Abstract
The setpoint of viral RNA concentration (viral load [VL]) during chronic human immunodeficiency virus type 1 (HIV-1) infection reflects a virus-host equilibration closely related to CD8+ cytotoxic T-lymphocyte (CTL) responses, which rely heavily on antigen presentation by the human major histocompatibility complex (MHC) (i.e., HLA) class I molecules. Differences in HIV-1 VL among 259 mostly clade C virus-infected individuals (137 females and 122 males) in the Zambia-UAB HIV Research Project (ZUHRP) were associated with several HLA class I alleles and haplotypes. In particular, general linear model analyses revealed lower log10 VL among those with HLA allele B*57 (P = 0.002 [without correction]) previously implicated in favorable response and in those with HLA B*39 and A*30-Cw*03 (P = 0.002 to 0.016); the same analyses also demonstrated higher log10 VL among individuals with A*02-Cw*16, A*23-B*14, and A*23-Cw*07 (P = 0.010 to 0.033). These HLA effects remained strong (P = 0.0002 to 0.075) after adjustment for age, gender, and duration of infection and persisted across three orders of VL categories (P = 0.001 to 0.084). In contrast, neither B*35 (n = 15) nor B*53 (n = 53) showed a clear disadvantage such as that reported elsewhere for these closely related alleles. Other HLA associations with unusually high (A*68, B*41, B*45, and Cw*16) or low (B*13, Cw*12, and Cw*18) VL were either unstable or reflected their tight linkage respecting disequilibria with other class I variants. The three consistently favorable HLA class I variants retained in multivariable models and in alternative analyses were present in 30.9% of subjects with the lowest (<10,000 copies per ml) and 3.1% of those with the highest (>100,000) VL. Clear differential distribution of HLA profiles according to level of viremia suggests important host genetic contribution to the pattern of immune control and escape during HIV-1 infection.
The specificity and diversity of human major histocompatibility complex (MHC) (i.e., HLA) products closely reflect the molecular interplay in infection and autoimmunity accumulated during human evolution and migration (10, 23, 25, 26, 36, 46). There is a growing consensus that specific HLA alleles and heterozygosity at HLA class I loci can collectively exert a strong impact on disease progression in human immunodeficiency virus type 1 (HIV-1) infection (6, 8, 21, 22, 30, 31, 35, 41, 62, 63); much of the effect appears to correlate with cytotoxic T-lymphocyte (CTL) responses directly restricted by class I allelic products in both humans (3, 16, 18, 34, 56) and chimpanzees (2). HLA markers associated with delayed HIV-1 disease progression may present epitopes found more frequently in various HIV-1 proteins (45), and the two most prominent alleles (B*27 and B*57) have been found experimentally to induce immunodominant CTL responses to conserved HIV-1 epitopes (15, 19, 20, 44). The same favorable HLA alleles were overrepresented in canarypox-HIV vaccine recipients with repeatedly detected CTL responses to certain viral proteins (32). These findings imply that HLA typing data may be useful in predicting the relative population coverage (58, 60) of epitope-rich HIV vaccine constructs (9, 37, 47).
On the other hand, the impact of HLA alleles on HIV-1 disease progression can differ in magnitude and consistency by cohort, ethnicity, and stage of chronic infection. Most of the work to date has been performed in populations infected with clade B viruses; information on differences by viral clade is sparse. Our analyses of HLA and HIV-1 viral load (VL) in 259 Zambians revealed a pattern of HLA associations with unusually high or low VL that is distinct from that reported in other populations. The findings now imply HIV-1 clade-specific differential antigen presentation.
(Data derived from this study were presented in part [abstr. 327-w] at the 9th Conference on Retroviruses and Opportunistic Infections in Seattle, Wash. [24 to 28 February 2002] and at the 27th Annual Meeting [abstr. 217] of the American Society for Histocompatibility and Immunogenetics in San Francisco, Calif. [13 to 17 October 2001]).
MATERIALS AND METHODS
Human subjects and HIV-1 identities.
From 1,022 cohabiting couples originally enrolled in the Zambia-UAB HIV Research Project (ZUHRP), 381 individuals (from 200 couples) were available for this study between February 1994 and November 2000. The procedures for recruitment, counseling, follow-up visits, and laboratory testing have been described elsewhere (12, 42). The 381 study subjects were classified into four categories: 108 transmitters (transmission pair donors [TPDs]) were HIV-1 positive at enrollment and subsequently transmitted virus to their heterosexual spouses or partners; 105 seroconverters (SCs) became seropositive after enrollment; 83 nontransmitters (nontransmission pair seropositives) were HIV-1 positive at enrollment but did not transmit virus during follow-up; and 85 discordant pair seronegatives remained uninfected during the entire prospective study period. A subgroup of 259 seropositive and antiretroviral therapy-naïve individuals (103 TPDs, 91 SCs, and 65 nontransmission pair seropositives) with both genetic and virologic data were analyzed in greater detail. HIV-1 isolates from all coupled TPDs and SCs were epidemiologically linked by phylogenetic analyses of HIV-1 subgenomic sequences corresponding to gag, env (gp120 and gp41), and the long terminal repeat regions (12, 66) against reference sequences representing a variety of viral isolates. Consistent with earlier results based on heteroduplex mobility assays (H. Sheppard et al., unpublished data) of randomly selected isolates from Zambia, the vast majority (95%) of HIV-1 sequences derived from ZUHRP participants belonged to subtype C, although other subtypes such as A, D, G, and J were also detected occasionally (66).
Immunological and virological measurements.
HIV-1 RNA level in plasma was measured by the Roche Amplicor 1.0 assay (Roche Diagnostics Systems Inc., Branchburg, N.J.) in a laboratory certified by the Virology Quality Assurance Program of the AIDS Clinical Trials Group. Direct comparison of four commercial viral assays (the Chiron Quantiplex HIV-1 branched DNA, Roche Amplicor versions 1.0 and 1.5, and nucleic acid sequence-based amplification HIV-1 RNA QT assays) revealed that each of them could successfully quantify plasma HIV-1 RNA in Zambians; the modified (new generation) Amplicor version 1.5 assay with additional primer sets targeting non-B subtype viruses showed slight (0.3 log10 copies per ml; P > 0.05) improvement compared with the first generation Amplicor 1.0 assay (24). For categorical analyses, HIV-1 RNA levels were stratified into three categories, <10,000 copies/mm3, 10,000 to 100,000 copies/mm3, and >100,000 copies/mm3; increasingly high levels of viremia are strongly predictive of HIV-1 transmission, especially from females to males (12). HIV-1 RNA level transformed to log10 was treated as a continuous variable in separate analyses. The unavailability of measurements of CD4+ T-cell levels in the majority of subjects precluded reliable analysis of corresponding levels of immunodeficiency in ZUHRP participants.
DNA extraction and HLA class I typing.
High-molecular-weight genomic DNA was extracted from whole blood by using the QIAamp blood kit and protocols recommended by the manufacturer (Qiagen Inc., Valencia, Calif.). All DNA samples were stored at 4°C in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0], 2 mM EDTA) before HLA typing. HLA class I alleles were initially typed by PCR with sequence-specific primers in a commercial kit (Pel-Freez Clinical Systems, Brown Deer, Wis.), which defined alleles largely to their two-digit specificities. Individual samples with apparent homozygosity at any class I locus and those carrying HLA-B*57 were further defined by sequencing-based typing (SBT) using locus-specific PCR amplification (7), followed by solid-phase direct sequencing (67) on the ALFexpress automated sequencer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Automated reference-strand conformation analyses (RSCA) (Pel-Freez Clinical Systems) were applied to resolve ambiguities in PCR-sequence-specific primers and SBT. Both RSCA and SBT detected common four-digit alleles in this cohort [J. Tang et al., abstr. 217, Hum. Immunol. 62(Suppl. 1):S148, 2001].
Assignment of HLA class I haplotypes.
In the absence of genetic data from families, the apparent two-locus haplotypes were assigned after linkage disequilibrium (LD) analyses. More specifically, χ2 and likelihood ratio (LR) tests were performed using a 2 × 2 table containing the numbers of chromosomes with both, one without the other, or neither of the two class I alleles in question. A significant P value (≤0.01) with an LR ≥ 2.0 was treated as indicative of positive LD. Additionally, two-locus haplotypes were deemed fully reliable when alleles at one or both loci remained homozygous after repeated typings that targeted separate regions of the allele-defining sequences (63). The class I extended haplotypes were determined by the pairwise frequency and the relative strength (LR and P values) of two-locus haplotypes.
