Abstract
Phosphorylation of hepatitis B virus (HBV) core protein has recently been shown to be a prerequisite for pregenomic RNA encapsidation into viral capsids, but the host cell kinases mediating this essential step of the HBV replication cycle have not been identified. We detected two kinases of 95 and 115 kDa in HuH-7 total cell lysates which interacted specifically with the HBV core protein and phosphorylated its arginine-rich C-terminal domain. The 95-kDa kinase was purified and characterized as SR protein-specific kinase 1 (SRPK1) by mass spectrometry. Based on this finding, the 115-kDa kinase could be identified as the related kinase SRPK2 by immunoblot analysis. In vitro, both SRPKs phosphorylated HBV core protein on the same serine residues which are found to be phosphorylated in vivo. Moreover, the major cellular HBV core kinase activity detected in the total cell lysate showed biochemical properties identical to those of SRPK1 and SRPK2, as examined by measuring binding to a panel of chromatography media. We also clearly demonstrate that neither the cyclin-dependent kinases Cdc2 and Cdk2 nor protein kinase C, previously implicated in HBV core protein phosphorylation, can account for the HBV core protein kinase activity. We conclude that both SRPK1 and SRPK2 are most likely the cellular protein kinases mediating HBV core protein phosphorylation during viral infection and therefore represent important host cell targets for therapeutic intervention in HBV infection.
Hepatitis B virus (HBV), a small DNA virus belonging to the family Hepadnaviridae, causes acute and chronic hepatitis in humans. Worldwide, an estimated 350 million persons are persistently infected with HBV (4). A significant subset of these HBV carriers progresses to severe liver disease, such as hepatocellular carcinoma, which is assumed to cause up to one million deaths per year (30). Current treatment of chronic HBV infections by the approved therapeutics alpha interferon and lamivudine has its limitations, and there is a clear medical need for new therapeutic strategies (33).
The mature HBV virion consists of an enveloped, spherical nucleocapsid which contains the viral DNA genome and is assembled from dimers of a single capsid protein, the 21-kDa HBV core protein (43). During the assembly process, viral polymerase mediates the specific encapsidation of pregenomic RNA and subsequently converts the pregenomic RNA to viral genomic DNA (3, 19; for reviews, see references 13 and 33).
Numerous studies have shown that the HBV core protein is phosphorylated in intact cells (27, 32, 41). The serine residues of three repeated SPRRR motifs in its arginine-rich C-terminal region were identified as phosphoacceptor sites in vivo (S155, S162, and S170 in strain ayw) (25). Core protein becomes phosphorylated prior to nucleocapsid assembly, and mutational analysis strongly suggests that phosphorylation of serines 162 and 170 is critical for subsequent pregenomic RNA packaging to occur (14, 24). As none of the viral proteins possesses intrinsic protein kinase activity, the enzyme catalyzing core protein phosphorylation must be of host cell origin. Several protein kinases, such as the cyclin-dependent kinase Cdc2, protein kinase C (PKC), and a 46-kDa serine kinase, have been found to phosphorylate HBV core protein in vitro (20, 23, 25). Notably, all three mapped serine phosphorylation sites of core protein represent consensus sites for Cdc2 or cyclin-dependent kinase 2 (Cdk2) phosphorylation (35), and both kinases are activated in cells overexpressing the HBV HBx protein (5).
PKC has been detected inside mature HBV virions. However, PKC-mediated core protein phosphorylation was shown to be dispensable for pregenomic RNA encapsidation and was instead proposed to be required for transport of the viral genome to the nucleus at a late step during the HBV replication cycle (20, 21). A 46-kDa serine kinase previously implicated in core protein phosphorylation was also found to associate with core particles. The identity of this kinase could not be determined, but it is noteworthy that the 46-kDa core particle-associated kinase was found to be distinct from PKC in this study (23). Thus, it is presently unclear whether these kinases or others not yet identified are responsible for HBV core protein phosphorylation during natural infection.
To address this open question, we set out to isolate protein kinases which specifically bound and phosphorylated recombinant HBV core protein. By this approach, we identified the SR protein-specific kinases 1 and 2 (SRPK1 and SRPK2) and could further demonstrate that these kinases indeed account for the cellular core protein kinase activity detected in total lysates from HuH-7 cells. Thus, SRPK1 and SRPK2 are responsible for HBV core protein phosphorylation and could therefore represent essential host factors for HBV replication.
MATERIALS AND METHODS
Plasmid constructs and adenoviruses.
Plasmid pGEX-HBV-C3 contains the entire coding sequence of HBV core protein (amino acid 1 to 183; subtype ayw) cloned in frame with the glutathione-S-transferase (GST) gene into the pGEX-5X (Pharmacia) vector (12). Plasmids pGEX-HBV-C1 and pGEX-HBV-C2 encode GST fusions with N-terminally truncated core proteins that start at amino acid positions 119 and 30, respectively. The insert from pGEX-HBV-C3 encoding full-length HBV core protein was cloned into the eukaryotic expression vector pcDNA3. In this construct, serines 155, 162, and 170 of HBV core protein were mutated to alanines to generate the pcDNA3-HBV-C3-AAA plasmid, and a mutant with an N-terminal truncation starting at amino acid position 119 was inserted into pGEX-5X for generation of GST-HVB-C1-AAA fusion protein. In addition, serines 176 and 178 were mutated to alanines in pcDNA3-HBV-C3 and pcDNA3-HBV-C3-AAA. The full-length SRPK1 coding sequence was PCR amplified from HeLa cell cDNA and fused to an N-terminal FLAG epitope by insertion into the pRK-FLAG vector (17). Plasmid pcDNA3-SRPK2-VSV expresses a functionally active vesicular stomatitis virus (VSV)-tagged version of human SRPK2 from a pcDNA3 vector background (38). The replication-competent HBV plasmid pSPT1.2xHBV (subtype adr4) was a kind gift from P. H. Hofschneider (Max Planck Institute for Biochemistry, Martinsried, Germany) (39).
Recombinant adenovirus vector construction and purification.
The adenoviruses used here were all E1/E3-defective derivatives of adenovirus type 5. Briefly, the cDNAs of SRPK1 and SRPK2 (VSV-tagged versions) were cloned into the transfer plasmid pPM7 between the cytomegalovirus immediate-early promoter-enhancer and the rabbit beta-globin intron-polyadenylation signal. This expression cassette was inserted into a bacterial plasmid-borne adenovirus genome by recombination in bacteria (8, 28). A cloned version of the novel genome was identified, the viral genome was released from the plasmid by restriction enzyme digestion, and virus replication was initiated by transfecting the genome into 293 cells by a modified polyethyleneimine transfection method (28). Virus was amplified in modified 293 cells (16) and purified from cell lysates by CsCl density gradient centrifugation as described (10). Virus was quantified by protein content by the conversion factor 1 mg of pure virion protein per ml = 3.4 × 1012 viral particles per ml (10). The control virus contains the same E1/E3-negative viral genome but no expression cassette.
Cell culture, transient expression, and cell lysis.
HuH-7 and COS-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. For plasmid transfection experiments in six-well dishes, COS-7 (HuH-7) cells were seeded at 3.5 × 105 (3.0 × 105) per well 20 h before transfection. Cells were incubated for 4 h in 1.0 ml of serum-free medium containing 8 μl (6 μl) of Lipofectamine (Gibco-BRL) and 1.5 μg of plasmid DNA per well. The transfection mixture was then supplemented with 1 ml of medium containing 20% fetal bovine serum, and 20 h later, cells were either lysed or incubated in phosphate-free medium in the presence of 100 μCi of [33P]orthophosphate per ml for 3 h prior to cell lysis in 150 μl of lysis buffer. Transfection experiments with the HBV plasmid pSPT1.2xHBV were performed in 10-cm dishes. HuH-7 cells were seeded at 1.5 × 106 per dish 20 h before transfection with 40 μl of Lipofectamine 2000 (Gibco-BRL) and 20 μg of plasmid DNA per dish according to the manufacturer"s instructions, and 48 h later, cells were lysed with 350 μl of lysis buffer per dish.
For adenoviral infections, HuH-7 cells were seeded at 2.5 × 105 per well into six-well dishes, followed by infection with 30,000 particles per cell on the following day. Cells were either lysed 24 h after infection or washed and incubated for a further 20 h prior to lysis in 150 μl of buffer.
Cells were harvested when about 80 to 90% confluent. For immunoprecipitation and in vitro association experiments, cells grown in 10-cm dishes (six-well dishes) were lysed in 500 μl (150 μl) of buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM EDTA, 2 mM MgCl2, and 10 mM sodium pyrophosphate plus additives (10 mM sodium fluoride, 1 mM orthovanadate, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 0.2 mM dithiothreitol [DTT]). When total cell lysates or supernatant fractions from binding experiments with chromatography media were assayed for HBV core protein kinase activity, cells cultivated in 10-cm dishes (six-well dishes) were lysed in 350 μl (100 μl) of buffer containing 20 mM HEPES (pH 7.5), 400 mM NaCl, 0.1% Nonidet P-40, 1 mM EGTA, 1.5 mM MgCl2, 5 mM β-glycerophosphate, 10 mM sodium fluoride, 1 mM orthovanadate, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 1 μM microcystin-LR, and 150 U of benzonase per ml. Where indicated, HEPES was replaced with 20 mM Tris-HCl (pH 7.5). All lysates were cleared by centrifugation at 13,000 rpm prior to further processing.
