 NLS (A-G) virus.
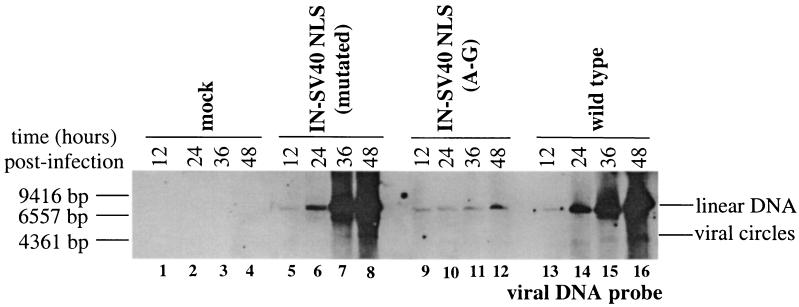
The level of Gag-Pol precursor packaged and the activity of RT within virions are critical for the generation of fully infectious particles. Intracellular Gag/Gag-Pol ratios of around 20:1 are found during the replication of all retroviruses examined (19-21, 37-40). Viral assembly and the subsequent viral infectivity decrease when this ratio is not conserved (18, 66). HIV-1 replication was drastically reduced even with 25% of the RT molecules active (42). Since alteration in the level of Gag-Pol or Pol activity within the virus can have profound effects on viral DNA replication, a time course of viral DNA synthesis was determined with the stable IN-SV40 NLS (A-G), the mutated IN-SV40 NLS, and the wild-type virus. For the initial round of infections, the virions were normalized to RT activity, allowing for a first round of infection at equivalent levels. Unintegrated viral DNA was isolated from cells 12, 24, 36, and 48 h postinfection and analyzed by Southern blots. In the initial round of infection, all three viruses yielded equivalent levels of linear viral DNA. For wild-type Mo-MuLV, the levels of unintegrated viral DNA increased with each 12-h time point as a result of multiple rounds of infection (Fig. 6, lanes 13 to 16). Interestingly, replicated viral DNA from the mutated IN-SV40 NLS virus, carrying a disrupted SV40 NLS sequence, accumulated with kinetics similar to that of the wild type (Fig. 6, lanes 5 to 8). In contrast to infection with wild-type virus, the IN-SV40 NLS (A-G) virus displayed a delay in the accumulation of replicated viral DNA (Fig. 6, lanes 9 to 12). These results indicate that although the IN-SV40 NLS (A-G) virus is capable of complete viral DNA synthesis, multiple rounds of infection are delayed by the presence of the SV40 NLS (A-G) sequence. When the virus preparations used to infect the cultures were normalized for the amount of CA present, the IN-SV40 NLS (A-G) virus showed a delay in the accumulation of newly synthesized viral DNA at the first 12-h time point (data not shown). These results are consistent with the decreased packaging of the IN-SV40 NLS (A-G) Gag-Pol precursor into virions in subsequent rounds of infection. The selective advantage of the IN-SV40 NLS (mutated) virus is thus explained by the return to the wild-type kinetics of infection.
FIG. 6.
Analysis of in vivo synthesis of viral DNA. Preintegrative viral DNAs were isolated from cells 12, 24, 36, and 48 h postinfection by the Hirt method of extraction (33). Equal volumes of isolated DNA were subjected to Southern blot analysis. DNA from the Hirt extraction was transferred to a nylon membrane (NEN Gene Screen) (64) and probed with biotin-labeled pNCA-C (nucleotides 31 to 8295; Mo-MuLV) (69) viral DNA probe. Hybridization was detected with chemiluminescence following the manufacturer's instructions (Renaissance nick translation biotin labeling kit and Renaissance nucleic acid chemiluminescence reagent; NEN). The specific virus stocks are indicated at the top, along with the time postinfection (hours) at which the samples were isolated. The locations of linear and circular viral DNA are indicated to the right. The positions of DNA standards and their molecular masses are shown to the left of each panel.
