Abstract
Assembly of retrovirus-like particles only requires the expression of the Gag polyprotein precursor. We have exploited this in the development of a model system for studying the virus particle assembly pathway for bovine leukemia virus (BLV). BLV is closely related to the human T-cell leukemia viruses (HTLVs), and all are members of the Deltaretrovirus genus of the Retroviridae family. Overexpression of a BLV Gag polyprotein containing a carboxy-terminal influenza virus hemagglutinin (HA) epitope tag in mammalian cells led to the robust production of virus-like particles (VLPs). Site-directed mutations were introduced into HA-tagged Gag to test the usefulness of this model system for studying certain aspects of the virus assembly pathway. First, mutations that disrupted the amino-terminal glycine residue that is important for Gag myristylation led to a drastic reduction in VLP production. Predictably, the nature of the VLP production defect was correlated to Gag membrane localization. Second, mutation of the PPPY motif (located in the MA domain) greatly reduced VLP production in the absence of the viral protease. This reduction in VLP production was more severe in the presence of an active viral protease. Examination of particles by electron microscopy revealed an abundance of particles that began to pinch off from the plasma membrane but were not completely released from the cell surface, indicating that the PPPY motif functions as a late domain (L domain).
The assembly of retrovirus particles requires the expression of the Gag polyprotein precursor (PrGag), which is used as a principal building scaffold for retrovirus assembly and budding from infected cells (42, 49). During or after the process of particle release, the action of the retroviral protease cleaves, except for the spumaviruses, PrGag into mature matrix (MA), capsid (CA), and nucleocapsid (NC) proteins (16, 42, 49).
The retrovirus Gag protein has all the necessary information to mediate intracellular transport to the cell membrane, to direct assembly of virus particles, and to catalyze the budding process (9). The events associated with Gag-mediated budding appear to be similar among different retroviruses. After synthesis of Gag in the cytoplasm, Gag-Gag interactions, Gag-RNA interactions, Gag-membrane interactions, and Gag-host protein interactions occur. The targeting of Gag to the cell membrane leads to a bulging out of the nascent virus particles. It is clear that each step of the virus assembly process requires a great degree of precision and involves highly specific macromolecular interactions, including protein-protein and protein-RNA interactions (9, 16).
The expression of retroviral PrGag alone can lead to the formation of virus-like particles (VLPs). The formation and release of VLPs occur in the absence of expression of the viral envelope glycoproteins, reverse transcriptase, or full-length viral RNA (46). Retroviral RNA can act as a scaffold to facilitate virus particle assembly (27, 38). In the absence of viral RNA, cellular RNAs are used and can be found in VLPs.
There are three functional assembly domains in PrGag (31). Each domain is responsible for a separate function in the assembly process. The three functional domains are the membrane-binding domain (M domain), the late domain (L domain), and the interaction domain (I domain). The M domain for most retroviruses resides primarily in the MA domain and contains a myristylation signal (2, 39), while the I domain resides in the CA and NC domains of Gag.
L domains have been observed to be critical for efficient pinching off of the virus particle. L domains have been identified by extensive mutational analysis in many retroviruses (3, 10, 11, 22, 34, 48, 50-52, 54). Different retroviruses may utilize different viral proteins and structural motifs to accomplish the same late budding function. Rous sarcoma virus (RSV), murine leukemia virus (MLV), and Mason-Pfizer monkey virus (MPMV) all have L domains that consist of a highly conserved PPPY motif as the core sequence and are located near the junction of the MA and CA domains in Gag (5, 31, 47, 50-52). A PPPY motif has been also found in the matrix protein of rhabdoviruses and can function as an L domain (5). In contrast, the L domains of lentiviruses are located at the C terminus of the PrGag and have distinct core motifs, PTAP in human immunodeficiency virus type 1 (HIV-1) and YXXL in equine infectious anemia virus (3, 11, 34). The retroviral L domains are protein-interaction domains and most likely function by binding to specific cellular proteins that facilitate the late stages of retroviral particle release (45).
Ubiquitination has been found to be involved in virus particle release from infected cells, extending earlier studies that had identified ubiquitin in retrovirus particles (36, 45). Recently, proteins associated with the ubiquitin pathway have been found that associate ubiquitin with virus particle release. Specifically, cellular factors that are associated with the ubiquitin pathway have been shown to interact with L domains (12, 20, 33, 35, 44). The Nedd4-like family of E3 ubiquitin protein ligases (specifically LDI-1) has been identified as the cellular protein that interacts with the PPPY motif (20, 33). TSG101, a putative ubiquitin regulator that is involved in trafficking of endosomal proteins, has been reported to interact with the PTAP motif and therefore to be involved in L domain function (6, 12, 44).