In vitro association and immunoprecipitation experiments.
After cell lysis in 10-cm dishes (six-well dishes), 300 μl (120 μl) of lysate per sample was subjected to in vitro association with either GST or GST-HBV-C1, -C2, or -C3 fusion protein (about 1 μg of each) bound to glutathione-Sepharose beads for 2.5 h at 4°C. For further immunoblot analysis or in-gel kinase assays, beads were washed three times with 500 μl (250 μl) of lysis buffer without additives, and the samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For immunoblotting, monoclonal anti-SRPK1, monoclonal anti-SRPK2 (both from Transduction Laboratories), monoclonal anti-FLAG (Sigma), or anti-VSV (Boehringer) antibodies were used. For in vitro kinase assays, beads were washed twice with 500 μl of lysis buffer without additives and twice with 300 μl of kinase buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl2). Kinase reactions were performed in 25 μl of kinase buffer supplemented with 50 μM ATP and 1 μCi of [γ-32P]ATP for 5 min at 37°C. Reactions were stopped by addition of 20 μl of SDS sample buffer (100 mM Tris-HCl [pH 6.8], 3% SDS, 30% glycerol, 5% β-mercaptoethanol). Samples were subjected to gel electrophoresis on 12.5% gels, and the Coomassie-stained gels were then autoradiographed.
Immunoprecipitations were performed with 120 μl (300 μl) of precleared lysate from six-well dishes (10-cm dishes) per sample by anti-FLAG, anti-VSV, or polyclonal anti-HBV core protein antibodies (Dako). For further immunoblot analysis, beads were washed three times with 200 μl (250 μl) of lysis buffer without additives prior to SDS-13% PAGE and immunoblotting. To detect HBV surface antigen expression, goat polyclonal anti-HBsAg antiserum was used (Dako). For measurement of specific kinase activity of transfected SRPKs, immunoprecipitates from six-well dishes were washed twice with 300 μl of lysis buffer without additives and twice with 200 μl of kinase buffer (50 mM HEPES [pH 7.5], 100 mM NaCl, 10 mM MgCl2). Kinase reactions were performed in 25 μl of kinase buffer supplemented with 50 μM ATP, 1 μCi of [γ-32P]ATP, and 1 μg of either GST-HBV-C1 or GST-HBV-C1-AAA for 5 min at 37°C. Reactions were stopped by addition of 20 μl of SDS sample buffer prior to SDS-12.5% PAGE.
Cell lysate fractionation and kinase assays.
For binding experiments with chromatography media, lysates prepared in HEPES- or Tris-HCl-containing buffers were first diluted with 1 volume of 20 mM HEPES (pH 7.5)-0.1% Nonidet P-40-1 mM EGTA-1.5 mM MgCl2-10 mM sodium fluoride for binding assays on SP or heparin-Sepharose FF beads (Amersham Pharmacia) or 1 volume of the same buffer containing 20 mM Tris-HCl (pH 7.5) instead of HEPES for Q Sepharose Fast Flow (Amersham Pharmacia) or phosphocellulose P11 beads (Whatman). The NaCl concentration was then adjusted to 0.2, 0.4, 0.6, or 0.8 M, and 200 μl per sample was added to 50 μl of packed beads of the different chromatography media which had been equilibrated to the same buffer and NaCl concentrations. Samples were rotated for 30 min at 4°C; control lysates were incubated without beads. Supernatants were then recovered and analyzed by immunoblotting or in-gel kinase assays.
In parallel, aliquots of the supernatants were normalized for NaCl concentration, and 20 μl of each was added to 50 μl of kinase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.5 mM DTT, 50 μM ATP, 1 μCi of [γ-32P]ATP) which had been mixed with approximately 2.5 μg of GST-HBV-C1 bound to washed beads. The final NaCl concentration was 150 mM. The kinase reaction was performed for 5 min at 25°C. Total cellular extracts from SRPK1-VSV- or SRPK2-VSV-overexpressing HuH-7 cells were tested in the same way for cellular HBV core kinase activity except that HEPES (pH 7.5) instead of Tris-HCl was included in the kinase buffer.
To examine cyclin-dependent kinase activity in total cell lysates, extracts from HuH-7 cells pretreated for 17 h with either solvent or 0.25 μg of nocodazole per ml were prepared and diluted with 1 volume of HEPES-containing buffer as described above. Aliquots (100 μl) were then subjected to either control incubations or incubations in the presence of 6 μl of packed p9CKShs1-agarose beads (Calbiochem) for 2 h. Supernatants were then recovered and analyzed by immunoblotting with anti-SRPK1, anti-SRPK2, anti-Cdc2 (Santa Cruz), and anti-Cdk2 (Santa Cruz) antibodies. In parallel, 20-μl aliquots were added to 50 μl of kinase buffer (50 mM HEPES [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 0.5 mM DTT, 50 μM ATP, 1 μCi of [γ-32P]ATP) which had been mixed with either approximately 2.5 μg of GST-HBV-C1 bound to washed beads or 5 μg of histone H1 (Calbiochem) as kinase substrates. Kinase reactions were performed for 3 min at 30°C.
When cellular PKC activity was measured, the HEPES-containing lysis and dilution buffers contained only 0.1 mM EGTA. Diluted lysates were preincubated with dimethyl sulfoxide 1 or 10 μM bisindolylmaleimide I (Calbiochem) on ice, and 20 μl was then added to 50 μl of different kinase reaction mixes (20 mM HEPES [pH 7.5], 10 mM MgCl2, 0.5 mM DTT, 50 μM ATP, 1 μCi of [γ-32P]ATP) containing the same inhibitor concentrations. Kinase reaction were performed for 3 min at 30°C in the presence or absence of PKC-specific cofactors (100 μg of phosphatidylserine per ml, 20 μg of diacylglycerol per ml, and 150 μM CaCl2) with either 2.5 μg of GST-HBV-C1 bound to washed beads or 5 μg of histone H1 as kinase substrates.
All kinase reactions were stopped by adding 40 μl of SDS sample buffer. In each experiment, control reactions were performed to ensure the linearity of phosphate incorporation into GST-HBV-C1. Samples were then analyzed by SDS-12.5% PAGE and autoradiography.
In-gel kinase assay.
Kinase assays in SDS-polyacrylamide gels were performed essentially as reported previously (7). Samples were prepared by incubation for 10 min at 50°C in SDS sample buffer. SDS-PAGE was performed on 10% minigels containing either approximately 75 μg of GST or GST-HBV-C1 protein per ml copolymerized in the separating gel. After sample resolution, gels were incubated twice for 30 min in 100 ml of 20% isopropanol-50 mM Tris-HCl (pH 8.0) and then washed for 1 h in 250 ml of 50 mM Tris-HCl (pH 8.0)-5 mM β-mercaptoethanol. To denature proteins, gels were incubated twice for 30 min in 100 ml of 6 M guanidine hydrochloride and then renatured for 16 h in 250 ml of 50 mM Tris-HCl (pH 8.0)-5 mM β-mercaptoethanol-0.04% Tween 40 at 4°C (five changes). Gels were then equilibrated for 1 h in 20 ml of 40 mM HEPES (pH 8.0)-100 mM NaCl-2 mM DTT-10 mM MgCl2. The kinase reaction was carried out for 1 h in 15 ml of 40 mM HEPES (pH 8.0)-100 mM NaCl-10 mM MgCl2-0.5 mM EGTA-75 μCi of [γ-32P]ATP-10 μM ATP. Gels were then washed extensively in 5% trichloroacetic acid-1% sodium pyrophosphate until washes were free of radioactivity (usually five changes). Gels were then Coomassie stained and dried, and autoradiography was performed.
16-BAC/SDS-PAGE.
Protein separation on 16-BAC/SDS-polyacrylamide gels was performed by modifications of procedures reported previously (18, 26). 16-BAC (benzyldimethyl-n-hexadecylammonium chloride) gels were cast as 0.75-mm-thick minigels. The 7.5% separating gel was prepared by mixing 1.8 g of urea, 2.5 ml of an acrylamide-N,N-methylenebisacrylamide solution (30%:0.8%, wt/vol), 0.3 ml of 2% N,N-methylenebisacrylamide (wt/vol), 5 ml of 2× separating gel buffer (KH2PO4, 2.05%, wt/vol; H3PO, 1%, vol/vol), 0.5 ml of 1.45% ascorbic acid (wt/vol, freshly prepared), and 16 μl of 0.14% FeSO4 (wt/vol, freshly prepared). The volume was adjusted with water to 9.6 ml, and after degassing the solution for 10 min, polymerization was initiated by adding 0.4 ml of H2O2 solution (1:1,200 dilution of 30% H2O2), and the top surface was overlaid with 1× separating gel buffer. The 4% stacking gel consisted of 1 g of urea, 1.3 ml of an acrylamide-N,N-methylenebisacrylamide solution (30%:0.8%, wt/vol), 1.16 ml of 2% N,N-methylenebisacrylamide (wt/vol), 2.5 ml of 4× stacking gel buffer (0.5 M KH2PO4, pH 4.1), 0.5 ml of 1.45% ascorbic acid (wt/vol), and 8.5 μl of 0.14% FeSO4 (wt/vol). The volume was adjusted with water to 9.5 ml, and after degassing the solution for 10 min, polymerization was initiated by adding 0.5 ml of H2O2 solution (1:750 dilution of 30% H2O2). After 1 h of polymerization at room temperature, gels were kept at 4°C and used the next day.
