Abstract
Chromatin remodeling complexes have been implicated in the disruption or reformation of nucleosomal arrays resulting in modulation of transcription, DNA replication, and DNA repair. Here we report the isolation of WCRF, a new chromatin-remodeling complex from HeLa cells. WCRF is composed of two subunits, WCRF135, the human homolog of Drosophila ISWI, and WCRF180, a protein related to the Williams syndrome transcription factor. WCRF180 is a member of a family of proteins sharing a putative heterochromatin localization domain, a PHD finger, and a bromodomain, prevalent in factors involved in regulation of chromatin structure.
The genome of eukaryotes is packaged into chromatin, the fundamental unit of which is the nucleosome. The wrapping of ≈146 bp of DNA around a histone octamer, two copies each histone H2A, H2B, H3, and H4, forms the nucleosome core. The arrangement of nucleosomes into an array forms the higher order chromatin structure. Although this packaging serves to compact the DNA in the nucleus, it also presents a barrier to cellular processes such as transcription, DNA replication, and DNA repair. The control of accessibility to the nucleosomal DNA therefore provides an important regulatory point for these processes (1).
Recent genetic and biochemical studies have identified a host of multisubunit complexes that, in an ATP-dependent manner, are able to alter the structure of the nucleosome. The first identified chromatin remodeling factor, the large 11-subunit SWI/SNF complex, was discovered through genetic studies in yeast. The catalytic subunit of this complex, SWI2/SNF2, was identified as the DNA-dependent ATPase (2–4). The SWI2/SNF2 enzyme is part of a larger family of enzymes whose members share a number of conserved domains including an ATP-binding and hydrolysis motif initially referred to as the helicase signature domain (5). Although the members of this family possess DNA-dependent ATPase activity, it is interesting that they do not appear to catalyze the unwinding of DNA (4). A complex similar to that of the SWI/SNF complex was recently identified in yeast (RSC), which, unlike the SWI/SNF complex, is essential for growth (6). The catalytic subunit has been identified as STH1, a homolog of SWI2/SNF2. Complexes homologous in polypeptide composition and biochemical activity to that of SWI/SNF have been identified in other organisms (7–12).
Additional chromatin remodeling complexes have been identified through biochemical studies in Drosophila. These complexes include the nucleosome remodeling factor (NURF) (13, 14), the chromatin accessibility complex (CHRAC) (15), and the ATP-utilizing chromatin assembly and remodeling factor (ACF) (16). The ATPase subunit of each of these complexes is the ISWI protein, a protein related to SWI2/SNF2 (17). Two human proteins (hSNF2L and hSNF2h) with high degree of homology to Drosophila ISWI have been described (18, 19). The homology between these proteins is not only in their highly conserved ATPase domain but also extends across the entire protein to include the SANT (SWI3, ADA2, N-COR, and TFIIB B′′ domain, which is conserved among many regulators of transcription and chromatin structure (20). Recent studies have also identified ISWI-containing chromatin remodeling complexes in both human and Saccharomyces cerevisiae (21, 22).
To gain further insight into the biochemical activity and polypeptide composition of chromatin remodeling complexes in human cells, we examined fractionated HeLa nuclear extract by using a high-resolution mononucleosome disruption assay. This analysis revealed a two-subunit (135 kDa and 180 kDa) chromatin remodeling complex displaying a qualitatively different mononucleosome DNase I cleavage pattern. Mass spectrometric analysis of the two subunits identified the 135-kDa subunit as human SNF2h. The 180-kDa subunit was identified through the analysis of expressed sequence tags (ESTs) as a protein closely related to Williams syndrome transcription factor (WSTF) (23, 24). The gene for WSTF is one of the many genes deleted in Williams syndrome, a complex developmental disorder marked by mental retardation, growth defects, cardiovascular disease, dysmorphic facial features, and a unique cognitive profile. This disorder is caused by a contiguous gene deletion (<1 Mb) at 7q11.23 (23, 24). Therefore it is not clear whether deletion of WSTF may result in any of the phenotypic changes seen in Williams syndrome. Analysis of the nonredundant GenBank database by using the blast algorithm (25) revealed the presence of another closely related protein to the 180-kDa subunit, indicating the presence of a family of related proteins.
Materials and Methods
Purification of the WCRF Complex.
