Abstract
Envelope glycoprotein M (gM) and the complex formed by glycoproteins E (gE) and I (gI) are involved in the secondary envelopment of pseudorabies virus (PrV) particles in the cytoplasm of infected cells. In the absence of the gE-gI complex and gM, envelopment is blocked and capsids surrounded by tegument proteins accumulate in the cytoplasm (A. R. Brack, J. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 73:5364-5372, 1999). Here we demonstrate by yeast two-hybrid analyses that the cytoplasmic domains of gE and gM specifically interact with the C-terminal part of the UL49 gene product of PrV, which represents a major tegument protein and which is homologous to VP22 of herpes simplex virus type 1. However, deletion of the UL49 gene from PrV had only minor effects on viral replication, and ultrastructural analyses of infected cells confirmed that virus maturation and egress, including secondary envelopment in the cytoplasm, were not detectably affected by the absence of UL49. Moreover, the UL49 gene product was shown to be dispensable for virion localization of gE and gM, and mutants lacking either gE or gM incorporated the UL49 protein efficiently into virus particles. In contrast, a PrV mutant with deletions of gE-gI and gM failed to incorporate the UL49 protein despite apparently unaltered intracytoplasmic UL49 expression. In summary, we describe specific interactions between herpesvirus envelope and tegument proteins which may play a role in secondary envelopment during herpesvirus virion maturation.
Pseudorabies virus (PrV; suid herpesvirus 1), the causative agent of Aujeszky's disease in pigs (41), has been classified as a member of the genus Varicellovirus within the subfamily Alphaherpesvirinae of the family Herpesviridae (51). All herpesvirus particles consist of an icosahedral capsid which encloses the double-stranded DNA genome and which in turn is surrounded by a tegument layer containing numerous viral gene products and a lipid envelope of cellular origin containing mostly glycosylated virus-encoded proteins (42, 51). For herpes simplex virus types 1 and 2 (HSV-1 and HSV-2, respectively), more than 10 different envelope proteins have been described and more than 15 viral gene products may be components of the tegument (42, 56). Whether this bulk of proteins is acquired during the budding of newly formed nucleocapsids at the inner nuclear membrane or during a secondary envelopment step in the trans-Golgi region of the cytoplasm has been a subject of research and debate, but recent genetic, biochemical, and ultrastructural studies of different alpha- and betaherpesviruses clearly support the secondary envelopment pathway (reviewed in references 16 and 42).
Most proteins required for viral DNA replication and capsid formation are highly conserved among all herpesviruses (23, 38). Similarly, homologs of the UL31 and UL34 gene products of HSV-1 and PrV, which physically interact with each other and which participate in virus egress from the nucleus (17, 32, 50), are found in all herpesvirus subfamilies. In contrast, of the envelope glycoproteins, only glycoproteins B, H, L, M, and N (gB, gH, gL, gM, and gN, respectively) are found in alpha-, beta- and gammaherpesviruses (56); among the genes encoding tegument proteins, the UL36 and UL37 homologs are conserved in all herpesvirus subfamilies (38). In HSV-1 and PrV, the UL36 and UL37 proteins are required for efficient virion formation (10, 11, 33) and presumably form the innermost layer of the tegument at the surface of the nucleocapsid (34, 66). However, many of the nonconserved tegument proteins of alphaherpesviruses play important roles during different steps of the viral replication cycle, such as the shutoff of host cell functions by the vhs protein encoded by UL41 (35, 53) and the transcriptional activation of viral immediate-early genes by the UL48 gene product VP16 (1). In addition, VP16 has been shown to be required for efficient virus egress (43). The tegument proteins VP11/12, VP13/14, and VP22, encoded by the adjacent UL46, UL47, and UL49 genes of HSV-1, have been shown to directly interact with VP16 (14) or to modulate its trans-activating functions (64). In addition, the UL49 gene products of HSV-1 and bovine herpesvirus 1 (BHV-1) exhibit virus-independent intercellular trafficking of unknown biological function (15, 22). Despite their prominence in the tegument, recent studies with BHV-1 (36), PrV (9), and HSV-1 (48; G. Elliott and A. Whiteley, 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001) proved that the respective UL49 homologs are dispensable for productive viral replication.
Viral envelope glycoproteins play important roles during the entry and egress of herpesviruses (42, 54). Glycoprotein E (gE) and glycoprotein I (gI) of alphaherpesviruses form a noncovalently linked complex in all alphaherpesviruses investigated so far (24). They are dispensable for the replication of most alphaherpesviruses, except for Marek's disease virus (MDV) (52) and varicella-zoster virus (VZV) (7, 37, 62). In PrV, the gE-gI complex is relevant for neuroinvasion (reviewed in reference 16) but is dispensable for replication in other animal tissues or in cell cultures (reviewed in reference 41). gM, a multiple membrane-spanning protein which is present in a complex with gN (30), is conserved in all herpesvirus subfamilies, despite not being required for efficient viral replication in cultured cells. However, simultaneous deletion of gM and gE-gI resulted in a drastic impairment of PrV maturation and egress (5). Ultrastructural analyses of cells infected with this mutant showed that capsids were able to exit the nucleus, but secondary envelopment in the trans-Golgi region was almost completely abolished. Remarkably, the cytoplasmic nucleocapsids of the gE-gI-gM triple mutant formed aggregates with electron-dense tegument material containing, among other tegument proteins, the UL49 protein (4). This phenotype could be reproduced with a PrV mutant which, in addition to the absence of gM, lacked the cytoplasmic tail of gE (4). These results suggested that interactions between gE or gM and viral tegument proteins may be a prerequisite for secondary envelopment. However, specific interactions between herpesvirus envelope and tegument proteins which are relevant for virion morphogenesis have not yet been characterized.
To identify viral proteins capable of interactions with gE and gM, yeast two-hybrid studies were performed in which the cytoplasmic domain of either gE or gM was used as bait. Screening of expression libraries of the PrV genome revealed that the UL49 protein is able to interact with both glycoproteins. To ascertain the biological relevance of these interactions, a UL49 deletion mutant of PrV was constructed and characterized, and virions of this mutant were tested for the incorporation of gE and gM. Conversely, gE-gI and gM mutants of PrV were analyzed for virion localization of the UL49 protein.
MATERIALS AND METHODS
Plasmids, viruses, and cells.