Aspirated beads from in vitro association experiments were mixed with 30 μl of freshly prepared sample buffer (0.6 g of urea, 1.25 ml of 10% 16-BAC, 200 μl of 87% glycerol, 90 μl of 1 M DTT, 12.5 μl of 2-mg/ml aprotinin) and incubated for 10 min at 37°C prior to sample loading. The electrode buffer used was 1.126% (wt/vol) glycine-0.35% (vol/vol) H3PO4, with 0.1% 16-BAC included in the upper electrode buffer. Electrophoresis was carried out towards the cathode with an initial current of 10 mA per gel, which was increased to 20 mA per gel after 20 min. Electrophoresis was terminated 10 min after the Schlieren line had run out of the bottom of the gel.
The gel was fixed for 1 h in 10% (vol/vol) acetic acid-40% (vol/vol) methanol and then Coomassie stained and destained. For equilibration, gels were incubated for 10 min in 100 mM Tris-HCl (pH 6.8), 10 min in a solution consisting of 50 mM Tris-HCl (pH 6.8), 30% (vol/vol) glycerol, 6 M urea, and 2% SDS, and then for a further 10 min in the same solution containing 10 mg of DTT per ml. The gel was cut into appropriate-sized strips with a glass plate, and the strips were then positioned on top of the flat stacking gel surfaces of 10% second-dimension SDS gels. Electrophoresis was carried out towards the anode with an initial current of 5 mA per gel, which was increased to 20 mA per gel upon protein entry into the separating gel. Electrophoresis was terminated when the Coomassie blue had eluted from the bottom of the gel, and gels were then Coomassie stained and destained.
For subsequent analysis by mass spectrometry, gels were washed three times for 10 min with water, and the spots of interest were excised from the gel. For analytical purposes, in vitro associations were performed essentially as described above. Protein elution was achieved by directly adding 16-BAC sample buffer to the aspirated beads. After 16-BAC-PAGE, SDS-PAGE was performed on gels containing copolymerized GST-HBV-C1 protein, and in-gel kinase assays were performed as described above. For preparative purposes, eight confluent 10-cm dishes of Huh-7 cells were lysed in 550 μl of lysis buffer, and six in vitro associations of 670 μl of lysate with about 10 μg of GST-HBV-C1 each were performed in parallel. Eluted proteins were resolved by 16-BAC/SDS-PAGE, and the six spots of the protein of interest were pooled and then subjected to analysis by mass spectrometry.
Mass spectrometry. (i) In-gel digestion.
The excised gel plugs were washed twice in 100 mM ammonium bicarbonate (NH4HCO3), pH 7.8, and twice in 100 mM ammonium bicarbonate-acetonitrile (60:40; vol/vol) and dried by vacuum centrifugation. For reduction, the plugs were reswollen in 100 mM ammonium bicarbonate containing 10 mM DTT and incubated for 45 min at 56°C. Thereafter the tubes were chilled to room temperature, and the liquid was replaced with roughly the same volume of 55 mM iodoacetamide-100 mM ammonium bicarbonate. Samples were incubated for 30 min in the dark. Iodoacetamide solution was removed, and the gel plugs were washed and dried as described above. Trypsin (15 ng/μl) was added to the dry gel pieces and incubated on ice for 1 h for reswelling. After this time, sufficient digestion buffer was added to cover the gel pieces, and digestion was continued at 37°C overnight. The supernatant was transferred to a sample cup and dried in a Speed Vac vacuum concentrator. The gel pieces were washed twice with 25 mM ammonium bicarbonate, twice with 25 mM ammonium bicarbonate-acetonitrile (40:60), and twice with acetonitrile (50%) containing 5% formic acid. All supernatants were collected and then dried in a Speed Vac.
(ii) Desalting and concentration.
The dried peptides were resolubilized in 20 μl of 0.5% formic acid, loaded onto a ZipTip (Millipore), and washed with 0.5% formic acid-5% methanol. The peptides were eluted with 60% methanol-0.5% formic acid.
(iii) ESI-MS.
Mass spectrometry/mass spectrometry (MS/MS) of peptides generated by in-gel digestion was performed by nano-electrospray ionization (ESI) on a Q-TOF mass spectrometer (Micro Mass, Manchester, United Kingdom) (6). The cone voltage was 50 V. The quadrupole analyzer was used to select precursor ions for fragmentation in the hexapole collision cell. The collision gas was argon at a pressure of 6 × 10−5 to 7 × 10−5 Torr. The collision energy was 20 to 30 V. The collision-induced dissociation products were analyzed by MassLynx MaxEnt 3 (Micromass, Ltd.) software. The deconvoluted MS/MS spectra were manually interpreted with the help of MassLynx software. Database search was performed with the MS blast function (http://dove.embl-heidelberg.de/Blast2/msblast.html) at EMBL or by database search programs in the Protein Prospector (http://prospector.ucsf.edu/).
RESULTS
Phosphorylation of HBV core protein by protein kinase(s) present in HuH-7 total cell lysate.
In a previous study, recombinant GST-HBV core fusion protein was found to serve as an interaction partner and substrate for an unidentified cellular protein kinase, which was detected in a cellular subfraction of ribosome-associated proteins (RAP) from HuH-7 cells (23). In efforts to reproduce these observations, we employed the analogous wild-type and deletion mutant GST-HBV core fusion proteins, but to avoid missing any cellular kinases, we used total cell lysate from HuH-7 cells instead of the RAP fraction as the cellular kinase source for in vitro association studies. As a control, in vitro association was done with GST alone.
After washing of the immobilized GST fusion proteins to remove unbound HuH-7 proteins, kinase reactions performed in buffer containing [γ-32P]ATP showed that all of the tested GST-HBV core fusion proteins were phosphorylated by associating cellular kinases, while no phosphate incorporation into GST itself could be detected (Fig. 1). Consistent with previous reports showing core protein phosphorylation in its arginine-rich C-terminal domain, the short GST-HBV-C1 fusion protein comprising only the C-terminal 65 amino acids of HBV core protein was efficiently phosphorylated in this assay (Fig. 1) (23, 25, 41). Moreover, the bound kinase(s) appeared to be highly active towards HBV core protein, as phosphate incorporation became clearly detectable after 1 h of exposure of the dried gels despite the relatively low specific activity of the [γ-32P]ATP used in this assay.
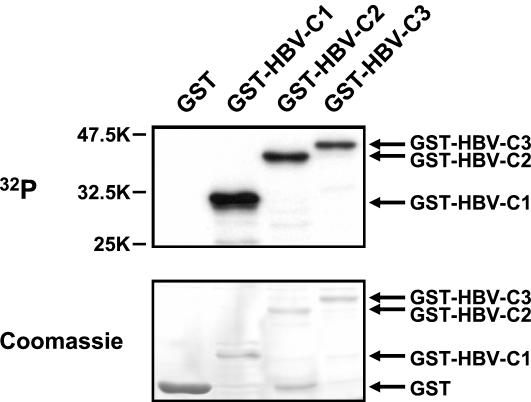
FIG. 1.
Phosphorylation of GST-HBV core protein by associating cellular kinases. Total cell lysates prepared from HuH-7 cells were incubated with either GST or different GST-HBV core fusion proteins. GST-HBV-C1 and GST-HBV-C2 are GST fusion proteins with N-terminally truncated core proteins that start at amino acid positions 119 and 30, respectively. GST-HBV-C3 contained wild-type HBV core protein. After in vitro association for 2.5 h, kinase reactions were performed on the washed beads containing bound GST fusion proteins as substrates for interacting cellular kinases. Samples were then resolved by SDS-12.5% PAGE. Upon Coomassie staining to visualize the different GST fusion proteins (lower panel), autoradiography was performed to detect the phosphorylated GST-HBV core fusion proteins (upper panel). Positions of GST fusion proteins are indicated on the right. In this and subsequent figures, sizes are shown in kilodaltons.
Two cellular protein kinases of 95 and 115 kDa catalyzing core protein phosphorylation interact specifically with HBV core protein.