WCRF was purified from 1.2 grams of HeLa nuclear extract (Fig. 2a). Nuclear extract was loaded on a 200-ml column of phosphocellulose (P11, Whatman) and fractionated stepwise by the indicated KCl concentrations in buffer A (20 mM Tris⋅HCl, pH 7.9/0.2 mM EDTA/10 mM β-mercaptoethanol/10% glycerol/0.2 mM PMSF). The P11 1.0-M KCl fraction (100 mg) was loaded on a 10-ml DEAE-Sephacel column (Pharmacia) and eluted with 0.35 M KCl. The 0.35 M KCl elution (50 mg) was dialyzed to 10 mM KxPO4 in buffer B (5 mM Hepes, pH 7.6/1 mM DTT/0.5 mM PMSF/10 μM CaCl2/10% glycerol/40 mM KCl) and loaded on a 20-ml BioGel HT column (Bio-Rad). The column was resolved by using a linear 10-column volume gradient of 10 to 600 mM KxPO4. Fractions containing WCRF (≈275 mM KxPO4, 5.0 mg) were dialyzed to 100 mM KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin, and loaded on Mono Q HR 5/5 (Pharmacia). The column was resolved by using a linear 10-column volume gradient of 100 to 500 mM KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin. WCRF containing fractions (≈300 mM KCl, 0.7 mg) were dialyzed to 100 mM KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin, and fractionated on a heparin-5PW (Toso-Haas, Montgomeryville, PA). The heparin column was resolved by using a linear 10-column volume gradient of 100 mM to 1 M KCl in buffer A containing 1 μg/ml aprotinin, leupeptin, and pepstatin. WCRF-containing fractions (≈600 mM KCl) were fractionated on a Superose 6 HR 10/30 (Pharmacia) equilibrated in 0.7 M KCl in buffer A containing 0.1% Nonidet P-40, 1 μg/ml aprotinin, leupeptin, and pepstatin. Purified WCRF was subjected to SDS/PAGE and stained with colloidal blue (NOVEX, San Diego) (Fig. 3a).
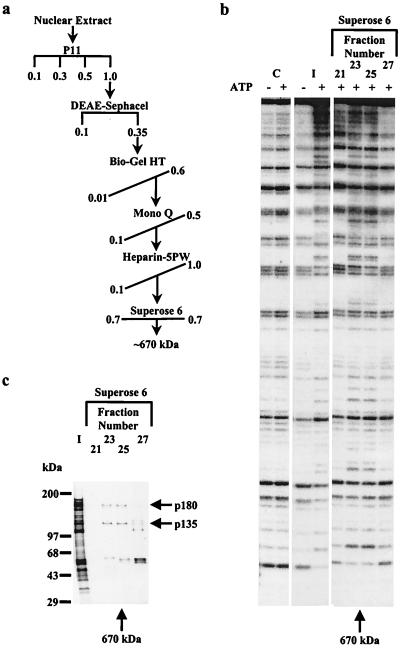
Figure 2.
Isolation of WCRF. (a) Purification scheme. HeLa nuclear extract was fractionated by chromatography as described in Materials and Methods. The horizontal and diagonal lines indicate stepwise and gradient elution, respectively. Concentrations are given in molars. (b) Assay of mononucleosome DNase I cleavage of Superose 6 fractions (6 μl, 50 nM) was conducted as described in Materials and Methods. C denotes the addition of 0.1 M potassium chloride. I is the 0.55-M potassium chloride eluate from Heparin-5PW. The arrow (Bottom) denotes the elution position of thyroglobulin. (c) Resolution of WCRF as two subunits on a Superose 6. The 0.55-M potassium chloride eluate from Heparin-5PW (I) was loaded on a Superose 6 column, and proteins were eluted with 0.7 M potassium chloride. Fractions (5 μl, 50 nM) were separated in an SDS-polyacrylamide (4–12%) gel, and proteins were visualized by silver staining. Molecular masses of marker proteins (Left) and WCRF subunits (Right) are indicated. (Bottom) The arrow denotes the elution position of thyroglobulin.
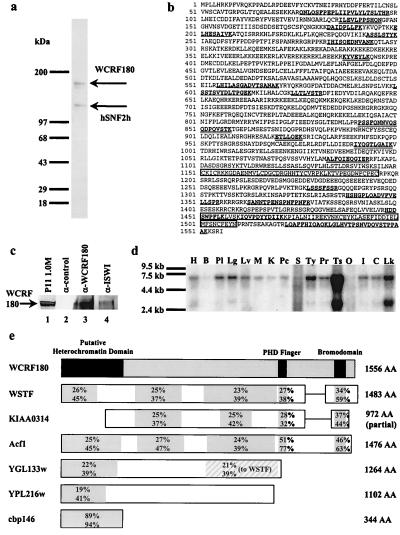
Figure 3.
