Abstract
In the present study, we describe a new simian immunodeficiency virus (SIV), designated SIVgsn, naturally infecting greater spot-nosed monkeys (Cercopithecus nictitans) in Cameroon. Together with SIVsyk, SIVgsn represents the second virus isolated from a monkey belonging to the Cercopithecus mitis group of the Cercopithecus genus. Full-length genome sequence analysis of two SIVgsn strains, SIVgsn-99CM71 and SIVgsn-99CM166, revealed that despite the close phylogenetic relationship of their hosts, SIVgsn was highly divergent from SIVsyk. First of all, they differ in their genomic organization. SIVgsn codes for a vpu homologue, so far a unique feature of the members of the SIVcpz/human immunodeficiency virus type 1 (HIV-1) lineage, and detailed phylogenetic analyses of various regions of the viral genome indicated that SIVgsn might be a mosaic of sequences with different evolutionary histories. SIVgsn was related to SIVsyk in Gag and part of Pol and related to SIVcpz in Env, and the middle part of the genome did not cluster significantly with any of the known SIV lineages. When comparing the two SIVgsn Env sequences with that of SIVcpz, a remarkable conservation was seen in the V3 loop, indicating a possible common origin for the envelopes of these two viruses. The habitats of the two subspecies of chimpanzees infected by SIVcpz overlap the geographic ranges of greater spot-nosed monkeys and other monkey species, allowing cross-species transmission and recombination between coinfecting viruses. The complex genomic structure of SIVgsn, the presence of a vpu gene, and its relatedness to SIVcpz in the envelope suggest a link between SIVgsn and SIVcpz and provide new insights about the origin of SIVcpz in chimpanzees.
AIDS is caused by two lentiviruses, human immunodeficiency viruses type 1 (HIV-1) and type 2 (HIV-2), both of zoonotic origin, which find their closest simian relatives in the common chimpanzee (Pan troglodytes) and the sooty mangabey (Cercocebus atys), respectively. In addition to these simian immunodeficiency viruses (SIVs), serological or molecular evidence for other SIVs has been reported in at least 30 African nonhuman primates (16, 29). Based on sequence similarity of the fully characterized viruses, SIVs are classified into six approximately equidistant phylogenetic lineages: (i) SIVcpz from chimpanzees (Pan troglodytes) together with HIV-1 (9, 12, 21, 43); (ii) SIVsm from sooty mangabeys (Cercocebus atys) (7, 19) together with HIV-2; (iii) SIVagm from four species of African green monkeys (members of the Chlorocebus aethiops superspecies) (1); (iv) SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis) (18); (v) SIVmnd from a mandrill (Mandrillus sphinx) (41) together with SIVlhoest from l'Hoest monkeys (Cercopithecus lhoesti) (17) and SIVsun from sun-tailed monkeys (Cercopithecus solatus) (3); and (vi) SIVcol from a guereza colobus (Colobus guereza) (10). Two other recently characterized SIV strains from wild mandrills in Cameroon (SIVmnd2) (35, 39) and from red-capped mangabeys in Nigeria (SIVrcm) (4) cannot be classified into one of the six lentivirus lineages because of a mosaic genome.
SIVs belong to the lentivirus subfamily of retroviruses and can display different genomic organizations. Viruses from all lineages have a similar basic genomic structure comprising the gag, pol, vif, vpr, tat, rev, env, and nef genes, but viruses from the SIVcpz/HIV-1 lineage have an additional gene, vpu, whereas viruses from the SIVsm/HIV-2 lineage have a vpx gene in the central part of the genome.
Phylogenetic relationships among the lentiviruses clearly indicate a common origin and provide evidence that some of the viruses have evolved in a host-dependent fashion, as is the case for African green monkeys (1, 22, 28) and within the Cercopithecus lhoesti superspecies (3). But there are also multiple examples of cross-species transmissions from simians to humans and between simians. Indeed, it appears now that the presence of human immunodeficiency viruses (HIV-1 and HIV-2) in the human population results from at least eight independent transmission events of viruses naturally infecting chimpanzees and sooty mangabeys (34). SIV infection of baboons and patas monkeys by viruses derived from the local sympatric species of African green monkeys confirms that simian-to-simian cross-species transmissions also occur in the wild (5, 23, 44). In addition, phylogenetic analyses also provided evidence for mosaic SIV genomes in at least three nonhuman primate species, West African sabaeus monkeys (SIVsab), red-capped mangabeys (SIVrcm), and mandrills (SIVmnd2) (4, 15, 22, 35), suggesting that recombination events have occurred between viruses in vivo. These observations indicate that both cross-species transmission and coinfection with highly divergent viral strains are possible.
For a better understanding of the evolutionary relationships of primate lentiviruses, which are becoming more and more complex, characterization of SIVs from other nonhuman primates species is essential. It is now widely accepted that HIV-1 came from a zoonotic transmission from SIVcpz to humans, but there is less evidence on whether chimpanzees are the natural reservoir for this group of lentiviruses or whether they became infected from another species. In this context, we initiated a large seroprevalence survey of wild-born monkeys in Cameroon (29) and identified several greater spot-nosed monkeys (Cercopithecus nictitans) whose sera contained antibodies cross-reacting with HIV-1 envelope gene products. The greater spot-nosed monkeys are found in lowland forest across the Congo Basin and as far west as Nigeria and are known to be hunted by chimpanzees for food.
This paper describes the genetic characterization of a novel primate lentivirus found in greater spot-nosed monkeys from Cameroon. Interestingly, this virus, designated SIVgsn, displays a typical lentivirus genomic structure but also encodes a vpu homologue, a gene which until now was found only among members of the SIVcpz/HIV-1 lineage.
MATERIALS AND METHODS
Animals and serologic testing.
Blood samples were obtained from 165 greater spot-nosed monkeys (C. nictitans) between January 1999 and March 2001. All the animals were caught in the wild in Cameroon. A total of 121 animals were sampled as bushmeat upon arrival at markets in Yaounde, in surrounding villages, or at logging concessions in southeastern Cameroon, and 44 animals were sampled as pets from the same areas. All primate samples were obtained with approval from the Cameroonian Ministry of Environment and Forestry. The bushmeat samples were obtained by employing a strategy specifically designed not to increase further the demand for bushmeat. Women preparing and preserving the meat for subsequent sale and hunters already involved in the trade were asked for permission to sample blood and tissues from carcasses, which were then returned to their owners.
For the bushmeat animals, blood was collected by intracardiac puncture. Information provided by the owners indicated that most of the animals had died 12 to 72 h prior to sampling. For the pet monkeys, blood was drawn by peripheral venipuncture after tranquilizing the animals with ketamine (10 mg/kg). Plasma and cells were separated on site by Ficoll gradient centrifugation. All samples, including peripheral blood mononuclear cells (PBMCs), plasma, whole blood, and other tissues, were stored at −20°C. Whole blood or plasma samples were tested for the presence of HIV/SIV antibodies with the INNO-LIA HIV confirmation test (Innogenetics, Ghent, Belgium). This test configuration includes HIV-1 and HIV-2 recombinant proteins and synthetic peptides that are coated as discrete lines onto a nylon strip. Five HIV-1 antigens are used, including synthetic peptides for the exterior envelope glycoprotein (sgp120) as well as recombinant proteins for the transmembrane envelope glycoprotein (gp41), integrase (p31), core (p24), and matrix (p17) proteins. HIV-1 group O envelope peptides are included in the HIV-1 sgp120 band. The HIV-2 antigens include synthetic peptides for the exterior envelope glycoprotein (sgp120) as well as recombinant gp36 protein. In addition to these HIV antigens, control lines are present on each strip: one sample addition line (3+) containing anti-human immunoglobulin (Ig) and two test performance lines (1+ and +/−) containing human IgG.