Although GST itself was not phosphorylated by interacting kinases, the experiment shown in Fig. 1 did not exclude that the GST moiety rather than the C-terminal part of HBV core protein was important for kinase binding. To address this issue and further characterize the kinases involved, proteins bound to GST or the three different GST-HBV core fusion proteins (GST-HBV-C1, -C2, and -C3) were either resolved on gels containing copolymerized GST-HBV-C1 as a test substrate or separated on control gels containing GST instead. In addition, total cell lysate from HuH-7 cells was applied to both types of gels. As shown in Fig. 2, this in-gel kinase assay of renatured proteins revealed two protein kinases of approximately 95 and 115 kDa which interacted with the different GST-HBV core fusion proteins but were absent from control associations in the presence of GST. Importantly, at this level of sensitivity, phosphate incorporation was detectable in GST-HBV-C1- but not in GST-containing gels (Fig. 2). These results demonstrated that the arginine-rich C-terminal domain of HBV core protein served as the substrate for the 95- and 115-kDa kinases, which also specifically interacted with this region of the core protein. A third major kinase of about 42 kDa could be visualized when total cell lysate was run on GST-HBV-C1-containing gels (Fig. 2). Although it is presently unclear whether this kinase is related to a candidate HBV core kinase of 46 kDa described in an earlier report, this kinase showed no detectable affinity for core protein under our assay conditions (23).
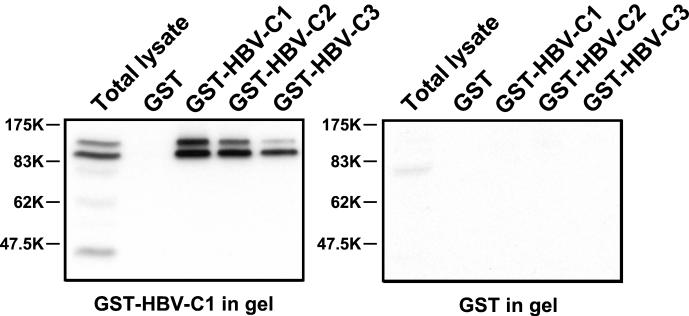
FIG. 2.
Detection of specifically associating HBV core protein kinases by an in-gel kinase assay. Total cell lysates prepared from HuH-7 cells were subjected to in vitro association with either GST or different GST-HBV core fusion proteins containing N-terminally truncated HBV core protein (GST-HBV-C1 and GST-HBV-C2) or wild-type HBV core protein (GST-HBV-C3). Bound proteins eluted with SDS sample buffer and total cell lysate were then resolved in parallel on gels containing either copolymerized GST-HBV-C1 (left panel) or GST protein (right panel). Upon denaturation and renaturation, in-gel kinase reactions were performed in the presence of [γ-32P]ATP. Gels were then washed and subjected to autoradiography.
Identification of two candidate HBV core protein kinases as SRPK1 and SRPK2.
The results presented above raised the possibility that GST-HBV core protein beads could be used for efficient affinity purification of the 95- and 115-kDa kinases. We reasoned that subsequent two-dimensional separation of HBV core protein-interacting proteins could provide us with sufficient resolving power to visualize distinct protein spots precisely comigrating with the in-gel HBV core kinase activities detected. For this purpose, we employed a two-dimensional technique which combines gel electrophoresis at an acidic pH in the presence of the cationic detergent 16-BAC with standard SDS-PAGE in the second dimension (18, 26). As shown in the upper panel of Fig. 3, the in-gel kinase assay of GST-HBV-C1-associating proteins resolved by 16-BAC/SDS-PAGE revealed two spots corresponding to the 95- and 115-kDa bands visualized in one-dimensional gels. These spots were not detected in GST-containing control gels (data not shown). Despite some background staining due to copolymerized GST-HBV-C1 protein, a Coomassie-stainable protein spot exactly comigrating with the lower HBV core kinase activity could be visualized in the same gel (Fig. 3, lower panel).
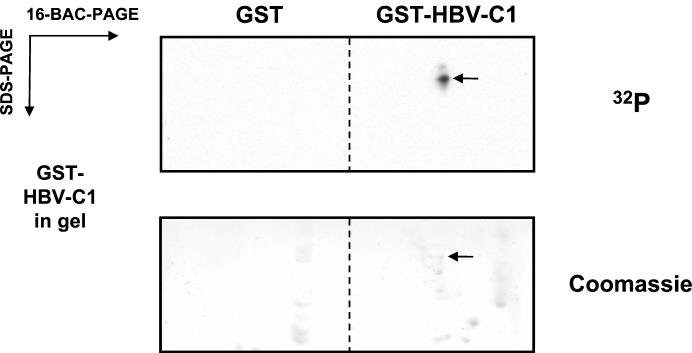
FIG. 3.
Purification of specifically associating GST-HBV core protein kinases. Total cell extracts from HuH-7 cells were incubated with either GST or GST-HBV-C1 immobilized on glutathione-Sepharose beads. After in vitro association, bound proteins were then resolved by 16-BAC-7.5% PAGE in the first dimension, and the two lanes containing the GST and GST-HBV-C1 in vitro association samples were then resolved on the same 10% SDS gel containing copolymerized GST-HBV-C1 in the second dimension as described under Materials and Methods. Upon denaturation and renaturation, an in-gel kinase reaction was performed in the presence of [γ-32P]ATP. Gels were then washed, Coomassie stained, and subjected to autoradiography. The arrows indicate the position of the faster-migrating HBV core protein kinase in the upper panel and its corresponding protein spot visualized by Coomassie staining in the lower panel. For subsequent mass spectrometry analysis, the same spot was excised from 16-BAC/SDS gels containing no copolymerized GST-HBV-C1 protein as described under Materials and Methods.
The same spots were excised from six preparative 16-BAC/SDS gels containing no copolymerized substrate protein, pooled, and digested with trypsin. Selected peptides were then subjected to MS/MS analysis by nano-ESI on a Q-TOF mass spectrometer. The spectra obtained allowed sequencing of two peptides (LEESSTIGQDQTLMER and EINCNGVLEVLNYTQNSNNETLR) which were identical with amino acids 330 to 345 and 353 to 375 in the human serine kinase SRPK1 (17). As shown in Fig. 4, this result was confirmed by immunoblot analysis of GST-HBV-C1-interacting proteins with anti-SRPK1 antibody. In addition, the 115-kDa kinase was identified as SRPK2 by immunoblotting. The highly related kinases SRPK1 and SRPK2 have been shown to phosphorylate serine/arginine-rich (SR) proteins, which play an important role in pre-mRNA splicing (17, 38). Moreover, transfection of either FLAG-SRPK1 or SRPK2-VSV expression plasmids into cells led to increased levels of specifically GST-HBV-C1-associating SRPK1 and SRPK2, and the transiently expressed kinases could be detected in anti-FLAG and anti-VSV immunoblots, respectively (Fig. 4).
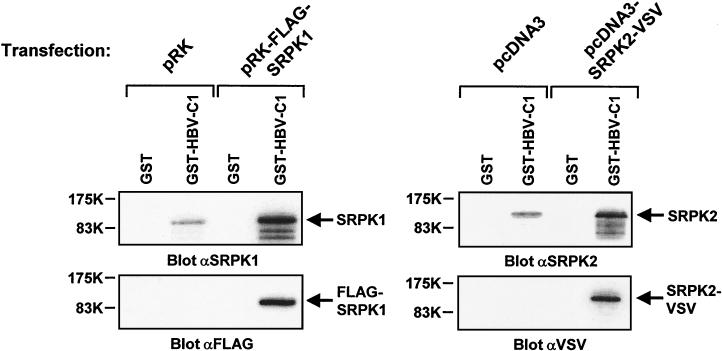
FIG. 4.
Specific association of endogenous and ectopically expressed SRPK1 and SRPK2 with GST-HBV core protein. COS-7 cells were transiently transfected with either control vectors or plasmids encoding FLAG-tagged SRPK1 (1.5 μg/well) or VSV-tagged SRPK2 (1.0 μg/well). After 24 h, cells were lysed, and extracts were subjected to in vitro association with either GST or GST-HBV-C1 protein. Samples were then resolved in duplicate by SDS-PAGE, followed by immunoblotting with anti-SRPK1 and anti-FLAG antibodies (left panels) or anti-SRPK2 and anti-VSV antibodies (right panels).
Similar in vivo phosphorylation and SRPK1/2-mediated in vitro phosphorylation of HBV core protein.