Statistical analyses.
Allele and marker (carrier) frequencies of HLA class I alleles and haplotypes were enumerated by direct counting. The observed and expected HLA class I genotypes (pairs of alleles at each locus) were tested for Hardy-Weinberg equilibrium, and the Ewens-Watterson neutrality test (11, 29, 68) was used to compare the observed and expected homozygosity frequencies. Both χ2 tests for trend in HLA distribution and general linear model (GLM) statistics were used to assess the association of individual HLA alleles and haplotypes (marker frequencies ≥ 3%) with HIV-1 RNA levels. For each univariate analysis, individuals carrying a specific HLA variant (marker positives) were compared with all others without the marker. Our analyses of HLA effects emphasized consistency, both internal (i.e., similarity between patient groups following different statistical approaches) and external (i.e., similarity to findings in earlier comparable studies). Univariate analyses have been reported for all possible HLA effects on HIV-1 VL with a nominal P ≤ 0.10, with or without adjustments for age, sex, and membership of patient groups. In the final multivariable model, only HLA variants with an adjusted P ≤ 0.05 were retained. Statistical routines available in Statistical Analysis Software, version 8.0 (SAS Institute, Cary, N.C.), were applied throughout these analyses. The P values shown throughout this work were calculated without Bonferroni correction (1, 38, 52) for multiple comparisons, mainly because the multifaceted analyses (by HLA allele, haplotype, and zygosity and by different VL categories) introduced considerable uncertainty about the number of independent tests performed or implied.
RESULTS
Distribution of major HLA class I variants in Zambians.
At the two-digit specificity level, PCR-based HLA typing detected 12 A, 18 B, and 11 Cw variants at a carrier frequency equal to or higher than 3% in the entire cohort (n = 381) and in the subset of 259 HIV-1 seropositive subjects with HIV-1 VL measurements (Table 1). RSCA and SBT were able to resolve major four (e.g., A*2301 and B*3501)- or five (e.g., B*07021 and B*57031)-digit alleles at each locus for about 20% of total subjects. Multiple four-digit alleles within A*02, A*30, A*68, B*14, B*15, B*39, B*44, B*58, Cw*02, Cw*06, Cw*07, and Cw*16 coexisted in this cohort (Table 1). The 259 HIV-1 seropositives showed HLA allelic distributions similar to those in the entire group (n = 381) (P = 0.327, 0.758, and 0.696 for the A, B, and C loci, respectively). The distribution of HLA genotypes (pairs of alleles at each locus; data not shown) in the cohort and its subgroups closely fit that required for Hardy-Weinberg equilibrium; i.e., common alleles produced common genotypes (e.g., A*30/A*68, B*42/B*58, and B*15/B*53) and accounted for the vast majority of the homozygotes (e.g., A*30/A*30 and Cw*04/Cw*04).
TABLE 1.
Major HLA class I allelic variants detected in Zambians
| Allelic variant(s)a | All subjects
|
Seropositive subject subset with HIV-1 RNA measurements
|
||
|---|---|---|---|---|
| 2N = 762 Alleleb | n = 381 Carriageb | 2N = 518 Alleleb | n = 259 Carriageb | |
| Major A | ||||
| A*01(01) | 18 (2.4)c | 16 (4.2)c | 10 (1.9)c | 8 (3.1)c |
| A*02(01,05) | 91 (11.9) | 86 (22.6) | 54 (10.4) | 51 (19.7) |
| A*03(01) | 27 (3.5) | 26 (6.8) | 16 (3.1) | 16 (6.2) |
| A*23(01) | 94 (12.3) | 90 (23.6) | 59 (11.4) | 57 (22.0) |
| A*29(01,02) | 48 (6.3) | 45 (11.8) | 40 (7.7) | 37 (14.3) |
| A*30(01,02,04) | 191 (25.1) | 164 (43.0) | 130 (25.1) | 111 (42.9) |
| A*33(01) | 15 (2.0) | 15 (3.9) | 8 (1.5) | 8 (3.1) |
| A*34(01) | 20 (2.6) | 20 (5.2) | 11 (2.1) | 11 (4.2) |
| A*3601 | 56 (7.3) | 53 (13.9) | 43 (8.3) | 41 (15.8) |
| A*6601 | 26 (3.4) | 26 (6.8) | 14 (2.7) | 14 (5.4) |
| A*68(01,02) | 100 (13.1) | 92 (24.1) | 80 (15.4) | 74 (28.6) |
| A*74(01,03) | 50 (6.6) | 47 (12.3) | 33 (6.4) | 31 (12.0) |
| Minor A | 26 (3.4) | 25 (6.6) | 20 (3.9) | 19 (7.3) |
| Major B | ||||
| B*07(021) | 50 (6.6) | 47 (12.3) | 33 (6.4) | 32 (12.4) |
| B*08(01) | 26 (3.4) | 26 (6.8) | 18 (3.5) | 18 (6.9) |
| B*13(01) | 14 (1.8) | 14 (3.7) | 10 (1.9) | 10 (3.9) |
| B*14(01,02) | 49 (6.4) | 48 (12.6) | 39 (7.5) | 38 (14.7) |
| B*15(01,03,10,17) | 123 (16.1) | 110 (28.9) | 73 (14.1) | 65 (25.1) |
| B*18(01) | 19 (2.5) | 19 (5.0) | 14 (2.7) | 14 (5.4) |
| B*35(01) | 23 (3.0) | 23 (6.0) | 15 (2.9) | 15 (5.8) |
| B*39(01,02) | 11 (1.4) | 11 (2.9) | 9 (1.7) | 9 (3.5) |
| B*41(01) | 12 (1.5) | 12 (3.1) | 7 (1.4) | 7 (2.7) |
| B*42(01,02) | 70 (9.2) | 66 (17.3) | 55 (10.6) | 51 (19.7) |
| B*44(02,03) | 38 (5.0) | 36 (9.4) | 26 (5.0) | 24 (9.3) |
| B*45(01) | 58 (7.6) | 57 (15.0) | 45 (8.7) | 44 (17.0) |
| B*49(01) | 9 (1.2) | 9 (2.4) | 6 (1.2) | 6 (2.3) |
| B*51(01) | 15 (2.0) | 15 (3.9) | 11 (2.1) | 11 (4.2) |
| B*53(01) | 77 (10.1) | 74 (19.4) | 56 (10.8) | 53 (20.5) |
| B*57(031) | 38 (5.0) | 36 (9.4) | 20 (3.9) | 18 (6.9) |
| B*58(01,02) | 84 (11.0) | 79 (20.7) | 48 (9.3) | 46 (17.8) |
| B*8101 | 34 (4.5) | 31 (8.1) | 25 (4.8) | 22 (8.5) |
| Minor B | 12 (1.6) | 12 (3.1) | 8 (1.5) | 8 (3.1) |
| Major Cw | ||||
| Cw*02(02,03) | 87 (11.4) | 82 (21.5) | 55 (10.6) | 53 (20.5) |
| Cw*03(02,03,04) | 53 (7.0) | 49 (12.9) | 36 (7.0) | 34 (13.1) |
| Cw*04(01) | 130 (17.1) | 125 (32.8) | 87 (16.8) | 85 (32.8) |
| Cw*05(01) | 11 (1.4) | 11 (2.9) | 5 (1.0) | 5 (1.9) |
| Cw*06(02,03) | 96 (12.6) | 87 (22.8) | 63 (12.2) | 55 (21.2) |
| Cw*07(01,02) | 113 (14.9) | 107 (28.1) | 80 (15.4) | 76 (29.3) |
| Cw*08(01) | 59 (7.8) | 58 (15.2) | 44 (8.5) | 43 (16.6) |
| Cw*12(02,03) | 9 (1.2) | 9 (2.4) | 8 (1.5) | 8 (3.1) |
| Cw*16(02,01) | 64 (8.4) | 62 (16.3) | 43 (8.3) | 41 (15.8) |
| Cw*17(01,03) | 63 (8.3) | 60 (15.7) | 48 (9.3) | 45 (17.4) |
| Cw*18(01) | 57 (7.5) | 51 (13.4) | 36 (7.0) | 32 (12.4) |
| Minor Cw | 18 (2.4) | 27 (7.1) | 13 (2.5) | 13 (5.0) |
Those present in ≥3% of subjects in any patient group. Identification of fully resolved four- or five-digit alleles (in parentheses) within each two-digit molecular specificity was based on selective typing by RSCA and SBT.
