Abstract
RelA-associated inhibitor (RAI) is an inhibitor of nuclear factor κB (NF-κB) newly identified by yeast two-hybrid screen as an interacting protein of the p65 (RelA) subunit. In this study, we attempted to examine the effect of RAI on transcription and replication of human immunodeficiency virus type 1 (HIV-1). We found that RAI inhibited gene expression from the HIV-1 long terminal repeat (LTR) even at the basal level. Upon in vitro DNA-binding reactions, RAI could directly block the DNA-binding of p65 subunit of NF-κB but not that of the p50 subunit or AP1. We found that RAI could also inhibit the DNA-binding of Sp1 and thus inhibit the basal HIV-1 promoter activity. We further examined the effects of RAI on Sp1 and found that RAI colocalizes with Sp1 in the nucleus and interacts with Sp1 in vitro and in vivo. Moreover, we found that RAI efficiently blocked the HIV-1 replication when cotransfected with a full-length HIV-1 clone. These findings indicate that RAI acts as an efficient inhibitor of HIV-1 gene expression in which both NF-κB and Sp1 play major roles.
Nuclear factor κB (NF-κB) and Sp1 are potent cellular activators of human immunodeficiency virus type 1 (HIV-1) gene expression (1, 9, 22, 33). In cells chronically infected with HIV-1, activation of NF-κB together with constitutive active Sp1 could trigger the transcription of viral genes including the trans-activator Tat, which would result in an explosive increase in HIV-1 replication (11, 19, 28; reviewed in reference 12). Thus, downregulation of NF-κB activity has long been sought to inhibit the HIV replication and prevent clinical development of AIDS in the HIV infected individuals (reviewed in references 1, 2, and 20).
The members of the NF-κB family in mammalian cells include p50/p105 (NF-κB1), p52/p100 (NFκB2), p65 (RelA), c-Rel, and RelB. These proteins share a highly conserved region in the N terminus, known as the Rel homology domain, which is responsible for DNA binding, dimerization, nuclear translocation, and inhibition by IκB proteins (1, 8). NF-κB is normally present in the cytoplasm in association with its inhibitor, IκB (1, 31). Upon stimulation with various stimuli including interleukin 1, tumor necrosis factor alpha (TNF-α), phorbol esters, radical oxygens, and UV irradiation, it is dissociated from IκB and translocated to the nucleus, where it activates target genes (1, 20, 22, 29). Since IκB proteins are susceptible to these extracellular signals, various approaches have been attempted in order to block HIV replication, such as using chemical inhibitors and dominant negative IκB mutants (13, 26, 28, 33). However, actions of these inhibitors are broad, and more specific inhibition of HIV gene expression is being sought after.
We have recently identified a novel inhibitor for NF-κB, RelA-associated inhibitor (RAI), by yeast two-hybrid screen using the central region of p65 as bait (39). RAI contains four tandem ankyrin repeats and an SH3 motif, structural features similar with other proteins interacting with NF-κB, such as IκB family proteins and 53BP2 (38). We showed that RAI is located in the nucleus and that overexpression of RAI inhibited the NF-κB DNA binding in the transfected cells, although it is not known whether RAI can directly block the DNA binding. In this report we demonstrate that RAI can strongly block HIV-1 replication and that it inhibits not only the DNA-binding activity of p65 but also that of Sp1, both of which play major roles in HIV-1 gene expression.
MATERIALS AND METHODS
Cell culture and transfection.
Human 293 cells were grown at 37°C in Dulbecco's modified Eagle's medium with 10% heat-inactivated fetal bovine serum, 1 mM glutamate, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were transfected using SuperFect transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's recommendations.
Construction of plasmids.
Plasmids were constructed by standard methods (27). Construction of pCMV-p65, pEBV-His-IκBα, pFLAG-RAI, and pEGFP-RAI was previously described (39). In order to create pGEX-RAI expressing a full-length RAI protein fused in-frame to glutathione-S-transferase (GST) in bacteria, the full-length RAI cDNA was amplified by PCR using pFLAG-RAI as a template with oligonucleotide primer pairs (forward, 5′-ACGCGAATTCGAATGTGGATGAAGGACCCT-3′, which contains an EcoRI site; reverse, 5′-GCCGCTCGAGTCTAGACTTTACTCCTTTG-3′, which contains an XhoI site) and was cloned into EcoRI-XhoI-digested pGEX-5X-2 vector (Amersham Pharmacia Biotech, Uppsala, Sweden). The construction of pMAL-p65 and pGEX-IκBα, expressing a full-length p65 protein fused to maltose-binding protein and a full-length human IκBα protein fused to GST, respectively, was described by Tetsuka et al. (32). HIV-1 LTR-based luciferase expression plasmids including CD12-Luc (containing the full-size LTR U3 and R), CD23-Luc (containing positions −117 to +80 of HIV-1 LTR), CD52-Luc (containing positions −65 to +80 of HIV-1 LTR), and CD54-Luc (containing positions −48 to +80 of HIV-1 LTR) were constructed from the original CD12, CD23, CD52, and CD54 chloramphenicol acetyltransferase reporter plasmids reported previously (20, 21). These chloramphenicol acetyltransferase reporter plasmids were digested by XhoI and HindIII, and the isolated DNA fragments containing the LTR regions were cloned into NheI-HindIII-digested pGL3-Basic vector (Promega, Madison, Wis.). All the constructs were confirmed by dideoxynucleotide sequencing using ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin Elmer, Foster City, Calif.) on Applied Biosystems 313 automated DNA sequencer. Mammalian expression vector pCMV-Sp1 was a generous gift from Stephen Smale (University of California—Los Angeles).
Antiviral assays.
Antiviral activity of RAI was evaluated based on the extent of inhibition of viral antigen expression in the culture supernatants of 293 cells transfected with a full-length HIV-1 molecular clone (pNL4-3). 293 cells were transfected with 0.2 μg of pNL4-3, together with various amounts of a plasmid coding for wild-type RAI or RAI mutants (RAIΔN or RAIΔC) (39) with Lipofectamine (19). The effect of RAI was examined in the presence or absence of transfection with pCMV-p65. Twenty-four hours after transfection, cell culture media were changed to new media, and the cells were incubated for an additional 48 h. The cells transfected without pCMV-p65 were suspended in a fixed amount (500 μl) of phosphate-buffered saline (PBS), sonicated using MICROSON Ultrasonic Homogenizer Model XL 2007 (Misonix Incorporated, Farmingdale, N.Y.), and centrifuged at 4,800 × g for 5 min, and the p24 antigen concentration in the culture supernatant was determined. The p24 antigen level was measured by p24 antigen capture ELISA assay using a commercial kit (RETRO-TEK HIV-1 p24 Antigen ELISA kit; Zepto Metrix Corp., Buffalo, N.Y.) as described previously (28).