WCRF180 contains a bromodomain and a PHD finger. (a) SDS/PAGE of WCRF. WCRF (Superose 6, 15 μl) was separated in an SDS-polyacrylamide (4–12%) gel, and proteins were visualized by colloidal blue staining. Molecular masses of marker proteins are indicated (Left). (b) The primary amino acid sequence of WCRF180. The single-underlined peptides correspond to peptides obtained from comparison of MS/MS spectra against the EST database. The double-underlined peptides correspond to sequences that were confirmed by comparison of the MS/MS spectra against the full length clone. The conserved PHD finger and bromodomain are boxed. (c) Immunoprecipitation of WCRF. P11 1.0-M fraction was incubated with control antibodies (α-CSA), polyclonal WCRF180 antibodies (α-WCRF180), or polyclonal ISWI antibodies (α-ISWI). Immunoprecipitates were subjected to SDS/PAGE and immunoblotted with affinity-purified WCRF180 antibodies. (d) Northern blot analysis of WCRF180. RNA size markers are indicated. Human tissues are as indicated: H, heart; B, brain; Pl, placenta; Lg, lung; Lv, liver; M, skeletal muscle; K, kidney; Pc, pancreas; S, spleen; Ty, thymus; Pr, prostrate; Ts, testis; O, ovary; I, small intestine; C, colon; Lk, peripheral blood leukocytes. (e) Diagram of WCRF180 showing regions of homology with WSTF, KIAA0314, Acf1, YGL133w, YPL216w, and cbp146. The numbers (Right) correspond to the number of amino acids in each protein. The numbers in the boxes denote the percent of identity (Top) and similarity (Bottom) of each protein to WCRF, with the exception shown in shaded lines.
For fractions used in Fig. 1, DEAE eluates from HeLa nuclear extracts/P-11 fractions were loaded on a calibrated Superose 6 column in 40 mM Hepes 7.5/350 mM NaCl/0.1% Tween-20/10 μM ZnCl2/0.5 mM DTT/20% glycerol plus 0.2 mM PMSF/1 μg/ml pepstatin, aprotinin, and leupeptin. Fractions (0.5 ml) were collected, and insulin was added to 100 μg/ml.
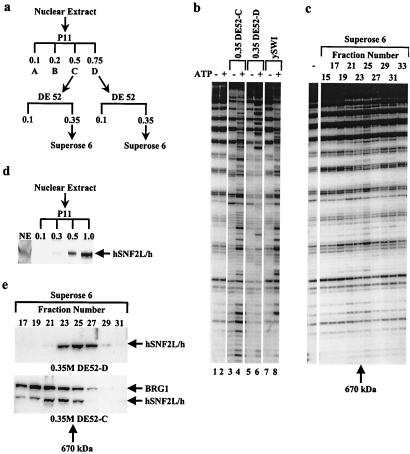
Figure 1.
Identification of a new chromatin remodeling activity. (a) HeLa nuclear extract was fractionated by using phosphocellulose (P11) chromatography. The 0.5- and 0.75-M KCl elutions were concentrated on a DEAE cellulose (DE52) column and further fractionated on a Superose 6 gel filtration. (b) P11 D-fraction concentrated on DE52 displays a unique mononucleosome DNase I cleavage pattern. Fractions (3 μl) corresponding to the 0.5-M potassium chloride elution of P11 (C-fraction) concentrated on a DE52 or the 0.75-M potassium chloride elution of P11 (D-fraction) concentrated on a DE52 were analyzed for mononucleosome disruption assay and compared with the disruption activity seen by using purified yeast SWI/SNF complex (3 μl) (ref. 4). (c) Mononucleosome DNase I disruption assay for the fractions (3 μl) of the Superose 6 derived from the P11 D-fraction, as described in a. The arrow (Bottom) denotes the elution position of thyroglobulin (670 kDa). Superose 6 calibration is as follows: fraction 16, void; fraction 25, 670 kDa; fraction 29, 440 kDa; fraction 32, 158 kDa. (d) Human ISWI predominantly elutes from P11 at potassium chloride concentration of 1 M. Equal volumes (5 μl) of either nuclear extract (NE, 30 μg) or fractions of P11 (3 μg of 0.1 M/1.5 μg of 0.3/M/1.0 μg of 0.5 M/0.5 μg of 1.0 M) were separated in an SDS-polyacrylamide (8%) gel, and proteins were immunoblotted with anti-human ISWI antisera. (e) The 0.35-M potassium chloride eluate of DE52 fraction derived from P11 C-fraction contains a human ISWI complex larger than WCRF. Fractions of the Superose 6 chromatography shown in a (12 μl) were separated in an SDS-polyacrylamide (8%) gel, and proteins were immunoblotted with either anti-human SNF2L or anti-BRG1 antisera. The hSNF2L/h immunoreactivity in the D-fraction closely coelutes with the disrupting activity present in the same fractions (c), indicating that the majority of hSNF2L/h is associated with WCRF.
Mass Spectrometric Peptide Sequencing.
Excised bands were subjected to in-gel reduction, carboxyamidomethylation, and tryptic digestion (Promega). Multiple peptide sequences were determined in a single run by microcapillary reverse-phase chromatography [a custom New Objective (Cambridge, MA) 50-μm column terminating in a nanospray 15-μm tip], directly coupled to a Finnigan LCQ quadrupole ion trap mass spectrometer (Finnigan-MAT, San Jose, CA). The ion trap was programmed to acquire successive sets of three scan modes consisting of: full-scan MS over alternating ranges of 395–800 m/z or 800–1,300 m/z, followed by two data-dependent scans on the most abundant ion in those full scans. These data-dependent scans allowed the automatic acquisition of a high-resolution (zoom) scan to determine charge state and exact mass and MS/MS spectra for peptide sequence information. MS/MS spectra were acquired with a relative collision energy of 30%, an isolation width of 2.5 Da, and dynamic exclusion of ions from repeat analysis. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by programs developed in the Harvard Microchemistry Facility (26) and by database correlation with the algorithm sequest (27).