The UL49 deletion mutant of PrV strain Kaplan (PrV-Ka) (31) was derived from the bacterial artificial chromosome (BAC) plasmid pPrV-K1 (17). To this end, the complete UL49 open reading frame (ORF) was amplified from cloned PrV DNA by PCR with Pfx DNA polymerase (Life Technologies) and primers PUL49-F (CCCACTCGCTCGCCATGTCCAG; initiation codon underlined) and PUL49-R (GCGGGGTGCCATTTGCAACG). The 787-bp PCR product was inserted into EcoRI-digested vector pcDNA3. The resulting plasmid, pcDNA-UL49 (Fig. 1B), was used for transient expression in transfected eucaryotic cells (19) as well as for in vitro transcription and translation (TNT-coupled reticulocyte lysate system; Promega). Furthermore, a radiolabeled antisense cRNA was transcribed (SP6/T7 transcription kit; Roche) and used as a probe for detection of the UL49 mRNA by Northern blot analysis of PrV-infected cells.
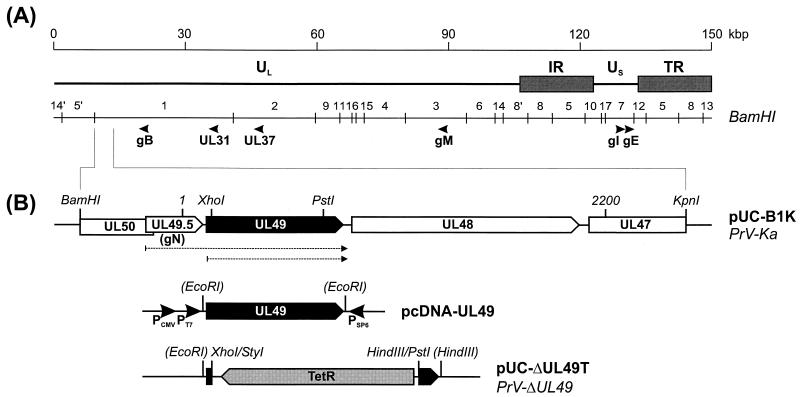
FIG. 1.
Structure of the PrV genome and construction of UL49 mutant viruses. (A) A schematic map of the PrV genome shows the long (UL) and short (US) unique regions, the inverted repeat sequences (IR and TR), the positions of BamHI restriction sites, and the coding regions of analyzed proteins. (B) Magnification of the BamHI-KpnI fragment encompassing the region under study. Pointed rectangles denote ORFs in transcriptional orientation. Relevant restriction sites and the extent of the sequenced region (nt 1 to 2200) are shown. Broken arrows indicate the UL49 and UL49.5-UL49 transcripts. Plasmid pcDNA-UL49 containing the SP6, T7, and human cytomegalovirus (CMV) immediate-early promoters was used for in vitro transcription and translation. Insertion of the tetracycline resistance gene (TetR) into a UL49 ORF with a partial deletion resulted in pUC-ΔUL49T and progeny virus PrV-ΔUL49.
For isolation of a UL49 deletion mutant, the PCR-amplified gene was inserted into EcoRI-HindIII-cleaved vector pUC19. Subsequently, a 632-bp XhoI-PstI fragment containing codons 4 to 214 of UL49 was replaced by the tetracycline resistance gene, which had been isolated as a 1,340-bp HindIII-StyI fragment from plasmid pBR322 (pUC-ΔUL49T; Fig. 1B). Noncompatible fragment ends were made blunt with the Klenow polymerase prior to ligation.
The mutated UL49 gene was amplified from pUC-ΔUL49T by PCR with primers PUL49-F and PUL49-R. The isolated PCR product was used for RecE- and RecT-mediated mutagenesis (65) of pPrV-K1 (17) in Escherichia coli as described previously (17, 52), except that recombinant bacteria were selected and propagated in medium containing 30 μg of chloramphenicol/ml and 10 μg of tetracycline/ml. This resulted in BAC vector pPrV-ΔUL49. Finally, the bacterial vector sequences were removed from the glycoprotein G (gG) gene locus by cotransfection of rabbit kidney (RK13) cells with pPrV-ΔUL49 and plasmid pU-6.3, which restores the authentic gG gene (17, 29), yielding virus mutant PrV-ΔUL49.
In addition, PrV strain Becker (PrV-Be) (2) and gE-gI (PrV-gE/I−) (40), gM (PrV-gM−) (12), or gE-gI-gM (PrV-gE/I/M−) (5) deletion mutants of PrV-Ka were used for this study. Viruses were propagated in porcine or RK13 cells or in gM-expressing RK13 cells (for PrV-gE/I/M−) (5) grown in minimum essential medium (Life Technologies) supplemented with 10% fetal calf serum. Extracellular virus particles were purified as described previously (32).
DNA sequencing.
From genomic BamHI fragment 1 of PrV-Ka, a 3,609-bp BamHI-KpnI fragment (Fig. 1) was subcloned into pUC19 (pUC-B1K). After digestion with suitable restriction enzymes, nested sets of deletion plasmids were generated (nested deletion kit; Amersham Pharmacia Biotech). DNA sequences of both strands of the insert fragment were determined with vector-specific M13 forward and reverse primers as described previously (32). DNA sequences were assembled and analyzed with the Wisconsin software package (Genetics Computer Group) and Unix version 10.2.
Western blot analysis.
For Western blot analyses of whole-cell lysates, RK13 cells were infected with PrV at a multiplicity of infection (MOI) of 2 and incubated for 16 h at 37°C. For PrV-gE/I/M−, virus stocks which had been grown on gM-expressing transcomplementing cells were used for infection. For virus purification, porcine cells were infected at an MOI of 0.1, and virions were isolated by sucrose gradient centrifugation as described previously (32). Samples were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and electrotransferred to nitrocellulose membranes (Schleicher & Schuell). Blots were blocked with 5% low-fat milk in phosphate-buffered saline and incubated for 1 h with monospecific polyclonal rabbit sera directed against UL49 (dilution, 1:105) (4), UL37 (dilution, 1:105) (33), UL31 (dilution, 1:105) (17), gN (dilution, 1:5,000) (28), gM (dilution, 1:5 × 105) (12), or the gE cytoplasmic tail (58) or with monoclonal antibodies against PrV gB (dilution, 1:100) (45). Bound antibodies were detected with peroxidase-conjugated secondary antibodies (Dianova) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
Electron microscopy.
