Abstract
The 3′-terminal ends of both the positive and negative strands of the hepatitis C virus (HCV) RNA, the latter being the replicative intermediate, are most likely the initiation sites for replication by the viral RNA-dependent RNA polymerase, NS5B. The structural features of the very conserved 3′ plus [(+)] strand untranslated region [3′ (+) UTR] are well established (K. J. Blight and C. M. Rice, J. Virol. 71:7345-7352, 1997). However, little information is available concerning the 3′ end of the minus [(−)] strand RNA. In the present work, we used chemical and enzymatic probing to investigate the conformation of that region, which is complementary to the 5′ (+) UTR and the first 74 nucleotides of the HCV polyprotein coding sequence. By combining our experimental data with computer predictions, we have derived a secondary-structure model of this region. In our model, the last 220 nucleotides, where initiation of the (+) strand RNA synthesis presumably takes place, fold into five stable stem-loops, forming domain I. Domain I is linked to an overall less stable structure, named domain II, containing the sequences complementary to the pseudoknot of the internal ribosomal entry site in the 5′ (+) UTR. Our results show that, even though the (−) strand 3′-terminal region has the antisense sequence of the 5′ (+) UTR, it does not fold into its mirror image. Interestingly, comparison of the replication initiation sites on both strands reveals common structural features that may play key functions in the replication process.
Hepatitis C virus (HCV), which is essentially transmitted by blood, represents a worldwide major health problem. The World Health Organization estimates that 170 million people are infected by this virus. Forty percent of infected people develop various liver diseases, such as chronic hepatitis, cirrhosis, and hepatocarcinomas (13, 35, 48). HCV is an RNA enveloped virus of positive polarity which is the sole member of the genus Hepacivirus within the family Flaviviridae (55). The HCV RNA genome is about 9,600 nucleotides (nt) in length, encodes a unique polyprotein of 3,030 amino acids, and is flanked at its 5′ and 3′ ends by two highly conserved noncoding regions involved in the translation and replication processes of the virus, respectively (11, 12, 29). The 5′ untranslated region (UTR) has been extensively studied and appears to be structurally very conserved among different HCV strains (5, 7, 21, 56). Its main feature consists of an internal ribosome entry site (IRES) driving cap-independent expression of the HCV polyprotein (19, 51, 61). This unique polyprotein is then processed by the host signal peptidase and viral proteases to generate 10 mature structural and nonstructural (NS) proteins (16, 20, 36, 57). Among them, NS5B, the RNA-dependent RNA polymerase, is a membrane-associated phosphoprotein (17). NS5B forms the replication complex, (22) together with other viral partners, including the protease-helicase NS3 (23). It was recently shown that the helicase activity of NS3 specifically recognizes the 3′ ends of both the minus [(−)] and plus [(+)] strands (1). NS3 is targeted to the perinuclear endoplasmic reticulum by an interaction with the viral protein NS4A (62). One hypothesis is that the NS3/NS4A complex may then associate with NS5B and subsequently recognize the 3′UTR of the coding RNA strand [(+) strand RNA], where initiation of (−) strand synthesis occurs. Accordingly, the RNA binding activity of NS5B was observed on homopolymeric RNA (24) but also, and with higher affinity, on the 3′ UTR of the coding strand (10). This activity was found to be associated with two distinct domains of NS5B. The 3′ UTR of the (+) strand RNA is approximately 200 nt in length and consists of a short nonconserved region, a poly(U) and/or poly(U-C) stretch of variable length, and the highly conserved 98-nt X region (32, 58). The presence of the 3′ UTR is absolutely required for in vivo replication (33, 64). Secondary-structure predictions, as well as structural probing experiments, have revealed that the X region folds into three stable stem-loop structures named SL-I, SL-II and SL-III (4, 25, 59).
Synthesis of full-length cRNA has been achieved in vitro, using full-length and truncated forms of NS5B (3, 9, 28, 37, 43, 63). However, the exact mechanism for initiation of RNA synthesis remains somewhat controversial. Several reports have shown that initiation of RNA synthesis by the RNA-dependent RNA polymerase of HCV and other flaviviruses was achieved through a “primer-dependent copy-back” mechanism or was associated with a terminal nucleotidyl transferase (TNT) activity of the same protein (3, 24, 49). In contrast, other studies (39, 68) have reported primer-independent de novo initiation of RNA synthesis supported by flavivirus polymerases devoid of TNT activity (27, 39, 68). Finally, primer dependency and template requirements, in particular secondary-structure requirements, were also extensively studied (44, 67). Recent data on the φ6 double-stranded RNA (dsRNA) polymerase have shed new light on the initiation mechanism of RNA-dependent RNA synthesis (8, 41). In this case, the RNA polymerase, which lacks helicase activity, initiates RNA synthesis on blunt-ended dsRNA (41). The proposed mechanism for initiation is through base stacking of two nucleotides against a priming aromatic ring of the protein, as deduced from the crystal structure of the protein (8). Although this mechanism could be generalized to HCV, it seems that HCV has evolved to use the NS3 helicase activity to perform strand separation within the replication complex and prior to initiation of RNA synthesis (52).
It might be assumed that once the replicative intermediate is produced, the replication complex recognizes the 3′ end of the (−) strand RNA and synthesizes the new HCV genomic RNA. Therefore, recognition of the 3′ end of HCV (−) strand RNA is a crucial step in viral replication. It is tempting to speculate that at least some features recognized for initiation of RNA synthesis on both the (+) and (−) strands are identical or have some homologies. However, little information is available concerning the structure of the 3′ end of the HCV (−) strand, which, notably, is in part the antisense sequence of the (+) strand IRES, known to be very structured.
In the present work, we have investigated in detail the conformation of the expected replication initiation site of HCV (−) strand RNA by chemical and enzymatic probing combined with computer prediction of secondary structure. The last 220 nt, where initiation of (+) strand RNA synthesis most likely occurs, are arranged in five stable stem-loops, forming domain I. Domain I is linked to a less stable structure, named domain II, containing the antisense sequence to the pseudoknot on the coding strand. Our results show that, even though the 3′ end of the (−) strand has the antisense sequence of the (+) strand 5′ UTR, it does not fold into its mirror image. Moreover, comparison of initiation sites of replication on both strands reveals common structural features that may play key roles in the replication process.