Transient luciferase assay.
293 cells were cultured in 12-well plates, and transfections were performed with SuperFect transfection reagent according to the manufacturer's recommendation. For each transfection, 100 ng of reporter plasmids (either CD12-Luc, CD23-Luc, CD52-Luc, or CD54-Luc) and 10 ng of the internal control plasmid, pRL-TK, expressing Renilla luciferase, were used. The relevant empty plasmid vector was used to adjust the total amount of plasmid DNA. Triplicate tissue culture dishes for each plasmid combination were transfected in each experiment. Forty-eight hours posttransfection, the cells were harvested for measurement of the luciferase activity as previously described (28, 32, 39). For experiments with TNF-α stimulation, the transfected cells were stimulated with TNF-α (5 ng/ml) after 24 h of transfection and cultured for further 24 h and harvested. The luciferase activity was normalized by Renilla luciferase activity used as an internal control for transfection efficiency.
Microscopic examination.
293 cells were cultured in chamber slides and transfected with a plasmid expressing GFP-RAI using Lipofectamine. After 24 h, cells were fixed with 4% (wt/vol) paraformaldehyde-PBS for 20 min at room temperature and then permeabilized by 0.5% Triton X-PBS for 10 min at room temperature. The cells were then incubated with goat anti-Sp1 (PEP 2) polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) for 1 h at 37°C and then at 4°C for 16 h. After washing with 0.05% Triton X-PBS, the cells were incubated with rhodamine conjugated anti-goat immunoglobulin G (IgG) (Chemicon International, Temecula, Calif.) and DAPI (4′,6-diamidino-2-phenylindole) (Sigma-Aldrich, Saint Louis, Mo.) for 1 h at 37°C.
Recombinant proteins and purification.
pGEX-RAI-full, pGEX-IκB-α, pGEX-5X-2 (expressing only GST), and pMAL-p65 were transformed in E. coli strain BL21(DE3)/pLysS following induction with 0.1 mM IPTG (isopropyl-1-thio-β-d-galactopyranoside) at 28°C overnight. Recombinant GST fusion proteins (GST, GST-RAI-full, and GST-IκBα) were purified by incubating the bacterial extracts in PBS with 10% Triton-X and protease inhibitors (200 μM phenylmethylsulfonyl fluoride, leupeptin [1 μg/ml], aprotinin [10 μg/ml], pepstatin [1 μg/ml]) and affinity purified with glutathione-Sepharose beads (Amersham Pharmacia Biotech) according to the manufacturer's recommendation. Recombinant maltose binding protein (MBP) fusion protein (MBP-p65) was affinity-purified with amylose resin (New England Biolabs, Beverly, Mass.) according to the manufacturer's recommendation. These affinity-purified MBP-p65 and GST-RAI proteins were further purified by column chromatography using Mono Q HR 5/5 columns and an ÄKTA purifier (Amersham Pharmacia Biotech). Briefly, the affinity-purified proteins were dialyzed against the starting buffer containing 70 mM Bis-Tris, 50 mM Tris-HCl [pH 7.8], 1 mM dithiothreitol, and 5% glycerol and loaded onto a Mono Q HR 5/5 column, and eluted by a continuous 0 to 500 mM NaCl gradient. Intact MBP-p65 and GST-RAI proteins were recovered in the 70 to 130 mM and the 15 to 25 mM NaCl eluted fractions, respectively. Intact GST-IκBα and the control GST proteins were obtained solely by affinity purification. These proteins were dialyzed against the electrophoretic mobility shift assay (EMSA) buffer containing 22 mM HEPES-KOH [pH 7.9], 80 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, leupeptin [1 μg/ml], aprotinin [10 μg/ml], pepstatin [1 μg/ml], 0.1% NP-40, and 5% glycerol and stored in aliquots at −80°C. Purified recombinant human Sp1, AP1 (c-Jun), and p50 subunit of NF-κB were purchased from Promega. The protein concentrations were measured by the DC Protein Assay (Bio-Rad, Hercules, Calif.).
EMSA.
EMSA was performed as described previously (37). The κB sequence was taken from HIV-1 LTR. The sequences of the κB wild-type and mutant oligonucleotides were described previously (39). The oligonucleotide probes for Sp1 and AP1 are Sp1 (5′-TTTCCCTTGGTGGGGGCGGGGCCTAAGCTG −3′ and 5′-TTTCAGCTTAGGCCCCGCCCCCACCAAGGG-3′) and AP1 (5′-TTTCGCTTG ATGAGTCAGCCGGAA-3′ and 5′-TTTTTCCGGCTGACTCATCAAGCG-3′). These oligonucleotide pairs were annealed and labeled using DNA polymerase Klenow fragment (Takara Biomedicals, Shiga, Japan) in the presence of [α-32P]dATP (3,000 Ci/mmol; ICN Pharmaceuticals Inc., Costa Mesa, Calif.). DNA binding reactions were performed at 30°C for 5 min in 10-μl reaction volume. Analysis of binding complexes was performed by electrophoresis in 7% polyacrylamide gels with 0.5× Tris-borate-EDTA buffer, followed by autoradiography. For DNA competition experiments, unlabeled double-stranded competitor oligonucleotides (cold-κBw and cold-κBm, for wild-type and mutated κB sites, respectively [9, 37]) were added into the reaction mixture at 30-fold molar excess over the probe.
In vitro protein interaction.
The protein-protein interaction in vitro was performed as described previously (32, 39). Briefly, an equal amount of the recombinant Sp1 or MBP-p65 was incubated with 5 μg of the GST-RAI or GST (as a negative control) bound to glutathione-Sepharose beads and in 250 μl of buffer A (39) at 4°C for 12 h. The beads were washed five times, and the bound proteins were eluted with an equal volume of 2× sodium dodecyl sulfate (SDS) loading buffer, boiled for 3 min, and resolved by SDS-8% polyacrylamide gel electrophoresis (SDS-8% PAGE) followed by Western blotting with goat anti-Sp1 (PEP 2) antibody or rabbit anti-p65 antibody (Santa Cruz) at a dilution of 1/1,000. Secondary antibody, horseradish peroxidase-conjugated anti-rabbit or anti-goat IgG antibody, was used at a dilution of 1/1,000, and protein bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech).
Coimmunprecipitation.