Allele refers to the number of occurrences of a selected allele divided by the number of alleles typed on two chromosomes per subject (2N); carriage refers to the number of subjects with a selected allele divided by the number of subjects typed (n). For the two groups, overall P = 0.327, 0.758, and 0.696 for the A, B, and C loci, respectively, based on Mantel-Haenszel χ2 tests.
Numbers correspond to n (%).
LD analyses revealed 42 A-B, 32 B-Cw, and 41 A-Cw haplotypes with an LR ≥ 2.0 and P ≤ 0.01. These two-locus haplotypes produced 27 extended haplotypes (Table 2) that collectively accounted for about 51% of the total. Most of these haplotypes were unique to Zambians; our earlier HLA class I typing in Caucasians (35) and Rwandans (64) detected only eight of them. Further comparison between Zambian HLA class I haplotypes and those in five major United States populations (5) also showed very limited interpopulation similarities (Table 2).
TABLE 2.
Extended HLA class I haplotypes observed in Zambians (n = 381)
| Haplotypea | Observed frequencyb | Other ethnic group(s) in which haplotype was detectedc |
|---|---|---|
| A*01-Cw*18-B*8101 | 0.013 | None |
| A*02-Cw*02-B*15 | 0.025 | None |
| A*02-Cw*07-B*07 | 0.010 | AA, C, H, NAN, & RWA |
| A*02-Cw*16-B*45 | 0.020 | AA & RWA |
| A*02-Cw*16-B*58 | 0.005 | None |
| A*23-Cw*02-B*15 | 0.024 | None |
| A*23-Cw*04-B*53 | 0.014 | None |
| A*23-Cw*08-B*14 | 0.017 | None |
| A*23-Cw*07-B*07 | 0.021 | AA |
| A*23-Cw*16-B*45 | 0.014 | None |
| A*30-Cw*03-B*15 | 0.022 | RWA |
| A*30-Cw*04-B*53 | 0.037 | RWA |
| A*30-Cw*07-B*08 | 0.018 | None |
| A*30-Cw*08-B*14 | 0.029 | None |
| A*30-Cw*17-B*42 | 0.052 | AA & RWA |
| A*30-Cw*18-B*57031 | 0.026 | None |
| A*34-Cw*04-B*44 | 0.010 | None |
| A*34-Cw*02-B*15 | 0.013 | None |
| A*3601-Cw*02-B*15 | 0.010 | None |
| A*66-Cw*06-B*58 | 0.017 | None |
| A*68-Cw*03-B*15 | 0.021 | RWA |
| A*68-Cw*04-B*35 | 0.015 | RWA |
| A*68-Cw*07-B*07 | 0.025 | RWA |
| A*68-Cw*08-B*14 | 0.025 | None |
| A*68-Cw*16-B*45 | 0.013 | None |
| A*74-Cw*02-B*15 | 0.021 | None |
| A*74-Cw*04-B*35 | 0.012 | RWA |
LR ≥ 2.0 (P ≤ 0.01) in all pair-wise 2 × 2 contingency tables.
Combined total = 0.517, but the actual haplotype frequency can be lower than the observed frequency.
Characteristics of HIV-1 viremia in Zambians with defined HLA class I variants.
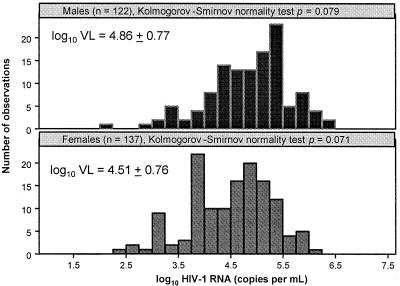
In the 259 seropositive and ambulatory Zambians, HIV-1 VL as a categorical variable and log10 VL as a continuous variable clearly differed by gender (P < 0.001), by patient group (TPDs, SCs, and DPSPs) (P < 0.0001 to 0.05), and by age (P < 0.006 to 0.007) (Table 3). The log10 VL measurements, stratified by gender, fit the normal distribution pattern in both females and males (P > 0.05) (Fig. 1) sufficiently well to justify the application of t tests in the GLM procedure. Compared with Caucasian and African-American SCs from the Multicenter AIDS Cohort Study (65), Zambian SCs here had slightly higher (by 0.1 to 0.2 log10) VL during a highly comparable period (i.e., within the first 42 months of infection) when measured by the same Amplicor assay.
TABLE 3.
Plasma HIV-1 RNA concentration in Zambians, stratified by gender, patient group, and age group
| Group (no.) | No. of subjects with a plasma HIV-1 RNA level (copies per ml) of:
|
Log10 VL
|
||||
|---|---|---|---|---|---|---|
| <10,000 (n = 55) | 10,000-100,000 (n = 106) | >100,000 (n = 98) | P value | Mean ± SD | P value | |
| Gender | ||||||
| Females (137) | 40 | 58 | 39 | Ref.b | 4.51 ± 0.76 | Ref. |
| Males (122) | 15 | 48 | 59 | <0.0001 | 4.86 ± 0.77 | 0.0004 |
| Patienta | ||||||
| TP donors (103) | 12 | 36 | 55 | <0.0001 | 4.93 ± 0.63 | <0.0001 |
| DP nontransmitters (65) | 21 | 27 | 17 | 0.005 | 4.45 ± 0.81 | 0.007 |
| TP seroconverters (91) | 22 | 43 | 26 | 0.055 | 4.54 ± 0.84 | 0.046 |
| Age (yr) | ||||||
| ≤40 (228) | 52 | 97 | 79 | Ref. | 4.63 ± 0.80 | Ref. |
| >40 (31) | 3 | 9 | 19 | 0.006 | 5.03 ± 0.58 | 0.007 |
P < 0.0001 (by F test) for the overall heterogeneity among the transmission pair (TP) donors, TP SCs, and discordant pair (DP) nontransmitters.
Ref., reference category.
FIG. 1.
Approximately normal distribution of log10 HIV-1 RNA concentration in Zambians with medium to high resolution of HLA class I profiles.
Differential distribution of HLA class I variants in Zambians according to HIV-1 VL categories.
With the 259 subjects divided into three categories of HIV-1 VL (12) (Table 4), overall (global) tests of HLA class I allelic distributions revealed genetic heterogeneities, mostly at the B (P = 0.022) and C (P = 0.013) loci, but not at the A locus (P = 0.274). Individually, 17 HLA class I allelic and haplotypic variants were differentially distributed across the VL categories in the univariate analyses without any adjustment for nongenetic factors (P = 0.003 to 0.088), all but two of them involving at least one B or C variant (Table 4). The number of differentially distributed HLA factors increased to 20 (P = 0.003 to 0.087) following adjustments for age, sex, and patient group membership that reflected differences in duration of HIV-1 infection (seroprevalents and SCs) and status of viral transmission (transmitters and nontransmitters). HLA variants known to be in LD accounted for many of these differences. For example, patients with low VL (<10,000) consistently had more B*57, along with A*30 and Cw*18 on the same extended haplotype, than did patients with higher (≥10,000 to >100,000) VL. Distribution of Cw*18 in the absence of B*57 showed a much smaller difference across the VL categories (P = 0.028 to 0.067). Similarly, the effect of B*45 alone entirely explained the increasing presence of A*02-Cw*16, B*45-Cw*16, and A*02-Cw*16-B*45 in patients with higher VL.
TABLE 4.