MATERIALS AND METHODS
Plasmid constructions and RNA synthesis.
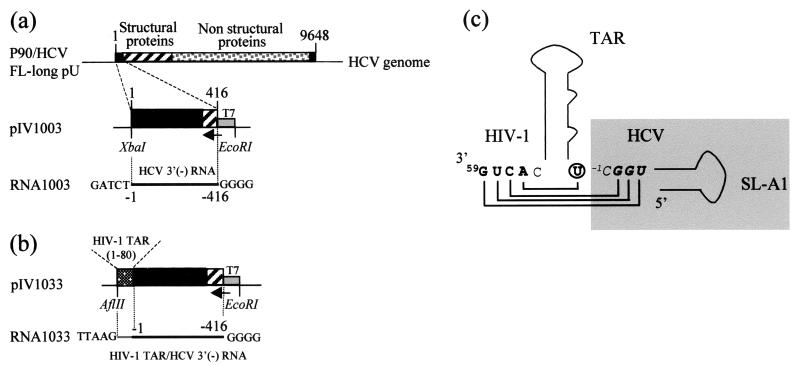
pIV1003 was obtained by PCR amplification of nt 1 to 416 of the original HCV consensus clone (HCV p90/HCV FL-long pU) (31) with primer 1 (5′GCTCTAGAGCCAGCCCCCTGATG3′) and primer 2 (5′GAGAATTCTAATACGACTCACTATAGGGGCGGGAACTTGACGTCCTG3′), containing the T7 promoter sequence (underlined), for the purpose of in vitro transcription. The PCR fragment was cut with EcoRI and XbaI and inserted into vector pUC19 cut with the same restriction enzymes (Fig. 1a). pIV1033 is a derivative of pIV1003 in which a PCR fragment encoding the human immunodeficiency virus type 1 (HIV-1) transactivating responsive element (TAR) sequence (Mal isolate) (2) was inserted in front of nt 1 of HCV. The HIV-1 TAR was obtained by PCR amplification of the 80 nt of TAR with primer 3 (5′GGTCTCTCTTGTTAGACC3′) and primer 4 (5′CAAGGCAAGCTTTATTGAGGCTT3′), using plasmid pJCB (45) as a template. The resulting fragment was ligated into the blunted XbaI site of pIV1003 (Fig. 1b) to generate pIV1033. The pIV1033 construct contains an extra U added by cloning (Fig. 1c). The DNA templates pIV1003 and pIV1033 were linearized with XbaI and AflII, respectively, and in vitro transcribed with T7 RNA polymerase. The resulting RNA1003 and RNA1033 contain five extra bases at their 3′ ends as a result of the enzymatic restriction and four G residues at the 5′ ends (Fig. 1a and b). RNA1003 and RNA1033, 423 and 491 nt in length, respectively, were purified according to standard procedures (42).
FIG. 1.
Plasmid constructions. (a) pIV1003 contains the first 416 nt of the HCV genotype 1a genome (clone p90/HCV FL-long pU) directly under the control of the T7 promoter. (b) pIV1033 contains the HIV TAR (2) sequence cloned in front of nt 1 of the HCV genome in pIV1003. (c) Sequence details of the junction between the extremity of the HCV terminus and the nucleotides of the HIV-1 TAR sequence that base-pair with the 3′ end of the HCV RNA. HCV nucleotides are in the shaded box (in italics and boldface), and HIV nucleotides are in boldface; an extra U (circled) was added by cloning.
RNA structure-probing experiments. (i) Buffers.
The buffers used were buffer M1 (10 mM sodium cacodylate [pH 7.5], 1 mM magnesium chloride, 60 mM potassium chloride), buffer M2 (10 mM sodium borate [pH 8.0], 1 mM magnesium chloride, 60 mM potassium chloride), and buffer M3 (50 mM HEPES-NaOH [pH 7.5], 5 mM magnesium acetate, 50 mM potassium acetate).
(ii) Chemical and enzymatic probing of in vitro-transcribed viral RNAs.
Standard RNA-probing assays were performed with 1 to 2 μg of in vitro-transcribed 1003 or 1033 RNA. The RNA was first denatured by heating it at 90°C for 2 min and then cooled on ice for 2 min. After the addition of buffer M1 for dimethylsulfate (DMS) modification and RNase V1 cleavage or buffer M2 for 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluene sulfonate (CMCT) modification experiments, the RNA was refolded for 15 min at 37°C. For lead cleavage experiments, refolding was obtained by incubation at room temperature for 10 min in buffer M3. Prior to modifications with DMS and CMCT, 1 μg of total yeast tRNA was added to the reaction mixture. Modifications with DMS were performed at room temperature for 5, 10, 15, and 20 min with 0.7 μl of DMS diluted 20-fold in ethanol. CMCT modifications were performed for 15, 30, and 45 min at room temperature with 2.5 μl of a CMCT solution (42 mg/ml). The reactions were stopped by ethanol precipitation. Lead cleavages were performed as follows. Refolded RNA, supplemented with 4 μg of total yeast tRNA, was incubated with lead acetate (12, 40, and 120 mM) for 5 min at 20°C. The reaction was stopped by addition of 2 μl of 0.1 M EDTA, and the RNA was ethanol precipitated. Cleavage with RNase V1 (0.035 U per reaction) was for 5, 10, or 15 min at 20°C, and the reaction was stopped by a phenol-chloroform extraction prior to ethanol precipitation.
Modified bases and cleavage sites were detected by primer extension with avian myeloblastosis virus reverse transcriptase as described previously (2) using various 5′-end 32P-labeled primers complementary to residues −77 to −91 (primer oIV1036), −186 to −203 (primer oIV1066), −237 to −253 (primer oIV1038), and −283 to −299 (primer oIV1025) for RNA1003. One additional primer, complementary to residues 22 to 40 of the HIV TAR region (primer oIV1057) (2) (Fig. 1c shows the nomenclature) was used to probe RNA1033. After reverse transcription, the cDNAs were separated by electrophoresis on 8% denaturing polyacrylamide gels. A dideoxynucleotide sequencing reaction was run in parallel to allow identification of the modified positions.
RESULTS
Experimental strategy. (i) Design of the RNAs.