In order to examine the protein-protein interaction in cultured cells in vivo, 293 cells were transfected with either pFLAG-RAI, pFLAG-RAIΔC, or pFLAG-RAIΔN and cultured for 24 h in 10-cm-diameter dishes; the total cell lysate was prepared by lysing the cells in 2 ml of ice-cold lysis buffer (50 mM Tris-HCl [pH 7.8], 300 mM KCl, 1 mM EDTA, 10% glycerol, 0.3% Nonidet P-40) for 15 min. The lysate was cleared by centrifugation. The supernatants were diluted by adding two times the volume of dilution buffer (50 mM Tris-HCl [pH 7.8], 1 mM EDTA, 10% glycerol, 0.3% Nonidet P-40). Then, 900 μl of the diluted lysate was incubated with 10 μl of anti-FLAG M2 affinity gel (Sigma-Aldrich) in PBS containing 0.1% bovine serum albumin at 4°C for 1 h. The beads were washed gently three times with diluted lysis buffer. The bound proteins were eluted with 25 μl of SDS loading buffer, boiled for 3 min, and resolved by SDS-8% PAGE. Western blotting was performed as described above using anti-Sp1 (PEP2) antibody.
RESULTS
Inhibition of gene expression from HIV-1 LTR by RAI.
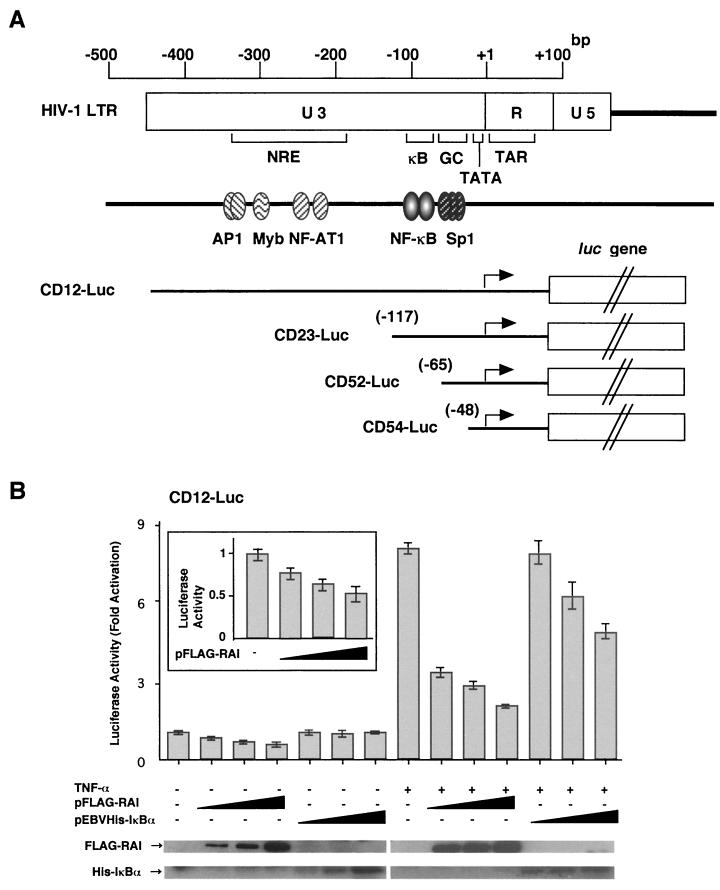
We first addressed whether RAI could inhibit gene expression from HIV-1 LTR (Fig. 1). As demonstrated in Fig. 1B, the basal transcription level from CD12-Luc containing the full-length HIV-1 LTR was inhibited by RAI in a dose-dependent manner. No such inhibition was observed with IkBa. Gene expression from other promoters including herpesvirus thymidine kinase (tk) promoter and p21 gene promoter was not inhibited by RAI as we previously reported (39) (data not shown). Upon stimulation of HIV-1 promoter by TNF-α, a physiological inducer of NF-κB (20), RAI could inhibit its gene expression greater than by IκBα (Fig. 1B).
FIG. 1.
Inhibition of HIV-1 LTR gene expression by RAI. 293 cells were transfected with reporter plasmid together with the various amounts of pFLAG-RAI or pEBV-His-IκBα plasmids. (A) Schematic diagram of HIV-1 LTR, position of various cis-elements for transcription factors, and reporter constructs. Binding sites for AP1 (position −343 to −350 and position −330 to −336), Myb (position −293 to −314), NFAT-1 (position −218 to −256), two tandem repeats of NF-κB-binding site (κB) (position −96 to −105 and position −82 to −91), and three tandem repeats of Sp1-binding sites (GC) (position −69 to −78, position −58 to −67, and position −47 to −56) are indicated. CD12-Luc contains the full sequence of both U3 and R regions. CD23-Luc, CD52-Luc, and CD54-Luc encompass the downstream sequence from position −117 (from the cap site of HIV-1 LTR) (containing both NF-κB and Sp1 sites), that from position −65 (containing only the two Sp1-binding sites), and that from position −48 (retaining no Sp1 site), respectively. (B to F) Results of transient luciferase assays. The luciferase activity was normalized by the Renilla luciferase activity produced from pRL-TK cotransfected as an internal control. In some experiments, cells were stimulated with TNF-α (5 ng/ml) after 24 h of transfection or cotransfected with pCMV-Sp1. Luciferase activities are indicated by the extent of increase in the luciferase activity obtained from the transfection with each reporter plasmid alone. The data are represented by the means and standard deviations (error bars) of three independent experiments. In order to indicate the amount of overexpressed proteins, the results of Western blot analysis of FLAG-RAI and His-IκBα are shown in the bottom of Fig. 1B. The inset in Fig. 1B indicates the effect of RAI on the basal level of gene expression from HIV-1 LTR (CD12-Luc).
In order to determine the target cis-regulatory element(s) within the HIV-1 LTR for the RAI-mediated transcriptional repression, we examined the effects of RAI on various 5′-deletion mutants: CD23-Luc (deleting the upstream sequences from position 2117 of HIV-1 LTR, including AP1-binding sites but still retaining the NF-κB and the Sp1 sites), CD52-Luc (deleting the upstream from position −65 but retaining the promoter-proximal two tandem repeats of Sp1 sites), and CD54-Luc (deleting the upstream from position −48 and retaining no Sp1 site) (30) (Fig. 1A). Similarly to the results with CD12-Luc, the TNF-α-stimulated gene expression from CD23 was suppressed either by overexpression of RAI or by IκBα, indicating that the upstream sequence from position −117 containing the cis-regulatory elements for AP1, Myb, and NFAT-1 is not involved in RAI-mediated repression (Fig. 1C). When CD52-Luc was used, neither TNF-α-mediated activation nor inhibition by IκBα was observed. However, RAI could still block the transcription from CD52-Luc (Fig. 1D). As demonstrated in Fig. 1E, overexpression of Sp1 augmented transcription from CD52-Luc by 3.5-fold. This Sp1-mediated transcription was completely abolished by expression of RAI. We also examined the effects of RAI with CD54-Luc. However, RAI could no longer inhibit the gene expression (Fig. 1F). These results indicate that RAI can block both the inducible transcription mediated by NF-κB and the constitutive transcription mediated by Sp1.
RAI directly inhibits the p65 and Sp1 DNA-binding in vitro.