All assays were performed according to the manufacturer's instructions, with alkaline phosphatase-labeled goat anti-human IgG used as the secondary antibody. We established the following working definition for SIV seropositivity: plasma samples were scored as INNO-LIA positive when they reacted with at least one HIV antigen and yielded a band intensity equal to or greater than that of the assay cutoff (+/−) lane; samples which reacted less strongly but still visibly with two or more HIV antigens were classified as indeterminant; and samples reacting with no bands or only one band with less than ± intensity were classified as negative.
A subset of samples were also tested for the presence of antibodies to HIV/SIV V3 loop peptides by an in-house indirect enzyme-linked immunosorbent assay (ELISA). Synthetic peptides corresponding to the V3 loop of HIV-1-M, -N and -O, SIVcpzANT, SIVsm, SIVagm, SIVsyk, and SIVmnd were synthesized, and the four N-terminal amino acids were truncated. Wells of microtiter plates were coated with 0,25 μg of each single peptide per well in 0.05 M bicarbonate buffer, pH 9.6, by incubation at 37°C for 20 h. After washing with phosphate-buffered saline (PBS) containing 0.5% Tween 20 (PBS-TW), unoccupied sites were blocked with PBS containing 5% fetal calf serum for 2 h at 37°C. Sera were diluted 1:100 in a hypertonic PBS solution (0.01 M sodium phosphate buffer [pH 7.4] containing 0.75 M NaCl, 10% fetal calf serum, and 0.5% Tween 20). After incubation for 30 min at room temperature, plates were washed and incubated with peroxidase-conjugated goat anti-human IgG for 30 min at room temperature. After washing, the reaction was revealed with hydrogen peroxide-o-phenylenediamine for 15 min at room temperature in the dark. Color development was stopped by adding 2 N H2SO4, and optical densities were read at 492 nm.
PCR amplification, cloning, and sequencing.
DNA was isolated from whole blood or PBMCs with the QIAamp blood kit (Qiagen), according to the manufacturer's instructions. PCR amplification was performed with an automated DNA thermal cycler (GeneAmp PCR system 2700).
We first amplified a fragment of 650 bp in the pol region with degenerate consensus primers designed to amplify this fragment from all known primate lentiviruses, DR1 and PolOR for the first round and Polis4/UNIPOL2 for the second round of amplification (8, 10, 26). PCR conditions were as reported previously (10). PCR products were cloned into the pGEM-TEasy vector (Promega) and sequenced. Specific primers were then designed to amplify the fragment between the two outer primers, cloned, and sequenced. We then amplified the full-length genome sequence of SIVgsn with two sets of specific primers designed based on the DR1/UNIPOL2 sequence from animal 99CM71: Ni1, 5′-ACCCGCAAAGTCAAGGGGTAGTAG-3′, and NI8, 5′-AATTCTTCATACAATGGTACACTG-3′, for the first round and Ni5, 5′-TCACATACTTAGAAACAGCAGTA-3′, and NI7, 5′-AGTTACGTGCTCTTTTTGTTCTAG-3′, for the second round of amplification. These primers were used to amplify the complete genome of both SIVgsn-99CM71 and SIVgsn-99CM166 by targeting unintegrated circular SIV DNA.
PCRs were performed with the Long Expand High Fidelity PCR kit (Roche Molecular Biochemicals), including a hot start (92°C for 3 min) with the following cycle conditions: 10 cycles of denaturation at 92°C for 10 s, annealing at 57°C for 30 s, and extension at 68°C for 7 min, followed by 20 cycles with extension at 68°C for 7 min with an increment of 20 s per cycle. Amplification was completed by a final extension at 68°C for 10 min. Then, 1/20 of the first PCR was used as the template in a nested amplification with the same cycling conditions. PCR amplification products were then purified and cleaved with EcoRI, and the resulting fragments were subcloned into pBluescript KS+ cleaved with EcoRI or with EcoRI and SmaI. Double-stranded recombinant plasmid DNA was sequenced by the cycle sequencing and dye terminator methods (ABI Prism Big Dye terminator cycle sequencing ready reaction kit with AmpliTaq FS DNA polymerase; PE Biosystems, Warrington, England) on an automated sequencer (ABI 373 model Stretch; Applied Biosystems) with genome priming system GPS-1 (New England BioLabs, Beverly, Mass.). To reconstitute the full-length genome sequence, overlapping sequences were joined with Sequencher software (Gene Codes Corp.).
To test for DNA degradation, a region of the glucose-6-phosphate dehydrogenase gene was amplified by single-round PCR. The following primers amplify on human DNA a fragment of 1,459 bp: G6PDH-1s, 5′-GACATCCGCAAACAGAGTGAG-3′, and G6PDH-2as, 5′-GAGCTGTGTTACCCCCTTGAA-3′. Amplification conditions were as follows: 94°C for 2 min, then 35 cycles at 94°C for 20 s, 58°C for 30 s, and 72°C for 1 min.
RNA folding.
RNA secondary structures of TAR were predicted with the RNA mfold version 3.1 by Zuker and Turner (http://bioinfo.math.rpi.edu/≈mfold/rna/form1.cgi), with the folding temperature fixed at 37°C (25, 48).
Sequence alignments.
The predicted protein sequences encoded by SIVgsn were compared with other published SIV sequences representing each of the six known nonhuman primate lentivirus lineages. Reference sequences were retrieved through the Internet from the Los Alamos sequence database (http://hiv-web.lanl.gov/).
Protein sequences were aligned by using the Clustal W software program (40) with subsequent manual adjustments. Sites containing a gap in any of the sequences were excluded from the analyses. The extent of sequence differences, along the genome, between SIVgsn and other primate lentiviruses was examined in a diversity plot in which protein (Gag, Pol, Vif, Env, and Nef) sequences were concatenated. Segments encoded by overlapping genomic regions (between Gag and Pol, Pol and Vif, and Env and Nef) were represented only once. The fractional amino acid sequence difference was calculated for a window of 200 residues, moved in steps of 20 residues. Diversity plots were performed with the Simplot program (32).
Nucleotide alignments were also constructed for the gag, pol, vif, env, and nef genes of SIVgsn-99CM71 and SIVgsn-99CM166 plus 23 SIV and HIV strains representative of the major lentivirus lineages available from the Los Alamos HIV sequence database: HIV1-ANT70 (accession no. L20587), HIV1-MVP5180 (L20571), HIV1-YBF30 (AJ006022), HIV1-U455 (M62320), HIV2-ALI (AF082339), HIV2-D205 (X61240), SIVsyk173 (L06042), SIVsun (AF131870), SIVlhoest (AF075269), SIVcpzUS (AF103818), SIVcpzGAB (X52154), SIVcol (AF301156), SIVagmVER155 (M29975), SIVagmGRI677 (M66437), SIVagmVER906 (L40990), SIVagmTAN1 (U58991), SIVmnd1 (M27470), SIVsmH4 (X14307), SIVagmSAB1C (U04005), SIVmnd2 (AF328295), SIVcpzCAM3 (AF115393), SIVcpzANT (U42720), and SIVdeb, isolated from a De Brazza monkey (Cercopithecus neglectus) from Cameroon (29). The nucleotide sequences from the different genes were first translated and then aligned with Clustal W (40) implemented in DAMBE (47) and translated back to nucleotides in order to get a more accurate alignment. Alignments were then edited manually where necessary. A concatenated nucleotide alignment was constructed, excluding the overlapping sequences between gag and pol, pol and vif, and env and nef. Gaps were removed from the final alignment.
Phylogenetic signal.