The Deltaretrovirus genus of the Retroviridae family includes bovine leukemia virus (BLV), human T-cell leukemia virus type 1 (HTLV-1), and HTLV-2. These viruses replicate to low titers in their natural hosts and are poorly infectious in cell culture. Cocultivation is typically used to infect permissive host cells. Because of these difficulties, information regarding the molecular details of their life cycles, including virus assembly, is limited. Aspects of the deltaretrovirus assembly process that have been studied in some detail include BLV RNA encapsidation, BLV and HTLV-1 Gag myristylation, and the role of basic residues in MA of HTLV-1 Gag membrane localization and virus production (1, 17, 21, 23, 24). The primary encapsidation signal of BLV consists of two stable RNA stem-loop structures (i.e., SL1 and SL2) located just downstream of the gag start codon (24).
To more easily study the steps involved in the virus assembly pathway of BLV, we have developed a VLP model system. In this system, the BLV Gag protein containing a C-terminal influenza virus hemagglutinin (HA) epitope tag was overexpressed in mammalian cells and led to the robust production of VLPs. A series of site-directed mutations were introduced to assess the validity of this model system. First, mutations that disrupted the myristylation signal led to a dramatic reduction in VLP production. Second, mutations in a PPPY motif in the MA domain reduced VLP production and resulted in particles that budded but were not released from cells, as determined by electron microscopy, indicating that this motif functions as an L domain.
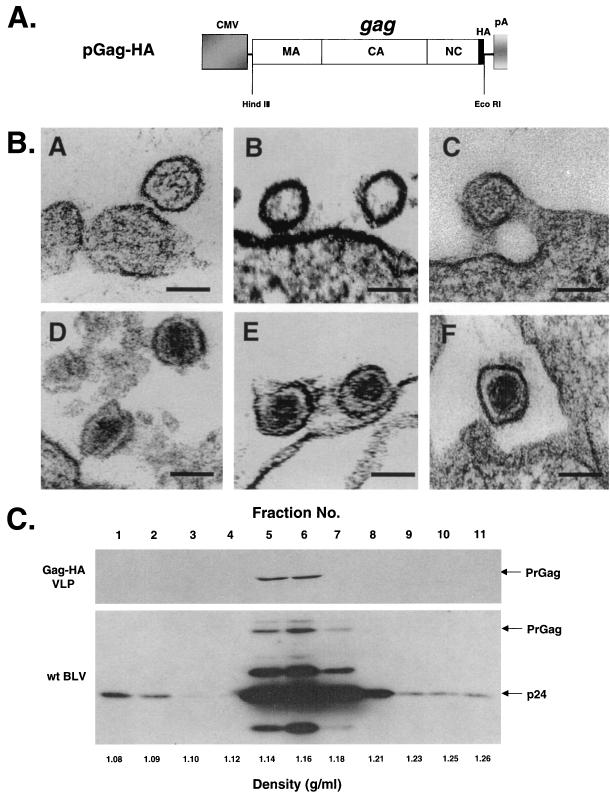
To create a BLV Gag expression vector, PCR amplification of the BLV gag gene from BLV-SVNEO (8) was performed to create a HindIII site immediately upstream of the start codon and an EcoRI site immediately downstream of the stop codon by using 5′-AAAAAAAAGCTTGATGGGAAATTCCCCCTCCTAT-3′ as the upstream primer and 5′-AAAAAAGAATTCTCGTTTTTTGATTTGAGGGTTGG-3′ as the downstream primer. The amplified fragment was digested with HindIII and EcoRI and cloned into a mammalian HA-tagged expression vector, pMH (Boehringer Mannheim, Indianapolis, Ind.). The resulting plasmid, pGag-HA, has a HA epitope tag fused to the C terminus of the BLV PrGag (Fig. 1A). In these studies, pGag-HA is the parental construct.
FIG. 1.
Overexpression of BLV PrGag leads to the production of VLPs. (A) Expression construct used for overexpression of PrGag. The gray box represents the cytomegalovirus promoter (CMV). The long white rectangular box represents the gag gene with the location of the corresponding MA, CA, and NC protein domains indicated. The influenza virus HA epitope tag is indicated by the thin black box. The bovine growth hormone polyadenylation signal (pA) is indicated. The locations of the HindIII and EcoRI restriction sites are indicated. (B) Visualization of VLPs by electron microscopy. Panels A to C show representative VLPs produced from COS-1 cells. The VLPs are immature and do not have core particles because the viral protease is absent. Panels D to F show representative wt virus particles produced from fetal lamb kidney cells chronically infected with BLV. The mature cores are readily visible. Bar, 100 nm. (C) Sucrose density gradient fractionation of VLPs. VLPs produced from COS-1 cells stably transfected with pGag-HA or wt BLV produced from fetal lamb kidney cells chronically infected with BLV were layered onto a sucrose gradient composed of 10, 20, 30, 40, 50, and 60% sucrose layers. The gradients were centrifuged, and 11 fractions were collected starting from the top of the centrifuge tubes. Fractionated samples were analyzed for BLV PrGag expression by Western blot analysis using either an anti-HA Ig (VLPs) or an anti-CA Ig (wt BLV). The locations of the 44-kDa Gag polyprotein (PrGag) and the 24-kDa capsid (p24) protein are indicated.