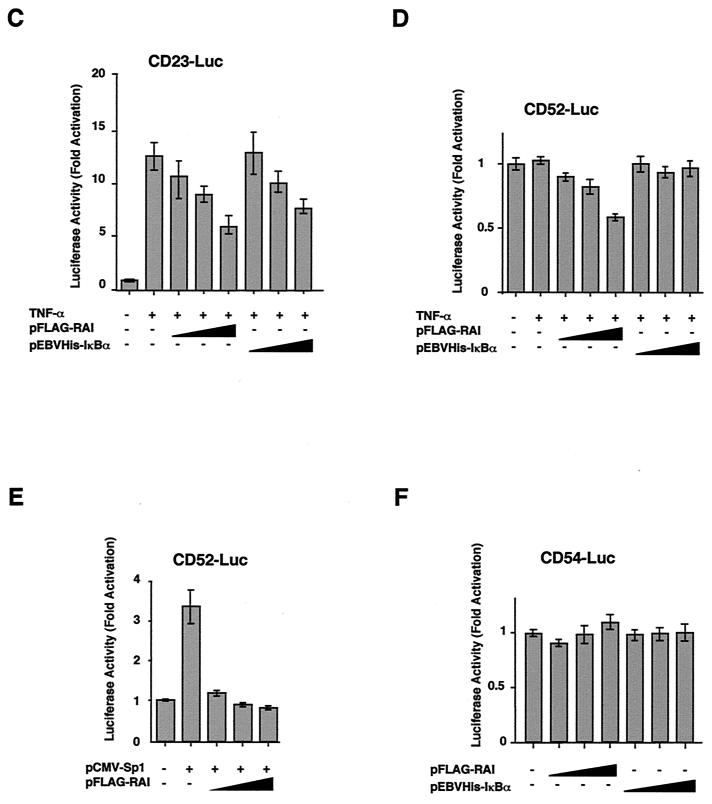
We have initially identified RAI as a nuclear protein interacting with NF-κB p65 subunit and demonstrated that RAI over-expression caused the inhibition of NF-κB DNA-binding in the transfected cells and the gene expression from the promoter containing only the NF-κB binding sites (39). However, it was not clear whether the RAI mediated inhibition of HIV-1 gene expression could be ascribed to the direct interaction between RAI and p65 or through other cellular factors that regulate HIV-1 transcription. We thus examined whether RAI could directly block the DNA-binding of NF-κB, either p65 or p50 subunit, AP1, or Sp1 (Fig. 2). In Fig. 2A, recombinant proteins of GST-RAI, GST-IκBα and MBP-p65 produced in E. coli were purified and the effects of GST-RAI and GST-IκBα on the DNA-binding of MBP-p65 were examined upon EMSA with κB DNA probe. When MBP-p65 was preincubated with GST-IκBα or GST-RAI, the DNA binding was inhibited in a dose-dependent manner (lanes 6 to 11). The control GST protein did not show such activity (lanes 3 to 5).
FIG.2.
Effects of RAI on the DNA-binding of p65, p50, AP1, and Sp1. (A) Direct inhibition of the p65 DNA binding by RAI. Purified recombinant proteins (GST-RAI, GST-IκBα, and GST) were prepared, and their activities in blocking the DNA binding of p65 were examined. The DNA-binding activity of MBP-p65 was analyzed by EMSA with a 32P-labeled oligonucleotide containing the NF-κB binding site. MBP-p65 was preincubated with the various amounts of GST-RAI, GST (negative control), or GST-IκBα (positive control) at onefold (1x) or fourfold (4x) molar excess. It was noted that the inhibitory activity of GST-IκBα was much greater than that of GST-RAI in vitro. For the cold DNA competition experiment, unlabeled double-stranded competitor oligonucleotides (wt and mut, wild-type and mutated κB sequences, respectively) were added into the reaction mixtures at 30-fold molar excess over the probe (lanes 12 to 14). (B to D) Effects of RAI on the DNA binding of other transcription factors. GST-RAI was added at onefold (1x) or fourfold (4x) molar excess to the purified p50 (B), AP1 (C), or Sp1 (D), and EMSA was carried out with the respective DNA probe. The positions of specific DNA-protein complex (filled arrowhead), the nonspecific band (asterisk), and the free probe (open arrowhead) are indicated.
Since results obtained in Fig. 2A did not sufficiently explain the effects of RAI on HIV-1 gene expression described in Fig. 1, in which RAI could block the gene expression from HIV-1 LTR even without TNF-α stimulation and that from CD52-Luc, lacking the κB site, we examined the effect of RAI on the DNA binding of other host transcription factors responsible for HIV-1 gene expression. In the experiments described in Fig. 2B to D, we examined the effect of GST-RAI on the DNA binding of p50 subunit of NF-κB, AP1 (c-Jun) and Sp1. The DNA-binding activities of p50 and AP1 were not significantly inhibited by RAI (Fig. 2B and C, respectively). This action of RAI is similar to that of IκBα and -β, which block the DNA binding of p65 but do not significantly block that of p50 (2, 6, 17). This feature of RAI is in contrast to Bcl-3, which is located in the nucleus but interacts with p50 and p52 without affecting their DNA-binding activity (6). More importantly, the DNA-binding activity of Sp1 was blocked by GST-RAI in a dose-dependent manner, as demonstrated in Fig. 2D. This effect was not due to the GST moiety, since Sp1 DNA-binding was not inhibited by GST or GST-IκBα (data not shown).
Direct interaction between RAI and Sp1 in vitro.