The presence of saturation at different codon positions (first plus second codon positions or third codon position) for the nonoverlapping genes gag, pol, vif, env, and nef was tested by comparing half of the theoretical saturation index expected in case of full saturation with the observed saturation index (46). Saturation indices were calculated with the program DAMBE. The phylogenetic signal was also investigated by analyzing all 4,845 possible groups of four sequences (quartets) in the data set with the likelihood-mapping method implemented in Tree-Puzzle (37). For a quartet, only three unrooted tree topologies are possible. The likelihood of each topology can be estimated with the maximum-likelihood method, and the three likelihoods can be reported as a dot in an equilateral triangle. Three main different areas in the triangle can be distinguished (37): (i) the tree corners, representing fully resolved tree topologies, i.e., the presence of tree-like phylogenetic signal in the data; (ii) the center, which is the area of star-like phylogeny, representing phylogenetic noise; and (iii) the three areas on the sides, where it is not possible to decide between two different tree topologies, representing net-like phylogeny. The percentage of dots belonging to each area can give an idea about the phylogenetic signal in the data set under investigation.
Bootscan analysis.
Bootscan analysis was performed on the concatenated nucleotide sequence alignment, and the following strains were used in the analysis: SIVsyk173, SIVsun, SIVlhoest, SIVcpzUS, SIVcpzGAB, SIVcol, SIVagmVER155, SIVagmTAN1, SIVmnd1, SIVsmH4, SIVagmSAB1C, SIVmnd2, SIVcpzANT, and SIVdeb. Bootscan analysis was performed with the Simplot program (32) on first and second codon position alignment, grouping the sequences per lineage with a sliding window of 500 nucleotides moved in steps of 20 nucleotides and 1,000 bootstrap replicates with Kimura's two-parameter model of evolution.
Phylogenetic analysis.
The nucleotide substitution model best fitting our data set was evaluated with the program Modeltest v3.06 (31). Phylogenetic analyses were performed with PAUP*4.0b8, written by David L. Swofford (38). Neighbor-joining and maximum-likelihood trees were calculated with the nucleotide substitution model selected and the parameters estimated through maximum likelihood. Bootstrap analysis (1,000 replicates) was applied to the neighbor-joining tree, whereas P values were obtained for the maximum-likelihood tree (11).
Nucleotide sequence accession numbers.
The complete sequences of SIVgsn-99CM71 and SIVgsn-99CM166 have been submitted to GenBank under accession numbers AF468658 and AF468659, respectively.
RESULTS
HIV/SIV cross-reactive antibodies in greater spot-nosed monkeys (C. nictitans).
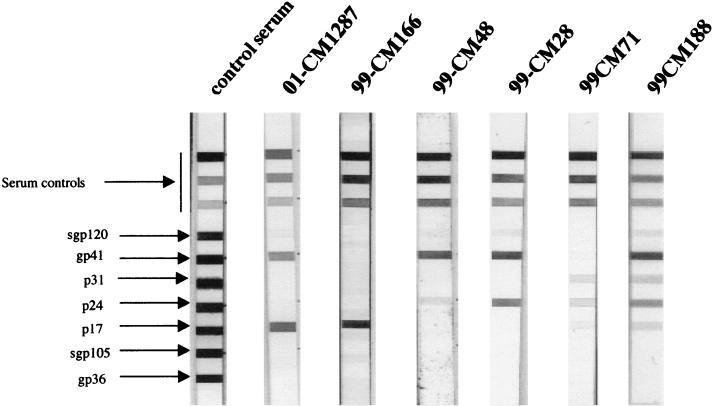
During a large serosurvey in Cameroon, a total of 165 wild-born greater spot-nosed monkeys were tested for HIV/SIV cross-reactive antibodies with a line-immunoassay (LIA) with multiple HIV antigens, the INNO-LIA HIV confirmation test (Innogenetics, Ghent, Belgium). Of 165 samples tested, 27 (16.4%) had antibodies that cross-reacted strongly with at least one HIV protein, and 3 of 165 (1.8%) were identified as indeterminate (i.e., weak but visible bands with two HIV antigens). An extensive variability was observed among all LIA profiles, as illustrated in Fig. 1: eight sera reacted only with HIV core proteins, one only with Pol, eight only with Env, nine with core and Env, and one with core, Pol, and Env. Among the three indeterminate samples, two reacted with core and Pol and one with core and Env. Interestingly, all sera showing cross-reactivity against Env proteins recognized HIV-1 Env (gp120 and/or gp41) with no cross-reactivity to HIV-2 Env proteins.
FIG. 1.
Antibody profiles observed in C. nictitans with a line immunoassay (INNO-LIA HIV confirmation; Innogenetics, Ghent, Belgium). The five HIV-1 antigens include synthetic peptides for the exterior envelope glycoprotein (sgp120) as well as recombinant proteins for the transmembrane envelope glycoprotein (gp41), integrase (p31), core (p24), and matrix (p17) proteins. The HIV-2 antigens include synthetic peptides for the exterior envelope glycoprotein (sgp105) as well as recombinant gp36 protein. Plasma samples which recognized at least one HIV antigen with an intensity equal to or greater than the assay cutoff line were scored as positive; samples which exhibited weaker but still visible reactivities with at least two HIV antigens were scored as indeterminant; and samples which yielded no reactivity or only a single band of less than ± intensity were scored as negative. Note that the 3+, 1+, and ± bands evident on the top portions of all test strips control for sample addition (presence of plasma immunoglobulin) and test performance (binding of secondary antibody).
We further screened 26 of the 27 LIA-positive, 3 LIA-indeterminate, and 43 LIA-negative samples for antibodies to different HIV/SIV V3 peptides. Interestingly, four samples (three LIA positives and one indeterminate) reacted exclusively with the SIVcpzANT peptide. Among these four samples, only one reacted simultaneously with HIV-1 Env proteins and SIVcpzANT, whereas none of the 43 LIA-negative samples showed reactivity with any of the V3 peptides tested.
In order to test whether the samples with serological cross-reactivity were infected with an SIV, we amplified with degenerate primers a fragment of the integrase gene in PBMCs from a subset of 20 LIA-positive and 1 indeterminate animal for which additional material was available. We successfully amplified two samples (animals 99CM-166 and 99CM-CN7) out of the LIA-positive monkeys as well as from the single indeterminate sample tested (animal 99CM-71). The PCR products were then further cloned and sequenced. Repeated attempts, with different PCR conditions and combinations of primers, to amplify the remaining positive samples were unsuccessful, due in part to degradation of the DNA (at least for 5 of the 18 PCR-negative samples) and probably also because of a combination of other factors, such as viral load and mismatches of primers. PCR was also performed on 61 LIA-negative samples, and all remained negative.
The amplified pol fragments were sequenced, and phylogenetic analysis with the neighbor-joining method showed that these three pol sequences clustered together on a separate branch compared with the existing SIV lineages. The pol fragments were closely related to each other, with an average of 85.5% nucleotide sequence identity (data not shown).
Sequence analysis of the full-length genome of SIVgsn: presence of a vpu gene.