Thus, our findings establish the importance of maintaining a normal ratio of mature Gag and Gag-Pol products in viral particles. Moreover, these data are in agreement with the model by which a balance between nuclear import of the PICs, through the utilization of NLS sequences, and proper viral assembly needs to be achieved. Thus, if NLS sequences within HIV-1 PICs participate in the active nuclear targeting of these complexes, these signals must be weak, masked, inactivated, or accompanied by NESs to maintain a supply of viral proteins at the plasma membrane during virion assembly. For HIV-1 Gag, an NES that partially overlaps a putative NLS of HIV-1 MA has been reported (13). During virus production, the NES may serve as the dominant signal and thus counteract the NLS to maintain proteins in the cytoplasm during assembly. Similarly, the accessory protein Vpr has been characterized as a nucleocytoplasmic shuttling (NS) protein, directing both nuclear import and nuclear export (67). The observation that the hnRNP M9 sequence, which can direct nucleocytoplasmic shuttling, was tolerated in our system is consistent with this model. Also consistent with this model is the observation that the ASV IN NLS and the 2X ASV IN NLS sequences, which are not basic in nature (44, 45), were both maintained during viral propagation. It is possible that the use of a nonclassical NLS sequence assists the virus in maintaining the crucial supply of viral proteins at the site of virion assembly. Further studies need to be performed to investigate this possibility.
Effects of NLS sequences on cell-cycle-independent viral entry of Mo-MuLV.
Studies were next performed to determine whether the presence of NLS sequences in Mo-MuLV IN could allow for cell cycle-independent viral infection. Five viruses shown to be capable of viral spreading in dividing cells [IN-HIV-1 MA NLS, IN-ASV IN NLS, IN-2X ASV IN NLS, IN-hnRNP M9 NLS, and IN-SV40 NLS (A-G)] were tested for their ability to infect nondividing cells. Despite the delay in DNA synthesis, the IN-SV40 NLS (A-G) virus is infectious and can be normalized for a single round of infection with wild-type virus. Mo-MuLVs that do not contain NLS sequences, namely wild-type MuLV, parental NCA-C-XN-SU8, and the IN-SV40 NLS (mutated) virus were included as controls. To facilitate measurement of infectivity, an Mo-MuLV-based retroviral vector (pGIP) encoding an EGFP reporter gene (10) (Ψ+EGFP-IRES-puro) was stably introduced into viral producer cells by calcium phosphate-mediated transformation (9, 75, 76). Individual clones were selected in puromycin. The relative ability of the viruses to enter the cells was determined by EGFP expression. The fluorescence intensity of cell samples was assayed by fluorescent microscopy or on a fluorescence-activated cell sorter (FACS) (Becton-Coulter, Fullerton, Calif., and Coulter EPICS XL-MCL). Infection of the various NLS viruses on cycling D17/pJET cells showed titers of at least 106 infectious units/ml (Table 2).
TABLE 2.
Effect of NLS sequences on Mo-MuLV infectivitya
| Virus | D17/pJET cells
|
Viral titer of 293T cells | EGFP expressionc
|
||||
|---|---|---|---|---|---|---|---|
| Viral titer | % EGFP expressionb
|
|
|
||||
| Cycling | G2 | M | U937 cells | Macrophages | |||
| Mock | <10 | 0.09 | 0.42 | 0.01 | <10 | − | − |
| Wild type | 2.3 × 107 | 97.7 | 0.28 | 7.96 | 2.1 × 104 | + | − |
| NCA-C-X/N-SU8 | 5.7 × 106 | 99.4 | 0.19 | 24.6 | NDd | + | − |
| IN-SV40 NLS (mutated) | 2.0 × 106 | 87.8 | 0.06 | 8.73 | ND | + | − |
| IN-SV40 NLS (A-G) | 2.8 × 107 | 97.2 | 0.20 | 12.0 | 3.7 × 103 | + | − |
| IN-HIV-1 MA NLS | 3.3 × 107 | 95.2 | 0.64 | 19.0 | 9.7 × 103 | + | − |
| IN-ASV IN NLS | 5.2 × 107 | 99.5 | 0.52 | 25.1 | 3.7 × 104 | + | − |
| IN-2X ASV IN NLS | 8.6 × 106 | 98.2 | 1.02 | 22.1 | 2.9 × 104 | + | − |
| IN-hnRNP M9 NLS | 2.1 × 107 | 97.5 | 0.24 | 16.4 | 6.8 × 103 | + | − |
Viral particles copackaging EGFP were used to infect the listed target cells and assayed by fluorescent microscopy and by FACS. Titers are presented as EGFP infectious units per milliliter of viral supernatant and were determined 24 (293T cells) and 48 (D17/pJET) h postinfection. EGFP expression was determined by FACS.
Percent EGFP expression in cycling cells was determined during the G2-arrest experiment. For G2 arrest, target cells were treated with 7 μg of mitomycin C per ml (Sigma) for 20 min followed by cultivation in fresh medium for 48 h. Target cells were then infected in the presence of Polybrene (8 μg/ml). M-arrested target cells were treated with 20 ng of nocodazole per ml (Sigma) of medium for 24 h. Medium with NZ was replaced every 12 h. Target cells were infected in the presence of nocadozole and 8 μg of Polybrene per ml.