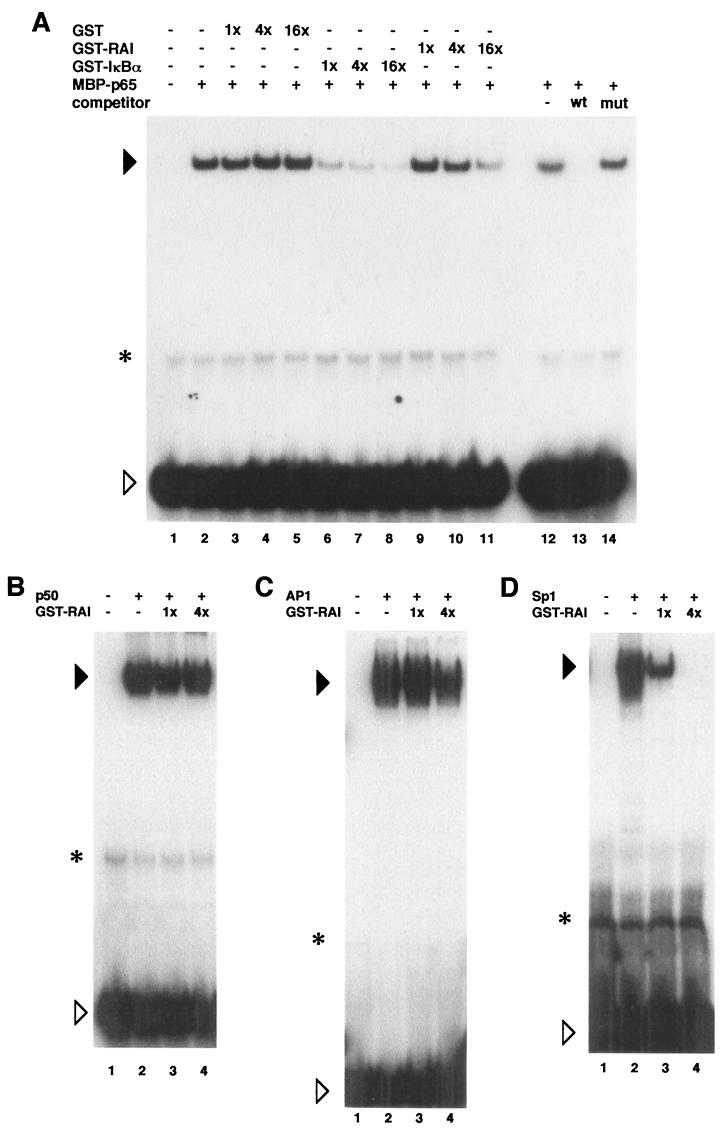
We then examined whether RAI directly binds to Sp1 in vitro. In Fig. 3A, the purified GST or GST-RAI proteins bound to glutathione-Sepharose beads were incubated with recombinant Sp1 or p65 proteins. The beads were washed extensively, and the bound proteins were analyzed by SDS-PAGE followed by Western blotting with specific antibodies to Sp1 or p65. As shown in Fig. 3A, Sp1 interacted with RAI in vitro. Binding between Sp1 and RAI was considered specific because no interaction was observed with the control GST protein. There was no binding between RAI and p50 or AP1 (data not shown).
FIG. 3.
(A) Binding of Sp1 and RAI in vitro. Either the recombinant Sp1 or p65 protein was incubated with GST-RAI or GST (negative control) bound to glutathione-Sepharose beads. After washing the beads, the bound proteins were analyzed by Western blotting with specific antibodies. The bands corresponding to the RAI-bound Sp1 (approximately 100 kDa) or MBP-p65 (110 kDa) are indicated by arrows. The interaction between RAI and p65 was observed as previously reported (39). The lower band detected by anti-Sp1 antibody is considered to be degraded Sp1. As input, one-fifth of the proteins used in each binding reaction was loaded. Positions of protein size markers are indicated to the left of each gel. (B) Colocalization of RAI with Sp1. 293 cells were transfected with a plasmid expressing GFP-RAI, and immunofluorescence microscopic examinations were carried out with anti-Sp1 antibody. The cells were counterstained with DAPI to visualize the nuclei.
Colocalization of RAI with Sp1 in the nucleus.
We have previously reported that RAI is localized in the nucleus. In order to examine the colocalization of RAI with Sp1 in the cells, we performed immunofluorescence microscopic examinations. After 24 h of transfection with pEGFP-RAI, cells were fixed and stained with anti-Sp1 antibody. As shown in Fig. 3B, RAI and Sp1 were colocalized in the nucleus. We also confirmed that RAI was colocalized with Sp1 in the nucleus with the stimulation of TNF-α (data not shown). These data shown in Fig. 3 suggested that RAI might interact with Sp1 in vivo.
Interaction between RAI and Sp1 in vivo.
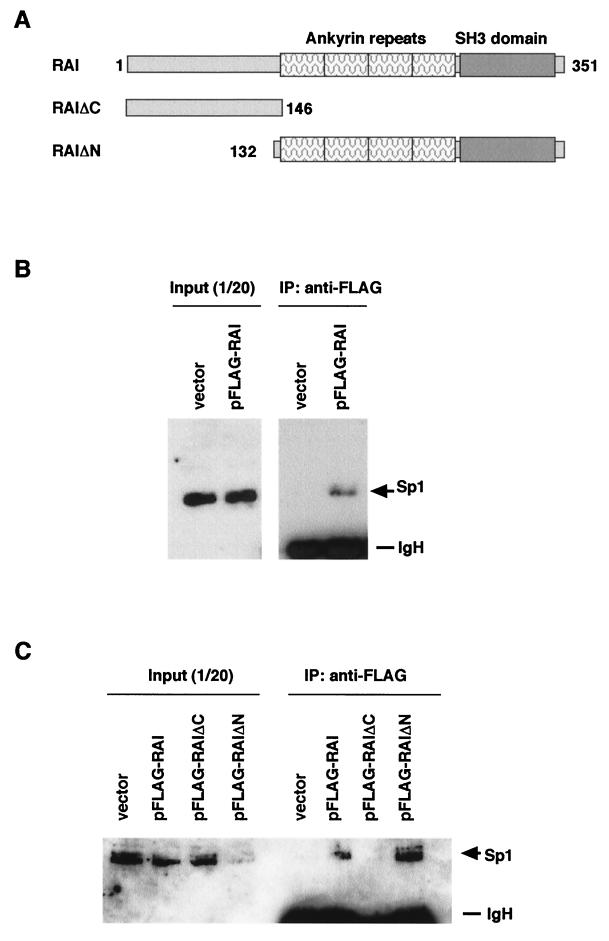
In order to examine whether Sp1 binds to RAI in vivo, 293 cells were transfected with a plasmid expressing RAI or its mutants (Fig. 4A), and the cell lysate was immunoprecipitated with anti-FLAG antibody. Immune complexes were collected and subjected to SDS-PAGE followed by Western blotting for detection of Sp1 using anti-Sp1 antibody. As Sp1 is abundant in 293 cells, it was not necessary to overexpress Sp1 by transfection. As shown in Fig. 4B, Sp1 was coimmunoprecipitated from the cell lysate expressing the wild-type RAI. We then examined which region of RAI was responsible for the interaction with Sp1 (Fig. 4C). When RAI (wild type) or the C-terminal (RAIΔC) or N-terminal (RAIΔN) RAI mutants were expressed, Sp1 was detected in the lysate from cells expressing either wild-type RAI or RAIΔN, but not in cells expressing RAIΔC. These results indicate that Sp1 interacts with RAI at its C terminus containing ankyrin repeats and SH3 domain.
FIG. 4.