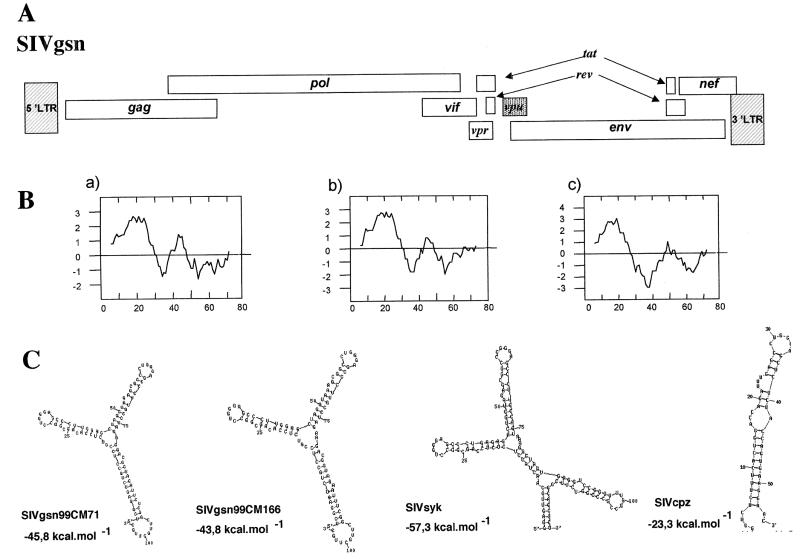
With a nested long-PCR method, we attempted to amplify full-length SIV genomes corresponding to the pol sequences amplified from animals 99CM-166 and 99CM-71, whose sera reacted with the SIVcpzANT V3 loop peptide. Complete genomes, SIVgsn-99CM71 and SIVgsn-99CM166, were successfully amplified by targeting unintegrated circular DNA. The long-PCR products were cleaved, and the different fragments were cloned. The two genomes were sequenced entirely and were 9,410 and 9,416 bp long, respectively. Surprisingly, the genomic organization of SIVgsn was similar to that of SIVcpz and HIV-1, i.e., it lacked the vpx gene present in the SIVsm/HIV-2 lineage and harbored the additional accessory gene vpu specific to members of the SIVcpz/HIV-1 lineage in addition to the gag, pol, and env genes and the accessory genes vif, vpr, tat, rev, and nef, common to all lentiviruses (Fig. 2A).
FIG. 2.
(A) Genomic organization of SIVgsn from greater spot-nosed monkeys (C. nictitans). (B) Hydrophobicity profiles of the SIVgsn-99CM71 (a), SIVgsn-99CM166 (b), and SIVcpzANT (c) Vpu proteins were calculated according to Kyte and Doolittle (24). Positive scores are hydrophobic, and the amino acid number is indicated on the x axis. (C) RNA secondary-structure predictions of TAR for SIVgsn-99CM71, SIVgsn-99CM166, SIVsyk, and SIVcpz as generated by the RNA MFOLD 3.0 program by M. Zuker and D. Turner at a 37°C folding temperature. The free energy is expressed in kilocalories per mole.
The product of the open reading frame (ORF) located at a position similar to that of the vpu gene in the HIV-1/SIVcpz lineage was difficult to recognize on the basis of amino acid similarities. The putative proteins encoded by these genes in SIVgsn-99CM71 and SIVgsn-99CM166 (76 and 78 amino acids, respectively) were closely related to each other (72% identity). A BLAST search against sequences from different databases (Los Alamos and GenBank) identified only six HIV-1 Vpu proteins with borderline identity (38 to 32,5% in a 55-amino-acid overlap). This was not really surprising, because even between Vpus from SIVcpz and HIV-1 group M or O, high variability has been documented. Nevertheless, the position in the genome and the length (consensus length for HIV-1 is 81 amino acids [26]) of this ORF fragment as well as the hydropathy profile displayed by its deduced protein, which is similar to Vpu (Fig. 2B), allowed us to identify this ORF as a vpu gene.
Long terminal repeat (LTR) sequences from both viruses contained all the characteristic features of other primate lentiviruses. LTRs include one predicted NF-κB site and two potential SP-1 binding sites. Despite some differences in the primary sequences, the predicted TAR secondary structures were very similar to each other for both SIVgsn strains. The energetically most stable structures predicted by RNA secondary-structure programs included the two stem loops (ΔG= −45.8 and −43.8 for SIVgsn-99CM71 and SIVgsn-99CM166, respectively) with the consensus sequence 5′-CUGGG(G/A)-3′. These TAR structures closely resembled those described for SIVsyk (Fig. 2C).
Relationship of the new SIVgsn strains to other known SIV/HIV lineages.
Amino acid identities between SIVgsn-99CM71 and SIVgsn-99CM166 as well as to representatives of the other primate lentivirus lineages were calculated. As shown in Table 1, SIVgsn-99CM71 and SIVgsn-99CM166 were closely related to each other and only distantly related to the other primate lentiviruses. Gag and Pol were the most closely related proteins, with average amino acid identities of 48.9 and 53%, respectively, followed by Env and Nef, with an average of 36.2 and 32.6%, respectively, Vif and Vpr being the most divergent proteins (24.5%). Comparisons to Tat and Rev were not included in the table because although these proteins present most of the features conserved among all nonhuman primate lentiviruses, their alignment with the corresponding genes showed only borderline similarities.
TABLE 1.
Amino acid identities between SIVgsn-99CM71 and other SIVs
| SIV | % Identity with protein from SIV gsn-99CM71 and other SIVs
|
|||||
|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Env | Nef | |
| SIVgsn-99CM166 | 89 | 91.5 | 86.9 | 86.7 | 79.9 | 80.7 |
| SIVsyk | 56.7 | 56.6 | 26.2 | 27.4 | 42.2 | 33.1 |
| SIVsmPBj | 53.5 | 55.4 | 29.1 | 27.4 | 37.3 | 33.8 |
| SIVmnd1 | 49.4 | 53.2 | 21.4 | 20.6 | 29.3 | 31.4 |
| SIVhoest | 44 | 50.8 | 25.6 | 24.3 | 29.6 | 33 |
| SIVsun | 45.6 | 50.9 | 19.2 | 25.9 | 29.6 | 35.3 |
| SIVagmGRI | 52.8 | 53.9 | 26.6 | 25.5 | 37.8 | 36.9 |
| SIVcpzGAB | 50.7 | 54.5 | 25.4 | 27.4 | 44.9 | 33.6 |
| SIVcpzANT | 48.6 | 52.7 | 27.6 | 22.2 | 45.2 | 32.9 |
| SIVcol | 40 | 48.2 | 21.3 | 19 | 29.9 | 24 |
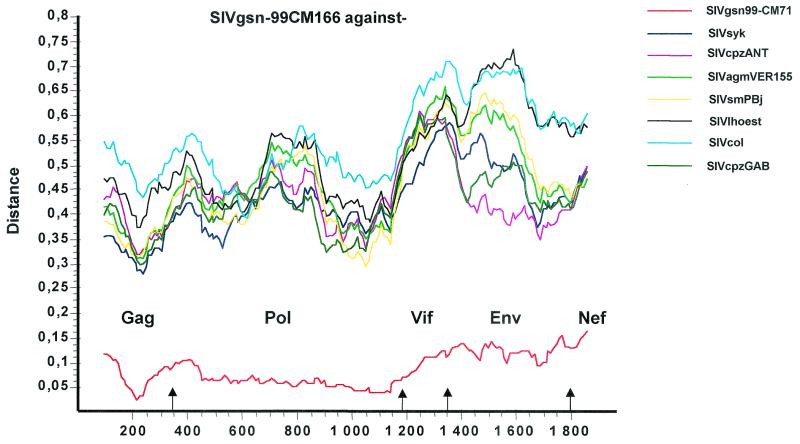
Diversity plots were then performed to further investigate the extent of sequence difference between SIVgsn and the other primate lentiviruses across the entire genome. A multiple alignment of concatenated predicted gene products was generated. Sites that could not be aligned unambiguously as well as sites containing a gap in any of the sequences were removed from the alignment in order to ensure that all comparisons were made across homologous positions. The percent sequence diversity between sequence pairs was then calculated for a window of 200 amino acids moved in steps of 10 amino acids along the alignment (Fig. 3). Table 1 and the diversity plot indicated that SIVgsn-99CM166 and SIVgsn-99CM71 were slightly more closely related to SIVsyk across the 5′ end of the genome and to SIVcpzANT in the envelope, with the latter similarity higher than the one observed with SIVsyk in the Gag-Pol region.
FIG. 3.