HLA variants with apparent associations with HIV-1 RNA level as a categorical variable
| HLA variant or variant statusd | Total no. of subjects | No. of subjects with a plasma HIV-1 RNA level (copies per ml) of:
|
P value
|
|||
|---|---|---|---|---|---|---|
| <10,000 (n = 55) | 10,000-100,000 (n = 106) | >100,000 (n = 98) | Unadjusted | Adjusteda | ||
| A*02-Cw*16 | 15 | 1 | 5 | 9 | 0.051 | 0.008 |
| A*02-B*45-Cw*16 | 11 | 1 | 4 | 6 | 0.193 | 0.040 |
| A*23-B*14 | 12 | 1 | 4 | 7 | 0.115 | 0.031 |
| A*23-Cw*08b | 12 | 1 | 5 | 6 | 0.237 | 0.106 |
| A*23-Cw*08-B*14b | 10 | 1 | 4 | 5 | 0.316 | 0.192 |
| A*23-Cw*07 | 18 | 1 | 6 | 11 | 0.023 | 0.084 |
| A*29-B*42 | 11 | 3 | 7 | 1 | 0.117 | 0.019 |
| A*30-B*57 | 11 | 5 | 6 | 0 | 0.005 | 0.001 |
| A*30-Cw*18-B*57 | 9 | 3 | 6 | 0 | 0.043 | 0.011 |
| A*30-Cw*03 | 12 | 5 | 6 | 1 | 0.018 | 0.008 |
| A*30-Cw*03-B*15 | 8 | 4 | 3 | 1 | 0.039 | 0.012 |
| A*3601b | 41 | 4 | 20 | 17 | 0.161 | 0.163 |
| A*68 | 74 | 12 | 28 | 34 | 0.076 | 0.087 |
| A*74-B*15b | 12 | 4 | 6 | 2 | 0.116 | 0.142 |
| B*13 | 10 | 4 | 4 | 2 | 0.116 | 0.129 |
| B*18 | 14 | 0 | 7 | 7 | 0.088 | 0.060 |
| B*39 | 9 | 6 | 2 | 1 | 0.003 | 0.012 |
| B*39-Cw*12 | 8 | 5 | 2 | 1 | 0.011 | 0.025 |
| B*41 | 7 | 0 | 2 | 5 | 0.050 | 0.019 |
| B*45 | 44 | 3 | 19 | 22 | 0.010 | 0.011 |
| B*45-Cw*16 | 26 | 1 | 11 | 14 | 0.017 | 0.020 |
| B*57 | 18 | 8 | 9 | 1 | 0.001 | 0.001 |
| B*57-Cw*18 | 15 | 6 | 9 | 0 | 0.003 | 0.002 |
| Cw*12 | 8 | 5 | 2 | 1 | 0.011 | 0.025 |
| Cw*16 | 41 | 4 | 17 | 20 | 0.037 | 0.037 |
| Cw*18 | 32 | 14 | 13 | 5 | 0.0003 | 0.0003 |
| Cw*18, no B*57 | 17 | 8 | 4 | 5 | 0.052 | 0.067 |
| Consistently favorablec | 37 | 17 | 17 | 3 | <0.0001 | <0.0001 |
| Unfavorablec | 43 | 3 | 15 | 25 | 0.004 | 0.0005 |
Adjusted for age, sex, and patient group membership (TPDs, transmission pair SCs, and discordant pair seropositives).
Became marginally significant in subsequent analyses (Table 5).
Favorable (A*30-Cw*03, B*39, and B*57) and unfavorable (A*02-Cw*16, A*23-B*14, and A*23-Cw*07) HLA variants show consistent associations with viral RNA levels in both categorical and general linear model statistics (Tables 4 and 5) as well as in alternative analyses based on sex or group membership (Table 7).
Indented entry(s) represents subset(s) of preceding entry.
Conversely, in other cases it was the HLA haplotype rather than its component alleles that showed differential distribution among categories of patients with contrasting VL. For example, A*30-Cw*03 (P = 0.005 to 0.019) and the closely related A*30-Cw*03-B*15 (P = 0.012 to 0.039) haplotype were associated with low VL, although neither A*30 alone (unadjusted P = 0.73), B*15 alone (unadjusted P = 0.43), nor Cw*03 alone (unadjusted P = 0.84) showed any appreciable differences. Weak and marginally significant effects were detected for several other haplotypes, including A*23-B*14 (P = 0.031 to 0.115) and A*23-Cw*07 (P = 0.023 to 0.084) (Table 4).
Univariate analyses of log10-transformed HIV-1 VL as a continuous variable.
GLM statistics (Table 5) largely supported findings based on categorical analysis. In particular, the mean VL was consistently about 0.9 to 1.1 log10 lower in B*13 (4.04 ± 1.03 log10)-, B*39 (3.89 ± 0.96)-, B*57 (4.11 ± 0.81)-, and A*30-Cw*03 (4.14 ± 0.74)-positive subjects than in those who tested positive for the presence of A*02-Cw*16 (5.00 ± 0.77 log10), A*23-B*14 (5.14 ± 0.61), and A*23-Cw*07 (5.11 ± 0.54). Two additional HLA variants, i.e., A*3601 and A*74-B*15, had marginally significant effects (P = 0.042 to 0.088) in the GLM analyses. Other HLA effects on mean log10 VL remained marginally significant, either alone or as a result of haplotypic combinations. Thus, HLA associations with various levels of HIV-1 VL were highly consistent in both categorical and GLM analyses.
TABLE 5.
HLA variants associated with HIV-1 RNA level as a continuous variable
| HLA variant or variant status | No. of carriers | Log10 VL (mean ± SD) | Unadjusted GLM P value | Adjusteda GLM P value |
|---|---|---|---|---|
| A*02-Cw*16 | 15 | 4.997 ± 0.769 | 0.010 | 0.025 |
| A*02-Cw*16-B*45 | 11 | 4.951 ± 0.870 | 0.230 | 0.081 |
| A*23-B*14 | 12 | 5.143 ± 0.608 | 0.033 | 0.009 |
| A*23-Cw*08 | 12 | 5.070 ± 0.576 | 0.072 | 0.030 |
| A*23-Cw*08-B*14 | 10 | 5.053 ± 0.621 | 0.118 | 0.078 |
| A*23-Cw*07 | 18 | 5.106 ± 0.540 | 0.015 | 0.075 |
| A*29-B*42 | 11 | 4.398 ± 0.704 | 0.234 | 0.094 |
| A*30-B*57 | 11 | 4.089 ± 0.603 | 0.011 | 0.004 |
| A*30-Cw*18-B*57 | 9 | 4.196 ± 0.595 | 0.062 | 0.025 |
| A*30-Cw*03 | 12 | 4.143 ± 0.738 | 0.016 | 0.017 |
| A*30-Cw*03-B*15 | 8 | 4.105 ± 0.710 | 0.036 | 0.018 |
| A*3601 | 41 | 4.881 ± 0.677 | 0.065 | 0.042 |
| A*68 | 74 | 4.836 ± 0.727 | 0.034 | 0.022 |
| A*74-B*15 | 12 | 4.298 ± 0.877 | 0.088 | 0.076 |
| B*13 | 10 | 4.038 ± 1.026 | 0.009 | 0.019 |
| B*18 | 14 | 4.986 ± 0.436 | 0.125 | 0.069 |
| B*39 | 9 | 3.891 ± 0.960 | 0.002 | 0.010 |
| B*39-Cw*12 | 8 | 3.995 ± 0.838 | 0.012 | 0.039 |
| B*41 | 7 | 5.052 ± 0.552 | 0.196 | 0.166 |
| B*45 | 44 | 4.885 ± 0.649 | 0.050 | 0.061 |
| B*45-Cw*16 | 26 | 4.957 ± 0.631 | 0.051 | 0.067 |
| B*57 | 18 | 4.111 ± 0.807 | 0.002 | 0.0002 |
| B*57-Cw*18 | 15 | 4.090 ± 0.782 | 0.003 | 0.0006 |
| Cw*12 | 8 | 3.995 ± 0.970 | 0.012 | 0.039 |
| Cw*16 | 41 | 4.867 ± 0.610 | 0.085 | 0.059 |
| Cw*18 | 32 | 4.276 ± 0.796 | 0.002 | 0.0006 |
| Cw*18, no B*57 | 17 | 4.415 ± 0.794 | 0.205 | 0.205 |
| Consistently favorableb | 37 | 4.110 ± 0.799 | <0.0001 | <0.0001 |
| Unfavorableb | 43 | 5.052 ± 0.625 | 0.0005 | 0.0002 |
Adjusted for age, sex, and patient group membership (TPDs, transmission pair SCs, and discordant pair seropositives).