To express BLV PrGag in mammalian cells, COS-1 cells were transfected with pGag-HA and placed under G418 selection to obtain stable cells expressing PrGag. Cells were grown in 100- or 60-mm-diameter dishes in Dulbecco's modified Eagle medium (GIBCO BRL, Gaithersburg, Md.) supplemented with 10% Fetal Clone III serum (HyClone, Logan, Utah). Superfect (Qiagen, Valencia, Calif.) was used for transfection of cells. Two days posttransfection, cells were placed under G418 selection until resistant colonies formed (∼3 weeks). Approximately 100 G418-resistant colonies were pooled and used for VLP analysis.
To confirm that the Gag detected in the pelleted material was an indication of VLP formation and release, the stably transfected COS-1 cells were examined by electron microscopy. For thin sectioning, cell pellets were fixed with 2.5% glutaraldehyde. After dehydration in a grade series of cold ethanol, the samples were embedded in Epon 812 resin. Ultrathin sections (90 nm) were stained with uranyl acetate and viewed with a Philips CM 12 electron microscope. Figure 1B (panels A to C) shows representative VLPs that were identified from the examination of thin sections. The VLPs do not have an electron dense core due to the absence of a viral protease. Immature retroviral particles can have an electron dense ring underneath the viral membrane. This is observed in Fig. 1B but is not easily observed in the other particles shown. These VLPs are similar in diameter to the wild-type (wt) BLV particles produced from fetal lamb kidney cells chronically infected with BLV (Fig. 1B, panels D to F). The electron dense cores in wt BLV particles are readily seen. Comparison of the diameter of the VLPs with that of wt BLV particles indicates that the VLPs are very similar in size to wt particles.
To further confirm that BLV Gag was being released from cells as VLPs, we used sucrose density gradient fractionation. VLPs or wt BLV preparations were pelleted from the culture supernatant and were resuspended in 0.1 ml of phosphate-buffered saline (PBS). Resuspended particles were placed onto a sucrose gradient composed of 10, 20, 30, 40, 50, and 60% sucrose layers in PBS (0.6 ml each). The gradients were centrifuged at 33,200 rpm at 4°C for 16 h in a Beckman SW60Ti rotor. Eleven gradient fractions were collected from top to bottom, and the amount of Gag in each fraction was determined by Western blotting (Fig. 1C). The majority of the Gag from wt BLV particles was identified in fractions 5 to 7, with the peak in fraction 6 (Fig. 1C, bottom panel). The Gag derived from the VLPs was detected only in fractions 5 and 6. This indicates that the VLPs had a density similar to that of wt BLV. In total, these data indicate that overexpression of PrGag in mammalian cells leads to the production of VLPs.
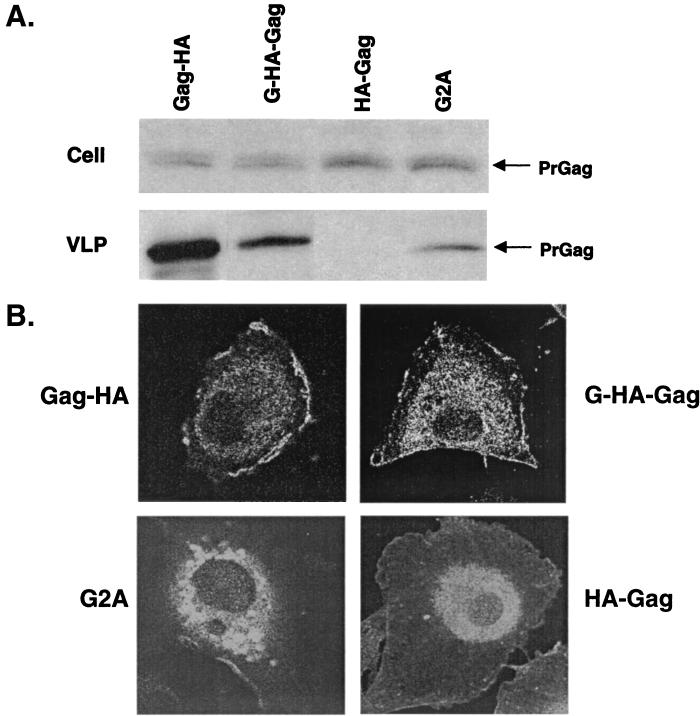
The Gag protein of most retroviruses, including BLV, is myristylated at the N-terminal glycine residue. The cotranslational addition of the 14-carbon fatty acid myristic acid to the N-terminal glycine residue plays an essential role in targeting the Gag precursor to the cell membrane and in the subsequent assembly, budding, and release of extracellular particles. The elimination or substitution of the glycine residue in some retroviruses abrogates particle production. To test this for BLV, three mutant constructs were made. First, the mutant G2A has the glycine residue immediately after the start codon changed to an alanine residue and would prevent myristylation of the BLV PrGag. Second, HA-Gag has an HA epitope tag fused to the N terminus of Gag and thereby blocks the exposure of the glycine residue to myristic acid, which would prevent the myristylation of the BLV PrGag. The plasmid pHA-Gag has an HA epitope tag fused to the N terminus of the BLV PrGag that was generated similarly by cloning the BLV gag gene into pHM6 (Boehringer Mannheim). Third, the mutant G-HA-Gag was constructed from HA-Gag by adding an extra glycine before the HA tag. The additional glycine was added to test if this residue could restore the myristylation signal. Site-directed mutations were added using the QuikChange XL kit (Stratagene) according to the manufacturer's instructions. All mutants made were sequenced to verify the correct introduction of the desired mutation and the absence of undesired mutations.