Diversity plot comparing SIVgsn-99CM166 with SIVgsn-99CM71 and representatives of the six major lineages of the primate lentiviruses, i.e., SIVcpz, SIVmnd, SIVsyk, SIVsm, SIVagm, and SIVcol. Protein sequence difference is plotted for windows of 200 amino acids moved in steps of 20.
These findings suggest that the SIVgsn genome has a mosaic structure which might result from a recombination event. To further investigate this mosaic structure, we performed bootscan analysis to define whether the different similarities observed in the similarity plots were supported by significant bootstrap values and to locate the putative recombination breakpoints. We therefore constructed a concatemer nucleotide alignment of the nonoverlapping regions of gag, pol, vif, env, and nef, including SIV strains representative of the six SIV lineages, SIVmnd2, SIVdeb from a De Brazza monkey (Cercopithecus neglectus) from Cameroon (unpublished), plus the two new SIVgsn strains. We tested the presence of saturation at different codon positions by comparing the half-saturation index expected with the observed saturation index.
As shown in Table 2, saturation always occurred at the third codon position for the five genes, while the first and second codon positions did not show evidence of saturation (gag, pol, and env) or borderline saturation (vif and nef). This implies that the first plus second codon position alignment was phylogenetically informative, and therefore, for the bootscan and subsequent analyses, we used only the first plus second codon position alignment. The bootscan analysis was performed by grouping the sequences per lineage, with a sliding window of 500 nucleotides moved in steps of 20 nucleotides, and 1,000 bootstrap replicates.
TABLE 2.
Saturation indicesa
| Gene | First plus second codon positions
|
Third codon position
|
||
|---|---|---|---|---|
| Half of expected saturation index | Observed saturation index (range) | Half of expected saturation index | Observed saturation index (range) | |
| gag | 0.96 | 0.64 (0.59-0.68) | 0.94 | 1.29 (1.24-1.34) |
| pol | 0.96 | 0.62 (0.59-0.65) | 0.90 | 1.22 (1.18-1.26) |
| vif | 0.97 | 1.08 (1.02-1.15) | 0.90 | 1.48 (1.38-1.59) |
| env | 0.96 | 0.88 (0.84-0.92) | 0.95 | 1.42 (1.39-1.46) |
| nef | 0.91 | 0.93 (0.85-1.01) | 0.94 | 0.94 (0.81-1.07) |
| vpu (all positions) | 0.83 | 0.84-1.16 | ||
Saturation indices were calculated with the program DAMBE at different codon positions on the concatenated nucleotide alignment for the nonoverlapping gene sequences, including representatives of the six major SIV lineages plus the SIV gsn-99CM71 and SIVgsn-99CM166 strains. Saturation positions for the different genes with a saturation index exceeding half of the expected saturation index are underlined.
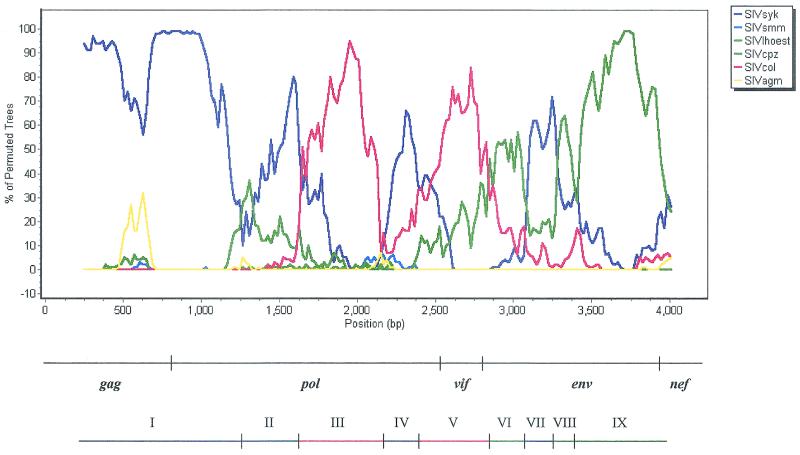
The bootscan plot of SIVgsn-99CM166 against representatives of the six major SIV lineages is shown in Fig. 4; the bootscan of SIVgsn-99CM71 gave similar results (data not shown). The bootscan analysis was also performed with a single sequence for each lineage, and similar results were obtained. In the bootscan analysis, only two fragments were significantly supported, one in the gag-pol region with SIVsyk and a second one in the env region with SIVcpz. The gap-stripped concatenated alignment of nucleotide sequences was then divided into nine regions according to different (significant as well as nonsignificant) lineage affinities (or putative recombinant breakpoints) revealed by the bootscanning profile (Fig. 4). Then we performed a likelihood mapping analysis implemented in the program Tree-Puzzle (37) in order to know the phylogenetic information contained in the first plus second codon position alignment of each region with different lineage affinities (putative recombinant fragment).
FIG. 4.
Bootscanning of SIVgsn-99CM166 against the six major SIV lineages. Bootscan analysis was performed on the concatenated nucleotide alignment with the nonoverlapping regions of gag, pol, env, vif, and nef with strains SIVsyk, SIVsun, SIVlhoest, SIVcpzUS, SIVcpzGAB, SIVcpzANT, SIVagmVER155, SIVagmTAN1, SIVmnd1, SIVsmH4, and SIVcol and by grouping them by lineage. Since saturation always occurred at the third codon position for all genes (see Table 2), the bootscan analysis was performed on first plus second codon positions with a sliding window of 500 nucleotides and a 20-nucleotide step increment and 1,000 bootstrap replicates. Below the graph, the bar at the top locates the gene regions, and the colored bar indicates the position of the putative recombination breakpoints in the SIVgsn genome.
The likelihood-mapping method identified in fragment I (region gag-pol) that 95.4% of quartets support the clustering of SIVgsn with SIVsyk and, for fragment IX (env), that 90.7% of quartets support the clustering of SIVgsn with SIVcpz. In fragment III, the phylogenetic signal was also significant, but no clear lineage affinity with any of the known lineages could be attributed. For the middle part of the genome, corresponding to fragments II to VIII, no clear assignment with any SIV strain has been found.
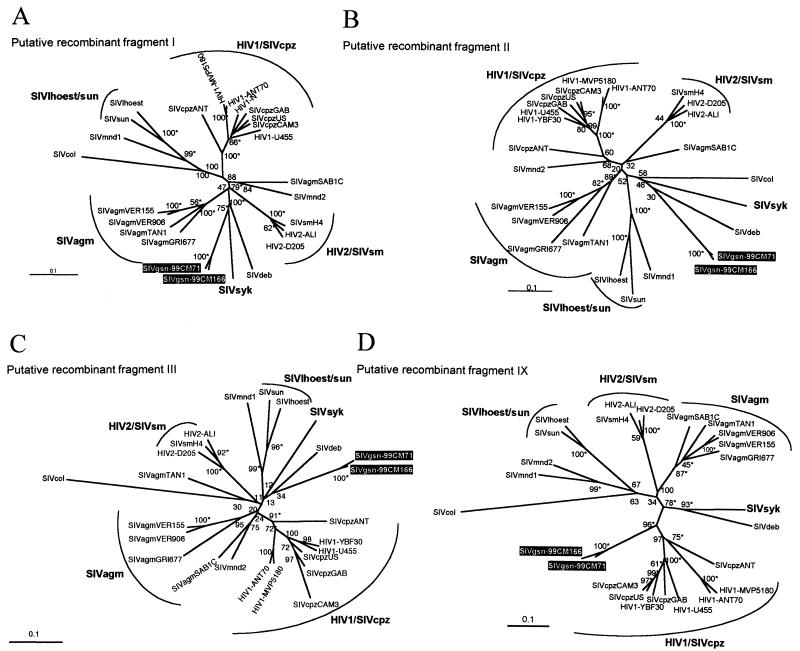
Neighbor-joining and maximum-likelihood tree topologies were performed on putative recombinant fragments I, II, III, and IX (Fig. 5A to D); fragments IV to VIII were too short to do reliable phylogenetic analysis, and Tree-Puzzle indicated a very low tree-like signal in these regions, suggesting a star-like evolution (Table 3). The best-fitting nucleotide substitution model for each fragment was evaluated with the Modeltest program (31). The model of evolution selected and the parameters used for each putative recombinant fragment alignment are shown in Table 4. As expected, the phylogenetic analysis of the different fragments revealed discordant topologies depending on the region of the genome analyzed. The phylogenetic analyses of fragment I showed that SIVgsn-99CM71 and SIVgsn-99CM166 clustered significantly with SIVsyk and SIVdeb (Fig. 5A), but in fragments II and III (Fig. 5B and C), SIVgsn-99CM71 and SIVgsn-99CM166 still clustered with SIVsyk or SIVdeb but not significantly (low bootstrap values). It is likely that in the central regions, including fragments IV to VIII, SIVgsn formed its own lineage. Finally, in the Env fragment IX, SIVgsn-99CM71 and SIVgsn-99CM166 clustered significantly with the SIVcpz lineage, branching off before SIVcpzANT (Fig. 5D).