U937 cells were grown in RPMI 1640 (Gibco RBL) supplemented with 10% fetal bovine serum and antibiotics. Human macrophages were generated from peripheral blood collected from healthy adult donors as previously described (34). +, EGFP expression in up to 10% of the cells; −, 10- to 1,000-fold decrease in EGFP expression compared to cycling U937 cells. HIV-1 titers from infection with M-tropic HIV-1Bal, shown as the amount of p24 per milliliter of viral supernatant, as measured 7 days postinfection by ELISA (Zeptometrix, Buffalo, N.Y.) are as follows: U937 cells, 8.4 pg/ml; macrophages, 16.6 pg/ml.
ND, not determined.
Cell cycle-independent Mo-MuLV viral entry was examined by challenging drug-arrested and nondividing cells with the various NLS viruses. D17/pJET cells were blocked at either the G2 or M phase of the cell cycle by treatment with mitomycin C or nocodazole, respectively. The block in cell cycle was confirmed by analyzing the DNA content distribution of treated D17/pJET cells at the time of infection and at the time of analysis (data not shown). Extrachromosomal, unintegrated viral DNA was detected 24 and 48 h postinfection by Southern blot analysis, confirming that each arrest did not block viral DNA synthesis (data not shown). Cycling cells showed EGFP expression in at least 88% of cells analyzed. For all viruses tested, however, there was no stimulation of EGFP expression over the controls in G2 and M growth-arrested cells 48 h postinfection (Table 2). With nocodazole treatment, target cells could reenter the cell cycle when the drug was removed from the media. Upon release of the block, which results in exit from metaphase and decondensation of chromosomes, high levels of EGFP expression were observed in all target cells (data not shown). Thus, insertion of an NLS onto the C terminus of Mo-MuLV IN does not facilitate transduction in both growth-arrested cells.
The engineered viruses were then assayed for infectivity on primary human macrophages. Since human cells lack the ecotropic receptor, Mo-MuLV was pseudotyped with vesicular stomatitis virus glycoprotein (VSV-G) to allow binding and entry. Two 150-mm-diameter plates of D17/pJET cells producing the various viruses harboring NLS fusions were transfected by the Lipofectamine (Gibco BRL/Invitrogen) method with 8 μg of pHIT-G (gift of M. Malim) and proportional levels of reagents. Transiently expressed viral particles were harvested approximately 44 h postaddition of DNA and filtered through a 0.45-μm-pore-diameter filter. Thirty milliliters of isolated virus was pelleted by spinning them at 18,000 rpm in an SW17 rotor for 2 h at 4°C in an SW17 swinging bucket rotor. The virus, within the bottom 3-ml portion, was stored in 1% bovine serum albumin in liquid nitrogen. One milliliter of virus was used to infect cells for 2 h and subsequently replaced with fresh medium. Infection of the VSV-G-pseudotyped viruses harboring NLS sequences yielded titers on the order of 104 infectious units/ml on cycling 293T cells (Table 2). The pseudotyped viruses were then used to infect primary human macrophages. The infection and subsequent expression were monitored by FACS. In addition, U937 cells, a proliferating human promonocytic cell line previously shown to be infected by MuLV (11, 71), were monitored as a control for the ability of the viral core to infect a macrophage cell line. On the day of infection, a portion of the differentiated macrophages was stained with propidium iodide for cell cycle analysis. More than 95% of the macrophages were in the G1/G0 phase of the cell cycle, compared to 39% in the proliferating U937 cell line. The various viruses listed in Table 2 showed EGFP expression upon infection of U937 cells. In contrast, EGFP expression in macrophages was observed to have a 10- to 1,000-fold decrease in transduction efficiency compared to cycling U937 cells, with controls showing similar efficiencies. Infection with HIV-1 on the primary human macrophages yielded a titer of 16.6 pg/ml, as measured by enzyme-linked immunosorbent assay (ELISA) for the presence of HIV-1 p24 CA, indicating the ability of these cells to be infected by a lentivirus. Furthermore, in this experiment, macrophages were infected more efficiently with HIV-1, as evidenced by lower titers on U937 cells. This is in contrast to what was observed for infection with the Mo-MuLV viruses harboring IN-NLS fusions.