Each vector was transfected into COS-1 cells in parallel with pGag-HA. To prepare VLP samples, the cell culture supernatant was clarified (5 min at 700 × g) and the VLPs were pelleted by centrifugation through a sucrose cushion for 1 h at 20,000 × g. The pelleted VLPs were resuspended in radioimmunoprecipitation assay buffer (1% IGEPAL CA-630, a nonionic detergent; 50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% deoxycholate; 5 mM EDTA; and 0.1% sodium dodecyl sulfate). Cell lysates were prepared by trypsinizing and pelleting VLP-producing cells and then resuspending them in radioimmunoprecipitation assay buffer. Western blot analysis was done using a primary antibody directed against the HA epitope tag (Covance, Berkeley, Calif.) and anti-mouse immunoglobulin (Ig) and a sheep horseradish peroxidase-linked whole antibody as the secondary antibody with the enhanced chemiluminescence Western analysis kit (Amersham, Arlington Heights, Ill.). Each vector was observed to express the Gag protein in cells at levels comparable to that of the parental pGag-HA (Fig. 2A). The efficiency of VLP production was analyzed by determining the percentage of VLP-associated Gag protein normalized to the amount of Gag in cells. Quantitation of band intensities was done using the Quantity One software package with the Chemi Doc 2000 Documentation System (Bio-Rad, Richmond, Calif.) according to the manufacturer's specifications. Analysis of VLP production indicated that the substitution of glycine for alanine in the G2A mutant drastically reduced VLP production to about one-fifth (19%) that of the parental vector (Fig. 2A). No BLV PrGag was detected for HA-Gag, indicating the complete loss of VLP production. Interestingly, by addition of the additional glycine in G-HA-Gag, VLP production could be detected at about half (56%) the level observed for the parental vector (Fig. 2A).
FIG. 2.
The amino-terminal glycine is required for efficient VLP production and PrGag membrane targeting. (A) VLP production. pGag-HA and mutants were stably transfected into COS-1 cells. Cell and virion lysates were analyzed by Western blot using an anti-HA antibody. The Western blots were subjected to quantitative fluorochemical analysis as described in the text. This experiment was repeated three times, with representative results shown. (B) Cellular distribution of PrGag. Cells stably transfected with VLP constructs were grown on coverslips, fixed, and incubated with an anti-HA Ig followed by incubation with Alexa Fluor 488-conjugated anti-mouse Ig. Images were collected using a confocal microscope.
To further examine the nature of the mutant phenotypes, we used confocal microscopy to analyze the subcellular localization of the Gag proteins. Transfected cells were grown on coverslips and fixed with 4% paraformaldehyde and permeabilized with Triton X-100, both diluted in PBS. The cells were then incubated with anti-HA antibody followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes, Eugene, Oreg.). Expression of Gag from Gag-HA yielded a stippled appearance along the plasma membrane (Fig. 2B). G2A was observed to have both perinuclear and cytoplasmic distribution (Fig. 2B). Very little plasma membrane staining was observed for G2A, indicating a severe defect in plasma membrane targeting. HA-Gag demonstrated staining that was primarily intracellular, with a predominant bright perinuclear halo (Fig. 2B). No peripheral staining was observed with HA-Gag, indicating that translocation of Gag to the plasma membrane was severely impaired. In contrast, the localization of G-HA-Gag led to a cytoplasmic distribution, though a stippled staining pattern along the plasma membrane was also observed (Fig. 2B).