RK13 cells were infected with PrV-ΔUL49 at an MOI of 1 and incubated for 14 h at 37°C. Fixation, embedding, and staining were done as described previously (32). Ultrathin sections were examined with an electron microscope (Philips 400T).
Yeast two-hybrid analyses.
Interactions between PrV proteins were investigated with two different yeast two-hybrid systems using the DNA binding domain (BD) of LexA (21) (distributed by Clontech) or Gal4 (25). To generate an expression library for the latter system, DNA from purified virions of PrV-Be was partially digested with MspI, and fragments ranging from 350 to 1,200 bp were isolated from agarose gels. The isolated DNA was ligated to ClaI-digested vectors pGAD-C1, pGAD-C2, and pGAD-C3, which permit the expression of fusion proteins with the Gal4 transactivation domain (AD) in all three translational frames. After electroshock transformation of E. coli DH10B cells, a library of ca. 2 × 104 plasmid clones of PrV genome fragments was obtained and further amplified. The creation of a similar PrV expression library for the LexA yeast two-hybrid system has been described elsewhere (17).
As bait, the 3′-terminal parts of the UL49, gE, gI, gM, and gN genes were expressed as fusion proteins with the Gal4 or LexA DNA BD in plasmid pgBDU-C2 (25) or pLexA (21), respectively. For that purpose, oligonucleotide primers were deduced from published DNA sequences to amplify the desired gene fragments by PCR, and suitable restriction sites were added to facilitate in-frame ligation with the vectors. The stable Gal4-inducible yeast reporter strain pJ69-4 (25) or the yeast strain EGY48 carrying the LexA-inducible reporter plasmid p8op-LacZ (Clontech) was subsequently transformed with the respective bait constructs and PrV expression libraries. Yeast clones exhibiting the amino acid and purine synthesis functions encoded by the plasmids or induced by interactions between the expressed DNA binding and transactivator fusion proteins were identified on selective agar plates and further tested for the induction of β-galactosidase activity by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining as described previously (17, 21, 25). The library plasmids of positive yeast clones were recloned in E. coli, again tested for specific two-hybrid interactions with the respective bait constructs, and characterized by DNA sequencing of the insert fragment with vector-specific primers.
Nucleotide sequence accession number.
The determined PrV-Ka DNA sequence was deposited in GenBank under accession number AJ437285.
RESULTS
Characterization of the UL49 gene of PrV-Ka.
The newly characterized genome fragment of PrV-Ka is 2,200 bp in size and closes a gap between two previously sequenced DNA fragments. Thus, nucleotides (nt) 1 to 127 correspond to the end of the UL49.5 (gN) gene sequence (28) (GenBank accession no. U38457), and nt 2072 to 2200 correspond to nt 1 to 129 of the UL47 and UL46 gene regions (6) (GenBank accession no. AJ010303). The DNA sequence contains two ORFs that encode homologs of the alphaherpesvirus UL49 and UL48 gene products. A comparison of the UL49 and UL48 homologs of PrV, equine herpesvirus 1 (EHV-1) (57), BHV-1 (36), VZV (8), and HSV-1 (38) is shown in Table 1. Database searches revealed no evidence for conservation of these genes in genomes of Betaherpesvirinae or Gammaherpesvirinae.
TABLE 1.
Properties of PrV UL48 and UL49 genes and gene products
| Gene | Nucleotide positionsa
|
mRNA size (kb)
|
Protein
|
% Related/% identical residues in homologous proteins from:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | TATA box | Poly(A) signal | Expected | Apparent | No. of amino acids | Mass (kDa)
|
||||||
| Expected | Apparent | EHV-1 | BHV-1 | VZV | HSV-1 | |||||||
| UL49 | 97-846 | 55-61 | 842-847 | >0.8 | 0.9 | 249 | 25.86 | 33 | 42.3/39.1 | 37.3/33.6 | 36.3/32.7 | 42.1/36.0 |
| UL48 | 910-2,151 | 838-844 | 6,759-6,764b | >5.9 | 5.6 | 413 | 45.11 | 53-57 | 60.6/52.5 | 57.3/51.0 | 59.7/49.4 | 46.6/40.8 |
The UL49 ORF of PrV is preceded by a putative TATA box element and followed by the polyadenylation signal consensus sequence AATAAA (Table 1). Northern blot analysis of total cellular RNA harvested 3 to 9 h after infection with PrV-Ka revealed increasing amounts of a UL49-specific transcript of 0.9 kb, which matches the calculated size (Table 1). The antisense cRNA probe of pcDNA-UL49 (Fig. 1B) detected an additional viral transcript of 1.3 kb which presumably represents the 3′-coterminal mRNA of the upstream UL49.5 gene (Fig. 1B).
The deduced UL49 protein of PrV-Ka contains 249 amino acids with a calculated molecular mass of ca. 26 kDa (Table 1). However, the electrophoretic mobility of the in vitro translation product of pcDNA-UL49 (data not shown) as well as of the UL49 protein in PrV-infected cells and virions (Fig. 2) indicates an apparent molecular mass of 33 kDa. At present it is unclear whether this discrepancy is due to posttranslational modifications of the UL49 gene product or to aberrant mobility in SDS-polyacrylamide gels. A fraction of the PrV UL49 protein was shown to be phosphorylated, but this modification had little effect on electrophoretic migration, and the nonphosphorylated form exclusively found in virions also had an apparent molecular mass of 33 kDa (9). A detailed characterization of the UL48 gene and protein is presented elsewhere (18), but basic properties are listed for comparison in Table 1.
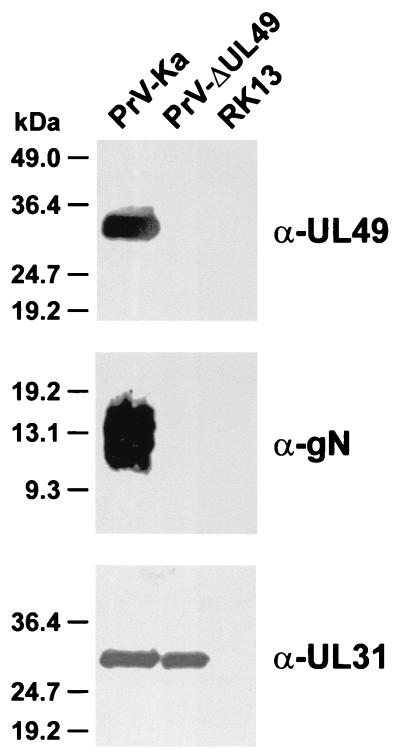
FIG. 2.