Favorable (A*30-Cw*03, B*39, and B*57) and unfavorable (A*02-Cw*16, A*23-B*14, and A*23-Cw*07) HLA variants show consistent associations with viral RNA levels in both categorical and general linear model statistics (Tables 4 to 5) as well as in alternative analyses based on sex group membership (Table 7).
Multivariable analyses of HIV-1 VL as a continuous variable.
We constructed a multivariable model which included each HLA variant with a nominally significant effect on HIV-1 VL (Table 6), plus B*35 and B*53, which have been reported as markers of rapid HIV-1 disease progression in other, primarily Caucasian cohort studies (6, 13, 21, 30). Age, sex, and patient group remained strong and significant (adjusted P, 0.0003 to 0.096) factors contributing to variability in HIV-1 VL. The effects of the presence of both B*35 and B*53 were negligible (P = 0.384 to 0.603), as were the effects of A*02-Cw*16, A*23-Cw*07, A*29-B*42, A*68, A*74, B*18, B*41, and B*45. The strongest associations with relatively low VL were those of A*30-Cw*03 (P = 0.014), B*13 (P = 0.035), B*39 (P = 0.007), and B*57 (P < 0.0001), while those of Cw*18 and no B*57 with low VL (P = 0.047) and A*23-B*14 with high VL (P = 0.064) were of borderline significance. In the second model (Table 6), five HLA factors retained significant effects on VL: A*30-Cw*03 (P = 0.012), B*13 (P = 0.003), B*39 (P = 0.003), B*57 (P < 0.0001), and Cw*18 without B*57 (P = 0.030).
TABLE 6.
Two multivariable models showing the contributions of HLA class I and nongenetic host factors to variation in HIV-1 RNA level
| Patient characteristic | GLM P value in model 1a | GLM P value in Model 2b |
|---|---|---|
| Age (per yr) | 0.008 | 0.005 |
| Sex (female vs male) | 0.079 | 0.096 |
| Groupc | 0.0003 | 0.002 |
| HLA markers | ||
| A*02-Cw*16 (n = 15) | 0.106 | NA |
| A*23-B*14 (n = 12) | 0.064 | 0.045 |
| A*23-Cw*07 (n = 18) | 0.185 | NA |
| A*29-B*42 (n = 11) | 0.199 | NA |
| A*30-Cw*03 (n = 12) | 0.014 | 0.010 |
| A*68 (n = 74) | 0.207 | NA |
| A*74-B*15 (n = 12) | 0.113 | NA |
| B*13 (n = 10) | 0.035 | 0.003 |
| B*18 (n = 14) | 0.116 | NA |
| B*39 (n = 9) | 0.007 | 0.003 |
| B*41 (n = 7) | 0.402 | NA |
| B*45 (n = 44) | 0.867 | NA |
| B*57 (n = 18) | <0.0001 | <0.0001 |
| Cw*18, no B*57 (n = 17) | 0.047 | 0.030 |
| B*35 (n = 17) | 0.990 | NA |
| B*53 (n = 58) | 0.473 | NA |
Based on all markers with an adjusted P < 0.10 in multivariable Model 1. NA, not applicable: dropped out of the fiNAl model when P > 0.05.
Three HIV-1 seropositive patient groups (TPDs, transmission pair SCs, and discordant pair seropositives).
Further dissection of favorable and unfavorable HLA variants.
Alternative analyses were performed to determine the consistency of HLA associations. The three favorable and three unfavorable HLA variants detected in earlier analyses were consistently associated with low and high HIV-1 viremia, respectively, in the three independent patient groups defined by the status of HIV-1 transmission instead of viremia (data not shown). Separate analyses of data in seropositive males and females revealed similar consistencies (Table 7). In addition, VL was slightly higher (P = 0.076) in B*53-positive than in B*53-negative males.
TABLE 7.
Alternative analyses showing consistent and inconsistent HLA associations with HIV-1 RNA levels in seropositive Zambians, stratified by sex
| Patient characteristic | Seropositive females (n = 137)
|
Seropositive males (n = 122)
|
||||
|---|---|---|---|---|---|---|
| Marker positive (no. of patients) | Marker negative (no. of patients) | Adjusted P valuea | Marker positive (no. of patients) | Marker negative (no. of patients) | Adjusted P valuea | |
| Consistent HLA markers | ||||||
| A*02-Cw*16 (n = 15) | 4.95 ± 4.48 (9) | 4.48 ± 0.75 (128) | 0.053 | 5.06 ± 0.55 (6) | 4.84 ± 0.78 (116) | 0.255 |
| A*23-B*14 (n = 12) | 4.91 ± 0.65 (6) | 4.49 ± 0.77 (131) | 0.076 | 5.38 ± 0.51 (6) | 4.83 ± 0.77 (116) | 0.056 |
| A*23-Cw*07 (n = 18) | 4.76 ± 0.44 (7) | 4.50 ± 0.78 (130) | 0.520 | 5.33 ± 0.49 (11) | 4.81 ± 0.77 (111) | 0.076 |
| A*30-Cw*03 (n = 12) | 3.86 ± 0.90 (6) | 4.54 ± 0.75 (131) | 0.050 | 4.42 ± 0.44 (6) | 4.88 ± 0.77 (116) | 0.175 |
| B*39 (n = 9) | 3.87 ± 1.15 (6) | 4.54 ± 0.74 (131) | 0.063 | 3.93 ± 0.59 (3) | 4.88 ± 0.76 (119) | 0.076 |
| B*57 (n = 18) | 4.00 ± 0.90 (10) | 4.55 ± 0.74 (127) | 0.011 | 4.25 ± 0.71 (8) | 4.90 ± 0.76 (114) | 0.008 |
| Other markers of interestb | ||||||
| B*35 (n = 15) | 4.49 ± 0.86 (10) | 4.51 ± 0.76 (127) | 0.943 | 5.15 ± 0.81 (5) | 4.84 ± 0.77 (117) | 0.784 |
| B*53 (n = 53) | 4.70 ± 0.63 (31) | 4.46 ± 0.80 (106) | 0.076 | 4.74 ± 0.98 (22) | 4.88 ± 0.71 (100) | 0.580 |
The three consistently favorable HLA markers (A*30-Cw*03, B*39, and B*57) were found in 17 out of 55 (30.9%) HIV-1 seropositives with VL below 10,000 (copies per milliliter), 17 out of 106 (16.0%) with VL between 10,000 and 100,000, and 3 out of 98 (3.1%) with VL exceeding 100,000 (P < 0.0001 in all analyses) (Tables 4 and 5). VL in subjects carrying at least one of these consistently favorable HLA variants (4.11 ± 0.80) (P < 0.0001, unadjusted or adjusted) was at least 1.0 log10 lower than VL in the remainder.
The three infrequent and consistently unfavorable HLA variants (A*02-Cw*16, A*23-B*14, and A*23-Cw*07) associated with high VL (P = 0.05 to 0.10) were detected in 3 out of 55 (5.5%) HIV-1 seropositives with VL of <10,000 (copies per milliliter), 15 out of 106 (14.2%) with VL between 10,000 and 100,000, and 25 out of 98 (25.5%) with VL of >100,000 (P < 0.0001 to 0.004 as a group in all analyses) (Tables 4 and 5). The mean VL in subjects carrying at least one of these unfavorable HLA variants (5.05 ± 0.63) differed by 0.45 log10 from the mean for the rest of the cohort (4.60 ± 0.79; P < 0.001 with or without adjustments). Thus, segregation of HLA class I profiles in Zambians with lowest (<10,000) and highest (>100,000) VL was mostly due to three consistently favorable and three unfavorable alleles and haplotypes.
HLA class I homozygosity and HIV-1 viremia.
At the two-digit specificity level, homozygosity at the A (n = 40), B (n = 27), or C (n = 28) locus showed no effect on log10 VL following standard adjustment for age, sex, and membership of patient groups (P = 0.491, 0.7211, and 0.744, respectively). Homozygosity at two (n = 9) or all three loci (n = 5) was rare in this cohort. Neither single nor multilocus (n = 73) homozygosity was associated with variability in VL (P = 0.359 to 0.663).