In this study, we have tested the accessibility of the last 416 nt of the HCV (−) strand RNA to chemical and enzymatic probes, using two in vitro-transcribed RNA fragments. In order to facilitate their positioning, the numbering of (−) strand nucleotides was the same as that of the (+) strand, except that they were preceded by a minus sign. For example nt −400 in the (−) strand RNA is complementary to nt 400 in the (+) strand RNA, and nt −1 in RNA1003 is the last nt of the HCV (−) strand RNA. RNA1003 contains the last 416 nt of the HCV (−) strand, which correspond to the antisense sequence of the (+) strand IRES (nt −1 to −342) and the N-terminal part of the core sequence (nt −343 to −416). This RNA also bears five and four additional nucleotides at the 5′ and 3′ ends of the molecule, respectively, introduced by cloning (Fig. 1a and b). In order to gain structural information on the extreme 3′ end of the HCV (−) strand RNA, RNA1033 was in vitro transcribed from plasmid pIV1033 (see Materials and Methods). This RNA contains the HIV-1 TAR sequence inserted immediately downstream of nt −1 of the HCV genome (Fig. 1b). This “tail” was used as a hybridization site for a primer, in order to detect RNA modifications very close to the 3′ end of the HCV RNA. It was specifically chosen because of its very stable structure (2). We assumed that a stable TAR structure would fold independently of the HCV sequence (see below).
(ii) Chemical probing and derivation of the secondary-structure model.
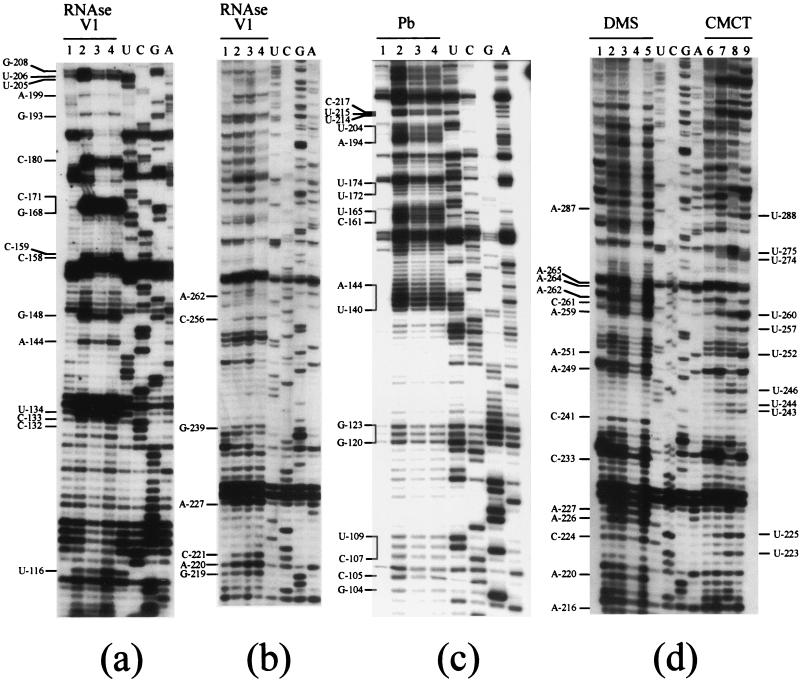
Prior to chemical and enzymatic modifications, RNA was denatured for 2 min at 90°C in order to open up all the possible secondary structures. It was then quickly cooled on ice to freeze the denatured conformation. Refolding buffer containing magnesium was then added, and refolding was allowed for 10 to 15 min at 37°C. During this step, the RNA adopts its thermodynamically most stable structure. Similar refolding strategies have been applied to ribozymes, where correct refolding could be monitored by the cleavage activity (53, 66). The chemical reagents we used allowed us to test the Watson-Crick (WC) positions of the bases [A(N-1) and C(N-3) with DMS; G(N-1) and U(N-3) with CMCT] or accessible positions in the ribose-phosphate backbone [lead(II) acetate]. Series of lead-induced cleavages are usually observed in single-stranded regions or poorly stable helices, while isolated strong lead-induced hydrolysis at specific positions is usually indicative of divalent cation binding sites (6). Finally, RNase V1, specific for double-stranded regions, provides a positive signal for the presence of helices or stacked nucleotides. The modified or cleaved positions were detected by primer extension with reverse transcriptase, using the primers described in Materials and Methods. In all cases, control experiments were run in parallel, in order to detect nicks in the RNA template or pausing of reverse transcription due to stable secondary structures. Probing experiments are shown below (see Fig. 3 to 6), and the results, representative of at least three independent experiments, were compiled (Fig. 2).
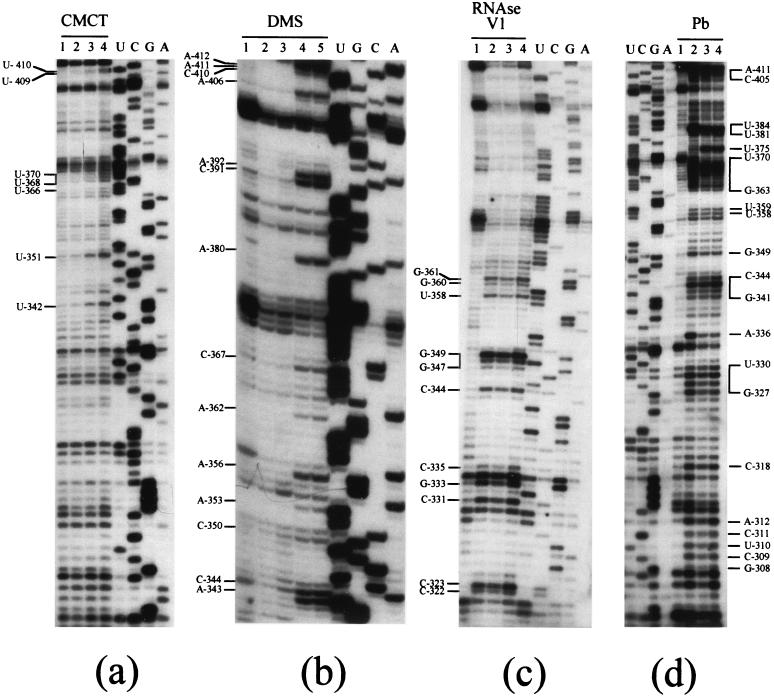
FIG. 3.