The results from multiple experiments are summarized in Table 2. Both growth-arrested cells and terminally differentiated cells, which do not progress through the cell cycle, were not transduced at significant levels by wild-type Mo-MuLV, as judged by expression of EGFP. The addition of various NLSs to IN did not result in an increase in EGFP expression over the level in the controls. These results are in sharp contrast with those of the initial studies, which used the isolated IN protein tagged with the SV40 NLS protein (Fig. 1), in which the presence of the NLS sequence was sufficient to transport the IN fusion protein to the nucleus. These results highlight the complexity of the viral system, in which viral assembly and association in higher-order protein-nucleic acid complexes are contributing factors.
Many possible explanations could account for lack of infection by the NLS viruses; all of which have important implications in developing murine-based retroviral vectors that can target PICs to the nucleus of nondividing cells. One possibility to account for the lack of infection in nondividing cells is that more than one NLS is required for efficient transport of the Mo-MuLV PIC into the nucleus. Of the proteins found within the HIV-1 PIC, NLSs have been identified within MA, Vpr, and IN, yielding karyophilic properties for each of these proteins (3, 6, 25, 30, 32, 54, 57). Potentially, the HIV-1 NLSs act cooperatively or in coordination with one another to allow for more efficient nuclear entry. This rationale may be needed to generate a cell cycle-independent Mo-MuLV.
Another possibility is that differences in the composition of the PICs influence nuclear import. The intracellular complexes shown to mediate Mo-MuLV reverse transcription (RTCs) are composed of CA, IN, and RT proteins (4, 16). In contrast, HIV-1 PICs minimally contain the viral RT, IN, MA, and Vpr proteins (7, 14, 15, 53). These differences in the composition of the PICs may influence the accessibility of NLS sequences within the complex. The possibility exists that, in our system, the NLS is buried within the PIC or that the presence of CA in MuLV PICs masks the NLS in IN. The size differences between HIV and MuLV PICs may also influence the ability of MuLV to transduce nondividing cells. Isolated at the same time after acute infection, HIV-1 RTCs are slower in sedimenting and presumably smaller than Mo-MuLV RTCs (15, 16). Furthermore, studies with electron microscopy showed that HIV cores are disrupted shortly after virus-cell fusion, in contrast to MuLV cores, which persist longer (29, 60). Thus, the uncoating or maturation of the PIC may be required before translocation through a nuclear pore complex (NPC) can take place. A similar scheme for import is utilized by adenovirus. Adenovirus targets its genome to the cell nucleus by a multistep process whereby ultimately the complex disassembles at the nuclear membrane before its viral DNA is transported into the nucleus (28). Alternatively, transient ruptures of the nuclear envelope may provide an unconventional route for nuclear entry that bypasses the size-restricted NPCs. Such a task of disrupting the nuclear envelope structure was recently described for the HIV-1 viral protein Vpr (12).
Host factors and chromatin structure may also play an important role in cell cycle-independent viral entry, making the cell cycle regulation of murine retroviruses complex and intriguing. It has been reported that nuclear import of Mo-MuLV DNA mediated by an adenoviral protein was not sufficient for efficient retroviral transduction in cells arrested in the G1/S phase of the cell cycle (47). The requirement for passage through M phase has a direct impact on the type of cells that can be infected by murine-based retroviral vectors. Through these studies, we have identified NLS sequences that can be incorporated into the MuLV IN protein and yield infectious virus. By investigating the different strategies and signals used by viruses to achieve subcellular localization, additional factors can now be incorporated into these IN-NLS viruses to develop murine-based vectors that render the safe delivery of genes encoding therapeutic proteins.
Acknowledgments
This work is supported by National Institutes of Health (NIH) grant R21-DK54374 and R01CA76545 to M.J.R. J.A.S. was supported in part by NIH Predoctoral Training grants T32-GM8360 and T32-AI7403.
We thank Christina DeCoste, Lucille O'Reilly, Jennifer Puglia, Michaela Stanton, and Cari Sadowski for technical assistance and Frank Ruscetti for helpful discussions. We also thank Keith Bupp and Chi-Wei Lu for their critical reading of the manuscript.