DISCUSSION
HLA class I polymorphism has been shown to be a major determinant of early HIV-1 RNA level (27, 57) as well as of later disease progression (6, 13, 21, 30, 41) in seropositive Caucasian populations where clade B virus dominates. In our analysis, the main influential HLA class I alleles and haplotypes showed more differences than resemblances between clade C virus-infected Zambians and clade B virus-infected Caucasians. These differences suggest that evolution of viral clades represents a response to the distinctive HLA class I population profiles and that this hypothesis provides an explanation for population-specific HLA-mediated effects. One particular difference between clade C- and clade B-infected groups is especially noteworthy. Despite the strong association of B*35 alleles (or a subset of those) and B*53 alleles in Caucasians and African-Americans with both rapid disease progression (6, 13, 21, 30) along with elevated early HIV-1 RNA level (R. A. Kaslow et al., unpublished data), those same alleles lacked any appreciable effect in Zambians. Although no Zambians carried the reportedly more unfavorable B*35 subtypes, subtle allelic heterogeneity in nucleotide sequence encoding residues surrounding the peptide binding groove could not entirely explain this discrepancy, because the only Zambian B*53 allele is B*5301, the one reported to account for rapid disease progression in clade B-infected persons and to prefer epitopes with the xPxxxxxxX motif (13). Conversely, several class I allele (B*13 and B*39) or haplotype (A*30-Cw*03 and A*23-B*14) associations observed in Zambians have not shown consistent effects in Caucasians, perhaps because their low frequency would have made detection difficult or again because relationships are clade specific. B*14, B*18, B*35, and B*53 alleles from another native African population have been reported to present a variety of HIV-1 epitopes for CTL responses (33); however, apparently only a subset of CTL epitopes were differentially recognized in highly exposed seronegative individuals showing some resistance to HIV-1 seroconversion. Thus, the timing of specific CTL responses, along with the presence of different competing CTLs induced by relatively favorable and unfavorable HLA class I alleles coexisting in the same individual, can add to the complexity of genetic effects.
Focusing attention on newly identified markers with the most statistically significant relationships can raise concern about potentially spurious associations generated by multiple comparisons. That concern is ever present in studies of numerous highly polymorphic loci, but particularly so in an assessment of previously unreported relationships in relatively small numbers of patients, as was the case in this clade C-infected Zambian population. General caution against overinterpreting the statistical significance of any original finding, pursuit of further comparable population studies in clade-C-infected cohorts, and demonstration of the functional importance of the putative markers are more appropriate ways to refine and clarify the findings described here than arbitrary correction of P values.
Besides the newly recognized cohort-specific differences noted above, certain consistencies between Zambians and other cohorts corroborated HLA allelic effects previously shown to transcend the boundaries of race and clade. For example, the presence of HLA-B*57 has already emerged as uniformly favorable in persons of both Caucasian and African ancestry and infected with both clade A and B viruses (8, 21, 30, 35, 44). Our work now extends those findings to native Africans bearing HIV-1C infection. Recognition of HLA-B*57 (specifically and exclusively, B*57031) as the single most favorable marker in Zambians, despite LD patterns that differ between this and all other cohorts, highlights B*57 itself rather than its haplotypic lineages as broadly capable of suppressing HIV-1 RNA to a relatively low level during the early stages of infection (44). B*5701- and B*5703-mediated immunodominant responses to conserved HIV-1 Gag epitopes have been documented in African-American adolescents (P. Goepfert and R. A. Kaslow et al., unpublished data). Cross-reactive HIV-1 p24 epitopes restricted by B*5701 and B*5703 have been simultaneously detected in seropositive European and Kenyan subjects defined as slow progressors (15). These new findings lend further weight to the importance of B*57 alleles beyond race- and clade-specific immune responses.
Although less impressive, the association of two A*23-related haplotypes (A*23-B*14 and A*23-Cw*07) in Zambians with high VL parallels the association of A*23 with accelerated course of disease progression in Caucasians with clade B infection (30, 35) and that of A23 with increased transmission of HIV-1A in Kenyans (39, 40). Two other comparisons are also worth mentioning: the association of Cw*16 on the B*45-Cw*16 haplotype with high VL here was consistent with its overrepresentation in rapid progressors in the GRIV cohort (21), whereas higher VL in Zambians carrying B*14-Cw*08 and A*23-B*14 contrasted with the associations of B*14 and Cw*08 with a favorable prognosis of disease progression reported elsewhere (21, 41). Ongoing efforts to evaluate CTL profiles in Zambians with both favorable and unfavorable HLA alleles and haplotypes should provide important clues to mechanisms of immunologic control and escape during HIV-1C infection.
The negligible risk of increased VL from homozygosity at individual class I loci with incomplete four-digit allele resolution was consistent with the minor effects seen in clade B-infected Caucasians with single locus homozygosity at similarly incomplete resolution (Kaslow et al., unpublished). The unequivocally strong influence of class I homozygosity reported in general for the progression of disease in both Africans (63) and Caucasians (6, 35, 63) may be due to an increasing importance of diverse alternatives for antigen presentation, as evolving viruses attempt to escape from initially effective HLA-mediated CTL control. Longitudinal comparison of rates of viral mutation and divergence (17) in homozygotes and heterozygotes could help verify or refute that explanation.
Several implications can be drawn from the diminishing number of relationships seen of both nongenetic host factors and HLA markers to variation in HIV-1 viremia as more parsimonious models were applied. First, nongenetic factors such as age, gender, and duration of infection may modulate genetic effects, but variable impact of those other factors in different settings may make it difficult to assess the multiple host influences on HIV-1 viremia. Second, because HLA effects may covary strongly as a result of LD or reciprocity (increased frequency of one allele compensated by a decrease of one or several others), neither univariate nor multivariable analysis can completely distinguish the colinear effects of closely related markers without comprehensive stratifications and subset comparisons. Finally, a clear host genetic contribution to variability in HIV-1 viremia may not always translate directly to an appreciable difference in later disease progression and vice versa (65). Complex genetic effects probably depend on differential regulation of events throughout the pathological process, not simply on relationships detectable only at early or late stages of HIV-1 infection.
HLA polymorphisms probably influence plasma HIV-1 RNA concentration relatively early in the infection process (28, 57). Evaluation of host genetic influence on HIV-1 RNA levels should prove increasingly useful, because early in the course of infection, those levels have profound implications not only for the rate of subsequent progression of both untreated (4, 28, 43, 50, 61) and treated (4, 53, 54) disease but also for the transmissibility of HIV-1 from infected to uninfected individuals (12, 14, 49, 51, 55, 59). If the three consistently favorable HLA markers (Table 7) prove more broadly protective in Zambians and other southern Africans, individuals carrying them should experience a relatively benign course of disease progression without treatment. Over successive generations of HIV-1C-infected native Africans with corresponding HLA profiles, favorable antigen-presenting molecules should accumulate as unfavorable ones diminish concomitantly. Such evolution may gradually alter the genetic composition and responsiveness of populations in greatest need of effective HIV-AIDS vaccines. Consensus through continuous monitoring of disease progression in diverse ethnic groups carefully characterized for their HLA profiles should improve the design of epitope-rich vaccine constructs (9, 37, 48) and help predict their relative population coverage (58, 60).
Acknowledgments
We are grateful to C. A. Rivers and D. Munfus for valuable contributions to various aspects of genotyping-related work and to G. Cloud, M. Schaen, and C. Flanigan for data management. We also thank B. Hahn, F. Gao, and S. A. Trask for sharing datasets related to HIV-1 genotyping and phylogenetic analyses of HIV-1 clusters.
The Zambia-UAB HIV Research Project (ZUHRP) was supported by a grant (R01 AI40951 to S.A.) from the National Institutes of Health. Additional funding (U01 AI41530) was channeled through the HIV Acute Infection and Early Disease Evaluation Research Network (AIEDRN; George Shaw, principal investigator) and provided from R01 AI41951 (R.A.K.).