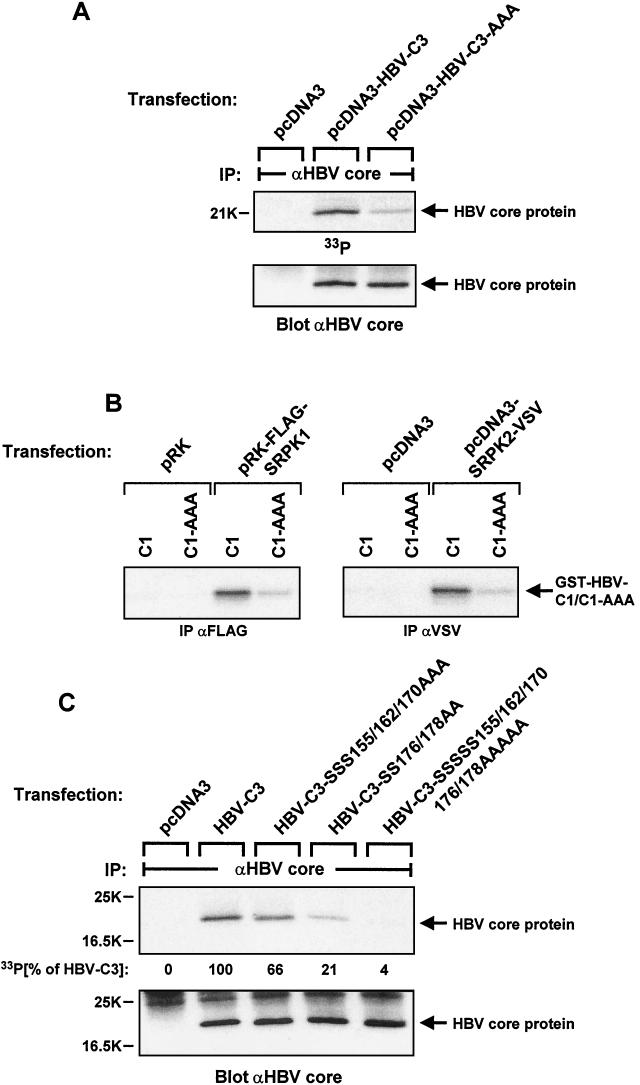
The HBV core protein has been shown to be phosphorylated in vivo in its arginine-rich C-terminal domain (41). Furthermore, three serine residues in this part of the protein (S155, S162, and S170 in subtype ayw) have been identified as the major in vivo phosphorylation sites (25). We cloned HBV core cDNA into the pcDNA3 expression plasmid and mutated the three reported phosphorylation sites to alanine. We transiently expressed the HBV core wild-type (HBV-C3) and mutant (HBV-C3-AAA) proteins in HuH-7 cells, labeled them with 33Pi in vivo, and isolated core protein by immunoprecipitation with specific antibody. As shown in Fig. 5A, the three serine-to-alanine mutations strongly reduced core protein phosphorylation in vivo. This result is quite similar to data reported previously by Liao and Ou (25), albeit the residual phosphate incorporation into mutant protein appeared to be more pronounced in our experiments.
FIG.5.
Analysis of in vivo phosphorylation and SRPK-mediated in vitro phosphorylation of HBV core protein. (A) HuH-7 cells were transiently transfected in six-well dishes with 1.5 μg of either empty expression vector, plasmid encoding wild-type HBV core protein, or plasmid encoding mutant HBV core protein devoid of the three reported C-terminal serine phosphorylation sites (serines 155, 162, and 170) per well. On the second day after transfection, cells were labeled with 50 μCi of 33Pi per ml for 3.5 h. Lysates were then subjected to immunoprecipitation (IP) with anti-HBV core antibody. After gel electrophoresis and transfer onto a nitrocellulose membrane, HBV core protein phosphorylation was first visualized by autoradiography (upper panel), followed by immunoblotting of the same filter with anti-HBV core protein antibody (lower panel). (B) COS-7 cells were transiently transfected in six-well dishes with either control plasmids or expression constructs encoding FLAG-tagged SRPK1 or VSV-tagged SRPK2 (1.5 μg/well each). After cell lysis and immunoprecipitation (IP) with anti-FLAG or anti-VSV antibodies, in vitro kinase reactions were performed in the presence of [γ-32P]ATP and GST-HBV-C1 core fusion protein with (C1) or without the three previously mapped C-terminal serine phosphorylation sites (C1-AAA, serines 155, 162, and 170 mutated to alanines). After gel electrophoresis, phosphate incorporation into substrate protein was visualized by autoradiography. (C) HuH-7 cells were transiently transfected in six-well dishes with plasmids encoding either wild-type HBV core protein or its mutants lacking serine phosphorylation sites in the C-terminal part as indicated (1.5 μg/well each). After metabolic labeling with 50 μCi of 33Pi per ml for 3.5 h, cell lysates were processed as described for panel A. HBV core protein phosphorylation was visualized by autoradiography (upper panel) and quantified by a PhosphorImager, and the same filter was then probed with anti-HBV core protein antibody (lower panel).
We next investigated SRPK-mediated HBV core protein phosphorylation in vitro. FLAG-tagged SRPK1 and VSV-tagged SRPK2 were transiently expressed in COS-7 cells and isolated by immunoprecipitation with antibodies recognizing the respective epitope tags. Immunoprecipitated SRPK1 and SRPK2 were then assayed for kinase activity towards either GST-HBV-C1 or the corresponding fusion protein lacking the three reported phosphorylation sites (GST-HBV-C1-AAA) as exogenous substrate proteins (Fig. 5B). Importantly, the reduced phosphorylation of mutant protein was similar to what we observed in vivo, indicating that SRPK1 and/or SRPK2 could be responsible for core protein phosphorylation in intact cells. To further analyze core protein phosphorylation in vivo, we mutated serines 176 and 178 to alanines. These mutations led to slightly reduced phosphorylation when introduced into wild-type HBV core protein, whereas the residual phosphate incorporation into HBV core protein already devoid of the three mapped serine phosphorylation sites was further reduced to barely detectable levels (Fig. 5C).
HBV core kinase activity in total cell lysate is biochemically indistinguishable from SRPK1 and SRPK2.
The extensive data presented above identified SRPK1 and SRPK2 as very good candidates for the cellular HBV core kinases, with the following caveats. First, other cellular protein kinases able to phosphorylate HBV core protein might simply not renature efficiently and would therefore be undetectable in an in-gel kinase assay. Second, cellular kinases might be involved which are only active when essential cofactors such as regulatory subunits are bound to them. As those complexes are disrupted during SDS-PAGE, the kinase subunits on their own would not regain activity. Third, kinases such as the 42-kDa protein kinase detected in total cell lysate might efficiently phosphorylate core protein in vivo even if the interaction is rather transient and not detectable in in vitro association experiments.
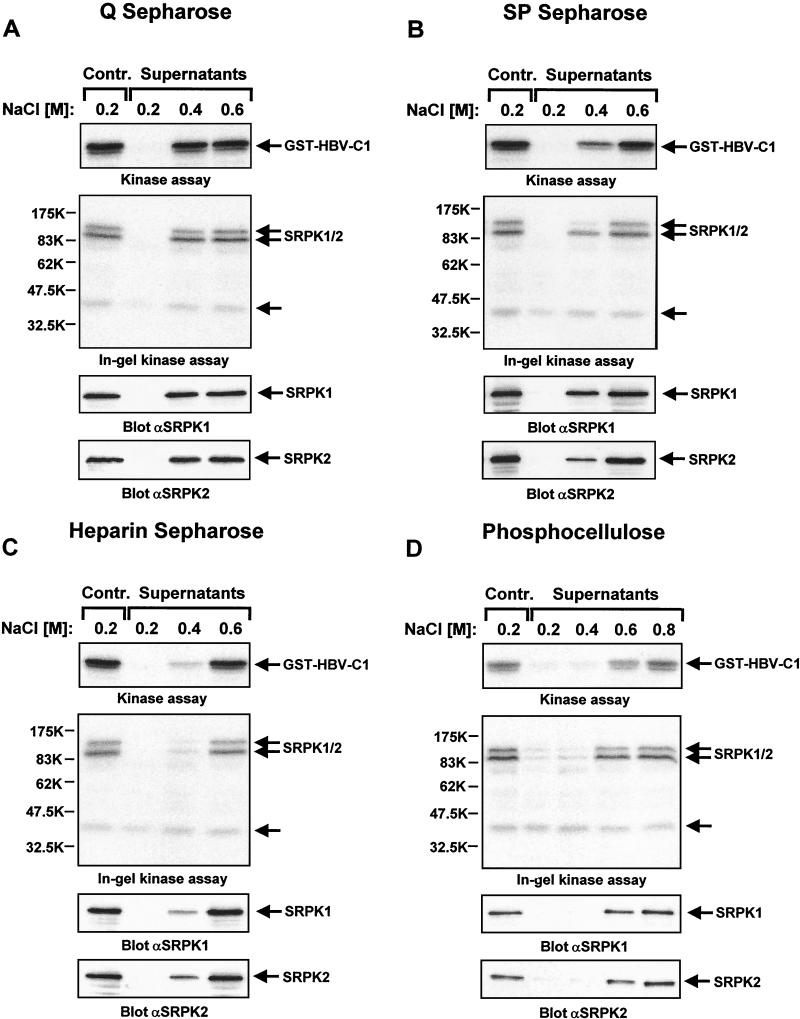
To address these limitations of the in-gel kinase detection, we set up an assay which enabled us to directly measure total cellular HBV core kinase activity in solution. For these experiments, cell lysis was performed in buffer devoid of kinase-inhibitory components at a salt concentration sufficient to disrupt cell nuclei. Subsequently, total cell extracts were incubated with beads of various chromatography media at different NaCl concentrations. Both the original total cell lysates and supernatants from these binding assays were then subjected to kinase reactions in the presence of GST-HBV-C1 fusion protein as an exogenous substrate. Furthermore, aliquots of the same cell lysates and supernatant fractions were run on parallel gels, which were then subjected to either in-gel kinase assays or SRPK1 and SRPK2 immunoblotting. This experimental approach enabled as to compare the biochemical properties of the cellular HBV core kinase(s) with those of SRPK1 and SRPK2. In this context, it is noteworthy that no specific antibodies capable of immunodepleting SRPK1 or SRPK2 from cell lysates are currently available.