Protein expression of PrV-ΔUL49. Noninfected RK13 cells as well as RK13 cells infected with wild-type PrV-Ka or the UL49 deletion mutant PrV-ΔUL49 were analyzed by Western blotting with monospecific antisera against the UL49 protein, gN, or the UL31 protein.
Isolation and characterization of a UL49 deletion mutant of PrV-Ka.
The UL49 deletion mutant PrV-ΔUL49 has a major part of the UL49 ORF deleted and substituted by the tetracycline resistance gene (Fig. 1B). The deleted viral DNA fragment encompasses codons 4 to 214 of the UL49 ORF, and the remaining 3′ end lacks an in-frame initiation codon. Thus, no UL49 protein expression should be possible. However, the polyadenylation signal common to the UL49 and UL49.5 (gN) genes was preserved.
Western blot analyses of infected cell lysates confirmed that PrV-ΔUL49 failed to express the 33-kDa UL49 protein or truncated gene products (Fig. 2, α-UL49). Unexpectedly, the UL49.5 gene product gN was also not expressed at detectable levels (Fig. 2, α-gN), whereas the expression of other viral genes, e.g., UL31 (Fig. 2, α-UL31), was not affected. In Northern blot analyses of PrV-ΔUL49-infected cells, no stable UL49.5-specific transcripts were detectable (data not shown). Presumably, this result is a consequence of the foreign gene insertion at the coterminally transcribed downstream UL49 locus (Fig. 1B).
A previous study showed that deletion of the gN gene does not significantly affect the replication of PrV in cultured cells (30), and PrV-ΔUL49, which does not express the UL49 and gN genes, also did not exhibit significant defects in either plaque formation or one-step growth (data not shown); these data correlate with previous results (9). Ultrastructural analyses of infected RK13 cells also demonstrated that maturation and egress of PrV-ΔUL49 were not detectably affected (Fig. 3). Similar to what is seen for virion maturation in cells infected with PrV-Ka (20), only a few perinuclear particles were detectable (Fig. 3B), and deenveloped cytoplasmic nucleocapsids were efficiently reenveloped (Fig. 3C and D) and released (Fig. 3E).
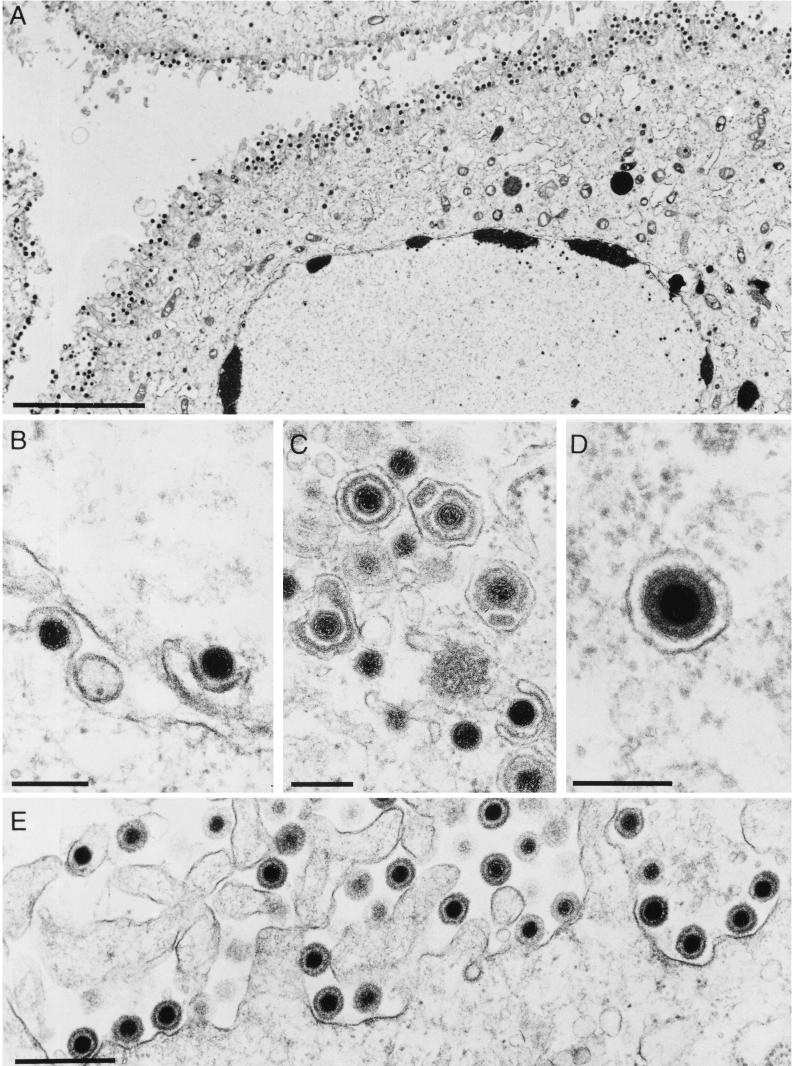
FIG. 3.
Electron microscopy of PrV-ΔUL49-infected cells. RK13 cells were infected with PrV-ΔUL49 at an MOI of 1 and analyzed 14 h postinfection. (A) Overall view of an infected cell with numerous extracellular virus particles lining the surface of the plasma membrane. (B) Nuclear egress, with primary envelopment by budding at the inner nuclear membrane and deenvelopment by fusion at the outer nuclear membrane. (C and D) Secondary envelopment in the trans-Golgi region by budding into vesicles (C) results in enveloped virions within cytoplasmic vesicles (D). (E) Larger magnification of extracellular complete virions at the plasma membrane. Bars, 5 μm in A, 200 nm in B to D, and 500 nm in E.
Yeast two-hybrid interactions of the UL49 gene product with gE and gM.