REFERENCES
- 1.Aickin, M., and H. Gensler. 1996. Adjusting for multiple testing when reporting research results: the Bonferroni vs Holm methods. Am. J. Public Health 86:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balla-Jhagjhoorsingh, S. S., G. Koopman, P. Mooij, T. G. M. Haaksman, V. J. P. Teeuwsen, R. E. Bontrop, and J. L. Neeney. 1999. Conserved CTL epitope shared between HIV-infected human long-term survivors and chimpanzees. J. Immunol. 162:2308-2314. [PubMed] [Google Scholar]
- 3.Bienzle, D., K. S. MacDonald, F. M. Smaill, C. Kovacs, M. Baqi, B. Courssaris, M. A. Luscher, S. L. Walmsley, and K. L. Rosenthal. 2000. Factors contributing to the lack of human immunodeficiency virus type 1 (HIV-1) transmission in HIV-1-discordant partners. J. Infect. Dis. 182:123-132. [DOI] [PubMed] [Google Scholar]
- 4.Bratt, G., A. Karlsson, A. C. Leandersson, J. Albert, B. Wahren, and E. Sandstrom. 1998. Treatment history and baseline viral load, but not viral tropism or CCR-5 genotype, influence prolonged antiviral efficacy of highly active antiretroviral treatment. AIDS 12:2193-2202. [DOI] [PubMed] [Google Scholar]
- 5.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygosity advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 7.Cereb, N., P. Maye, S. Lee, Y. Kong, and S. Y. Yang. 1995. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, and -C alleles. Tissue Antigens 45:1-11. [DOI] [PubMed] [Google Scholar]
- 8.Costello, C., J. Tang, C. Rivers, E. Karita, J. Meizen-Derr, S. Allen, and R. A. Kaslow. 1999. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS 13:1990-1991. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, D. V., M. Ozgur, K. Sari, M. Ghanayem, and D. D. Kostyu. 2001. Ramifications of HLA class I polymorphism and population genetics for vaccine development. Genet. Epidemiol. 20:87-106. [DOI] [PubMed] [Google Scholar]
- 10.Dupont, B. 1995. Immunobiology of HLA. Springer, New York, N.Y.
- 11.Ewens, W. J. 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3:87-112. [DOI] [PubMed] [Google Scholar]
- 12.Fideli, U. S., S. Allen, R. Musunda, S. Trask, B. Hahn, J. Mulenga, F. C. Kasolo, S. H. Vermund, and G. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 (HIV-1) in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, P. M., L. A. Kalish, J. Pitt, H. Minkoff, T. C. Quinn, S. K. Burchett, J. Kornegay, B. Jackson, J. Moye, C. Hanson, C. Zorrilla, J. F. Lew, and Women and Infants Transmission Study Group. 1999. Maternal levels of plasma human immunodeficiency virus type 1 RNA and the risk of perinatal transmission. N. Engl. J. Med. 341:394-402. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie, G. M., R. Kaul, T. Dong, H. B. Yang, T. Rostron, J. J. Bwayo, P. Kiama, T. Peto, F. A. Plummer, A. J. McMichael, and S. L. Rowland-Jones. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16:961-972. [DOI] [PubMed] [Google Scholar]
- 16.Goulder, P., D. Price, M. Nowak, S. Rowland-Jones, R. Phillips, and A. McMichael. 1997. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol. Rev. 159:17-29. [DOI] [PubMed] [Google Scholar]
- 17.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulder, P. J., A. K. Sewell, D. G. Lalloo, D. A. Price, J. A. Whelan, J. Evans, G. P. Taylor, G. Luzzi, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1997. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)—identical siblings with HLA-A*0201 are influenced by epitope mutation. J. Exp. Med. 185:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulder, P. J. R., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. McMichael. 1996. Novel, cross-restricted, conserved and immunodominant epitopes. I. Slow progressors in HIV infection. AIDS Res. Hum. Retrovir. 12:1691-1698. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J. R., A. Edwards, R. E. Phillips, and A. J. McMichael. 1997. Identification of a novel HLA-B*2705-restricted cytotoxic T-lymphocyte epitope within a conserved region of HIV-1 Nef. AIDS 11:536-538. [PubMed] [Google Scholar]
- 21.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942-6946. [PubMed] [Google Scholar]
- 22.Hendel, H., N. Henon, H. Lebuanec, A. Lachgar, H. Poncelet, S. Caillat-Zucman, C. A. Winkler, M. W. Smith, L. Kenefic, S. O'Brien, W. Lu, J. M. Andrieu, D. Zagury, F. Schachter, J. Rappaport, and J. F. Zagury. 1998. Distinctive effects of CCR5, CCR2, and SDF1 genetic polymorphisms in AIDS progression. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:381-386. [DOI] [PubMed] [Google Scholar]
- 23.Hill, A. V., S. N. Yates, C. E. Allsopp, S. Gupta, S. C. Gilbert, A. Lalvani, M. Aidoo, M. Davenport, and M. Plebanski. 1994. Human leukocyte antigens and natural selection by malaria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 346:379-385. [DOI] [PubMed] [Google Scholar]
- 24.Hoesley, C. J., S. Allen, J. L. Raper, R. Musonda, Y. F. Niu, F. Gao, K. E. Squires, and G. M. Aldrovandi. Comparative analysis of commercial assays for the detection and quantification of plasma HIV-1 RNA in patients infected with HIV-1 subtype C. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 25.Hughes, A. L., and M. K. Hughes. 1995. Natural selection on the peptide-binding regions of major histocompatibility complex molecules. Immunogenetics 42:233-243. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, A. L., M. Yeager, and M. Carrington. 1996. Peptide binding function and the paradox of HLA disease associations. Immunol. Cell Biol. 74:444-448. [DOI] [PubMed] [Google Scholar]
- 27.Ioannidis, J. P., P. G. McQueen, J. J. Goedert, and R. A. Kaslow. 1998. Use of neural networks to model complex immunogenetic associations of disease: human leukocyte antigen impact on the progression of human immunodeficiency virus infection. Am. J. Epidemiol. 147:464-471. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis, J. P. A., J. J. Goedert, P. G. McQueen, C. Enger, and R. A. Kaslow. 1999. Comparison of viral load and human leukocyte antigen statistical and neural network predictive models for the rate of HIV-1 disease progression across two cohorts of homosexual men. J. Acquir. Immune Defic. Syndr. 20:129-136. [DOI] [PubMed] [Google Scholar]
- 29.Just, J. J., M. C. King, G. Thomson, and W. Klitz. 1997. African-American HLA class II allele and haplotype diversity. Tissue Antigens 49:547-555. (Corrected and republished article originally printed in Tissue Antigens 48: 636-644, 1996). [DOI] [PubMed] [Google Scholar]
- 30.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. L. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 31.Kaslow, R. A., R. Duquesnoy, M. VanRaden, L. Kingsley, M. Marrari, S. Su, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1990. Combinations of A1, Cw7, B8, DR3 HLA antigens associated with rapid decline of T-helper lymphocytes in HIV-1 infected homosexual men: a report from the Multicenter AIDS Cohort Study. Lancet 335:927-930. [DOI] [PubMed] [Google Scholar]
- 32.Kaslow, R. A., C. R. Rivers, Tang, J., T. J. Bender, P. A. Goepfert, R. El Habib, K. Weinhold, M. J. Mulligan, and NIAID AIDS Vaccine Evaluation Group. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 75:8681-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaul, R., T. Dong, F. A. Plummer, J. Kimani, T. Rostron, P. Kiama, E. Njagi, E. Irungu, B. Farah, J. Oyugi, R. Chakraborty, K. S. MacDonald, J. J. Bwayo, A. McMichael, and S. L. Rowland-Jones. 2001. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Investig. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaul, R., F. A. Plummer, J. Kimani, T. Dong, P. Kiama, T. Rostron, E. Njagi, K. S. MacDonald, J. J. Bwayo, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 35.Keet, I. P., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and class II and transporter gene products with progression of human immunodeficiency virus-1 infection in homosexual men. J. Infect. Dis. 180:299-309. [DOI] [PubMed] [Google Scholar]