As shown in the upper panels of Fig. 6A to D, the cellular HBV core kinase activity remained bound to Q, SP, and heparin-Sepharose as well as phosphocellulose beads when lysates had been adjusted to 0.2 M NaCl. An identical biochemical fractionation was observed with SRPK1 and SRPK2 (Fig. 6A to D, lower two panels). For all four matrices tested, increasing ionic strength led to partitioning of HBV core kinase activity into the supernatant fractions in a way that precisely correlated with the SRPK1 and SRPK2 fractionation. Importantly, we did not observe such a correlation when we probed the same filters with specific anti-Cdc2 and anti-Cdk2 antibodies (data not shown). Moreover, in-gel kinase assays revealed that the 42-kDa protein kinase detected in HBV core protein-containing gels only partially bound to Q Sepharose at the lowest ionic strength tested but to none of the other materials that bound SRPK1 and SRPK2 (Fig. 6A to D, second panels from top). Thus, we conclude that SRPK1 and SRPK2 have the same biochemical properties as the major cellular HBV core kinase activity, which is clearly not the case for the 42-kDa protein kinase.
FIG. 6.
Comparison of binding characteristics of HBV core kinase activity and SRPKs for different chromatography media. Total cellular extracts of HuH-7 cells were adjusted to different NaCl concentrations and then either control incubated without chromatography media orincubated in the presence of Q Sepharose (A), SP Sepharose (B), heparin-Sepharose (C), or phosphocellulose beads (D). Supernatants were then assayed for HBV core kinase activity in an in vitro kinase assay with GST-HBV core protein C1 as a substrate. After gel electrophoresis, phosphate incorporation into substrate protein was visualized by autoradiography (upper panels). In parallel, the same supernatants were resolved on GST-HBV-C1-containing SDS gels and subjected to an in-gel kinase assay. Phosphorylation of copolymerized GST-HBV-C1 by renatured cellular kinases was detected by autoradiography (second panels from the top). In addition, identical samples were subjected to immunoblotting with either anti-SRPK1 or anti-SRPK2 antibodies (lower two panels). Positions of GST-HBV-C1, SRPK1, and SRPK2 are indicated on the right. The position of an unidentified 42-kDa kinase detected in the in-gel kinase assay is marked by an arrow.
Neither the cyclin-dependent kinases Cdc2 and Cdk2 nor PKC account for major cellular HBV core kinase activities.
The three mapped serine phosphorylation sites of HBV core protein (S155, S162, and S170) are all part of SPRRR motifs, which have been characterized as consensus sequences for Cdc2- and Cdk2-mediated substrate phosphorylation (25, 35). As Cdc2 has been reported to phosphorylate HBV core protein in vitro and the HBV protein HBx was found to stimulate both Cdc2 and Cdk2 activity in transfected cells, these kinases could account for major cellular HBV core kinase activities (5, 25).
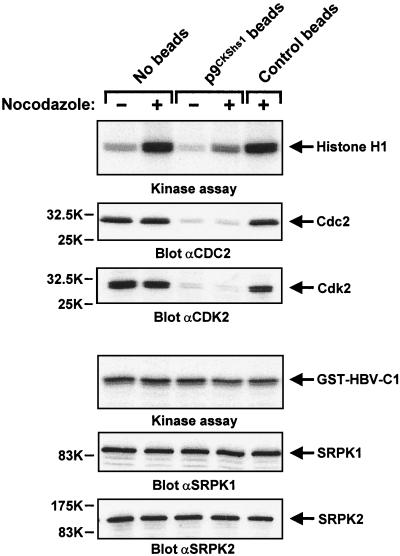
To test this, we prepared total cell lysates from solvent- and nocodazole-treated cells. Under our experimental conditions, nocodazole led to M-phase arrest in about 50% of the cells. Prior to performing in vitro kinase assays and immunoblotting, these lysates were either control incubated or depleted of Cdc2 and Cdk2 by incubating them in the presence of p9CKShs1 beads (2). As shown in the upper panel of Fig. 7, lysates from nocodazole-treated cells showed the expected pronounced increase in histone H1 kinase activity, which was strongly reduced when lysates had been preincubated with p9CKShs1 beads. This result indicates that Cdc2-mediated histone H1 phosphorylation could be detected in our extracts, as this p9CKShs1-interacting kinase is known to be activated during M phase (11). Importantly, neither nocodazole treatment nor Cdc2 and Cdk2 depletion had any effect on HBV core kinase activity and SRPK levels in the total cell extracts (Fig. 7). Thus, both biochemical fractionation and biological manipulation support our conclusion that these two CDKs are not important cellular HBV core protein kinases.
FIG. 7.
Cyclin-dependent kinases Cdc2 and Cdk2 are not major cellular HBV core protein kinases. HuH-7 cells were either mock treated or incubated for 17 h in the presence of 0.25 μg of nocodazole per ml prior to lysis. Total cellular extracts were then incubated without beads or in the presence of either p9CKShs1-agarose or control beads. Supernatants were then assayed for either histone H1 or HBV core kinase activity. After gel electrophoresis, phosphate incorporation into histone H1 or GST-HBV-C1 was visualized by autoradiography (first and fourth panel from top, respectively). In parallel, identical samples were analyzed by immunoblotting with either anti-Cdc2, anti-Cdk2, anti-SRPK1, or anti-SRPK2 antibodies. Positions of kinases and kinase substrates are indicated on the right.
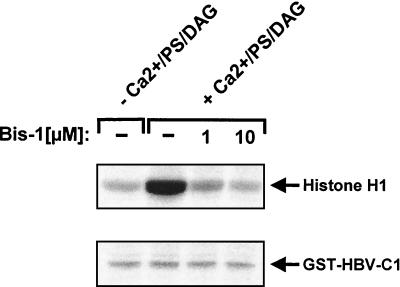
Furthermore, earlier studies have also implicated PKC in HBV core protein phosphorylation (20, 22). To assess whether members of the PKC family might contribute to cellular HBV core protein kinase activity, the effects of PKC activation and its inhibition in total cell extracts were measured in in vitro kinase assays with either histone H1 or GST-HBV-C1 as exogenous substrates. As shown in Fig. 8, addition of calcium, phosphatidylserine, and diacylglycerol to fully activate PKC strongly enhanced histone H1 phosphorylation, and this stimulation was reduced to basal levels when the PKC inhibitor bisindolylmaleimide I was included (37). In stark contrast, these reagents had no effect on HBV core kinase activity in total cell lysate. Thus, PKC does not account for a major cellular HBV core kinase activity.
FIG. 8.
Cellular PKC activity does not mediate HBV core protein phosphorylation. Total cell lysates from HuH-7 cells were incubated with either dimethyl sulfoxide or the indicated amounts of bisindolylmaleimide I (Bis-1). Aliquots were then added to kinase reaction mixes containing the same concentrations of bisindolylmaleimide I, which were supplemented with calcium, phosphatidylserine (PS), and diacylglycerol (DAG) where indicated. Kinase reactions were performed in the presence of either histone H1 (upper panel) or GST-HBV-C1 protein (lower panel). After gel electrophoresis, substrate phosphorylation was visualized by autoradiography. Positions of kinase substrates are indicated on the right.
Exclusive formation of stable HBV core protein-SRPK2 complexes in vivo correlates with stronger SRPK2- than SRPK1-mediated core protein phosphorylation in vitro.
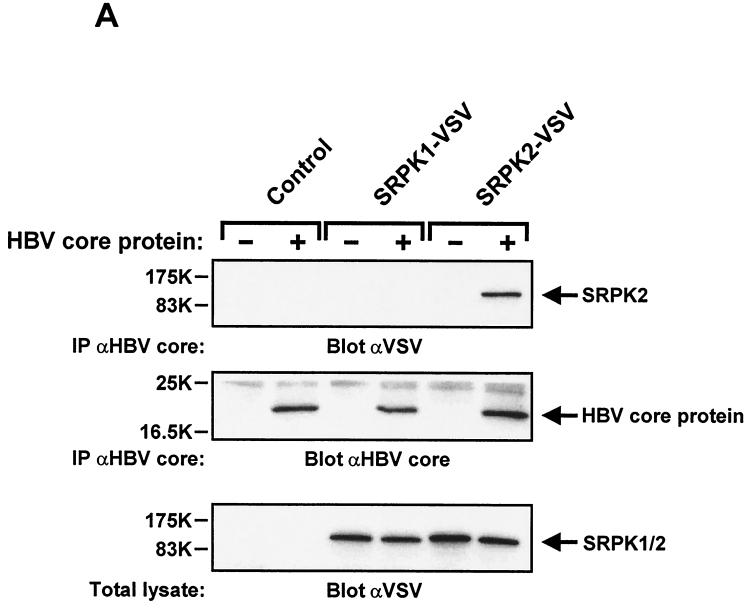
To study the interaction of HBV core protein with SRPKs in vivo, we used adenovirus-mediated gene transfer for transient protein expression in HuH-7 cells. The virus titers used were calibrated to express similar amounts of C-terminally tagged SRPK1 and SRPK2, as verified by anti-VSV immunoblotting of total cell extracts (Fig. 9A, lower panel). When HBV core protein was coexpressed and isolated by immunoprecipitation (Fig. 9A, middle panel), we could clearly detect specifically coprecipitating SRPK2, whereas SRPK1 was not found to associate under these experimental conditions (Fig. 9A, lower panel). Thus, when compared at similar expression levels, SRPK2 shows stronger affinity for HBV core protein than SRPK1. However, it is conceivable that core protein-SRPK1 complexes present in intact cells might dissociate when transferred into a larger volume upon cell lysis. Thus, we think that this finding does not exclude SRPK1 as a physiological HBV core protein kinase.