REFERENCES
- 1.Adam, S. A., and L. Gerace. 1991. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66:837-847. [DOI] [PubMed] [Google Scholar]
- 2.Albritton, L. M., L. Tweng, D. Scadden, and J. M. Cunningham. 1989. A putative murine retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 3.Bouyac-Bertoia, M., J. D. Dvorin, R. A. M. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 5.Bukrinsky, M. I., and O. K. Haffar. 1999. HIV-1 nuclear import: in search of a leader. Front. Biosci. 4:772-781. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buvoli, M., G. Biamonti, P. Tsoulfas, M. T. Bassi, A. Ghetti, S. Riva, and C. Morandi. 1988. cDNA cloning of human hnRNP protein A1 reveals the existence of multiple mRNA isoforms. Nucleic Acids Res. 16:3751-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C.-C., A. Rivera, N. Ron, J. P. Dougherty, and Y. Ron. 2001. A gene therapy approach for treating T-cell-mediated autoimmune diseases. Blood 97:886-894. [DOI] [PubMed] [Google Scholar]
- 11.Ch'ng, J. L., R. C. Mulligan, P. Schimmel, and E. W. Holmes. 1989. Antisense RNA complementary to 3′ coding and noncoding sequences of creatine kinase is a potent inhibitor of translation in vivo. Proc. Natl. Acad. Sci. USA 86:10006-10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.deNoronha, C. M. C., M. P. Sherman, H. W. Lin, M. V. Cavrois, R. D. Moir, R. D. Goldman, and W. C. Greene. 2001. Dynamic disruptions in nuclear envelope architecture and integrity by HIV-1 vpr. Science 294:1105-1108. [DOI] [PubMed] [Google Scholar]
- 13.Dupont, S., N. Sharova, C. DeHoratius, C.-M. A. Virbasius, X. Zhu, A. G. Bukrinskaya, M. Stevenson, and M. R. Green. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402:681-685. [DOI] [PubMed] [Google Scholar]
- 14.Farnet, C. M., and W. A. Hazeltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 65:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 75:3626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felkner, R. H., and M. J. Roth. 1992. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J. Virol. 66:4258-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, K. M., and S. P. Goff. 1988. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J. Virol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein, K. M., and S. P. Goff. 1992. Mutational analysis of the gag-pol junction of Moloney murine leukemia virus: requirements for expression of the gag-pol fusion protein. J. Virol. 66:6601-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, Y.-X., D. L. Hatfield, A. Rein, and J. G. Levin. 1989. Translational readthrough of the murine leukemia virus gag gene amber codon does not require virus-induced alteration of tRNA. J. Virol. 63:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng, Y.-X., J. G. Levin, D. L. Hatfield, T. S. Schaefer, R. J. Gorelick, and A. Rein. 1989. Suppression of UAA and UGA termination codons in mutant murine leukemia viruses. J. Virol. 63:2870-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouchier, R. A. M., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 23.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goff, S., P. Traktman, and D. Baltimore. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 38:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246-248. [DOI] [PubMed] [Google Scholar]
- 28.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grewe, C., A. Beck, and H. R. Gelderblom. 1990. HIV: early virus-cell interactions. J. Acquir. Immune Defic. Syndr. 3:965-974. [PubMed] [Google Scholar]
- 30.Haffar, O. K., S. Popov, L. Dubrovsky, I. Agostini, H. Tang, T. Pushkarsky, S. G. Nadler, and M. Bukrinsky. 2000. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J. Mol. Biol. 299:359-368. [DOI] [PubMed] [Google Scholar]
- 31.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 75:9526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-371. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman, P. M., S. Dhib-Jalbut, J. A. Mikovits, D. S. Robbins, A. L. Wolf, G. K. Bergey, N. C. Logrey, O. S. Weislow, and F. W. Ruscetti. 1992. Human T-cell leukemia virus type I infection of monocytes and microglial cells in primary human cultures. Proc. Natl. Acad. Sci. USA 89:11784-11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-30012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imamoto, N., T. Shimamoto, T. Takao, T. Tachibana, S. Kose, M. Matsubae, T. Sekimoto, Y. Shimonishi, and Y. Yoneda. 1995. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 14:3617-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacks, T., H. D. Madhani, F. R. Masiarz, and H. E. Varmus. 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacks, T., M. C. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 39.Jacks, T., K. Townsley, H. E. Varmus, and J. Majors. 1987. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl. Acad. Sci. USA 84:4298-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacks, T., and H. E. Varmus. 1985. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 232:1237.. [DOI] [PubMed] [Google Scholar]