Binding of Sp1 and RAI in vivo. (A) Schematic diagram of RAI plasmids: wild-type RAI (containing ankyrin repeats and the SH3 domain), RAI ΔN (containing 132 to 351 amino acids), and RAI ΔC (containing 1 to 146 amino acids). (B and C) The lysates prepared from 293 cells transfected with pFLAG (empty vector), pFLAG-RAI (wild type), pFLAG-RAIΔC, or pFLAG-RAIΔN were used for coimmunoprecipitation. Whole-cell lysates were prepared from 293 cells 24 h after transfection and were precipitated with anti-FLAG affinity gel. Immune complexes were collected and subjected to SDS-PAGE followed by Western blotting with anti-Sp1 antibody. The position of the Sp1 proteins and Ig heavy chain (IgH) proteins are indicated. Sp1 was detected in the lysate overexpressing FLAG-RAI wild type and FLAG-RAI ΔN. As input, 1/20 of the proteins used in each reaction was loaded.
Suppression of HIV-1 replication by RAI.
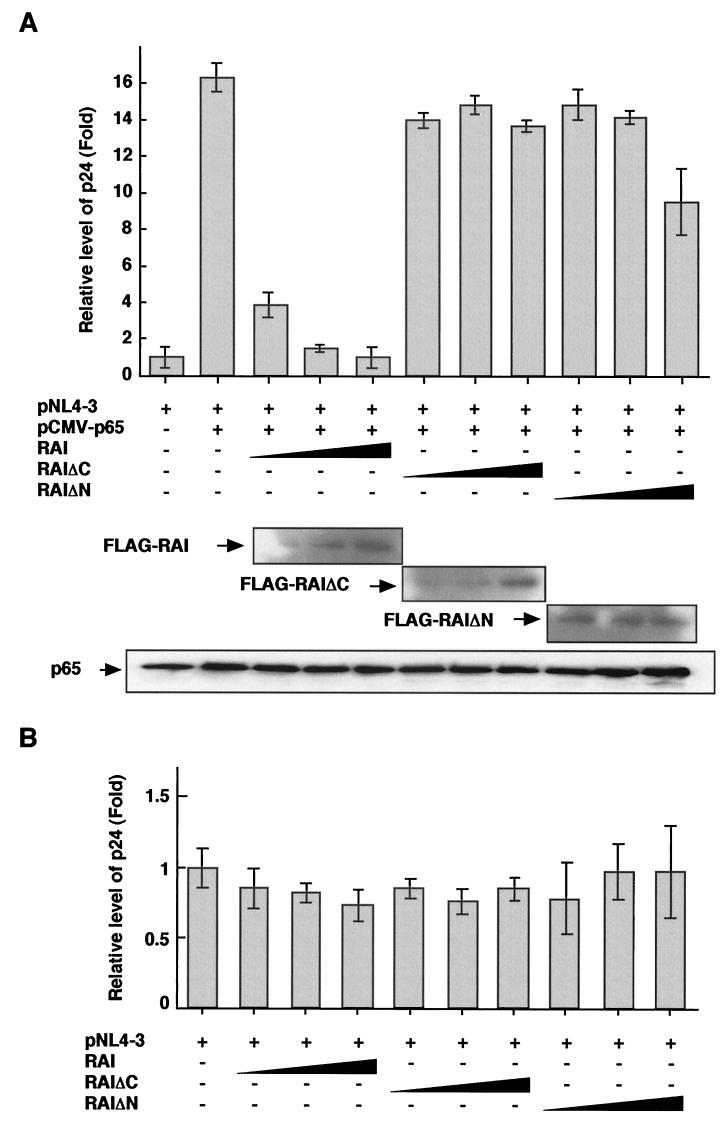
In order to examine the effects of RAI on HIV-1 replication, 293 cells were transfected with a replication-competent full-length HIV-1 clone (pNL4-3) and various amounts of RAI-expressing plasmids (either wild type, RAIΔC, or RAIΔN). Since 293 cells have no relevant HIV-1 receptors, the HIV-1 produced from the full-length clone could not reinfect other cells. Thus, this assay solely represents the level of HIV-1 replication. After infection of HIV-1, the proviral DNA is synthesized and is integrated into the host genome, and the viral replication is initiated by extracellular stimuli such as proinflammatory cytokines, UV wavelengths, and radical oxygens by inducing NF-κB (10). Thus, we examined the effects of RAI on HIV-1 replication when its gene expression is induced by NF-κB. When pNL4-3 was cotransfected with pCMV-p65, approximately 16-fold increase of HIV-1 production was observed in the culture supernatant (Fig. 5A). When wild-type RAI was expressed, a dramatic suppression of HIV-1 production was observed in a dose-dependent manner. However, when RAI mutants were expressed, no significant suppression of HIV-1 production was observed except at the highest amount of RAIΔN retaining the ankyrin repeats and the SH3 domain. As assessed by Western blotting, expression of wild-type RAI or its mutants did not significantly affect the level of p65 expression. We also examined the effect of RAI on the HIV-1 replication in cells transfected with pNL4-3 alone. However, the level of p24 in the culture supernatant for these cells was too low to evaluate the effect of RAI. Therefore, we measured the p24 level in the cell lysate prepared from each transfectant. As shown in Fig. 5B, wild-type RAI could suppress the HIV-1 production in a dose-dependent manner. When RAI mutants, RAIΔC, and RAIΔN, were cotransfected, no detectable suppression were observed.
FIG.5.
Effects of RAI on HIV-1 replication. (A) 293 cells were transfected with pNL4-3, pCMV-p65, and various amounts of plasmids expressing wild-type RAI, RAIΔC, or RAIΔN. These transfected cell cultures were maintained for 72 h, and the culture supernatants were analyzed for the determination of p24 antigen level by enzyme-linked immunosorbent assay. Western blots for FLAG-RAI, FLAG-RAIΔC, FLAG-RAIΔN, and p65 indicate the amount of proteins expressed in each transfectant. (B) 293 cells were transfected with pNL4-3 and various amounts of plasmids expressing wild-type RAI, RAIΔC, or RAIΔN. These transfected cell cultures were maintained for 72 h, the cell lysates were prepared by sonication, and the p24 antigen level was determined similarly. The amount of p24 is indicated by comparison of the increases in the amount of p24 obtained from the transfection with pNL4-3 alone. The data are represented by the means and standard deviations (error bars) of three independent experiments.