Cloning and Sequencing of WCRF180.
To identify the WCRF180 ORF, multiple overlapping EST cDNA fragments which encode the tryptic peptide sequences generated from the purified 180-kDa WCRF protein were identified in dbEST by using the blast algorithm (25). These tags were used to perform subsequent blast analysis of a larger database of over one million ESTs generated at Human Genome Sciences from approximately 750 independent human cDNA libraries to identify cDNA sequences that encoded the predicted start methionine. Complete sequence analysis of these putative full length clones and multiple independent overlapping ESTs yielded an ORF of 1,558 amino acids with a predicted molecular mass of 178.7 kDa and estimated pI of 6.42. The presence of an in-frame upstream stop codon indicates a full length clone was identified.
DNA-Dependent ATPase Assays.
ATPase reactions were performed as described (28). Briefly, 25 μl of reaction mixture containing 20 mM Tris⋅Cl, pH 7.4/8 mM MgCl2/0.1 mM DTT/50 mM KCl/2% glycerol/50 μg/ml of BSA/0.5 μCi [γ-32P]ATP and where indicated 100 ng of double-stranded plasmid DNA, core histones, or mono- or oligonucleosomes purified from HeLa (29). Reactions were incubated for 30 min at 30°C, stopped by the addition of 25 μl of 2× formamide loading buffer (90% deionized formamide/0.1% bromophenol blue and xylene cyanol), and 10 μl was resolved on a 12% polyacrylamide gel (19:1 acrylamide to bisacrylamide) containing 7 M urea for 1.0 h at 150 V. Wet gels were autoradiographed for 10 min at −80°C. Autoradiographs were quantified by using imagequant software, Ver. 5.0 (Molecular Dynamics).
Mononucleosome DNase I Cleavage Assay.
Labeled nucleosomes were reconstituted by octamer transfer from HeLa H1-depleted oligonucleosomes to a 172-bp 32P end-labeled DNA fragment as described (29). Binding reactions were conducted in 20 μl of final volume and included 25 ng of total nucleosomes, 1 mM ATP where indicated, and the indicated protein fraction for 1 hr at 30°C in 20 mM Hepes 7.5/3 mM MgCl2/50 mM NaCl/1 μM ZnCl2/2 mM DTT/0.2 mM PMSF/5% glycerol/200 μg/ml BSA. DNase I, 0.5 units, was added, and samples were further incubated for 1 min at room temperature. The reactions were stopped with 1 volume of 20 mM Tris⋅HCl, pH 7.5/50 mM EDTA/2% SDS/0.2 mg/ml proteinase K/0.25 mg/ml yeast tRNA. Samples were then incubated at 50°C for 1 h, precipitated with ammonium acetate, and washed with 80% EtOH. Pellets were suspended in 2 μl of water and 3 μl of loading buffer (95% formamide/10 mM EDTA/0.1% xylene cyanol/0.1% bromophenol blue). After incubation at 90°C, samples were run on an 8% polyacrylamide (19:1 acrylamide to bisacrylamide) gel with 1× TBE (100 mM Tris-borate/2 mM EDTA) and 8 M urea. Gels were exposed wet. Purified yeast SWI/SNF complex was prepared as described (4).
Helicase Assay.
Helicase assays were performed exactly as described by Marinoni et al. (30). The template was generated by annealing the M13/pUC −20 sequencing primer to ssM13 mp18 and then extending to 24 nucleotides with the Klenow fragment in the presence of dATP, dGTP, and [α-32P]dCTP (800 Ci/mmol, NEN).
Immunoblot Analysis and Immunoprecipitation.
Anti-ISWI antibodies were a gift from W. Gu and R. G. Roeder. The anti-ISWI antibodies were raised against the human SNF2L protein. Anti-BRG1 antibodies were described (31). Affinity-purified anti-WCRF180 antibodies were raised against keyhole limpet hemocyanin-conjugated peptides (Research Genetics, Huntsville, AL) corresponding to residues 10–29 of WCRF180. Immunoprecipitations and immunoblotting with alkaline phosphatase were performed as described (32).
Northern Blot Analysis.
The human multiple tissue Northern blots I and II (CLONTECH) were probed with a 0.9-kb fragment of WCRF180 that spans amino acids 358 to 666. The filters were hybridized overnight at 42°C with 1.5 × 106 cpm probe/ml Hybrizol I (Intergen, Purchase, NY). Filters were washed twice at 65°C for 20 min in 0.2× SSC/0.1% SDS and exposed to x-ray film for 7 days. Similar concentration of RNA for each tissue was confirmed by analysis by using a ubiquitously expressed cDNA.