Secondary envelopment of PrV is blocked when gE-gI and gM are deleted simultaneously (4, 5). To identify possible interactions of these proteins with other viral gene products, yeast two-hybrid studies were performed. In a first set of experiments, DNA fragments encoding the cytoplasmic tail of either gE or gM were inserted into host shuttle plasmids that permitted the expression of fusion proteins with the LexA or Gal4 DNA BD in yeast cells (Fig. 4). Both proteins are components of glycoprotein complexes. Whereas gE forms a noncovalently linked complex with gI (63, 68), gM is disulfide linked to gN (30). Therefore, the cytoplasmic domains of gI and gN were also expressed as LexA BD fusion proteins (data not shown). During characterization of these expression plasmids, we found that the C-terminal domains of gI of PrV-Ka and gI of PrV strain Rice are different (47) (GenBank accession no. M14336). As a consequence of a frameshift after codon 313, the deduced gI of PrV-Ka, like that of a recently sequenced Asian PrV strain (S. B. Xiao et al., unpublished data; GenBank accession no. AF306511), is 366 amino acids long; the predicted strain Rice protein is 350 amino acids long. The C-terminal domains of gE are identical in PrV strain Rice (47) (GenBank accession no. M14336) and PrV-Be (T. del Rio and L. W. Enquist, personal communication), but the cytoplasmic tail of gE in PrV-Ka contains one amino acid substitution at position 499 (Val → Asp). As expected, the inserts of the gM and gN expression plasmids used specified the published sequences of the respective genes of PrV-Ka (12, 28).
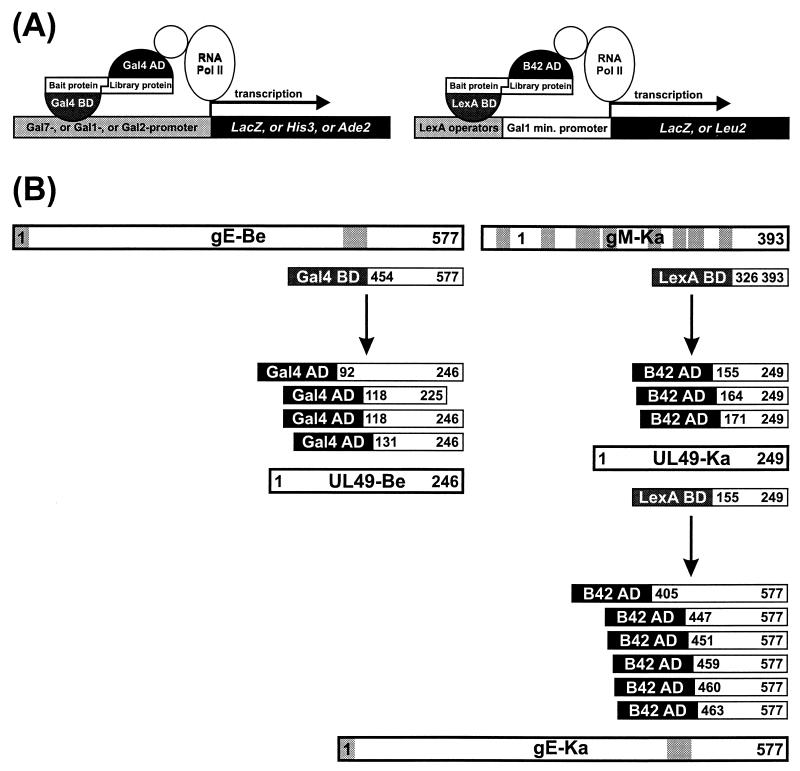
FIG. 4.
Yeast two-hybrid interactions between gE and gM and the UL49 protein. (A) Schematic depiction of the principles of the two different yeast two-hybrid systems used. min., minimal. (B) Observed interactions. Bars represent proteins with hydrophobic signal sequences (for gE) and transmembrane domains (for gE and gM), highlighted by grey shading. Also indicated are the different portions of gE, gM, and UL49 found in the interacting fusion proteins from the PrV expression libraries. Proteins from PrV-Be (gE-Be and UL49-Be) and PrV-Ka (gM-Ka, UL49-Ka, and gE-Ka) were analyzed.
The DNA binding bait constructs were used to screen expression libraries of randomly cleaved genomic PrV DNA fused to genes encoding transcriptional activators (B42 AD for PrV-Ka and Gal4 AD for PrV-Be; Fig. 4A). Interactions between bait and library proteins in yeast cells were detected by induction of selectable marker genes that permit leucine-, histidine-, or adenine-independent growth and by trans-activation of the lacZ reporter gene (Fig. 4A). Whereas no specific interactions of the C-terminal domains of gI and gN with other PrV proteins were detectable, several independent but overlapping in-frame expression clones which all contained the 3′-terminal part of the UL49 gene were selected from the PrV genome libraries with the LexA-gM and Gal4-gE bait proteins (Fig. 4B). After recloning of the identified library plasmids in E. coli, the specificity of the two-hybrid interactions was verified by cotransfection of yeast cells with individual plasmids. Only combinations of transcription-activating UL49 and DNA binding LexA-gM or Gal4-gE fusion constructs induced LacZ expression. Expression was abolished when one component was substituted by the corresponding vector plasmid (data not shown). However, expression of the C-terminal part of gE as a LexA fusion protein caused strong trans-activation of the marker genes in yeast cells, even in the absence of any B42 AD construct; therefore, no two-hybrid screening was possible in this system.
In a reciprocal set of experiments, the UL49 proteins of PrV-Ka and PrV-Be were recloned from the prey plasmids into bait vectors and expressed as LexA BD or Gal4 BD fusion proteins in yeast cells (Fig. 4B). The expression of PrV-Be UL49-Gal4 BD constructs yielded nonspecific activation, prohibiting useful library screening (data not shown). However, this result was not observed with PrV-Ka UL49-LexA BD constructs, and after library screening, six positive yeast clones were selected; all contained different but overlapping parts of the 3′ terminus of the gE gene fused in frame to the B42 AD gene sequence (Fig. 4B). Thus, physical interactions between the C-terminal parts of the UL49 protein and gE could be demonstrated in two independent library screens with either of the proteins as bait. However, interactions between the same parts of the UL49 protein and gM could not be confirmed in reciprocal experiments with BD-UL49 and AD-gM fusions. We hypothesize that proper folding of the bait-prey fusion protein complex is required to permit simultaneous DNA binding and transcriptional initiation. It is unlikely that the different results obtained with the UL49 fusion constructs of PrV-Be and PrV-Ka were caused by sequence variations within the viral gene, since all observed alterations were restricted to the N-terminal parts of the protein and did not affect the interacting C-terminal domains (data not shown).