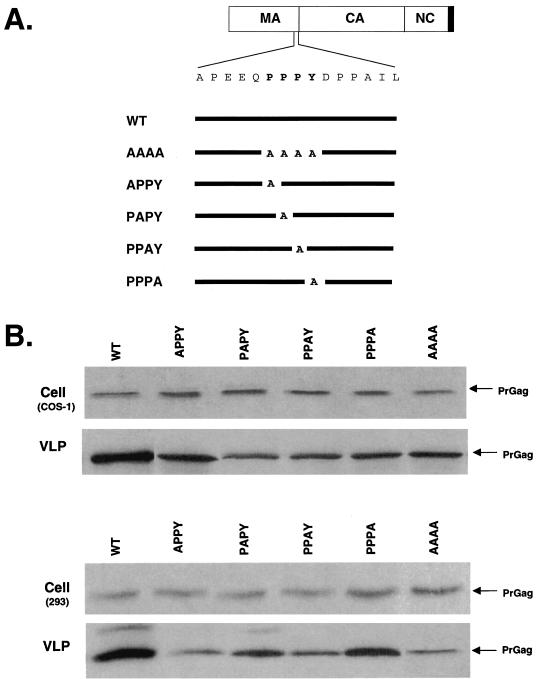
A functional L domain is required for the budding and release of many retroviruses. The RSV, MLV, and MPMV Gag L domain maps specifically to a proline-rich sequence, PPPY, located near the MA and CA protein domain junction in Gag (48, 51, 52). Examination of the protein-coding sequence of BLV indicates that a similar PPPY motif is located in the MA domain near the boundary with CA. To examine the significance of the PPPY motif in the process of particle budding and release, a series of mutations were introduced into pGag-HA (Fig. 3A). In the APPY, PAPY, PPAY, and PPPA mutants, each residue within the PPPY motif was individually replaced with alanine. In the mutant AAAA, all four residues in the PPPY motif replaced alanine residues.
FIG. 3.
Mutation of the PPPY motif reduces VLP production. (A) Amino acid substitutions to the conserved PPPY sequence. The PrGag is shown as described for Fig. 4. The amino acid sequence of PPPY domain and its flanking sequence are expanded. The full-length black line represents the wt Gag sequence. The alanine substitutions are indicated below and are aligned with the expanded sequence. The name of each mutant is indicated on the left. (B) Analysis of VLP production. COS-1 cells or 293 cells were stably transfected with pGag-HA or the indicted mutants. Cell and VLP lysates were analyzed by Western blot using an anti-HA Ig. The Western blots were subjected to quantitative fluorochemical analysis as described in the text. This experiment was repeated three times, with representative results shown.
To test the influence of the PPPY mutants on VLP production, COS-1 cells were transfected in parallel with the parental construct and each mutant. Analysis of Gag expression in cells and subsequent VLP production indicated that, for each of the alanine substitution mutants, there was a general decrease in the release of VLPs (Fig. 3B). Again, the efficiency of VLP production was analyzed by determining the percentage of VLP-associated Gag protein normalized to the amount of Gag in cells. The reduction in VLP production was most pronounced for APPY, PAPY, and AAAA, where there was a reduction in VLP production that was approximately half (42, 45, and 42%, respectively) that of the parental construct. These results indicate that mutations in the PPPY motif can significantly reduce VLP production and suggest that this motif may function as an L domain.
Recent observations have indicated that L domain function can be cell type dependent (7). To test this for BLV, the panel of PPPY mutants was tested in parallel in 293 cells for expression in cells and for production of VLPs. As was observed with COS-1 cells, there was a general decrease in VLP release for each of the mutants tested (Fig. 3B). In particular, the APPY and AAAA mutants were most pronounced, reducing VLP production to approximately one-fourth (26 and 28%, respectively) that of the parental construct. These results provide further evidence in support of the PPPY motif functioning as an L domain.