FIG. 9.
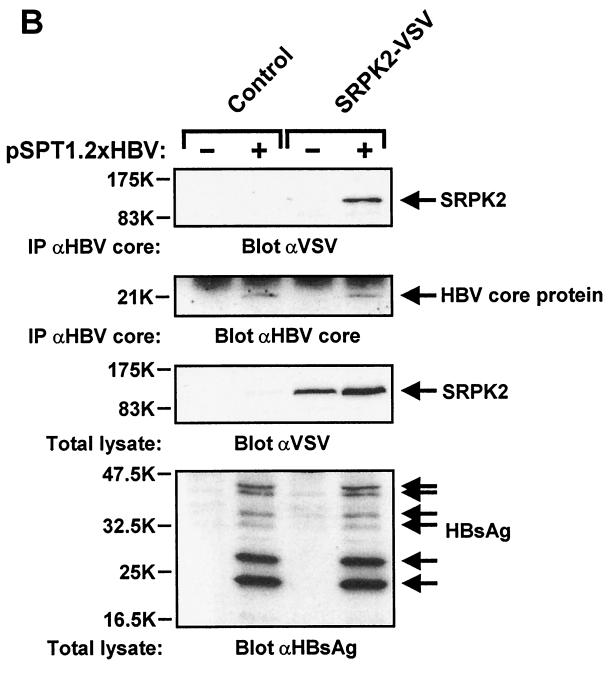
Isolation of stable HBV core protein-SRPK2 complexes from HuH-7 cells. (A) HuH-7 cells were infected in six-well dishes with adenovirus at 5,000 particles of AdHBVcore per cell plus either control adenovirus (no expression cassette), 25,000 particles of AdSRPK1-VSV per cell, or 15,000 particles of AdSRPK2-VSV per cell as indicated. Control adenovirus was added to give a total particle number of 30,000/cell for each of the infections. On the second day after infection, cell lysates were prepared and subjected to immunoprecipitation (IP) with polyclonal anti-HBV core protein antibody. Samples were then resolved by SDS-13% PAGE and analyzed by immunoblotting with anti-VSV antibody (upper panel) or anti-HBV core protein antibody (middle panel). In parallel, total cell lysates were analyzed by immunoblotting with anti-VSV antibody (lower panel). Positions of SRPKs and HBV core protein are indicated on the right. (B) HuH-7 cells were transiently transfected in 10-cm dishes with control plasmid, replication-competent HBV plasmid pSPT1.2xHBV (15 μg/dish), or plasmid encoding SRPK2-VSV (5 μg/dish), as indicated. After 48 h, cell lysates were prepared and processed as described for panel A. In addition, total cell lysates were analyzed by immunoblotting with anti-HBsAg antibody (lower panel). Positions of SRPK2, HBV core protein, and the different forms of HBsAg are indicated on the right.
To verify that stable HBV core protein-SRPK2 complexes are also formed under physiologically relevant conditions in intact cells, we cotransfected SRPK2 plasmid DNA together with a replication-competent HBV plasmid encoding the four major HBV open reading frames and demonstrated to support authentic HBV replication (39). As shown in Fig. 9B, SRPK2 was only detected in anti-HBV core protein immunoprecipitates when HBV proteins were expressed from viral genomic DNA. Correct expression of HBV proteins was further confirmed by immunoblotting of total lysates with anti-HBsAg antiserum, which showed that the different forms of HBsAg were present at their expected molecular weights (Fig. 9B, lower panel) (39).
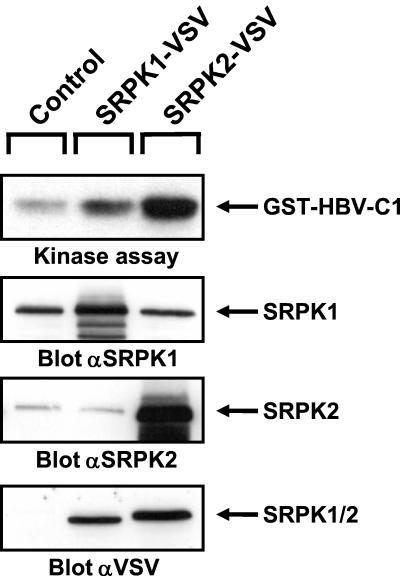
We next investigated how overexpression of SRPKs affects HBV core protein phosphorylation in vitro. As shown in Fig. 10, total lysates from SRPK2-overexpressing HuH-7 cells showed a more pronounced increase in HBV core kinase activity than extracts from SRPK1-overexpressing cells, although comparable amounts of both VSV-tagged kinases were detected. Interestingly, parallel immunoblots performed with either SRPK1- or SRPK2-specific antibody revealed that, relative to endogenous kinase levels, SRPK2 was overexpressed to a much higher degree than SRPK1. Thus, we conclude that endogenous SRPK1 is present at a significantly higher copy number than endogenous SRPK2 in parental HuH-7 cells. As higher endogenous protein expression should compensate for weaker affinity and activity, both SRPKs should be considered candidate host kinases mediating HBV core protein phosphorylation during viral infection.
FIG. 10.
Comparison of SRPK1- and SRPK2-mediated HBV core protein phosphorylation. HuH-7 cells were infected with adenovirus as described for Fig. 9A. Total cellular extracts were assayed for HBV core kinase activity in an in vitro kinase assay with GST-HBV core protein C1 as a substrate. After gel electrophoresis, phosphate incorporation into substrate protein was visualized by autoradiography (upper panel). In parallel, identical samples were analyzed by immunoblotting with either anti-SRPK1, anti-SRPK2, or anti-VSV antibodies (lower three panels). Positions of kinases and kinase substrates are indicated on the right.
DISCUSSION
Our results presented here demonstrate that the major cellular HBV core protein kinase activity can be attributed to the highly related protein kinases SRPK1 and SRPK2. These kinases display both cytoplasmic and nuclear localization (38). They have previously been reported to specifically phosphorylate SR proteins in their arginine/serine-rich (RS) domains, which regulate splice site selection and spliceosome assembly in a phosphorylation-dependent manner (9, 17, 38, 40). Remarkably, phosphorylation of RS domains was also shown to prevent nonspecific RNA binding (36), which is reminiscent of specific encapsidation of pregenomic HBV RNA mediated by core protein phosphorylated in its arginine- and serine-rich C-terminal part. Thus, an interesting functional similarity exists between core protein and SR proteins.
The host cell kinases Cdc2, Cdk2, and PKC have been implicated previously as HBV core kinases (20, 25). In this study, we clearly demonstrate that the major cellular HBV core protein kinase activity is biochemically distinguishable from Cdc2, Cdk2, and PKC activities detectable in a total cell lysate. Close inspection of the amino acid sequence of HBV core protein adds a further line of evidence and gives further support to our hypothesis that SRPKs account for HBV core kinase activity not only in vitro but also in vivo. The substrate specificity of SRPK2 has been determined previously by a peptide selection approach. SRPK family members belong to the group of arginine-directed protein kinases (34, 38), and SRPK2 was found to prefer arginines near the phosphorylation site, especially in the P−3 position. Moreover, SRPK2 also selects for proline in the P+1 position, and SRPK1-mediated phosphorylation of the SPRY peptide derived from the SR protein ASF/SF2 has been observed in vitro (9, 38, 42).
Importantly, inspection of the three mapped serine phosphorylation sites of HBV core protein (S155, S162, and S170 in strain ayw) reveals that all of them are in perfect agreement with the critical determinants reported for SRPK2 substrate specificity. The same should apply to SRPK1, because these SRPK family members are highly related in sequence and have been shown to phosphorylate the same substrate proteins in vitro (38). Interestingly, when we compared core protein phosphorylation in vivo with SRPK-mediated phosphorylation in vitro, we found a similar reduction to significantly lower but still detectable levels of phosphate incorporation into the core protein mutant devoid of serines 155, 162, and 170. Consistent with the residual mutant phosphorylation observed both in vivo and in vitro, two additional consensus SRPK phosphorylation sites are located in the very C-terminal part of HBV core protein (S176 and S178 in subtype ayw) (38, 42). Importantly, when we further mutated these two putative phosphorylation sites lying within an RRRRSQSREP sequence to alanines, cellular phosphate incorporation into the resulting HBV core protein mutant lacking five consensus SRPK phosphorylation sites was barely detectable.