Nucleosome Array Disruption Assay.
The 2.6-kb G5E4–5S fragment (Asp-718-ClaI) used in nucleosome array reconstitution was produced from plasmid pIC208G5E4 as described (33). Nucleosomal array reconstitution was performed by octamer transfer and analyzed by DNase I digestion followed by migration on a native agarose gel (34). Binding reactions were performed and analyzed as for mononucleosome disruption assays, except that 10 ng of nucleosomes was used.
Results
A Nucleosome Disruption Activity in Human Cells.
To identify different nucleosome remodeling activities in human cells, we analyzed HeLa nuclear extract after fractionation by phosphocellulose (P11) chromatography (Fig. 1a) by using a high-resolution mononucleosome disruption assay. Analysis of fractions displaying ATP-dependent disruption activity revealed that the 0.35-M potassium chloride eluate of DEAE cellulose (DE52) chromatography, which was derived from the high salt eluate (D-fraction) of the P11, displayed a qualitatively different ATP-dependent DNase I cleavage pattern (Fig. 1b; compare lanes 5 and 6). This chromatin remodeling activity was different from that of either the equivalent DE52 eluate derived from the P11 C-fraction (compare lanes 4 and 6) or the yeast SWI/SNF complex (lane 8). To gain insight into this distinct disruption activity, the DE52 eluate derived from the P11 D-fraction was further chromatographed on a gel filtration column (Superose 6). Analysis of the Superose 6 column fractions in a mononucleosome disruption assay revealed that the chromatin remodeling activity eluted with the apparent molecular mass of 600–700 kDa, peaking in fraction 25 (Fig. 1c). These experiments indicated that the chromatin remodeling activity contained in the P11 D-fraction exhibits a distinct molecular mass compared with that of the 2-MDa SWI/SNF complex. Moreover, immunodepletion of this fraction by using anti-SWI/SNF antibodies did not affect the nucleosome remodeling activity (data not shown).
A New Human Chromatin Remodeling Complex.
By using ATP-dependent DNase I cleavage of mononucleosomes to assay for the new chromatin remodeling activity, we fractionated HeLa nuclear extract following the scheme indicated in Fig. 2a. Analysis of the final chromatographic step on Superose 6 gel filtration indicated that the chromatin remodeling activity eluted with an apparent molecular mass of 600–700 kDa (Fig. 2b), similar to the elution profile of the complex from a partially purified fraction described above (Fig. 1c). Nucleosome remodeling activity coeluted with a complex composed of two polypeptides, 135 kDa and 180 kDa (Fig. 2c). The 55- to 60-kDa polypeptides did not coelute with the chromatin remodeling activity. Moreover, analysis of the chromatin remodeling complex by using a colloidal blue-stained SDS-polyacrylamide gel revealed an equal stoichiometry for only two polypeptides (Fig. 3a). These results suggest that the chromatin remodeling activity behaves as a tetramer containing two each of subunits of 135 kDa and 180 kDa, resulting in an apparent molecular mass of 600–700 kDa for the complex. To identify the two subunits of the complex, the bands corresponding to each polypeptide were cut from a colloidal blue-stained SDS-polyacrylamide gel, digested in-gel, and sequenced by microcapillary HPLC directly coupled to an ion trap mass spectrometer. Fifteen peptide sequences revealed that the 135-kDa subunit is hSNF2h, a human homolog of Drosophila ISWI (18, 19). Tryptic peptide sequences derived from the 180-kDa polypeptide were used to identify overlapping cDNA clones corresponding to an ORF that possesses a high degree of homology to WSTF (Fig. 3b). We have therefore named the chromatin remodeling complex WCRF for Williams syndrome transcription factor-related chromatin remodeling factor. We confirmed the association of WCRF180 and hSNF2h by using immunoprecipitation experiments. Polyclonal anti-ISWI and anti-WCRF180 antibodies specifically precipitate WCRF180 from the P11 1.0-M potassium chloride eluate (Fig. 3c) verifying the association of these polypeptides. Northern blot analysis of WCRF180 revealed the presence of a 6.8-kb transcript that is expressed highly in testis and to a detectable level in most other adult human tissues examined. The notable exception was undetectable levels of the 6.8-kb transcript in the brain and ovaries (Fig. 3d). We also observed a smaller transcript of about 3.0 kb, indicating the possible occurrence of an alternative spliced message (Fig. 3d).
WCRF180 Is a Williams Syndrome-Related Transcription Factor.
WCRF180 is a protein similar in amino acid sequence and protein architecture to WSTF (Fig. 3e). Moreover, analysis of the nonredundant GenBank database by using the blast algorithm (25) revealed the presence of another human protein (KIAA0314) with a high homology to WCRF180 (Fig. 3e). The homology between the three proteins extends over the entire ORF. Two conserved motifs in the C terminus were identified in all three proteins. Similar to WSTF, WCRF180 and KIAA0314 also contain a zinc-finger-like motif known as the PHD finger. The PHD finger is a cysteine-rich structure containing a consensus four cysteine–histidine–three cysteine (C4HC3) motif that spans 50 to 80 residues (35). These domains have been shown to bind Zn2+ in vitro (36). The second domain identified in WCRF180 is a bromodomain (Fig. 3b). Bromodomains, a motif that contains 60 amino acids, are thought to mediate interactions with histones (37, 38).