Probing experiments on RNA1003 and RNA1033. Modification experiments were performed as described in Materials and Methods. (a) Modifications of A (N-1) and C(N-3) with DMS. Lanes 1 and 6, incubation control; lanes 2 and 7, 5 min; lanes 3 and 8, 10 min; lanes 4 and 9, 15 min; lanes 5 and 10, 20 min. Modifications were done on RNA1003 (lanes 1 to 5) and RNA1033 (lanes 6 to 10). (b) Modifications of U(N-3) and G(N-1) with CMCT. Lanes 1 and 5, incubation control; lanes 2 and 6, 15 min; lanes 3 and 7, 30 min; lanes 4 and 8, 45 min. Modifications were done on RNA1003 (lanes 1 to 4) and RNA1033 (lanes 5 to 8). (c) Modification experiments were performed as for panel b.
FIG. 6.
Probing experiments on nt −308 to −416 (domain II, SL-BII, SL-CII, and SL-DII). Modifications with CMCT (a) and DMS (b) and cleavages with RNase V1 (c) and lead (d) were as for Fig. 4.
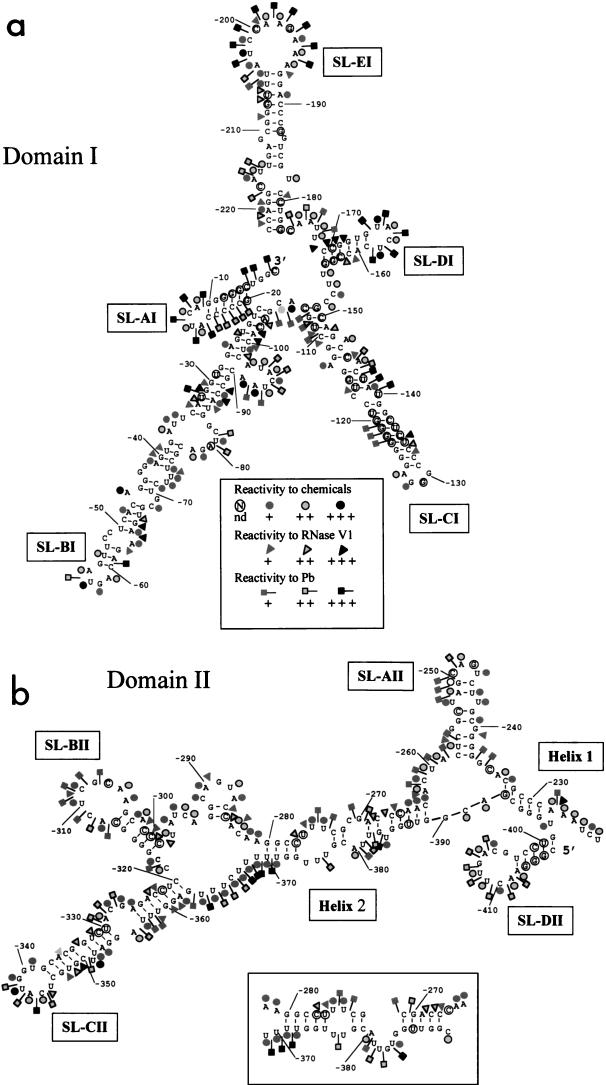
FIG. 2.
Secondary-structure model of the 3′ end of HCV genotype 1a (−) strand RNA. Domain I (a) and domain II (b) are shown. The DMS and CMCT reactivities are represented by circles, RNase V1 sensitivity is represented by triangles, and lead cleavages are represented by squares. The reactivity and sensitivity codes are indicated in panel a; nonreactive nucleotides are not labeled, and the reactivities of the circled nucleotides were not determined (nd). A possible alternative structure for region −283 to −264 interacting with region −371 to −389 is shown in the inset to panel b.
RNA-folding computer programs (40) were used to predict the thermodynamically most stable secondary structures of RNA1003. Thirteen different structures were proposed, with ΔG varying from −159.12 to −168.71 kcal/mol. All but two proposed secondary structures presented two main domains that we named domain I and domain II. Domain I, for which the same secondary structure was found in 45% of the structures, encompasses nt −230 to −1 and folds into five very stable stem-loop structures (Fig. 2a). Domain II, spanning nt −416 to −230, is more versatile. One of its possible foldings was predicted in 30% of the cases, whereas the others were almost unique. The various foldings were screened, and the secondary-structure motifs consistent with our experimental data were selected. The folding program was then run again, forcing these secondary-structure elements to be integrated. A general secondary-structure model, integrating both experimental data and computer predictions, can thus be proposed (Fig. 2).
Domain I folds into five stable stem-loop structures.
Domain I, spanning nt −230 to −1, most likely includes the starting point of (+) strand RNA synthesis. Domain I was predicted to be very structured, and indeed, the experimental data confirmed the existence of five stem-loops, named SL-AI, SL-BI, SL-CI, SL-DI, and SL-EI (Fig. 2a).
RNA 1033, containing the HIV-1 TAR sequence, was used to investigate the conformation of the last 40 nt of the HCV (−) strand RNA. Comparative probing of RNA1003 and -1033 confirmed that both RNAs mostly adopt the same conformation, thus validating the use of RNA1033 to study the structure of the extreme 3′ end of the HCV (−) strand. Figure 3 shows an example of comparative chemical probing with DMS (Fig. 3a) and CMCT (Fig. 3b and c) of RNA1003 and -1033, revealing that similar reactivities are observed on both RNAs. In Fig. 2a, nucleotides present in the linker region between SL-BI and SL-CI (A−106), in the loop of SL-CI (A−127), or bulged in the SL-CI stem (A−147 and A−144) were reactive in both RNAs (Fig. 3a). Similarly, nucleotides involved in the loops of SL-DI (nt −165 to −162) (Fig. 3a and b) and SL-EI (nt −215 to −202) (Fig. 3c) were reactive in both RNAs. The same conclusions hold true for domain II, where the reactivities are similar in both RNAs (see nt −275 and −274 in helix 2) and nucleotides of SL-CII (U−351 and U−342) (Fig. 3c).