FIG. 5.
Unrooted neighbor-joining trees of fragments I, II, III, and IX (see Table 2 and Fig. 4), including 23 SIV/HIV strains of the six major known SIV lineages and the new SIVgsn-99CM166 and SIVgsn-99CM71 isolates (boxed in black). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacement per site. Numbers along the branches represent percentage of bootstrap replicates (out of 1,000) for which the sequences at one end of the branch are monophyletic in the neighbor-joining tree. The maximum-likelihood method gives a similar tree topology: an asterisk along a branch indicates that the branch has a P value of <0.001 in the maximum-likelihood analysis. The trees were inferred with first plus second codon position with the TN plus Inv plus G, HKY plus Inv plus G, HKY plus G, or GTR plus G model (see Table 4).
TABLE 3.
Likelihood mapping analysis of the 4,845 possible groups of four sequences (quartets) for the putative recombinant fragments according to the SIVgsn-99CM71 genome identified on the bootscan analysis
| Fragment | Nucleotide positionsa | Gene region | Phylogenetic signalb (%) | Assignmentc |
|---|---|---|---|---|
| I | 455-2754 (1,250) | gag-pol | 95.4 | SIVsyk |
| II | 2755-3330 (380) | pol | 84.2 | N.A. |
| III | 3331-4101 (500) | pol | 92.1 | N.A. |
| IV | 4102-4536 (290) | pol | 78.3 | N.A. |
| V | 4537-5464 (360) | pol-vif | 72.9 | N.A. |
| VI | 6147-6761 (280) | env | 76.7 | N.A. |
| VII | 6762-7076 (189) | env | 81.4 | N.A. |
| VIII | 7077-7454 (160) | env | 70.8 | N.A. |
| IX | 7455-8360 (544) | env | 90.7 | SIVcpz |
| X | 8361-9255 (316) | env-nef | 70.7 | N.A. |
The number of nucleotides effectively used in the phylogenetic analysis after excluding gaps and third codon positions from the alignment is given in parentheses. The putative recombinant breakpoints were inferred from the bootscanning profile in Fig. 4.
Values indicate the percentage of quartets that are compatible with tree-like phylogeny (phylogenetic signal) in the likelihood maps obtained with Tree-Puzzle (37). Values of >90% are in boldface. Only first plus second codon positions were used in the likelihood mapping analysis because the third positions of each gene are always saturated (see Table 2).
Deduced from the bootscan analysis. N.A., not assignable.
TABLE 4.
Best-fitting nucleotide substitution model for putative recombinant fragments identified in the bootscan of the SIVgsn-99CM71 and SIVgsn-99CM166 strains against the known SIV lineages
| Putative recombinant fragmenta | Model selectedb | Ti/Tvc | αd | fie |
|---|---|---|---|---|
| I | TN+Inv+G | 1.04 (0.65) | 1.4 | 0.26 |
| II | HKY+Inv+G | 0.88 | 2.05 | 0.24 |
| III | HKY+G | 0.81 | 0.51 | |
| IV-VIIIf | TN+Inv+G | 0.81 (0.52) | 1.66 | 0.18 |
| IX | GTR+G | 2.3492 | 0.94 | |
| 2.3389 | ||||
| 0.9659 | ||||
| 1.6196 | ||||
| 2.2660 | ||||
| 1.0000 |
The putative recombinant fragments are those inferred from the bootscanning profile (see Fig. 4 and Table 3).
Best-fitting substitution models and their parameters were estimated through maximum likelihood with Modeltest v3.04 (31). TN, Tamura and Nei model; HKY, Hasegawa-Kishino-Yano model; GTR, general time reversible. Inv indicates models allowing for invariable sites. G indicates models allowing Γ-distributed rates across sites.
Ti/Tv, transition-transversion ratio. The pyrimidine transition-purine transition ratio is given for the Tamura and Nei model in parentheses. For the GTR model, the six different instantaneous substitution rates are shown.
Shape parameter α of the Γ distribution of the rates across sites estimated via maximum lilkelihood.
Fraction of invariable sites estimated via maximum likelihood.
Fragments IV to VIII were too short to perform a maximum-likelihood analysis. Because of their unclear assignment in the bootscanning, they were analyzed together.
Similarities between SIVgsn and SIVcpz in Vpu and Env.
Given the similarity observed in the diversity plot and phylogenetic analysis between SIVgsn and SIVcpz envelope proteins and that the vpu and env ORFs overlap, we investigated whether the acquisition of vpu (which is not present in SIVsyk) by SIVgsn might have occurred during a process of recombination. No reliable phylogenetic analyses could be performed on Vpu because of the extensive saturation observed at the first plus second and third codon positions (Table 2), and a DNA dot matrix of vpu sequences between SIVgsn-99CM166 and SIVcpzANT showed clearly that the similarity is too low to allow unambiguous alignment (data not shown). Despite the high variability observed among Vpu sequences, the EDS GNESXG(E/D) domain is highly conserved among HIV-1 M and O and SIVcpz (GAB1, US, and CAM3 strains). Noticeably, this domain is absent in SIVcpzANT and our newly described SIVgsn. On the other hand, a WDINDL motif, located at the C terminus of the protein, is conserved in HIV-1 group M and corresponds to a GFA/DNP motif located at the same position in HIV-1 group O and SIVcpz isolates. In SIVgsn, it is the latter motif which we have found.