- 41.Jonsson, C. B., G. A. Donzella, E. Gaucan, C. M. Smith, and M. J. Roth. 1996. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 70:4585-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz, R. A., J. G. Greger, K. Darby, P. Boimel, G. F. Rall, and A. M. Skalka. 2002. Transduction of interphase cells by avian sarcoma virus. J. Virol. 76:5422-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kukolj, G., K. S. Jones, and A. M. Skalka. 1997. Subcellular localization of avian sarcoma virus and human immunodeficiency virus type 1 integrases. J. Virol. 71:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kukolj, G., R. A. Katz, and A. M. Skalka. 1998. Characterization of the nuclear localization signal in the avian sarcoma virus integrase. Gene 223:157-163. [DOI] [PubMed] [Google Scholar]
- 46.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 11:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lieber, A., M. A. Kay, and Z.-Y. Li. 2000. Nuclear import of Moloney murine leukemia virus DNA mediated by adenovirus preterminal protein is not sufficient for efficient retroviral transduction in nondividing cells. J. Virol. 74:721-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobel, L. I., M. Patel, W. King, M. C. Nguyen-Huu, and S. P. Goff. 1985. Construction and recovery of viable retroviral genomes carrying a bacterial suppressor transfer RNA gene. Science 228:329-332. [DOI] [PubMed] [Google Scholar]
- 49.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 50.McCutchan, J. H., and J. S. Pagano. 1968. Enhancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J. Natl. Cancer Inst. 41:351-357. [PubMed] [Google Scholar]
- 51.Michael, W. M. 2000. Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 10:46-50. [DOI] [PubMed] [Google Scholar]
- 52.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 53.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nadler, S. G., D. Tritschler, O. K. Haffar, J. Blake, A. G. Bruce, and J. S. Cleaveland. 1997. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J. Biol. Chem. 272:4310-4315. [DOI] [PubMed] [Google Scholar]
- 55.O'Reilly, L., and M. J. Roth. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 74:899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parveen, Z., A. Krupetsky, M. Engelstadter, K. Cichutek, R. J. Pomerantz, and R. Dornburg. 2000. Spleen necrosis virus-derived C-type retroviral vectors for gene transfer to quiescent cells. Nat. Biotechnol. 18:623-629. [DOI] [PubMed] [Google Scholar]
- 57.Pluymers, W., P. Cherepanov, D. Schols, E. D. Clercq, and Z. Debyser. 1999. Nuclear loczalization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 58.Radu, A., G. Blobel, and M. S. Moore. 1995. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA 92:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Risco, C., L. Menendez-Arias, T. D. Copeland, P. P. da Silva, and S. Oroszlan. 1995. Intracellular transport of the murine leukemia virus during acute infection of NIH 3T3 cells: nuclear import of nucleocapsid protein and integrase. J. Cell Sci. 108:3039-3050. [DOI] [PubMed] [Google Scholar]
- 61.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roth, M. J. 1991. Mutational analysis of the carboxyl terminus of the Moloney murine leukemia virus integration protein. J. Virol. 65:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roth, M. J., P. Schwartzberg, N. Tanese, and S. P. Goff. 1990. Analysis of mutations in the integration function of Moloney murine leukemia virus: effects on DNA binding and cutting. J. Virol. 64:4709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 65.Seamon, J. A., M. Adams, S. Sengupta, and M. J. Roth. 2000. Differential effects of C-terminal molecular tagged integrase on replication competent Moloney murine leukemia virus. Virology 274:412-419. [DOI] [PubMed] [Google Scholar]
- 66.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherman, M. P., C. M. C. de Noronha, M. I. Heusch, S. Greene, and W. C. Greene. 2001. Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol. 75:1522-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin, C.-G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shinnick, T. M., R. A. Lerner, and J. G. Sutcliffe. 1981. Nucleotide sequence of Moloney murine leukaemia virus. Nature 293:543-548. [DOI] [PubMed] [Google Scholar]
- 70.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sundstrom, C., and K. Nilsson. 1976. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 17:565-577. [DOI] [PubMed] [Google Scholar]
- 72.Tanese, N., M. J. Roth, and S. P. Goff. 1986. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J. Virol. 59:328-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.vonSchwedler, U., R. S. Kornbluth, and D. Trono. 1994. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc. Natl. Acad. Sci. USA 91:6992-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weinberg, J. B., T. J. Matthews, B. R. Cullen, and M. H. Malim. 1991. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J. Exp. Med. 174:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wigler, M., A. Pellicer, S. Silverstein, and R. Axel. 1978. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell 14:725-731. [DOI] [PubMed] [Google Scholar]
- 76.Wigler, M., R. Sweet, G. K. Sim, B. Wold, A. Pellicer, E. Lacy, T. Maniatis, S. Silverstein, and R. Axel. 1979. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell 16:777-785. [DOI] [PubMed] [Google Scholar]
- 77.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]