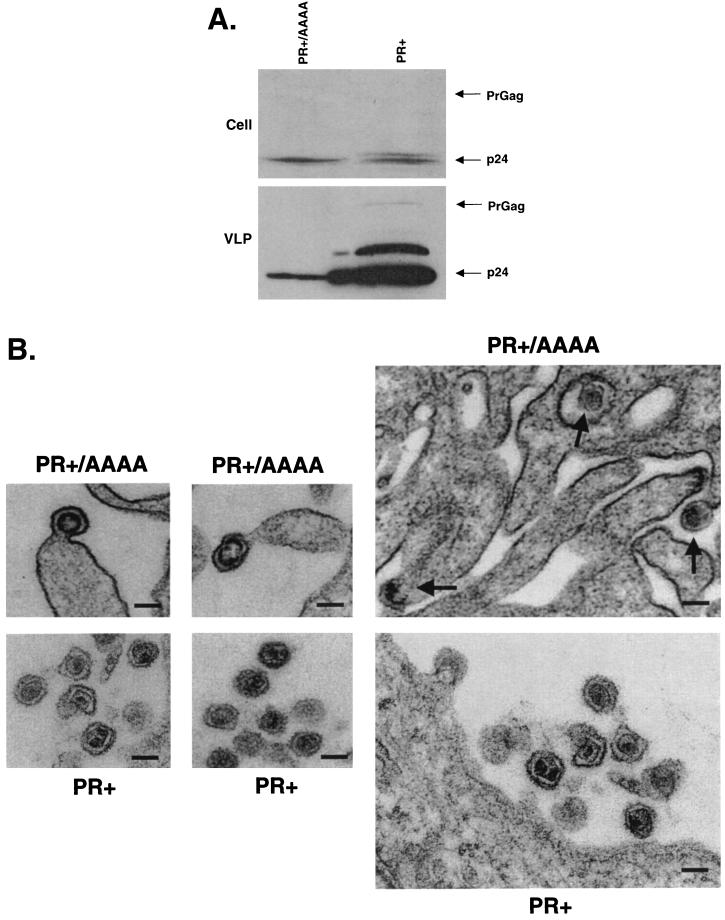
Previous studies have indicated that perturbation of Gag precursor processing can severely impair the assembly of infectious virions (18, 19). In previous studies with HIV-1, a less severe L domain defect was observed when protease function was absent (15). To determine if the presence of protease activity can enhance the VLP production defect observed by mutation of the PPPY motif, we examined the phenotype of the PPPY domain mutant in the context of an active viral protease (Fig. 4). The construct expressing Gag and the viral protease (PR+) was transfected into 293 cells in parallel with PR+/AAAA, a derivative with the PPPY motif mutated to AAAA. The production of VLPs was monitored by Western blotting using an anti-BLV CA monoclonal antibody (i.e., BLV3) (VMRD, Pullman, Wash.). As anticipated for a PR+ clone, the completely processed BLV CA was detected in both PR+ and PR+/AAAA, demonstrating the expression of the viral protease (Fig. 4A). Although not readily observed from cell lysates, PrGag was observed with longer exposures (data not shown). VLP production for PR+/AAAA was reduced to about 1/20 (5%) that of PR+. An incompletely processed Gag intermediate and a small amount of unprocessed Gag precursor could also be readily observed with PR+ but not in PR+/AAAA. This was presumably due to the relative abundance of VLPs produced by PR+ to that of PR+/AAAA. These observations in total indicate that the effect of the PPPY motif on VLP production was more pronounced in the context of an active protease.
FIG. 4.
The PPPY motif functions as an L domain. (A) An active protease enhances the L domain defect. 293 cells were transfected with PR+ or PR+/AAAA. Two days posttransfection, cell and VLP lysates were made and analyzed by Western blot using an anti-CA Ig. The Western blots were subjected to quantitative fluorochemical analysis as described in the text. This experiment was repeated three times, with representative results shown. (B) Visualization of the L domain defect. Representative VLPs budding but not released from 293 cells are shown (PR+/AAAA). The VLPs appear to be undergoing core maturation, presumably due to the activation of the viral protease. Representative particles released from 293 cells are also shown for the parental BLV vector construct (PR+). Arrows point to the location of budding particles. Bar, 100 nm.
Finally, we used electron microscopy to determine if the reductions in VLP production that we had observed correlated with defects in virus release. By comparing VLP production of wt BLV with that of PR+/AAAA, we observed a very high proportion of budding virus particles that did not release from cells compared to that of wt BLV (Fig. 4B). Interestingly, VLPs with donut-shaped (immature) cores were observed for PR+/AAAA. The high proportion of VLPs observed for PR+/AAAA that did not release from cells provide further support that the PPPY motif functions as an L domain. It is interesting that no long “strings” of particles were observed like those seen with HIV-1 and MLV L domain mutants.
We have described a model system that overexpresses the BLV PrGag in mammalian cells and have demonstrated that this leads to the assembly, budding, and release of VLPs. Our observations that overexpression of Gag leads to the production of VLPs is similar to observations made with other retroviruses, including BLV, that relied on the use of recombinant vaccinia viruses (14) or recombinant baculovirus systems (1, 17, 37, 43).