Virion localization of gE, gM, and the UL49 protein.
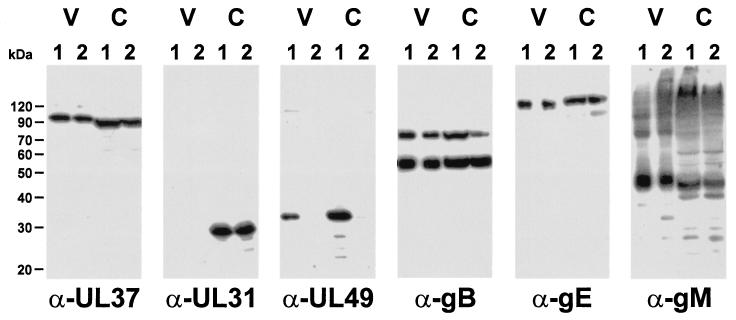
To investigate whether efficient incorporation of gE, gM, or other membrane proteins into the viral envelope depends on the presence of the UL49 protein, virions of PrV-Ka (Fig. 5, V, lanes 1) and PrV-ΔUL49 (Fig. 5, V, lanes 2) were purified from the supernatants of infected cells by sucrose gradient centrifugation. In addition, whole-cell lysates of cells infected with PrV-Ka (Fig. 5, C, lanes 1) or PrV-ΔUL49 (Fig. 5, C, lanes 2) were also analyzed. Equal amounts of protein were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analyses. Reactions of an antiserum against the tegument protein encoded by UL37 (Fig. 5, α-UL37) served as a loading control, and the absence of the UL31 protein, which is not a constituent of mature extracellular virions (17) (Fig. 5, α-UL31), verified the purity of the virion preparations. Both UL37 and UL31 proteins were present in the lysates of cells infected with either virus. As expected, the UL49 gene product (Fig. 5, α-UL49) was detectable in virions of PrV-Ka but was absent from PrV-ΔUL49. It was also absent from PrV-ΔUL49-infected cells. However, virion gB (Fig. 5, α-gB), glycoprotein C, glycoprotein D, gH, and gL (data not shown), gE (Fig. 5, α-gE), and gM (Fig. 5, α-gM) were detected at similar levels in wild-type and mutant PrV virions as well as in lysates of infected cells. Thus, the UL49 gene product of PrV is not required for virion localization of any of the investigated viral envelope glycoproteins.
FIG. 5.
The incorporation of gE and gM into virions is not affected by the absence of the UL49 protein. Western blot analysis of purified wild-type PrV-Ka (V, lanes 1) and PrV-ΔUL49 (V, lanes 2) virions compared to whole-cell lysates of cells infected with PrV-Ka (C, lanes 1) or PrV-ΔUL49 (C, lanes 2) was carried out with monospecific antisera against the tegument proteins UL37 (33) and UL49 (5), the UL31 protein (17), and envelope glycoproteins M (12) and E (58) as well as a monoclonal antibody against gB (45). Neither the intracellular nor the virion-associated amounts of proteins differ between the wild-type and the UL49 deletion mutant, other than the absence of UL49 in the mutant virus. As a component of primary enveloped virions but not mature virus particles, the UL31 protein is present in whole-cell lysates but absent from mature virions. Note that gM forms higher-molecular-weight oligomers in wild-type and mutant virus infections.
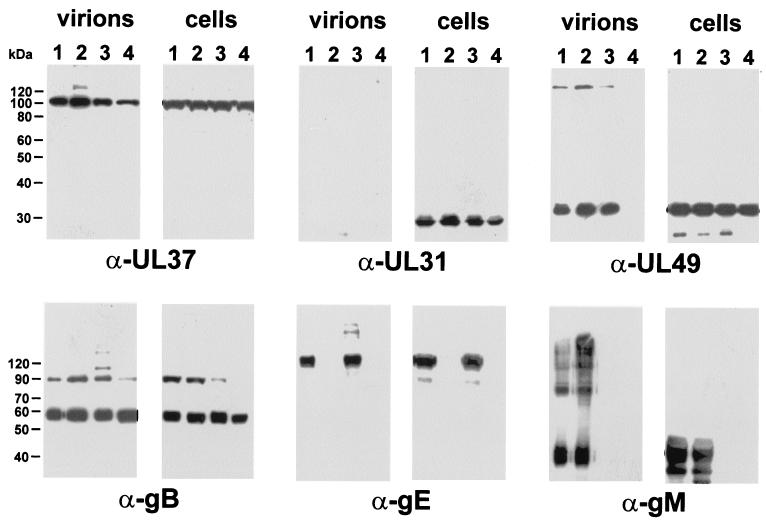
Alternatively, gE or gM might be required for virion incorporation of the PrV UL49 protein. To test this hypothesis, purified virions of wild-type PrV-Ka (Fig. 6, lanes 1), PrV-gE/I− (Fig. 6, lanes 2), PrV-gM− (Fig. 6, lanes 3), and the triple mutant PrV-gE/I/M− (Fig. 6, lanes 4) as well as infected cell lysates were analyzed. The presence of the tegument protein UL37 served as a positive control, whereas the absence of the UL31 protein from virions verified the purity of the preparations. The reactions of antisera directed against the C-terminal parts of these glycoproteins (Fig. 6, α-gE and α-gM) confirmed that these virus recombinants indeed lacked the deleted gene products or at least their UL49-interacting domains. However, similar amounts of the UL49 protein were detected in virions of PrV-gE/I− or PrV-gM− and in particles of wild-type PrV-Ka (Fig. 6, α-UL49). Apparently, the presence of either gE or gM is sufficient to permit the incorporation of the UL49 protein. In contrast, simultaneous deletion of gE-gI and gM, which resulted in a drastic impairment of secondary envelopment (5), also led to a lack of UL49 protein incorporation into virions, despite the unaltered presence of this protein in whole-cell lysates of cells infected with PrV-gE/I/M− (Fig. 6, α-UL49, lanes 4). Replication of PrV-gE/I/M− on either gE-gI- or gM-expressing cells restored the incorporation of the UL49 protein (data not shown).