The only interference we could detect was base-pairing between four single-stranded nucleotides at the 3′ end of the TAR sequence with an extra U (at the junction between TAR and the HCV sequence) added by cloning and three nucleotides (−2GGU−4) of the 3′ end of the HCV RNA, otherwise predicted to be single stranded (Fig. 1c). Indeed, we found that, in the context of RNA1033, the last 5 nt of the RNA were protected from chemicals, even though they were predicted to be single stranded within the original RNA1003 (Fig. 4a and b).
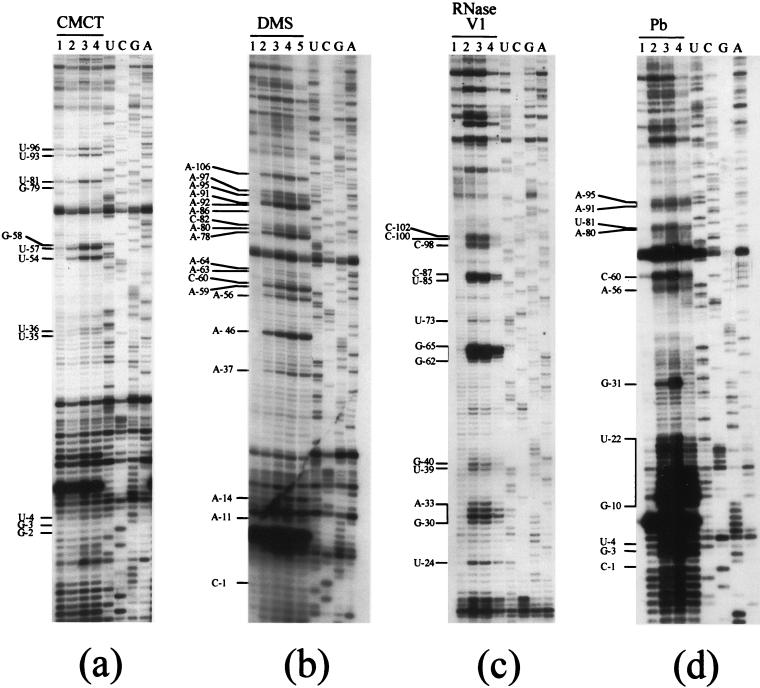
FIG. 4.
Probing experiments on nt −1 to −106 (domain I; SL-AI and SL-BI). (a) Modifications of U(N-3) and G(N-1) with CMCT. Lane 1, incubation control; lane 2, 15 min; lane 3, 30 min; lane 4, 45 min. (b) Modifications of A (N-1) and C(N-3) with DMS. Lane 1, incubation control; lane 2, 5 min; lane 3, 10 min; lane 4, 15 min; lane 5, 20 min. (c) Cleavage of double-stranded and stacked RNA regions by RNase V1. Incubation was with 0.035 U per reaction at 20°C. Lane 1, incubation control; lane 2, 5 min; lane 3, 10 min; lane 4, 15 min. (d) Cleavages of accessible RNA regions by lead. Incubation was for 5 min at 20°C. Lane 1, incubation control; lane 2, 12 mM lead acetate; lane 3, 40 mM lead acetate; lane 4, 120 mM lead acetate. Sequencing reactions (U, G, C, and A) were run in parallel.
Residues −5 to −20 of the HCV (−) strand RNA fold into a short hairpin, SL-AI, comprising a 6-bp stem, in which most nucleotides are nonreactive at their WC positions (Fig. 2b). Conversely, the 4-nt loop of SL-AI was highly reactive to chemicals (Fig. 4b) and strongly cleaved by lead acetate (Fig. 4d), further proving strong accessibility. Surprisingly, the 5′ part of the stem was also cut by lead acetate (Fig. 4d), and we have no explanation for this observation.
Nucleotides −115 to −21 fold into a long stem-loop, SL-BI, which was proposed in 12 out of the 13 predicted secondary structures (Fig. 2a). The basis of the stem is very stable, and most of its residues were protected from modifications by DMS, CMCT, and lead acetate (Fig. 4a, b, and d). Likewise, RNase V1, which generally cleaves close to the ends of helices and in the vicinity of bulges or internal loops (6), cleaved at the basis of SL-BI, from residues C−102 to C−100 and C−98, as well as immediately after the bulge, from A−33 to G−30 and C−87 to U−85 (Fig. 4c), confirming the presence of the double-stranded regions. The internal loops in the upper part of SL-BI were moderately reactive to chemicals (Fig. 2a and 4a and b, nt A−80, A−78, and A−37 to U−35), together with the bulged residues −97 to −91 (Fig. 4a and b), which were incidentally moderately cut by lead acetate (Fig. 4d). Even though marginal reactivities were observed in this part of the stem, the existence of the two short base-paired regions in the upper part of SL-BI was confirmed by mild RNase V1 cleavages at nt G−40 and U−39 (Fig. 4c) and stronger cuts in the −65GAAG−62 sequence (Fig. 4c). Residue A−46 displayed a strong reactivity to DMS (Fig. 4a), consistent with its bulging out in the secondary-structure model. Finally, nucleotides in the SL-BI apical loop also displayed strong reactivity at their WC positions (Fig. 4a and b). Residue G−31 exhibited a high reactivity to lead acetate (Fig. 4d) that could reflect the presence of an ion binding site (46).
The third stem-loop of domain I, SL-CI, is highly stable (Fig. 2a shows its reactivity to chemicals, and Fig. 5a shows RNase V1 cleavages). Within SL-CI, nt A−113, A−144, and A−147 were accessible to DMS modifications (Fig. 2a), suggesting that they were exposed to the solvent. They are indeed bulging out in our model. The apical loop of SL-CI displayed a modification pattern typical of the well-known GNRA tetraloops (26): the first G residue of the loop was nonreactive at its WC position, the last A was highly reactive, and the residue immediately upstream showed only marginal reactivity. GNRA tetraloops are usually involved in tertiary interactions, as found in rRNAs, self-splicing introns, and other highly structured RNA molecules, where the GNRA sequences are captured by helical stems (14). However, among those tetraloops, GGGA sequences are rare, and no strong receptor has been identified so far. Finally, lead cleavage experiments revealed an unexpected high accessibility of residues −144 to −140 (Fig. 5c).