Gag proteins of many retroviruses are myristylated at the N-terminal glycine residue, and the covalent attachment of myristic acid is required for efficient membrane association and virion formation (2, 30, 32, 39-41). We found that mutation of the N-terminal glycine led to a dramatic reduction in VLP production, similar to previous observations made with BLV Gag expressed from a baculovirus vector in insect cells (17). The mutation of the N-terminal glycine also led to a dramatic reduction in the ability of PrGag to localize at the plasma membrane. When an HA epitope tag was fused to the N terminus of PrGag, N-terminal myristylation of Gag was blocked, and VLP production along with localization of Gag to the plasma membrane was severely impaired. However, addition of a glycine in front of HA-Gag restored VLP production.
Two observations suggest that the N-terminal glycine residue alone was not sufficient for efficient PrGag membrane binding and VLP production. First, the mutant G2A severely impaired but did not completely eliminate VLP production. It is worth noting that, in previous studies with BLV and HTLV-1, mutation of the N-terminal glycine eliminated detectable particle production (1, 17, 21). It is possible that the level of Gag expression in our system was high enough that low-level VLP production was detected, whereas it was not in previous reports. Second, the G-HA-Gag mutant restored VLP production, presumably by restoration of PrGag myristylation, though this has not been formally tested. Studies with HIV-1 have demonstrated that hydrophobic residues within the first 14 amino acids of HIV-1 Gag can enhance the interaction with the plasma membrane (28, 29, 54). Extensive mutagenesis of the N-terminal residues of the HIV-1 Gag has indicated that the first five residues blocked or impaired Gag myristylation (29), while mutation of residues 6, 7, and 8 reduced membrane binding without affecting N-terminal myristylation (28). In light of these observations with HIV-1, the restored VLP production phenotype observed with the G-HA-Gag may be due to the introduction of an N-terminal glycine residue but with the proviso that the HA sequence disrupted the myristylation signal and prevented restoration to wt levels. The nature of the restored VLP production phenotype for G-HA-Gag is presently not completely understood and needs to be further investigated. The results with HIV-1 support the myristyl switch model for the regulation of Gag membrane binding, which proposes that membrane binding is determined by the degree of exposure or sequestration of the N-terminal myristate moiety. The electrostatic interaction between a cluster of basic amino acids positioned between residues 15 and 31 in HIV-1 MA and acidic membrane phospholipids can stabilize the protein-lipid association and can potentially contribute more binding energy than the hydrophobic myristate moiety (13, 28, 32, 54). This is further supported by the fact that the PrGag of several retroviruses, including RSV, visna virus, equine infectious anemia virus, and caprine arthritis-encephalitis virus, is fully capable of membrane targeting and budding without the need of myristylation (26, 49). Therefore, the proximity of myristate with downstream residues can be important for membrane targeting and VLP production. In total, analysis of the affects of site-directed mutations for all the residues near the N terminus of the BLV PrGag will help establish the roles of these residues in Gag membrane targeting.
Basic residues in the BLV MA domain likely play a role in PrGag membrane binding. The majority of the basic residues in BLV MA are not concentrated at the N terminus but appear on one side of the molecule within the three-dimensional structure (4, 25). This could create a basic surface exposed to the solvent that is ideally situated for the interaction with acidic phospholipid headgroups of the inner face of the membrane. We are presently investigating the role of the basic residues in BLV MA in PrGag membrane binding.
We have shown that mutagenesis of the PPPY motif in the MA domain of PrGag plays an important role in VLP release. PPPY mutants displayed a reduced level of VLP production from COS-1 cells that appears more pronounced when 293 cells are used. The PPPY mutant phenotype also appeared more pronounced in the presence of the viral protease. An abundance of budding particles that had not been released by cells was commonly seen with the PPPY mutants, which is in stark contrast to that observed with the wt particles. Interestingly, most of the budding particles for the PR+/AAAA mutant that had not released had developing core particles. This indicates the presence of viral protease activity prior to release of the VLP. This is in contrast to what has been observed for HIV-1 (6). However, our observations could be influenced by the overexpression of Gag in the VLP-producing cells.
In this study, the individual exchange of an alanine residue with a residue in the PPPY motif led to a significant reduction in VLP production, as did the simultaneous exchange of the PPPY motif with four alanine residues. With MPMV, each residue in the PPPY motif was found to be critical for late domain function, but the tyrosine residue appeared to be more important (51). The individual proline residues in the PPPY motif of RSV were found to equally influence virus budding when mutated to glycine (50). The simultaneous exchange of the PPPY motif in MLV with alanine residues resulted in a significant defect in virus budding (53).
Our finding that the viral protease enhanced the phenotype of the PPPY mutants is similar to an observation made with HIV-1, where the inactivation of the viral protease alleviated the requirement of the PTAP motif in virus release (15). Interestingly, in both RSV and MPMV, protease inactivation did not alter the requirement of the late domains for budding (48, 51).