- 36.Knight, J. G., and D. D. Adams. 1982. The genetic basis of autoimmune disease. Ciba Found. Symp. 90:35-56. [DOI] [PubMed] [Google Scholar]
- 37.Longmate, J., J. York, C. La Rosa, R. Krishnan, M. Zhang, D. Senitzer, and D. J. Diamond. 2001. Population coverage by HLA class-I restricted cytotoxic T-lymphocyte epitopes. Immunogenetics 52:165-173. [DOI] [PubMed] [Google Scholar]
- 38.Ludbrook, J. 1998. Multiple comparison procedures updated. Clin. Exp. Pharmacol. Physiol. 25:1032-1037. [DOI] [PubMed] [Google Scholar]
- 39.MacDonald, K. S., J. E. Embree, N. J. Nagelkerke, J. Castillo, S. Ramhadin, S. Njenga, J. Oyug, J. Ndinya-Achola, B. H. Barber, J. J. Bwayo, and F. A. Plummer. 2001. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J. Infect. Dis. 183:503-506. [DOI] [PubMed] [Google Scholar]
- 40.MacDonald, K. S., K. R. Fowke, J. Kimani, V. A. Dunand, N. J. D. Nagelkerke, T. B. Ball, J. Oyugi, E. Njagi, L. K. Gaur, R. C. Brunham, J. Wade, M. A. Luscher, P. Hrausa, S. Rowland-Jones, E. Ngugi, J. J. Bwayo, and F. A. Plummer. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:1581-1589. [DOI] [PubMed] [Google Scholar]
- 41.Magierowska, M., I. Theodorou, P. Debre, F. Sanson, B. Autran, Y. Riviere, D. Charron, and D. Costagliola. 1999. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 93:936-941. [PubMed] [Google Scholar]
- 42.McKenna, S. L., G. K. Muyinda, D. Roth, M. Mwali, N. Ng'andu, A. Myrick, C. Luo, F. H. Priddy, V. M. Hall, A. A. von Lieven, J. R. Sabatino, K. Mark, and S. A. Allen. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS 11(Suppl. 1):S103-S110. [PubMed] [Google Scholar]
- 43.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 44.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson, G. W., J. J. Goedert, R. A. Kaslow, and D. L. Mann. 1997. Frequency of HLA allele-specific peptide motifs in HIV-1 proteins correlates with the allele's association with relative rates of disease progression after HIV-1 infection. Proc. Natl. Acad. Sci. USA 94:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nepom, G. T., and H. Erlich. 1991. MHC class-II molecules and autoimmunity. Ann. Rev. Immunol. 9:493-525. [DOI] [PubMed] [Google Scholar]
- 47.Novitsky, V., P. O. Flores-Villanueva, P. Chigwedere, S. Gaolekwe, H. Bussman, G. Sebetso, R. Marlink, E. J. Yunis, and M. Essex. 2001. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum. Immunol. 62:146-156. [DOI] [PubMed] [Google Scholar]
- 48.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific Elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Operskalski, E. A., D. O. Stram, M. P. Busch, W. Huang, M. Harris, S. L. Dietrich, E. R. Schiff, E. Donegan, J. W. Mosley, and Transfusion Safety Study Group. 1997. Role of viral load in heterosexual transmission of human immunodeficiency virus type 1 by blood transfusion recipients. Am. J. Epidemiol. 146:655-661. [DOI] [PubMed] [Google Scholar]
- 50.O'Shea, S., T. Rostron, A. S. Hamblin, S. J. Palmer, and J. E. Banatvala. 1991. Quantitation of HIV: correlation with clinical, virological, and immunological status. J. Med. Virol. 35:65-69. [DOI] [PubMed] [Google Scholar]
- 51.Pedraza, M. A., J. del Romero, F. Roldan, S. Garcia, M. C. Ayerbe, A. R. Noriega, and J. Alcami. 1999. Heterosexual transmission of HIV-1 is associated with high plasma viral load levels and a positive viral isolation in the infected partner. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 21:120-125. [PubMed] [Google Scholar]
- 52.Perneger, T. V. 1998. What's wrong with Bonferroni adjustments. BMJ 316:1236-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips, A. N., S. Staszewski, R. Weber, O. Kirk, P. Francioli, V. Miller, P. Vernazza, J. D. Lundgren, and B. Ledergerber. 2001. HIV viral load response to antiretroviral therapy according to the baseline CD4 cell count and viral load. JAMA 286:2560-2567. [DOI] [PubMed] [Google Scholar]
- 54.Polis, M. A., I. A. Sidorov, C. Yoder, S. Jankelevich, J. Metcalf, B. U. Mueller, M. A. Dimitrov, P. Pizzo, R. Yarchoan, and D. S. Dimitrov. 2001. Correlation between reduction in plasma HIV-1 RNA concentration 1 week after start of antiretroviral treatment and longer-term efficacy. Lancet 358:1760-1765. [DOI] [PubMed] [Google Scholar]
- 55.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, R. H. Gray, and Rakai Project Study Group. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. N. Engl. J. Med. 342:921-929. [DOI] [PubMed] [Google Scholar]
- 56.Rowland-Jones, S. L., T. Dong, L. Dorrell, G. Ogg, P. Hansasuta, P. Krausa, J. Kimani, S. Sabally, K. Ariyoshi, J. Oyugi, K. S. MacDonald, J. Bwayo, H. Whittle, F. A. Plummer, and A. J. McMichael. 1999. Broadly cross-reactive HIV-specific cytotoxic T-lymphocytes in highly-exposed persistently seronegative donors. Immunol. Lett. 66:9-14. [DOI] [PubMed] [Google Scholar]
- 57.Saah, A. J., D. R. Hoover, S. Weng, M. Carrington, J. Mellors, C. R. J. Rinaldo, D. Mann, R. Apple, J. P. Phair, R. Detels, O. B. S., C. Enger, P. Johnson, R. A. Kaslow, et al. 1998. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. AIDS 12:2107-2113. [DOI] [PubMed] [Google Scholar]
- 58.Schipper, R. F., C. A. van Els, J. D'Amaro, and M. Oudshoorn. 1996. Minimal phenotype panels. A method for achieving maximum population coverage with a minimum of HLA antigens. Hum. Immunol. 51:95-98. [DOI] [PubMed] [Google Scholar]
- 59.Semba, R. D., N. Kumwenda, D. R. Hoover, T. E. Taha, T. C. Quinn, L. Mtimavalye, R. J. Biggar, R. Broadhead, P. G. Miotti, L. J. Sokoll, L. van der Hoeven, and J. D. Chiphangwi. 1999. Human immunodeficiency virus load in breast milk, mastitis, and mother-to-child transmission of human immunodeficiency virus type 1. J. Infect. Dis. 180:93-98. [DOI] [PubMed] [Google Scholar]
- 60.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50:201-212. [DOI] [PubMed] [Google Scholar]
- 61.Shearer, W. T., T. C. Quinn, P. LaRussa, J. F. Lew, L. Mofenson, S. Almy, K. Rich, E. Handelsman, C. Diaz, M. Pagano, V. Smeriglio, L. A. Kalish, and Women and Infants Transmission Study Group. 1997. Viral load and disease progression in infants infected with human immunodeficiency virus type 1. N. Engl. J. Med. 336:1337-1342. [DOI] [PubMed] [Google Scholar]
- 62.Steel, C. M., C. A. Ludlam, D. Beatson, J. F. Peutherer, R. J. G. Cuthbert, H. Morrison, M. Jones, and P. Simmonds. 1988. HLA haplotype A1 B8 DR3 as a risk factor for HIV-related disease. Lancet i:1185-1188. [DOI] [PubMed]
- 63.Tang, J., C. Costello, I. P. M. Keet, C. Rivers, S. LeBlanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 15:317-324. [DOI] [PubMed] [Google Scholar]
- 64.Tang, J., E. Naik, C. Costello, E. Karita, C. Rivers, S. Allen, and R. A. Kaslow. 2000. Characteristics of HLA class I and class II polymorphisms in Rwandan women. Exp. Clin. Immunogenet. 17:185-198. [DOI] [PubMed] [Google Scholar]
- 65.Tang, J., B. Shelton, N. J. Makhatadze, Y. Zhang, M. Schaen, L. Louie, J. J. Goedert, E. C. Seaburg, J. B. Margolick, J. Mellors, and R. A. Kaslow. 2002. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J. Virol. 76:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trask, S. A., C. A. Derdeyn, U. Fideli, Y. Chen, S. Meleth, F. Kasolo, R. Musonda, E. Hunter, F. Gao, S. Allen, and B. H. Hahn. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J. Virol. 76:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner, S., M. E. Ellexson, H. Hickman, D. A. Sidebottom, M. Fernandez-Vina, D. L. Confer, and W. H. Hildebrand. 1998. Sequence-based typing provides a new look at HLA-C diversity. J. Immunol. 161:1406-1413. [PubMed] [Google Scholar]
- 68.Watterson, G. A. 1978. The homozygosity test of neutrality. Genetics 88:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]