FIG. 5.
Probing experiments on nt −104 to −288 (domain I, SL-CI, SL-DI, and SL-EI; domain II, SL-AII). Cleavages with RNase V1 (a and b) and lead (c) were as for Fig. 4. (d) Modifications with CMCT and DMS. Lane 1, incubation control with buffer M1; lanes 2 to 5, incubation with DMS for 5 (lane 2), 10 (lane 3), 15 (lane 4), and 20 min (lane 5); lane 6, incubation control with buffer M2; lanes 7 to 9, incubation with CMCT for 15 (lane 7), 30 (lane 8), and 45 min (lane 9).
Stem-loop DI is made up of a 6-bp stem, protected from chemical modification, and a 4-base loop with all residues fully reactive at their WC positions (Fig. 2a). The existence of this short hairpin was confirmed by strong RNase V1 cleavages on the 5′ side of the stem (nt C−171 to G−168) (Fig. 5a).
The last hairpin of domain I, SL-EI, consists of two stems of 5 and 10 bp, interrupted by an internal loop (Fig. 2a). Residues in both stems were mainly protected from chemical modification (Fig. 2a), and RNase V1 recognized sequences in both stems (C−180, −206UU−205, and G−208 [Fig. 5a] and −221CCG−219 [Fig. 5b]). Residues U−182, U−215 (Fig. 2a), and A−216 (Fig. 5d) were modified at their WC positions, and their corresponding ribose-phosphate backbone supported lead cleavage (Fig. 5c). Accordingly, these residues are part of an internal loop in SL-EI. In contrast, G residues at positions −186 and −211 were unreactive to CMCT (Fig. 2a), even though they are bulging out in our secondary-structure model. Such reactivity patterns can most likely be explained by stacking interactions within the stem. Finally, the large apical loop of SL-EI (nt A−203 to A−194) was highly reactive to CMCT and DMS (Fig. 2a) and lead acetate (Fig. 5c).
Stem-loops BI, CI, DI, and EI are linked by single-stranded regions varying in length from 2 to 5 nt that are highly reactive to chemicals (nt −106 to −105, −155 to −152, and −175 to −172 [Fig. 2a]).
Domain II (nt −416 to −230) folds into a more versatile secondary structure.
Domain II is closed up by a 5-bp helix, named helix 1 (nt −232 to −229 and nt −396 to −393), and connected to domain I by a 5-nt linker region (−227AAUCU−223) highly reactive to DMS and CMCT (Fig. 2b and 5d). Within domain II, which appears considerably less stable than domain I, the most clear-cut results concern the existence of SL-CII. This stable structure is divided into two stems of 7 and 9 bp connected by an internal loop. The existence of the two stems was supported by the low level of reactivity to chemicals of the paired nucleotides, as well as by several RNase V1 cleavage sites (−323CC−322, C−331, G−333, C−335, −349GUG−347, U−358, and −361GG−360) (Fig. 6c). Residue U−351 was strongly reactive at its WC position (Fig. 6a), consistent with it bulging out of the helix (Fig. 2b). Residues within the internal and apical loops of SL-CII were highly reactive at their WC positions, and lead cleavages were observed at residues −330UACG−327 and −341UAC−344 (Fig. 6a, b, and d), in keeping with the proposed secondary-structure model.
The rest of domain II is organized in a series of loops and short metastable helices (SL-AII and SL-BII) connected by helix 2, which encompasses residues −280 to −263 and −388 to −371.
SL-AII is rather unstable, as demonstrated by the level of reactivity at the WC positions C−241, −244UU−243, −251UA−252, and U−257, with GC pairings alternating with labile AU and UG pairings (Fig. 5d). On the other hand, the WC positions of G residues −240 and −255 were protected, even though they were predicted to be unpaired, most likely because these bases were stacked inside the helix (Fig. 2b).
An internal loop, comprising −265AACACUA−259 and −392AC−391, highly reactive to chemicals (Fig. 5d and 6b) and cleaved by lead acetate (Fig. 2b), bridges helix 2 and SL-AII and -DII within domain II. Helix 2 forms an irregular stem structure that can fold into two alternative conformations (Fig. 2b). We have observed RNase V1 cleavages at positions −268 to −266, confirming the starting point of the stem region predicted in the alternative conformation. In this conformation, the unreactive −271CG−270 would be paired with −387UG−386. This helical segment would be preceded by an extended bulge (from nt −384 to −381) strongly cut by lead (Fig. 6d). In both conformations, −275UU−274 are single stranded, consistent with their accessibility to chemical reagents (Fig. 5d). Finally, the strong lead-mediated hydrolysis at nt U−375 was most likely due to a specific cation binding site (Fig. 6d).
Helix 2 is linked to the stable SL-CII by a weakly structured region, as revealed by the general high level of reactivity (Fig. 2b). However, the presence of mild RNase V1 cleavage sites suggests that the two helices predicted in this region, and mainly the stem of SL-BII, do exist. Nucleotides −312ACUCG−308 in the loop of SL-BII were cleaved by lead acetate (Fig. 6d), even though they exhibited only mild or no reactivity to modification agents (Fig. 2b).
The last hairpin (SL-DII), encompassing residues −416 to −399, comprises a 5-bp stem and an 8-nt loop. With the exception of G−407, the nucleotides forming the loop were reactive to the modification agents (Fig. 6a and b).
DISCUSSION
dsRNA viruses are able to perform replication from blunt-ended dsRNAs. However, the HCV RNA polymerase, even though structurally very close to the φ6 RNA polymerase (41), has diverged from a common ancestor. As a result, flaviviruses require a distinct helicase activity in order to separate the two RNA strands prior to replication, which consequently takes place on a single-stranded molecule (34). Determination of the secondary structure of the 3′ end of HCV (−) strand RNA is therefore an obligatory step to understand the replication process of this virus.