DISCUSSION
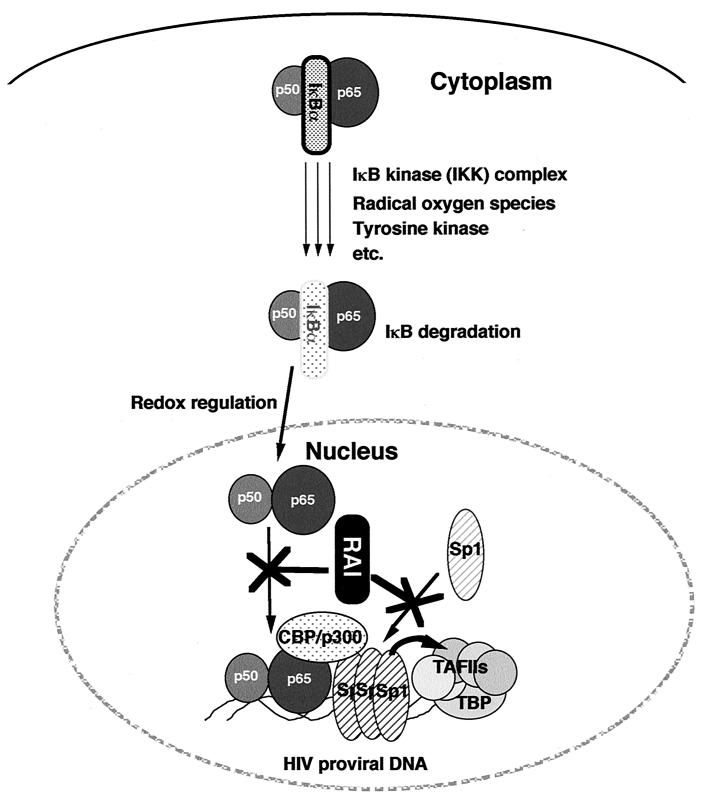
Our observations indicate that RAI inhibits not only the action of NF-κB but also that of Sp1 by direct interaction and blocks their DNA-binding activities. Because of these actions, RAI could efficiently block the gene expression from a promoter such as HIV-1 in which both NF-κB and Sp1 play major roles (Fig. 6). The action of RAI on Sp1 was unexpected, because RAI does not appear to contain any region that shares homology with other transcription factors known to interact with Sp1 such as steroidogenic factor-1 (15), p53 (3), and E2 (14). Regarding the inhibitory factors for Sp1, there are some precedents. For example, Vallian et al. (35) reported that promyelocytic leukemia protein directly interacted with Sp1 and inhibited its transcriptional activity. Similarly, Chen et al. (4) reported that the Sp1 DNA binding and its transcriptional activity were blocked by Rb-associated 20-kDa protein, called Sp1-I. Two other negative Sp1 regulators, p107 (5) and the von Hippel-Lindau gene product (18), are known to block Sp1 transcription activity but without inhibiting the DNA-binding activity. These findings raise the possibility that Sp1 is tightly controlled by the interaction with multiple regulators including promyelocytic leukemia protein, Sp1-I, the von Hippel-Lindau gene product, p107, and RAI.
FIG. 6.
RAI represses HIV-1 transcription by blocking the actions of NF-κB and Sp1. The signal transduction pathway for the NF-κB activation involves the IκB kinase (IKK) complex, tyrosine kinase, radical oxygens, and a redox control mechanism. RAI effectively blocks HIV-1 replication by blocking the DNA-binding activity of NF-κB and Sp1. Several studies have indicated the functional synergy between NF-κB and Sp1. RAI may also interrupt this synergy.
It was previously noted that effects of Sp1 and NF-κB are cooperative in positively regulating the HIV-1 gene expression and replication (24, 25). In fact, Perkins et al. (25) demonstrated the direct interaction between Sp1 and p65 at least in vitro. As the synergistic cooperation between these transcription factors in HIV-1 gene expression is also considered to be based on the adjacent binding of both transcription factors in the promoter context, it is suggested that RAI may also interrupt the synergy. Other studies revealed synergistic activation of Sp1 with other transcription factors (3, 14, 15, 16, 23). For example, the synergy between Sp1 and the sterol-responsive element-binding proteins required CREB-binding protein (CBP) for the transcriptional activation from the low-density lipoprotein receptor promoter (23). A similar observation was reported with TNF-α promoter (34). NF-κB also requires CBP/p300 for its transcriptional activity (7). It is possible that the synergism between NF-κB and Sp1 may also involve CBP/p300.
RAI exerts a strong negative effect on expression of genes that are controlled by NF-κB and Sp1. Thus, the target genes of its transcriptional inhibition are rather limited. As RAI gene expression is relatively restricted in tissues such as heart and placenta (39), negative effects of RAI may also be tissue specific. For example, the promoters for TNF-α (34, 40), a proinflammatory cytokine, and cyclooxygenase-2 (36), one of the effector molecules of inflammation, contain binding sites for NF-κB and Sp1 and are controlled by these transcription factors. Thus, it is conceivable that inflammatory responses mediated by TNF-α and cyclooxygenase-2 can be strongly suppressed in these tissues, which may have physiological significance.
Although RAI is not constitutively expressed in the cells normally susceptible to HIV-1 infection, such as CD4 T-lymphocytes, macrophages, and microglial cells (39), transduction of the RAI gene should block HIV-1 transcription selectively and render the cell resistant to HIV-1 replication. Because RAI represses the HIV-1 promoter activity more efficiently than IκBα, RAI can be used as a candidate replacement gene for the attenuated live HIV-1 vaccine (26) in which the nef gene was replaced by the IκBα gene. Furthermore, although the promoter sequence of the RAI gene has not yet been elucidated, identification of a specific regulator(s) for RAI gene expression and its selective activation may overcome the tissue-specific barrier for the ectopic expression of endogenous RAI gene in order to inhibit the HIV-1 gene expression in the natural host cells for the viral replication.
Acknowledgments
This work was supported in part by grants-in-aid from the Ministry of Health, Labor, and Welfare; the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and the Japanese Health Sciences Foundation.