FIG. 6.
The incorporation of UL49 into virions is blocked in the simultaneous absence of gE-gI and gM. Western blot analysis of purified virions of wild-type PrV-Ka (lanes 1), PrV-gE/I− (lanes 2), PrV-gM− (lanes 3), and PrV-gE/I/M− (lanes 4) compared to whole-cell lysates of virus-infected cells was carried out. Blots were probed as outlined in the legend to Fig. 5.
DISCUSSION
In this report, we demonstrate specific interactions between the carboxy-terminal domain of the major tegument protein UL49 of PrV and the cytoplasmic domains of virion envelope glycoproteins E and M. Our data suggest that these interactions are relevant for the formation of mature PrV virions, in particular, for the incorporation of the major tegument protein UL49 during secondary envelopment of intracytoplasmic capsids (42). We also describe the DNA sequences of the UL48 and UL49 genes of PrV-Ka and the construction and characterization of a PrV-Ka UL49 deletion mutant. The deduced protein encoded by the UL49 gene of PrV-Ka is 249 amino acids long and has a calculated molecular mass of 26 kDa. Interestingly, the PrV UL49 gene product is not required for efficient viral replication in cultured cells or for neuroinvasion, spread, or virulence in rodents (9), and no phenotype could be assigned to the absence of UL49 (9; this study).
gE complexes with gI, and gM forms a complex with gN (30, 68). The functions of the individual proteins and the complexes are not well understood. In fact, mutants lacking any one of these four proteins grow relatively normally in cultured cells. However, the simultaneous absence of gE-gI and gM results in a striking impairment of virion maturation in the cytoplasm. Secondary envelopment is drastically reduced, and capsids enclosed in tegument are found in large aggregates in the cytoplasm (5). The absence of only the cytoplasmic tail of gE had no effect on the expression and virion incorporation of the remaining part of the gE-gI complex (59) but, in combination with a deletion of gM, yielded the same phenotype as did deletion of gE-gI and gM (4). We hypothesized that in the absence of gE and gM, the recruitment of capsids with teguments to the envelopment site for budding is strongly compromised; we suggested that these two glycoproteins are involved in the envelopment process, presumably by interacting via their cytoplasmic domains with tegument proteins of nascent virions.
We demonstrated here by yeast two-hybrid analyses that the cytoplasmic tails of gE and gM indeed interact with a major tegument protein, the product of the UL49 gene. Yeast two-hybrid screening of a PrV expression library with the C-terminal domain of gE resulted in the identification of four different clones which all expressed approximately the carboxy-terminal half of the UL49 protein. A common overlap between these clones mapped the interacting domain to between amino acids 131 and 225 of the UL49 protein of PrV-Be. Moreover, with a different yeast two-hybrid system and an independent PrV expression library, amino acids 155 to 249 of the PrV-Ka UL49 protein used as bait selected six independent clones which all expressed the C-terminal part of gE with a common overlap between amino acids 463 and 577. Thus, the carboxy-terminal half of the UL49 protein contains sequences necessary and sufficient for interactions with the C terminus of gE. When the C terminus of gM was used as bait, three independent clones of UL49 of PrV-Ka were selected which also expressed sequences from the C terminus with a common overlap between amino acids 171 and 249. Whether the parts of the UL49 protein which interact with gE and gM are indeed identical or overlapping or can be separated in a more detailed analysis remains to be demonstrated.
It is interesting that with the two yeast two-hybrid assays, different but complementing interactions could be demonstrated. Whereas the C terminus of gE of PrV-Ka in the LexA system produced only nonspecific transactivation, an identical clone of PrV-Be was successfully used in the Gal4 system for the identification of UL49 interactions. In contrast, bait clones expressing UL49 sequences were helpful only in the identification of interactions with the C terminus of gE in the LexA system and failed to yield specific interactions in the Gal4 system. One possibility is that the conformation of the fusion proteins used as bait and/or prey profoundly influences the outcome of the analyses. This notion may also explain why a UL49 bait clone did not select clones expressing the C terminus of gM from the library, while a bait clone of the gM C-terminal domain readily identified UL49 expression clones.
The UL49 protein is not required for the replication of BHV-1 (36, 49) or HSV-1 (G. Elliott and A. Whiteley, Abstr. 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001). It is also nonessential for PrV replication in cell culture (9; this study) and for neuroinvasion, retrograde or anterograde spread, or virulence in the rat model (9). Since it seems unlikely that an abundant tegument protein would have no function, the lack of a phenotype most likely indicates that other proteins can substitute for UL49 (42). This situation has already been shown for envelope gE-gI and gM, which can be deleted individually with only a slight impairment of viral replication, whereas deletion of all three results in a drastic inhibition of virion maturation (5). Thus, it is likely that the required interactions involve not only UL49 but also other tegument proteins (3, 55, 67). We are currently testing PrV mutants lacking other tegument proteins for their phenotypes in virus morphogenesis.
In fact, there may be differences in the requirements for secondary envelopment not only between but also within herpesvirus subfamilies. For example, the UL49 protein appears to be essential for the productive replication of MDV (13), as is gE-gI (52). Moreover, gI is apparently essential for VZV replication (7, 37, 62). These data could mean that the observed redundancy of functions of these proteins in the envelopment process of PrV may not apply for these other viruses. Interestingly, MDV and VZV are highly cell associated in culture, whereas PrV is efficiently released from infected cells.
The redundancy of interactions between PrV envelope gE and gM and the UL49 protein complicates analyses of their in vivo importance. However, we were able to purify enough extracellular particles from noncomplementing cells infected with gM-transcomplemented PrV-gE/I/M− to perform Western blot analyses of these samples in comparison with whole-cell lysates. The results demonstrated that in the simultaneous absence of gE-gI and gM, the incorporation of the UL49 protein into virions is indeed blocked despite its unaltered presence in infected cells. Thus, interactions of the UL49 protein with either gE-gI or gM appear to be required for it to become a component of the viral tegument. We hypothesized that similar interactions between other viral tegument and envelope glycoproteins are important for virion formation and that the redundancy of these interactions is a hallmark of herpesvirus virion morphogenesis. These additional interactions may involve the tegument protein UL48 and gB, gD, and gH (67), although this possibility remains to be established unequivocally. Moreover, in HSV-1, the UL49 protein interacts with the UL48 protein (14) which, in turn, contacts the UL41 protein (53). The UL13 protein kinase, also a constituent of the tegument, is known to interact with gE, resulting in phosphorylation of the gE-gI complex (44). Whether this interaction is also relevant for virion morphogenesis, e.g., for inclusion of the UL13 protein into nascent virions, has not been analyzed. Therefore, a network of interactions between tegument proteins appears to secure their inclusion into nascent virions (reviewed in reference 42).