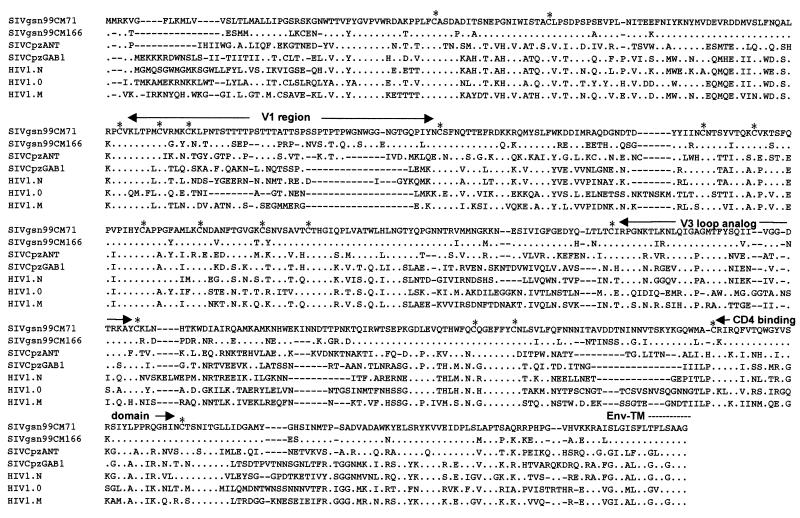
Figure 6 shows an alignment of the gp120 sequences of SIVgsn-99CM71, SIVgsn-99CM166, SIVcpzANT, SIVcpzGAB, and HIV-1 groups M, O, and N. The amino acid sequences corresponding to the surface unit of the envelope protein of SIVgsn show a structure similar to that of the previously described SIV gp120. A high level of conservation between SIVgsn-99CM166 and SIVgsn-99CM71 Env sequences was observed, as illustrated by 18 conserved cysteines, 19 conserved N-linked glycosylation sites, and only four and two amino acid changes in the V3 loop analog and the CD4 binding domain, respectively. When comparing the two SIVgsn Env sequences with SIVcpzANT, for example, conservation of the same 18 cysteine residues and 11 N-linked glycosylation sites was observed. A remarkable conservation is also seen in the V3 loop. In the central part of the loop, SIVgsn contains the QIGAGMTFYS motif, which is almost identical to the QIGPGMTFYN motif found in the SIVcpzANT V3 sequence and is known to be highly conserved among other SIVcpz isolates and the YBF30 HIV-1 N strain. The V3 loop sequences from SIVgsn were overall more related to that from SIVcpzANT than to that from other SIVcpz isolates (10, 14, 15, and 16 amino acid differences with SIVcpzANT, -GAB, -CAM3, and -US, respectively), explaining why we observed a reactivity against the SIVcpzANT V3 loop peptide in the V3-based peptide assay. Consistent with the bootscan analyses, and given that the V3 loop is not a hypervariable region in SIVcpz isolates as it is in HIV-1, this similarity could indicate a common origin for the envelopes of these two viruses or that SIVgsn and SIVcpz have a common ancestor, at least for the env gene, in the not so distant past.
FIG. 6.
Alignment of gp120 of SIVgsn-99CM71, SIVgsn-99CM166, SIVcpzANT, SIVcpzGAB, HIV1-YBF30, HIV1-ANT70, and HIV1-U455. The amino acid sequence of SIVgsn-99CM71 is shown on the top, with variable regions analogous to those observed in previously described SIVs indicated. The CD4 binding domain is shown. An asterisk represents a conserved cysteine residue. Dots indicate amino acid identity at a residue, and dashes indicate gaps introduced to optimize alignment.
DISCUSSION
Our understanding of the complex picture of primate lentiviral evolution gains from characterization of novel SIVs. In this study, we describe a new primate lentivirus (designated SIVgsn) that naturally infects greater spot-nosed monkeys (C. nictitans) in Cameroon. Among the six known SIV lineages, only two are derived from Cercopithecus species, the SIVsyk lineage and the SIVlhoest/SIVsun lineage. Together with SIVsyk, SIVgsn, described here, represents the second lentivirus isolated from a monkey belonging to the C. mitis group or superspecies of the Cercopithecus genus. However, despite the close phylogenetic relationship of their hosts, the molecular characterization of the two full-length SIVgsn sequences revealed that SIVgsn is highly divergent from SIVsyk.
A unique feature that distinguishes SIVgsn from SIVsyk and from all the other SIVs isolated previously from lower nonhuman primate species is the presence of a vpu gene in their genome. So far, this gene has been found only in lentiviruses isolated from chimpanzees and humans, i.e., the SIVcpz/HIV-1 lineage. Detailed phylogenetic analyses of different regions of the genome with known SIV lineages indicated that SIVgsn is most likely a mosaic of sequences with different evolutionary histories and which presented the following characteristics. SIVgsn was related to the SIVsyk lineage in Gag and part of Pol and had a similar TAR secondary structure; it was related to the SIVcpz lineage in Env, but the central part of its genome did not cluster significantly with any known SIV lineage, indicating a species-specific lineage for this portion of the genome. SIVgsn is therefore the first and only known SIV isolated from a lower monkey species to have a vpu gene and significant sequence homology with the SIVcpz lineage in the env gene.
Retroviruses are known to be highly recombinogenic (20, 45), and HIV-1, HIV-2, and SIV are all quite prone to recombination. Indeed, several examples of mosaic genomes have been described within HIV-1, HIV-2 (13, 14), SIVsm (7), and SIVlhoest and SIVagm (2). There is now evidence for recombination between diverse SIV lineages in at least four nonhuman primate species, and recombinant lentiviruses have been already identified in sabaeus monkeys (SIVsab), red-capped mangabeys (SIVrcm), mandrills (SIVmnd2), and drills (SIVdrl) (4, 15, 22, 35). All that indicates that cross-species transmissions and recombinations have existed since the beginning of the evolution of primate lentiviruses. Important to note is that identification of “pure lineages” or “recombinants” is mainly a function of chronological findings. Thus, if one virus previously determined to be “pure” has actually been derived from early recombinations between ancestral viruses, “parental” viruses characterized afterward will be identified as recombinants. With this regard, our observations suggest that SIVgsn is a complex recombinant, likely resulting from ancient recombination events involving ancestors of the current SIVsyk and SIVcpz viruses.
The habitats of the two subspecies of chimpanzees infected by SIVcpz (Pan troglodytes troglodytes and Pan troglodytes schweinfurthii) (12) have overlapping geographic ranges with greater spot-nosed monkeys and other monkey species, potentially allowing recombination between coinfecting viruses. Based on the behavior of these primates, it seems more plausible that chimpanzees acquired SIV infection from other species, because they are known to hunt and eat several small monkeys (6, 36, 42). Thus, this characterization of SIVgsn as a putative recombinant can be envisaged otherwise.
An alternative hypothesis may be proposed which is supported by the following observations. In the envelope tree presented in Fig. 5D, SIVgsn clusters with the SIVcpz/HIV-1 lineage; it branches off before SIVcpzANT, which itself is an outgroup of the SIVcpz/HIV-1 lineage. When considering this envelope tree without the SIVcpz/HIV-1 lineage, SIVgsn clusters then more tightly with SIVsyk/SIVdeb, in a pattern similar to that observed with the gag-pol tree (Fig. 5A). These observations were also confirmed by a diversity plot analysis performed without the SIVcpz lineage (data not shown). Therefore, it cannot be excluded that SIVgsn is a pure lineage related to SIVsyk or a recombinant between vpu-harboring SIVs related to SIVsyk. In this case, it remains possible that vpu was acquired during the evolution of SIVgsn or from another SIV from monkeys belonging to the C. mitis group or the Cercopithecus genus sharing the same habitat rather than resulting from recombination between a C. mitis SIV and SIVcpz. This acquisition of a vpu gene may have conferred a selective advantage on SIVgsn, which was then successfully spread into the greater spot-nosed monkey population. Screening and characterization of SIVs from other monkeys from the Cercopithecus genus will allow us to establish whether, among lower nonhuman primate species, Vpu is unique to SIVgsn or not. It will also be important to study the functionality of this protein.
If we assume that SIVgsn is a pure lineage, that implies that SIVcpz occurred by recombination rather than being a pure SIV lineage. Since the current prevalence of SIVcpz infection among chimpanzees seems low in captivity as well as in their natural habitat in Africa (9, 30, 33), it has been suggested that another, as yet unidentified primate species could be the natural host for SIVcpz. Obviously, SIVgsn is not the immediate SIVcpz progenitor, since the close relationship of SIVcpz and SIVgsn is restricted to Env. But given that chimpanzees, as mentioned above, are more susceptible to exposure to infection from other species, it is also conceivable that chimpanzees have been coinfected by different viruses and that this recombinant virus then successfully spread among chimpanzees and therefore in humans.