The 3′ end of the HCV (−) strand RNA is the strict antisense sequence of the (+) strand IRES. It is therefore of particular interest to compare both structures, which could adopt mirror image conformations. The first stem-loop in the (−) strand RNA, SL-AI, is indeed the mirror image of domain in the IRES. The same observation holds true for the loops of SL-DI and SL-EI, corresponding to loops IIIa and IIIb in the IRES structure, respectively, as well as for the nucleotides forming the external loop of SL-BI, which are also exposed in domain II of the IRES sequence. In contrast, the organization of nt −105 to −20, forming SL-BI, displays no homology with the IRES structure, in which a large proportion of the corresponding nucleotides are single stranded. Finally, it should be stressed that residues −331UACGAG−325 and −311CUCGCA−306, which correspond to the sequences involved in the pseudoknot structure of the IRES, are undoubtedly accessible in the 3′-end HCV (−) strand RNA as stressed by the lead cleavages. In contrast with the situation in the (+) strand RNA, our experimental data do not suggest any involvement of those sequences in a tertiary structure, such as a pseudoknot. In conclusion, we found that the 3′ end of the HCV (−) strand RNA does not adopt a strict mirror image conformation of its antisense 5′ UTR (+) sequence.
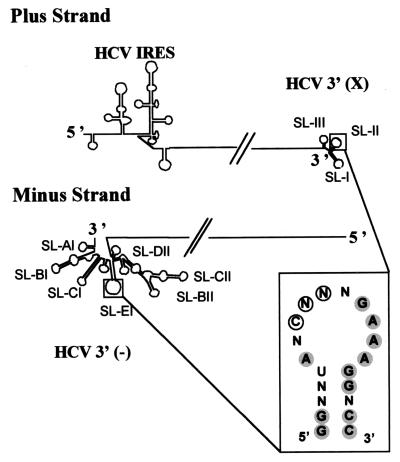
Another goal of our study was to investigate the possible existence of common structural characteristics between RNA regions involved in the replication process of the (+) and (−) RNA strands of HCV in particular and of flaviviruses in general. Comparison of the secondary structures of the 3′ X region of the HCV coding strand (4) and those of the (−) strand presented in this paper allows some features to be highlighted (Fig. 7). Interestingly, both regions are highly structured and assemble into long stem-loops. As a result, the SL-I stem at the 3′ end of HCV (+) strand RNA is comparable to SL-CI, both in length and stability. Another striking feature is the 60% homology between the loops of SL-EI and SL-II (Fig. 7, inset). SL-II was found to be involved in polypyrimidine tract binding (PTB) protein binding (25). However, the nucleotides in SL-II involved in PTB protein binding (Fig. 7, inset) are not homologous in both loops. Moreover, no homology with the PTB protein consensus binding site was found, either in this region or in any other region of the 3′-end HCV (−) strand RNA, disproving the hypothesis that the SL-EI loop would be involved in PTB protein binding. Accordingly, it was shown that the PTB protein binds solely to the 3′ end of the (+) strand and not to the 3′ end of the (−) strand (60). In spite of this, and given their strong sequence homology, we can still speculate that the two loops might share a common unknown functional role. In support of this hypothesis, an in vitro study using recombinant NS5B showed that, although efficient in vitro transcription was obtained using a template with SL-II deleted, the resulting fragment was not of the expected size (43). Thus, SL-II, as well as its counterpart on the HCV 3′ (−) strand RNA, SL-E1, could be involved somehow in the specificity of the replication process.
FIG. 7.
Schematic representation of the HCV (+) and (−) strands. The inset summarizes the conserved nucleotides between SL-II and SL-EI, which are shaded. The nucleotides involved in PTB binding are circled.
Investigation of template requirements for RNA synthesis by recombinant NS5B revealed that a stable secondary structure, together with at least one single-stranded 3′-end cytidine, is necessary for optimal initiation of replication (28). In agreement with this study, the structure we propose suggests the existence of an unpaired cytosine at the 3′ end of the (−) strand. This result could provide an explanation for higher efficiency of (−) strand than (+) strand replication (38, 50), since the extreme 3′ end of the HCV (+) strand RNA is a uridine, involved in a U-G base pair (32). In this case, RNA synthesis is initiated at the single-stranded region closest to the 3′ end of the X region, i.e., at a specific site within the loop of SL-I (44).
Even though our study is the first describing the secondary structure of the 3′ end of the (−) strand RNA of a flavivirus, the 3′ ends of the (+) strands of several flaviviruses have been investigated (47, 54). In particular, the 3′ UTR of the West Nile virus has been shown to contain two stem loops interacting with each other to form a pseudoknot (54, 54). Our study provides no evidence for the existence of such a tertiary interaction. Several studies also pointed to the existence of cyclization sequences involved in the flavivirus replication process (18, 30, 65). Such sequences may not exist in the HCV (+) strand, since there were no significant differences between nucleotide accessibilities whether tested on a full-length genome or on the 3′ (+) end alone (4). Whether cyclization sequences exist in the nonreplicative strand is still unknown and is under investigation.
It is noteworthy that in vitro replication studies performed with recombinant NS5B showed that the minimal RNA fragment required for efficient replication of the (−) strand spans nt −239 to −1 (43), which corresponds to the domain I identified in the present study. This strongly suggests that the replication complex may recognize a stable structure at the 3′ end of the RNA molecule. This hypothesis is further supported by the fact that unstructured homopolymeric RNAs are poor templates for de novo initiation of RNA synthesis (24). In addition, a specific interaction between domain I and the HCV nonstructural protein NS3 was reported last year (1). The first stem-loop, SL-AI, and in particular the run of guanosines in the stem, were absolutely required for this interaction. Recently, Friebe et al. (15), using an adapted replicon system, developed the first test allowing replication in vivo to be followed. They showed that the presence of the entire IRES region was required for maximal replication efficiency. In these experiments, the involvement of the IRES in translation was dissociated from its role in the replication process. Once transcribed to give rise to the 3′ (−) end of the minus strand, the nucleotides encoded by the IRES sequence are involved only in the replication process of the virus. In addition, the authors showed that the region encompassing nt −125 to −1 also supported significant replication. Accordingly, a functional subdomain encompassing SL-AI and SL-BI may represent the minimal initiation site for (+) strand RNA synthesis. Interestingly, in this study, truncated constructs bearing deletions of nt −96 to −72 or −104 to −61 totally abolished replication under transient-transfection conditions. In our model, these two regions are located in SL-BI, suggesting the involvement of this stem-loop in the mechanism of initiation of replication.