Notably, both Cdc2 and Cdk2 are unlikely to exhibit kinase activity towards the RRRRSQSREP sequence, as these cyclin-dependent kinases strictly require proline in the P+1 position (35). Conversely, PKC family members are arginine-directed kinases and might phosphorylate S176 and S178 but are unlikely to phosphorylate the three repeated SPRRR motifs (S155, S162, and S170) under physiological conditions, since they strictly select against proline in the P+1 position (29). Taken together, it appears that SRPK activity but neither CDK nor PKC activity is sufficient to account for HBV core protein phosphorylation in vivo.
Recently, Gazina et al. reported that HBV core protein is phosphorylated prior to assembly into nucleocapsids and that its phosphorylation is apparently not changed during capsid formation (14). This finding also suggests that core protein encounters the viral polymerase-pregenomic RNA complex subsequent to its phosphorylation by cellular kinases, while the capsid assembly is in process (3, 13, 31). As we demonstrated for SRPK2, specific cellular kinase binding to core protein occurred in both the absence and the presence of other viral factors, indicating that our identification of SRPKs is also likely to apply to the physiological situation when viral polymerase and pregenomic RNA are expressed in cells.
It has been known for more than two decades that a cellular kinase is present in mature, liver-derived HBV virions and displays moderate core protein-specific kinase activity in vitro (1, 15). There is some discrepancy in the literature as to whether either PKC or a 46-kDa serine kinase could account for this kinase activity (20, 23). Currently, it cannot be excluded that other cellular kinases such as SRPKs are located inside mature virions as well. We have not addressed this issue. Moreover, it is certainly conceivable that the kinases phosphorylating dimeric HBV core protein precursors prior to capsid assembly in intact cells are distinct from those which are encapsidated together with polymerase and pregenomic RNA into core particles. Based on this hypothesis, SRPK activity could prime the specific encapsidation step during which either SRPKs or distinct host cell kinases get trapped inside the core particle. The encapsidated kinase could then play a role at a later step of the HBV life cycle and, for example, be required for nuclear import of the viral genome, as reported by Kann and colleagues (21).
To establish the role of SRPKs during HBV infection, specific inhibition of their function has to be achieved in intact cells. Novel tools such as SRPK-specific inhibitors and dominant-interfering kinase mutants have to be generated to validate SRPK-mediated core protein phosphorylation as an essential step during virus assembly. These future studies could then define SRPK family members as promising targets for therapeutic intervention and will fuel the development of novel antiviral agents to treat HBV infections.
Acknowledgments
We are grateful to Peter Hans Hofschneider, Alan Hall, and Claus-Hobe Schröder for kindly providing us with plasmids and RNA. We thank our colleagues Dorian Bevec and Christian Wallasch for stimulating discussions. We are also grateful to Uli Elben and Rainer Wessel for managerial support.
REFERENCES
- 1.Albin, C., and W. S. Robinson. 1980. Protein kinase activity in hepatitis B virus. J. Virol. 34:297-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzi, L., L. Meijer, A. C. Ostvold, J. Lew, and J. H. Wang. 1994. Purification of a 15-kDa cdk4- and cdk5-binding protein. J. Biol. Chem. 269:13279-13288. [PubMed] [Google Scholar]
- 3.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, R. P. 1988. The hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 5.Benn, J., and R. J. Schneider. 1995. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. USA 92:11215-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borchers, C., J. F. Peter, M. C. Hall, T. A. Kunkel, and K. B. Tomer. 2000. Identification of in-gel digested proteins by complementary peptide mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem. 72:1163-1168. [DOI] [PubMed] [Google Scholar]
- 7.Cano, E., C. A. Hazzalin, and L. C. Mahadevan. 1994. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol. Cell. Biol. 14:7352-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwill, K., L. L. Feng, J. M. Yeakley, G. D. Gish, J. F. Cáceres, T. Pawson, and X.-D. Fu. 1996. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271:24569-24575. [DOI] [PubMed] [Google Scholar]
- 10.Cotten, M., A. Baker, M. L. Birnstiel, K. Zatloukal, and E. Wagner. 1996. Adenovirus polylysine DNA conjugates, p. 12.3.1-12.3.33. In N. C. Dracopoli, J. L. Haines, B. R. Korf, D. T. Moir, C. C. Morton, C. E. Seidman, J. G. Seidman, and D. R. Smith (ed.), Current protocols in human genetics. John Wiley and Sons, Inc., New York, N.Y.
- 11.Draetta, G., and D. Beach. 1988. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell 54:17-26. [DOI] [PubMed] [Google Scholar]
- 12.Galibert, F., E. Mandart, F. Fitoussi, P. Tiollais, and P. Charnay. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646-650. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlich, W. H., U. Goldmann, R. Müller, W. Stibbe, and W. Wolff. 1982. Specificity and localization of the hepatitis B virus-associated protein kinase. J. Virol. 42:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 17.Gui, J.-F., W. S. Lane, and X.-D. Fu. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678-682. [DOI] [PubMed] [Google Scholar]
- 18.Hartinger, J., K. Stenius, D. Högemann, and R. Jahn. 1996. 16-BAC/SDS-PAGE: A two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal. Biochem. 240:126-133. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, R. C., J. E. Lavine, L.-J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature (London) 344:552-555. [DOI] [PubMed] [Google Scholar]
- 20.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear core complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kann, M., R. Thomssen, H. G. Kochel, and W. H. Gerlich. 1993. Characterisation of the endogenous protein kinase activity of the hepatitis B virus. Arch. Virol. 8(Suppl.):52-62. [DOI] [PubMed] [Google Scholar]
- 23.Kau, J.-H., and L.-P. Ting. 1998. Phosphorylation of the core protein of hepatitis B virus by a 46-kilodalton serine kinase. J. Virol. 72:3796-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan, Y. T., J. Li, W.-Y. Liao, and J.-H. Ou. 1998. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 25.Liao, W., and J.-H. Ou. 1995. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 69:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macfarlane, D. E. 1989. Two-dimensional benzyldimethyl-n-hexadecyl-ammonium chloride → sodium dodecyl sulfate preparative polyacrylamide gel electrophoresis: a high capacity high resolution technique for the purification of proteins from complex mixtures. Anal. Biochem. 176:457-463. [DOI] [PubMed] [Google Scholar]
- 27.Machida, A., H. Ohnuma, F. Tsuda, A. Yoshikawa, Y. Hoshi, T. Tanaka, S. Kishimoto, Y. Akahane, Y. Miyakawa, and M. Mayumi. 1991. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J. Virol. 65:6024-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michou, A.-I., H. Lehrmann, M. Saltik, and M. Cotten. 1999. Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J. Virol. 73:1399-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikawa, K., A. Toker, F.-J. Johannes, Z. Songyang, and L. C. Cantley. 1997. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272:952-960. [DOI] [PubMed] [Google Scholar]
- 30.Parkin, D. M., P. Pisani, and J. Ferlay. 1999. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 80:827-841. [DOI] [PubMed] [Google Scholar]
- 31.Pollack, J. R., and D. Ganem. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roossinck, M. J., and A. Siddiqui. 1987. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J. Virol. 61:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, et al. 1996. A structural basis for the substrate specificities of protein ser/thr kinases: primary sequence preference of casein kinase I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Songyang, Z., S. Blechner, N. Hoagland, M. F. Hoekstra, H. Piwnica-Worms, and L. C. Cantley. 1994. Use of an oriented library to determine optimal substrates of protein kinases. Curr. Biol. 4:973-982. [DOI] [PubMed] [Google Scholar]
- 36.Tacke, R., Y. Chen, and J. L. Manley. 1997. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl. Acad. Sci. USA 94:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 38.Wang, H.-Y., W. Lin, J. A. Dyck, J. M. Yeakley, Z. Songyang, L. C. Cantley, and X.-D. Fu. 1998. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 140:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss, L., A. S. Kekule, U. Jakubowski, E. Burgelt, and P. H. Hofschneider. 1996. The HBV-producing cell line HepG2-4A5: a new in vitro system for studying the regulation of HBV replication and for screening anti-hepatitis B virus drugs. Virology 216:214-218. [DOI] [PubMed] [Google Scholar]
- 40.Yeakley, J. M., H. Tronchère, J. Olesen, J. A. Dyck, H.-Y. Wang, and X.-D. Fu. 1999. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 145:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh, C.-T., and J.-H. Ou. 1991. Phosphorylation of hepatitis B virus precore and core proteins. J. Virol. 65:2327-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun, C. Y., and X.-D. Fu. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150:707-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, S., and D. N. Standring. 1992. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc. Natl. Acad. Sci. USA 89:10046-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]