Among all the species known so far to harbor an SIV, none carries an SIV that is closely related to SIVcpz across the entire genome. According to our seroprevalence survey of wild-born monkeys in Cameroon, the partial characterization of two new viruses from the Cercopithecus genus (C. mona and C. cephus) indicates that they are not related to SIVcpz in the pol gene (29). The recent identification of SIVrcm and SIVmnd2, which are closely related to SIVcpz in pol, and the present identification of SIVgsn, which is closely related to SIVcpz in env, support our hypothesis that the present SIVcpz may have been generated by recombination between ancestral SIVs from the Cercopithecinae family. According to that, it is important to mention that Old World monkeys are the most numerous and the most geographically dispersed primates in Africa as well as the most commonly SIV infected. Furthermore, bootscan analysis of SIVcpzANT against representatives of the six major SIV lineages, including SIVmnd2 and SIVgsn, is more consistent with this hypothesis.
Chimpanzees are a natural host of SIVcpz in the wild; otherwise, members of this group of viruses would not be found throughout equatorial Africa (33). The questions are when, where, and how chimpanzees became infected and with which and how many species. Additional characterizations of SIVs from other monkeys are needed to answer these questions.
Since Sykes' monkeys and greater spot-nosed monkeys belong to the C. mitis group but live in geographically nonoverlapping regions in east central and west central Africa, respectively, our results may also suggest an ancient host-dependent evolution among viruses infecting monkeys from the C. mitis group. Thus, the amount of divergence found between SIVsyk and SIVgsn might reflect coevolution of virus and host within hosts geographically isolated for an extended period of time. Other examples of SIV host-dependent evolution have been described for the related SIVagm, present in the Chlorocebus genus, and SIVlhoest and SIVsun, isolated from the l'Hoest superspecies (1, 3, 22, 28). Screening of other members of the C. mitis group, such as blue monkeys, silver monkeys, and golden monkeys, which all inhabit different geographic regions of Africa, should allow us to investigate the hypothesis that ancestral SIVs infected ancestral monkeys before the speciation events occurred within the C. mitis group.
The factors that determine the capacity of the viruses to spread in humans have not been fully elucidated. Newly described viruses such as SIVlhoest, SIVrcm, and SIVmnd2 replicated efficiently in human PBMCs and therefore pose a potential health risk to humans. SIVsyk does not replicate in PBMCs from humans and chimpanzees; hence, it will be interesting to study the ability of a molecular infectious clone of SIVgsn to replicate in chimpanzee and human PBMCs, given that Env of SIVgsn and in particular the V3 loop are closely related to SIVcpz. Furthermore, the only SIVs actually known to have been transmitted to humans carried either a vpu or a vpx gene. In Cameroon, greater spot-nosed monkeys along with other monkeys are regularly hunted by the human population, and the risks for other zoonoses cannot be excluded (29).
Acknowledgments
We thank Christelle Butel and Florian Liegeois for technical assistance and Marc Sitbon for comments and critical reading of the manuscript.
This work was supported in part by grants from the National Institutes of Health (RO1 AI 50529) and the Agence National de Recherche sur le SIDA (ANRS). This work was partially supported by the Flemish Fonds voor Wetenschappelijk onderzoek (FWO grants G.0288.01 and KAN2002 1.5.193.02, Postodoctoraal Onderzoeker contract 530).
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. S. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. [DOI] [PubMed] [Google Scholar]
- 6.Boesch, C., and H. Boesch-Acherman. 1998. Dim forest, bright chimps, p. 74-79. In R. L. Ciochon and R. A. Nisbett (ed.), The primate anthology—essays on primate behaviour, ecology and conservation from natural history. Prentice Hall, Princeton, N.J.
- 7.Chen, Z., P. Telfier, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewley, J. P., J. C. Lewis, D. W. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbet, S., M. C. Muller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP: Phylogenetic Inference Package, version 3.5c. Department of Genetics, University of Washington, Seattle, Wash.
- 12.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 13.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for nonsubtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus lhoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., G. A. Dapolito, S. Goldstein, H. McClure, P. Emau, P. N. Fultz, M. Isahakia, R. Lenroot, G. Myers, and P. R. Johnson. 1993. A distinct African lentivirus from Sykes' monkeys. J. Virol. 67:1517-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 20.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 21.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 22.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 68:8454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 25.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 26.McCormick-Davis, C., S. B. Dalton, D. K. Singh, and E. B. Stephens. 2000. Comparison of Vpu sequences from diverse geographical isolates of HIV type 1 identifies the presence of highly variable domains, additional invariant amino acids, and a signature sequence motif common to subtype C isolates. AIDS Res. Hum. Retrovir. 16:1089-1095. [DOI] [PubMed] [Google Scholar]
- 27.Miura, T., J. Sakuragi, M. Kawamura, M. Fukasawa, E. N. Moriyama, T. Gojobori, K. Ishikawa, J. A. Mingle, V. B. Nettey, H. Akari, et al. 1990. Establishment of a phylogenetic survey system for AIDS-related lentiviruses and demonstration of a new HIV-2 subgroup. AIDS 4:1257-1261. [DOI] [PubMed] [Google Scholar]
- 28.Muller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. Herve, J. P. Durand, P. Legal-Campodonico, M. C. Lang, J. P. Digoutte, A. J. Georges, et al. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeters, M., C. Honore, T. Huet, L. Bedjabaga, S. Ossari, P. Bussi, R. W. Cooper, and E. Delaporte. 1989. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3:625-630. [DOI] [PubMed] [Google Scholar]
- 31.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 32.Ray, S. 1999. SIMPLOT version 2.5 software. Distributed by the author at http://sray.med.som.jhmi.edu/RaySoft/Simplot.
- 33.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465.. [DOI] [PubMed] [Google Scholar]
- 34.Sharp, P. M., E. Bailes, R. R. Chaudhuri, C. M. Rodenburg, M. O. Santiago, and B. H. Hahn. 2001. The origins of acquired immune deficiency syndrome viruses: where and when? Phil. Trans. R. Soc. Lond. B Biol. Sci. 356:867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanford, C. 1998. To catch a colobus, p. 84-87. In R. L. Ciochon and R. A. Nisbett (ed.), The primate anthology—essays on primate behaviour, ecology and conservation from natural history. Prentice Hall, Princeton, N.J.
- 37.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swofford, D. L. 1998. PAUP*: phylogenetic analysis with parsimony (* and other methods), version 4.0b5. Sinauer Associates, Sunderland, Mass.
- 39.Takehisa, J., Y. Harada, N. Ndembi, I. Mboudjeka, Y. Taniguchi, C. Ngansop, S. Kuate, L. Zekeng, K. Ibuki, T. Shimada, B. Bikandou, Y. Yamaguchi-Kabata, T. Miura, M. Ikeda, H. Ichimura, L. Kaptue, and M. Hayami. 2001. Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 17:1143-1154. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 11:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 42.Tutin, C. E. 2000. Ecology and social organization of African tropical forest primates: aid in understanding retrovirus transmission. Bull. Soc. Pathol. Exot. 93:157-161. [PubMed] [Google Scholar]
- 43.Vanden Haesevelde, M. M., M. Peeters, G. Jannes, W. Janssens, G. van der Groen, P. M. Sharp, and E. Saman. 1996. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology 221:346-350. [DOI] [PubMed] [Google Scholar]
- 44.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 79:1809-1814. [DOI] [PubMed] [Google Scholar]
- 45.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 46.Xia, X., M. Salemi, Z. Xie, C. Lu, and Y. Wang. An index of substitution saturation and its applications. Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 47.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 48.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. NATO ASI Series. Kluwer Academic Publishers, Dordrecht, The Netherlands.