WCRF Displays Nucleosome-Dependent ATPase but Not Helicase Activity.
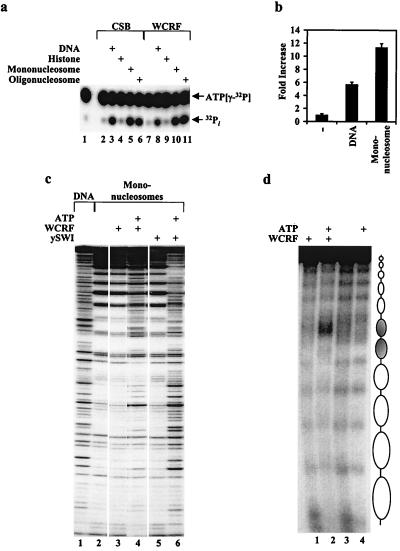
To further characterize the ATP requirement for the nucleosome remodeling activity of the WCRF complex, we analyzed the ability of the complex to hydrolyze ATP in the presence of equal amounts of either plasmid DNA, mononucleosomes, or oligonucleosomes purified from HeLa cells. In addition, we compared the ATPase activity of WCRF to the Cockayne syndrome B (CSB) protein, another SNF2 family member that contains the signature helicase domain (5). This analysis revealed that the ATPase activity of CSB is stimulated to the same extent by either naked DNA or nucleosomes (Fig. 4a, lanes 1–6). In contrast to CSB, although the ATPase activity of WCRF is stimulated by naked DNA (lane 8 and Fig. 4b), addition of the nucleosomes to the ATPase reaction further stimulated the ATPase activity by about 2-fold (lanes 10–11 and Fig. 4b). The ATPase activity of neither CSB nor WCRF was stimulated by core histones alone (lanes 4 and 9), and nucleosomes alone did not display any ATPase activity (data not shown). The DNA-dependent stimulation of the ATPase activity of WCRF prompted us to analyze whether WCRF displayed ATP-dependent DNA helicase activity. We compared the ATP-dependent DNA helicase activity of transcription factor IIH (TFIIH) to that of the WCRF complex. In contrast to TFIIH, WCRF does not display a detectable ATP-dependent DNA helicase activity (data not shown).
Figure 4.
Functional analysis of WCRF. (a) DNA-dependent ATPase assays of WCRF. Fractions from Superose 6 (2 μl, 50 nM) corresponding to WCRF (fraction 23) or CSB (2 μl, 50 nM; fraction 27) were assayed for stimulation of ATPase activity, as described in Materials and Methods by using 100 ng of either double-stranded plasmid DNA, mononucleosomes or oligonucleosomes. (b) Quantification of fold stimulation of WCRF ATPase activity by DNA and oligonucleosomes. (c) Mononucleosome DNase I disruption assay comparing the mononucleosome disruption activity of yeast SWI/SNF (3 μl, 100 nM) to WCRF (heparin fraction, 3 μl, 70 nM). (d) WCRF remodels nucleosome arrays in an ATP-dependent fashion. Nucleosome array was reconstituted on a DNA fragment containing a dinucleosome length sequence with a synthetic GAL4-E4 promoter flanked by on either side by five repeats of a nucleosome-positioning sequence from sea urchin 5S rDNA. Purified WCRF (heparin fraction, 3 μl; 70 nM) was then incubated with the 12-nucleosome array in the presence or the absence of ATP, as indicated. Nucleosomal structure was then analyzed by DNase I digestion and migration on a 2% native agarose gel. Positions of the 5S and G4E4 nucleosomes are shown (Right) (opened and shaded ovals respectively).
To analyze further the DNase I cleavage pattern of mononucleosomes by WCRF, we compared the activity of the purified WCRF complex to that of yeast SWI/SNF. Addition of the yeast SWI/SNF complex to the mononucleosome changed the DNase I digestion pattern to nearly that observed with naked DNA (Fig. 4c, compare lanes 1 and 6). In contrast, addition of nearly 1:1 molar ratio of WCRF complex to nucleosome results in a DNase I protection pattern that resembles nucleosomal DNA much more than naked DNA, but that clearly includes several distinct alterations (compare lanes 3 and 4). To characterize further WCRF for chromatin remodeling, we tested its activity on nucleosomal arrays. The array was constructed from a DNA template placing two nucleosomes on a G4-E4 promoter (Fig. 4d, shaded ovals) in phase with a spaced array of 5S rDNA (208-bp repeats), a strong nucleosome positioning sequence. Interestingly, in the absence of Gal4 protein, WCRF strongly disrupted the nucleosomes over the G4-E4 sequences in an ATP-dependent manner (Fig. 4d, increased DNase I digestion) without altering the nucleosomes on the 5S rDNA positioning sequences (Fig. 4d). The strong DNase I hypersensitivity seen after WCRF treatment is indicative of the appearance of nucleosome-free DNA in a large region of the G4-E4 promoter.