In summary, we have described a BLV-like particle model system to study virus assembly and release. The data presented indicate that aspects of Gag membrane targeting and virus release can be analyzed. Given the difficulties in studying the molecular biology of the deltaretroviruses, this model system will greatly facilitate future molecular analyses.
Acknowledgments
We thank Melodie Knowlton for technical assistance, Kathy Wolken for assistance with confocal microscopy and electron microscopy, and Akira Ono for very helpful advice regarding sucrose density gradient ultracentrifugation. We also thank Eric Freed and Anette Schneemann for critical reviews of the manuscript.
This work was supported by the American Cancer Society (RPG 0027801).
REFERENCES
- 1.Bouamr, F., L. Garnier, F. Rayne, A. Verna, N. Rebeyrotte, M. Cerutti, and R. Z. Mamoun. 2000. Differential budding efficiencies of human T-cell leukemia virus type I (HTLV-I) Gag and Gag-Pro polyproteins from insect and mammalian cells. Virology 278:597-609. [DOI] [PubMed] [Google Scholar]
- 2.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conte, M. R., and S. Matthews. 1998. Retroviral matrix proteins: a structural perspective. Virology 246:191-198. [DOI] [PubMed] [Google Scholar]
- 5.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derse, D., and L. Martarano. 1990. Construction of a recombinant bovine leukemia virus vector for analysis of virus infectivity. J. Virol. 64:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 10.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnier, L., L. J. Parent, B. Rovinski, S. X. Cao, and J. W. Wills. 1999. Identification of retroviral late domains as determinants of particle size. J. Virol. 73:2309-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, S. A., and J. L. Affranchino. 1998. Substitution of leucine 8 in the simian immunodeficiency virus matrix protein impairs particle formation without affecting N-myristylation of the Gag precursor. Virology 240:27-35. [DOI] [PubMed] [Google Scholar]
- 14.Hertig, C., A. D. Pye, A. D. Hyatt, and D. B. Boyle. 1994. Retrovirus-like particles produced by vaccinia viruses expressing gag-pro-pol region genes of bovine leukaemia virus. J. Gen. Virol. 75:2213-2221. [DOI] [PubMed] [Google Scholar]
- 15.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, E. 1994. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin. Virol. 5:71-83. [Google Scholar]
- 17.Kakker, N. K., M. V. Mikhailov, M. V. Nermut, A. Burny, and P. Roy. 1999. Bovine leukemia virus Gag particle assembly in insect cells: formation of chimeric particles by domain-switched leukemia/lentivirus Gag polyprotein. Virology 265:308-318. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 67:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 20.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Blanc, I., A. R. Rosenberg, and M. C. Dokhelar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansky, L. M., A. E. Krueger, and H. M. Temin. 1995. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J. Virol. 69:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansky, L. M., and R. M. Wisniewski. 1998. The bovine leukemia virus encapsidation signal is composed of RNA secondary structures. J. Virol. 72:3196-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews, S., M. Mikhailov, A. Burny, and P. Roy. 1996. The solution structure of the bovine leukemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO J. 15:3267-3274. [PMC free article] [PubMed] [Google Scholar]
- 26.McDonnell, J. M., D. Fushman, S. M. Cahill, W. Zhou, A. Wolven, C. B. Wilson, T. D. Nelle, M. D. Resh, J. Wills, and D. Cowburn. 1998. Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus. J. Mol. Biol. 279:921-928. [DOI] [PubMed] [Google Scholar]
- 27.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono, A., M. Huang, and E. O. Freed. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart, J.-C., and H. G. Göttlinger. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parent, L. J., C. B. Wilson, M. D. Resh, and J. W. Wills. 1996. Evidence for a second function of the MA sequence in the Rous sarcoma virus Gag protein. J. Virol. 70:1016-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen, L., J. K. Battles, W. H. Ennis, K. Nagashima, and M. A. Gonda. 1990. Characterization of virus-like particles produced by a recombinant baculovirus containing the gag gene of the bovine immunodeficiency-like virus. Virology 178:435-451. [DOI] [PubMed] [Google Scholar]
- 38.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 39.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz, A. M., and S. Oroszlan. 1983. In vivo modification of retroviral gag gene-encoded polyproteins by myristic acid. J. Virol. 46:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 43.Takahashi, R. H., K. Nagashima, T. Kurata, and H. Takahashi. 1999. Analysis of human lymphotropic T-cell virus type 2-like particle production by recombinant baculovirus-infected insect cells. Virology 256:371-380. [DOI] [PubMed] [Google Scholar]
- 44.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss, R., N. Teich, H. Varmus, and J. Coffin. 1982. Molecular biology of tumor viruses: RNA tumor viruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Weldon, R. A., and J. W. Wills. 1993. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J. Virol. 67:5550-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wills, J. W., and R. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 50.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]