The gE-gI complex plays an important role in directed spread of HSV-1 in cultured polarized cells by sorting nascent virions to epithelial cell junctions, thus promoting infection of adjacent cells (26, 27, 39). Although the mechanism of this directed transfer is unclear, our data raise the possibility that secondary envelopment driven by interactions between envelope glycoproteins and tegument proteins may play a role. A similar directed transfer results in transneuronal infection of synaptically connected neuronal networks by HSV-1 and PrV (reviewed in reference 16). In these specialized, highly polarized cells, final virus assembly appears to occur at or close to the synapse (46, 61), with viral subassemblies being transported separately to the final envelopment site (61). For HSV-1, the tegument protein US11 has been implicated in this transport process (R. J. Diefenbach, E. Szabados, M. Miranda, P. Armatt, and A. L. Cunningham, Abstr. 24th Int. Herpesvirus Workshop, abstr. 7.001, 1999), and for PrV, the membrane protein US9 has been demonstrated to play a role in axonal transport (60). It will be interesting to analyze within the context of the described tegument-envelope interactions the specific functions of these viral proteins.
In summary, we describe specific interactions between herpesvirus envelope and tegument constituents that are relevant for virion morphogenesis. The presence of either gE-gI or gM is required for the incorporation of the tegument protein UL49 into nascent virions. Although the roles of these proteins in virion morphogenesis apparently are redundant in cell cultures, they may be important at specific stages of the viral life cycle in vivo as well as in related alphaherpesviruses, which may rely more heavily on their functions.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Me 854/4-2) to T.C.M., from the National Institutes of Health (NS33506) to L.W.E., and from the American Cancer Society (postdoctoral fellowship award PF00-167-01-MBC) to C.H.
We thank Charlotte Ehrlich, Uta Hartwig, Kathrine Kuipers, Petra Meyer, and Nadine Müller for expert technical assistance.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, C. H. 1967. Zur primären Schädigung vegetativer Ganglien nach Infektion mit dem Herpes suis Virus bei verschiedenen Tierarten. Experientia 23:209-217. [DOI] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane-targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bras, F., S. Dezélée, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, J., and H. Nguyen. 1997. Varicella-zoster virus glycoprotein I is essential for growth of virus in Vero cells. J. Virol. 71:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai, P., G. Sexton, J. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the UL37 polypeptide of herpes simplex virus type 1 abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49 is indispensable for virus growth. J. Virol. 75:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 16.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not of mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. A new technique for assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 20.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 22.Harms, J., X. Ren, S. Oliveira, and G. Splitter. 2000. Distinctions between bovine herpesvirus 1 and herpes simplex virus type 1 tegument protein subcellular associations. J. Virol. 74:3301-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homa, F., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, L. 1994. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch. Virol. 137:209-228. [DOI] [PubMed] [Google Scholar]
- 25.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. C., and M. T. Huber. 2001. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jöns, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jöns, A., and T. C. Mettenleiter. 1997. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. J. Virol. Methods 66:283-292. [DOI] [PubMed] [Google Scholar]
- 30.Jöns, A., J. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. The pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong, A., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, X., B. Chow, Y. Li, C. Raggio, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallory, S., M. Sommer, and A. Arvin. 1997. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J. Virol. 71:8279-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 39.McMillan, T., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C., L. Zsak, A. S. Kaplan, T. Ben-Porat, and B. Lomniczi. 1987. Role of a structural glycoprotein of pseudorabies in virus virulence. J. Virol. 61:4030-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mossman, K., R. Sherburne, C. Lavery, J. Duncan, and J. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng, T., W. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 45.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penfold, M. E., P. Armati, and A. L. Cunningham. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. USA 91:6529-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrovskis, E., J. Timmins, and L. E. Post. 1986. Use of λgt11 to isolate genes for two pseudorabies virus glycoproteins with homology to herpes simplex virus and varicella-zoster virus glycoproteins. J. Virol. 60:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomeranz, L., and J. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren, X., J. Harms, and G. A. Splitter. 2001. Tyrosine phosphorylation of bovine herpesvirus 1 tegument protein VP22 correlates with the incorporation of VP22 into virions. J. Virol. 75:9010-9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roizman, B., and P. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 52.Schumacher, D., B. K. Tischer, S. Reddy, and N. Osterrieder. 2001. Glycoproteins E and I of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Virol. 75:11307-11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 55.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. 17(Suppl.):71-79. [DOI] [PubMed] [Google Scholar]
- 56.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 57.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 58.Tirabassi, R. S., and L. W. Enquist. 2000. Role of the pseudorabies virus gI cytoplasmic domain in neuroinvasion, virulence, and posttranslational N-linked glycosylation. J. Virol. 74:3505-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tirabassi, R. S., R. A. Townley, M. G. Eldridge, and L. W. Enquist. 1997. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J. Virol. 71:6455-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomishima, M., and L. W. Enquist. 2001. A conserved α-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomishima, M., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 62.Wang, Z.-H., M. Gershon, O. Lungu, Z. Zhu, S. Mallory, A. Arvin, and A. Gershon. 2001. Essential role played by the C-terminal domain of glycoprotein I in envelopment of varicella-zoster virus in the trans-Golgi network: interactions of glycoproteins with tegument. J. Virol. 75:323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whealy, M. E., J. P. Card, A. K. Robbins, J. R. Dubin, H.-J. Rziha, and L. W. Enquist. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., D. Sirko, and J. L. C. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, Z., D. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, W., and R. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]
- 68.Zuckermann, F., T. C. Mettenleiter, C. Schreurs, N. Sugg, and T. Ben-Porat. 1988. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J. Virol. 62:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]