WCRF Is a Predominant Human ISWI-Containing Complex.
To determine the abundance of WCRF and the number of hISWI complexes in HeLa cells, we analyzed the purification of WCRF by Western blotting by using anti-human SNF2L polyclonal antibodies. We first confirmed that the anti-hSNF2L antibodies recognize the hSNF2h derived from the last step of our purification (data not shown). We then analyzed the immunoreactivity for hSNF2L/h after the fractionation of nuclear extract on the phosphocellulose column (Fig. 1d). This analysis revealed the majority of hSNF2L/h immunoreactivity is in P11 D-fraction, consistent with the presence of WCRF in this fraction (see Figs. 1c and 2 b and c). However, the P11 C-fraction also contained a smaller quantity of hSNF2L/h immunoreactivity. To analyze further the hSNF2L/h complexes, P11 fractions were concentrated on a DE 52, and the 0.35-M potassium chloride eluate was subjected to gel filtration by using Superose 6. Consistent with the remodeling activity shown in Figs. 1b and 2b, Western blot analysis of Superose 6 fractions derived from the P11 D-fraction revealed the hSNF2L/h complex is eluting with the apparent molecular mass of 600–700 kDa (Fig. 1e, fractions 23–27). Furthermore, Western blot analysis by using anti-SNF2L/h antibodies of every chromatographic step in WCRF purification revealed only a single peak of SNF2L/h immunoreactivity coeluting with WCRF (data not shown). However, in contrast to the P11 D-fraction, hSNF2L/h immunoreactivity for Superose 6 fractions derived from the P11 C-fraction eluted with a larger apparent molecular mass (Fig. 1e, fractions 19–25) partially overlapping the fractions containing the immunoreactivity for BRG-1/hBrm, the SWI2/SNF2 components of human SWI/SNF complex. These results are consistent with WCRF representing one of the smaller and more abundant human ISWI-containing complexes in HeLa cells and with HeLa nuclear extract containing at least one other larger hISWI-containing complex, consistent with a previous report (10).
Discussion
We have purified and characterized WCRF, a human ISWI-containing complex that exhibits an ATP-dependent chromatin-remodeling activity, which includes a protein closely related to WSTF. The gene for WSTF is one of the many genes deleted in the human developmental disorder Williams syndrome (23, 24). The originality of this work lies in the following. First, it presents a complete molecular definition of a new human ISWI-containing complex. Second, it shows that WCRF is a predominant ISWI-containing complex in HeLa nuclear extract. Third, it presents evidence indicating that WCRF ATPase activity is stimulated by double-stranded DNA. However, in contrast to the ATPase activity of the SWI/SNF and RSC complexes (3, 6), the ATPase activity of WCRF is further stimulated by nucleosomal DNA. Fourth, through the homology between WCRF180 and WSTF, it provides a possible link between a human developmental disorder and chromatin remodeling machinery.
The ISWI-containing chromatin remodeling complexes were first isolated from Drosophila (13–16). Interestingly, the large subunit of the ACF complex in Drosophila (39) also displays similarity to WSTF and WCRF180 (Fig. 3e). In addition, two ISWI-containing complexes, a four-subunit ISW1p complex and a two-subunit ISW2p complex, were isolated from Saccharomyces cerevisiae (22). The WCRF complex may correspond to the two-subunit ISW2p-containing complex because blast analysis, by using the human WCRF180 protein sequence as a query, reveals a previously uncharacterized protein of theoretical mass of 145 kDa, corresponding to the ORF YGL133w. The molecular mass of YGL133w is consistent with the size (140 kDa) of the large subunit of ISW2 complex (22) (Fig. 3e). The yeast protein is 40% identical to another yeast ORF (YPL216 w; see Fig. 3e). Interestingly, both proteins do not possess a C-terminal PHD finger or bromodomain. The presence of the PHD finger and the bromodomain in WCRF180 may underlie a unique function that WCRF serves in higher organisms.
A functional distinction between SWI/SNF-related and ISWI-related remodeling complexes corresponds to the differences in their ATPase-containing remodeling engines. Although the ATPase activity of SWI/SNF-related complexes is stimulated by either naked or nucleosomal DNA to the same extent, the ATPase activity of ISWI complexes responds to a greater extent to nucleosomal DNA (15, 22). Furthermore, it was also observed that NURF, a Drosophila ISWI-containing complex, perturbs the DNase I digestion pattern of mononucleosomes differently than either SWI/SNF or the RSC complex (13, 14). The difference in the DNase I cleavage pattern most likely reflects changes in the induced physical state of nucleosome after the interaction of each complex with the nucleosome.