REFERENCES
- 1.Baeuerle, P. A., and V. R. Baichwal. 1997. NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv. Immunol. 65:111-137. [PubMed] [Google Scholar]
- 2.Baldwin, A. S., Jr. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 3.Borellini, F., and R. I. Glazer. 1993. Induction of SP1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor dependent proliferation in human erythroleukemia cell line TF-1. J. Biol. Chem. 268:7923-7928. [PubMed] [Google Scholar]
- 4.Chen, L. I., T. Nishinaka, K. Kwan, I. Kitabayashi, K. Yokoyama, Y.-H. Fu, S. Grunwald, and R. Chiu. 1994. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol. Cell. Biol. 14:4380-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta, P. K., P. Raychaudhuri, and S. Bagchi. 1995. Association of p107 with Sp1: genetically separable regions of p107 are involved in regulation of E2F- and Sp1-dependent transcription. Mol. Cell. Biol. 15:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita, T., G. P. Nolan, H. C. Liou, M. L. Scott, and D. Baltimore. 1993. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev. 7:1354-1363. [DOI] [PubMed] [Google Scholar]
- 7.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, and Y. Shi. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh, G., G. van Duyne, S. Ghosh, and P. B. Sigler. 1995. Structure of NF-κB p50 homodimer bound to a κ B site. Nature 373:303-310. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi, T., Y. Ueno, and T. Okamoto. 1993. Oxidoreductive regulation of nuclear factor κB. Involvement of a cellular reducing catalyst thioredoxin. J. Biol. Chem. 268:11380-11388. [PubMed] [Google Scholar]
- 10.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral approprication of the NF-κB pathway. J. Clin. Investig. 107:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blockes the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 12.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 13.Kwan, H., N. Pelletier, C. Duluca, P., Genin, S. Cisternas, R. Lin, M. A. Wainberg, and J. Hiscott. 1998. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J. Biol. Chem. 273:7431-7440. [DOI] [PubMed] [Google Scholar]
- 14.Li, R., J. D. Knight, S. P. Jackson, R. Tjian, and M. R. Botchan. 1991. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell 65:493-505. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Z., and E. R. Simpson. 1999. Molecular mechanism for cooperation between Sp1 and steroidogenic factor-1 (SF-1) to regulate bovine CYP11A gene expression. Mol. Cell. Endocrinol. 153:183-196. [DOI] [PubMed] [Google Scholar]
- 16.Look, D. C., M. R. Pelletier, R. M. Tidwell, W. T. Roswit, and M. J. Holtzman. 1995. Stat1 depends on transcriptional synergy with Sp1. J. Biol. Chem. 270:30264-30267. [DOI] [PubMed] [Google Scholar]
- 17.Malek, S., T. Huxford, and G. Ghosh. 1998. IκBα functions through direct contacts with the nuclear localization signals and the DNA binding sequences of NF-κB. J. Biol. Chem. 273:25427-25435. [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, D., B. Knebelmann, H. T. Cohen, S. Ananth, and V. P. Sukhatme. 1997. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol. Cell. Biol. 17:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto, H., C. Sheline, J. Corden, K. Jones, and B. M. Peterlin. 1996. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA 93:11575-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto, T., T. Matsuyama, S. Mori, S., Hamamoto, N. Kobayashi, N. Yamamoto, S. F. Josephs, F. Wong-Staal, and K. Shimotohno. 1989. Augmentation of human immunodeficiency virus type 1 gene expression by tumor necrosis factor α. AIDS Res. Hum. Retrovir. 5:131-138. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, T., T. Benter, S. F. Josephs, M. R. Sadaie, and F. Wong-Staal. 1990. Transcriptional activation from the long terminal repeat of human immunodeficiency virus in vitro. Virology 177:606-614. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, T., S. Sakurada, J.-P. Yang, and J. P. Merin. 1996. Regulation of NF-κB and disease control: identification of a novel serine kinase and thioredoxin as effectors for signal transduction pathway for NF-κB activation. Curr. Top. Cell. Regul. 3:149-161. [DOI] [PubMed] [Google Scholar]
- 23.Oliner, J. D., J. M. Andresen, S. K. Hansen, S. Zhou, and R. Tjian. 1996. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 10:2903-2911. [DOI] [PubMed] [Google Scholar]
- 24.Pazin, M. J., P. L. Sheridan, K. Cannon, Z. Cao, J. G. Keck, J. T. Kadonaga, and K. A. Jones. 1996. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 10:37-49. [DOI] [PubMed] [Google Scholar]
- 25.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 14:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinto, I., M. Mallardo, F. Baldassarre, G. Scala, G. Englund, and K. T. Jeang. 1999. Potent and stable attenuation of live-HIV-1 by gain of a proteolysis-resistant inhibitor of NF-κB (IκB-αS32/36A) and the implications for vaccine development. J. Biol. Chem. 274:17567-17572. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sato, T., K. Asamitsu, J.-P. Yang, N. Takahashi, T. Tetsuka, A. Yoneyama, A. Kanagawa, and T. Okamoto. 1998. Inhibition of human immunodeficiency virus type 1 replication by a bioavailable serine/threonine kinase inhibitor, fasudil hydrochloride. AIDS Res. Hum. Retrovir. 14:293-298. [DOI] [PubMed] [Google Scholar]
- 29.Siebenlist, U., K. Brown, and G. Franzoso. 1995. NF-κB: mediator of pathogen and stress responses, p. 93-141. In P. A. Baeuerle (ed.), Inducible gene expression, vol. 1. Birkhauser, Boston, Mass.
- 30.Starcich, B., L. Ratner, S. F. Josephs, T. Okamoto, R. C. Gallo, and F. Wong-Staal. 1985. Characterization of long terminal repeat sequences of HTLV-III. Science 227:538-540. [DOI] [PubMed] [Google Scholar]
- 31.Sun, S.-C., P. A. Ganchi, C. Beraud, and D. W. Ballard. 1994. Autoregulation of the NF-κ B transactivator RelA (p65) by multiple cytoplasmic inhibitors containing ankyrin motifs. Proc. Natl. Acad. Sci. USA 91:1346-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tetsuka, T., H. Uranishi, H. Imai, T. Ono, S. Sonta, N. Takahashi, K. Asamitsu, and T. Okamoto. 2000. Inhibition of nuclear factor-κB-mediated transcription by association with the amino-terminal enhancer of split, a groucho-related protein lacking WD40 repeats. J. Biol. Chem. 275:4383-4390. [DOI] [PubMed] [Google Scholar]
- 33.Traber, K. E., H. Okamoto, C. Kurono, M. Baba, C. Saliou, T. Soji, L. Pecker, and T. Okamoto. 1999. Anti-rheumatic compound aurothioglucose inhibits tumor necrosis factor-α-induced HIV-1 replication in latently infected OM10.1 and Ach2 cells. Int. Immunol. 11:101-108. [DOI] [PubMed] [Google Scholar]
- 34.Tsai, E. Y., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor α promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallian, S., K.-V. Chin, and K.-S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Q., Y.-S. Ji, and J. F. Schmedtje, Jr. 2000. Sp1 increases expression of cyclooxygenase-2 in hypoxic vascular endothelium. J. Biol. Chem. 275:24583-24589. [DOI] [PubMed] [Google Scholar]
- 37.Yang, J.-P., J. P. Merin, T. Nakano, T. Kato, Y. Kitade, and T. Okamoto. 1995. Inhibition of the DNA-binding activity of NF-κB by gold compounds in vitro. FEBS Lett. 361:89-96. [DOI] [PubMed] [Google Scholar]
- 38.Yang, J.-P., M. Hori, N. Takahashi, T. Kawabe, H. Kato, and T. Okamoto. 1999. NF-κB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene 18:5177-5186. [DOI] [PubMed] [Google Scholar]
- 39.Yang, J.-P., M. Hori, T. Sanda, and T. Okamoto. 1999. Identification of a Novel inhibitor of nuclear factor-κB, RelA-associated inhibitor. J. Biol. Chem. 274:15662-15670. [DOI] [PubMed] [Google Scholar]
- 40.Yao, J., N. Mackman, T. S. Edgington, and S.-T. Fan. 1997. Lipopolysaccharide induction of the tumor necrosis factor-α promoter in human monocyte cells. J. Biol. Chem. 272:17795-17801. [DOI] [PubMed] [Google Scholar]