The aim of our study was to gain insight into the structure of the replication initiation site on the HCV (−) strand RNA and to further compare it with the HCV (+) strand. Interesting hypotheses could be drawn with respect to rules that might govern the replication process. Our study also provides a secondary-structure model that should prove useful for the study of RNA-protein interactions within the replication complex, for instance, by site-directed mutagenesis. It therefore constitutes an essential approach to a better understanding of the replication mechanism, in parallel with analysis of quasispecies sequences selected from patients with high viral titers. Indeed, Proutski et al. (47) demonstrated that polymorphism of the 3′ UTR of yellow fever virus, another flavivirus, was associated with either virulence or attenuation. Selected mutations in the IRES sequence which do not affect translation of the viral genome could affect the structural organization of the 3′ end of the HCV (−) strand RNA and therefore affect the replication efficiency.
The next step of our work will be to test the relevance of the secondary structure we propose in cell cultures expressing the viral replication complex. This study will be conducted in permissive and nonpermissive cell lines, in order to identify the cell-specific partners of the replication process. Such partners probably exist, since the HCV replicons replicate only in the Huh-7 cell line (38).
Acknowledgments
Thanks are due to C. M. Rice for providing the HCV p90FL-pU clone, ATM no. PH 13149611755, and to P. Romby for helpful advice.
We thank the Association pour la Recherche contre le Cancer for a grant to one of us (I.I.). This work was supported by the following grants: contract no. INE 20000407028/1 from the Fondation pour la Recherche Médicale, contract no. CR300/7603 from the Association pour la Recherche contre le Cancer, and contract no. 1A133C from the Réseau National Hépatite C.
REFERENCES
- 1.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudin, F., R. Marquet, C. Isel, J. L. Darlix, B. Ehresmann, and C. Ehresmann. 1993. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 229:382-397. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. A., H. Zhang, L. H. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunel, C., and P. Romby. 2000. Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol. 318:3-21. [DOI] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. USA 89:4942-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J. C., M. F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, and P. J. Barr. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colombo, M. 1999. Natural history and pathogenesis of hepatitis C virus related hepatocellular carcinoma. J. Hepatol. 31(Suppl. 1):25-30. [DOI] [PubMed] [Google Scholar]
- 14.Costa, M., and F. Michel. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 16:3289-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagedorn, C. H., E. H. van Beers, and C. De Staercke. 2000. Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase). Curr. Top. Microbiol. Immunol. 242:225-260. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 19.Hellen, C. U., and T. V. Pestova. 1999. Translation of hepatitis C virus RNA. J. Viral Hepat. 6:79-87. [DOI] [PubMed] [Google Scholar]
- 20.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda, M., L. H. Ping, R. C. Rijnbrand, E. Amphlett, B. Clarke, D. Rowlands, and S. M. Lemon. 1996. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 222:31-42. [DOI] [PubMed] [Google Scholar]
- 22.Hwang, S. B., K. J. Park, Y. S. Kim, Y. C. Sung, and M. M. Lai. 1997. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology 227:439-446. [DOI] [PubMed] [Google Scholar]
- 23.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 24.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 25.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jucker, F. M., H. A. Heus, P. F. Yip, E. H. Moors, and A. Pardi. 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264:968-980. [DOI] [PubMed] [Google Scholar]
- 27.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 28.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 32.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong, A. D., J. L. Kim, and C. Lin. 2000. Structure and function of hepatitis C virus NS3 helicase. Curr. Top. Microbiol. Immunol. 242:171-196. [DOI] [PubMed] [Google Scholar]
- 35.Lavanchy, D. 1999. Hepatitis C: public health strategies. J. Hepatol. 31(Suppl. 1):146-151. [DOI] [PubMed] [Google Scholar]
- 36.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins. J. Hepatol. 24:11-19. [PubMed] [Google Scholar]
- 37.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 39.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyngso, R. B., M. Zuker, and C. N. Pedersen. 1999. Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics 15:440-445. [DOI] [PubMed] [Google Scholar]
- 41.Makeyev, E. V., and D. H. Bamford. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi6. EMBO J. 19:124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marquet, R., F. Baudin, C. Gabus, J. L. Darlix, M. Mougel, C. Ehresmann, and B. Ehresmann. 1991. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 19:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 45.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486-27493. [PubMed] [Google Scholar]
- 46.Pan, T., D. M. Lond, and O. C. Uhlenbeck. 1993. Divalent metal ions in RNA folding and catalysis, p. 271-302. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Proutski, V., M. W. Gaunt, E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′-untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J. Gen. Virol. 78:1543-1549. [DOI] [PubMed] [Google Scholar]
- 48.Purcell, R. 1997. The hepatitis C virus: overview. Hepatology 26:11S-14S. [DOI] [PubMed] [Google Scholar]
- 49.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reigadas, S., M. Ventura, L. Sarih-Cottin, M. Castroviejo, S. Litvak, and T. Astier-Gin. 2001. HCV RNA-dependent RNA polymerase replicates in vitro the 3′ terminal region of the minus-strand viral RNA more efficiently than the 3′ terminal region of the plus RNA. Eur. J. Biochem. 268:5857-5867. [DOI] [PubMed] [Google Scholar]
- 51.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]
- 53.Russell, R., and D. Herschlag. 2001. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 308:839-851. [DOI] [PubMed] [Google Scholar]
- 54.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 55.Simmonds, P. 1999. Viral heterogeneity of the hepatitis C virus. J. Hepatol. 31(Suppl. 1):54-60. [DOI] [PubMed] [Google Scholar]
- 56.Smith, J. W. 1995. Medical manifestations of alcoholism in the elderly. Int. J. Addict. 30:1749-1798. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki, R., T. Suzuki, K. Ishii, Y. Matsuura, and T. Miyamura. 1999. Processing and functions of Hepatitis C virus proteins. Intervirology 42:145-152. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolk, B., D. Sansonno, H. G. Krausslich, F. Dammacco, C. M. Rice, H. E. Blum, and D. Moradpour. 2000. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 64.Yanagi, M., M. St Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 66.Zarrinkar, P. P., and B. A. Sullenger. 1998. Probing the interplay between the two steps of group I intron splicing: competition of exogenous guanosine with omega G. Biochemistry 37:18056-18063. [DOI] [PubMed] [Google Scholar]
- 67.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]