Many questions remain concerning the function of WCRF and related ISWI-containing complexes, including their physiological function, regulation, and mechanism of remodeling activity. For example, the precise relationship of WCRF to RSF, the other reported SNF2h-containg chromatin remodeling complex, is not clear. However, the close homology of WCRF180 to WSTF, a gene deleted in the human developmental disorder Williams syndrome, not only predicts a similar function for WSTF in chromatin remodeling but also points to a role for WCRF in developmental control. Consistent with this notion, a recent report using a gene trap approach describes the localization of the N terminus of a putative mouse homolog of WCRF180 (see Fig. 3e) to the centromeric heterochromatin of the mouse chromosomes (40). A fusion of the first 344 N-terminal amino acids of mouse WCRF180 (cbp146; see Fig. 3e) was sufficient for localization of the protein to the heterochromatin (40). Therefore, through modulation of heterochromatin structure, WCRF may play a critical role in developmental control because higher order chromatin not only is involved in epigenetic control of gene expression but also is essential for the correct folding and segregation of mitotic and meiotic chromosomes (41) and for centromere (42, 43) and telomere activity (44). Moreover, because gene expression in eukaryotes is governed by positional information and higher order chromatin structure, the localization of WCRF to the heterochromatin may reveal a novel mechanism by which ISWI-containing remodeling complexes modulate gene expression. This is consistent with a recent report indicating that mutations in an Arabidopsis thaliana gene highly similar to hSNF2h result in predominant hypomethylation in the heterochromatin (45). Future studies by using biochemical as well as genetic approaches should reveal the function of WCRF and other ISWI-containing complexes in the regulation of gene expression and developmental control.
Acknowledgments
We thank Drs. W. Gu and R. G. Roeder for anti-SNF2L antiserum. The contribution of the sequencing facility at Human Genome Sciences is acknowledged. We also thank W. S. Lane and the expert staff of the Harvard Microchemistry Facility for expertise in the microcapillary HPLC/MS. Part of this work was supported by a Canadian Medical Research Council (MRC) operating grant (to J.C.). J.C. is a Canadian MRC scholar. R.S. is a V-foundation scholar and was supported by the startup fund of the Wistar Institute.
Abbreviations
- WSTF
Williams syndrome transcription factor
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF221130).
References
- 1.Kornberg R D, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Laurent B C, Yang X, Carlson M. Mol Cell Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Côté J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 5.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 7.Imbalzano A N, Kwon H, Green M R, Kingston R E. Nature (London) 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 8.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, et al. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 11.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukiyama T, Wu C. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 14.Tsukiyama T, Daniel C, Tamkun J, Wu C. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 15.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Nature (London) 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 17.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okabe I, Bailey L C, Attree O, Srinivasan S, Perkel J M, Laurent B C, Carlson M, Nelson D L, Nussbaum R L. Nucleic Acids Res. 1992;20:4649–4655. doi: 10.1093/nar/20.17.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aihara T, Miyoshi Y, Koyama K, Suzuki M, Takahashi E, Monden M, Nakamura Y. Cytogenet Cell Genet. 1998;81:191–193. doi: 10.1159/000015027. [DOI] [PubMed] [Google Scholar]
- 20.Aasland R, Stewart A F, Gibson T. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 21.LeRoy G, Orphanides G, Lane W S, Reinberg D. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 22.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Meng X, Morris C A, Keating M T. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- 24.Peoples R J, Cisco M J, Kaplan P, Francke U. Cytogenet Cell Genet. 1998;82:238–246. doi: 10.1159/000015110. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chittum H S, Lane W S, Carlson B A, Roller P P, Lung F D, Lee B J, Hatfield D L. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 27.Eng J K, McCormick A L, Yates J R., III J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Tantin D, Kansal A, Carey M. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utley R T, Owen-Hughes T A, Juan L J, Côté J, Adams C C, Workman J L. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 30.Marinoni J C, Rossignol M, Egly J M. Methods. 1997;12:235–253. doi: 10.1006/meth.1997.0476. [DOI] [PubMed] [Google Scholar]
- 31.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 32.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Nature (London) 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 33.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 34.Owen-Huges T, Utley R T, Cote J, Peterson J L, Workman J L. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 35.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 36.Mazo A M, Huang D H, Mozer B A, Dawid I B. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ornaghi P, Ballario P, Lena A M, Gonzalez A, Filetici P. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 38.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Nature (London) 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 39.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tate P, Lee M, Tweedie S, Skarnes W C, Bickmore W A. J Cell Sci. 1998;111:2575–2585. doi: 10.1242/jcs.111.17.2575. [DOI] [PubMed] [Google Scholar]
- 41.Dernburg A F, Sedat J W, Hawley R S. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 42.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 43.Williams B C, Murphy T D, Goldberg M L, Karpen G H. Nat Genet. 1998;18:30–37. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- 44.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 45.Jeddeloh J A, Stokes T L, Richards E J. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]