Abstract
It has previously been shown that upon infection of HeLa cells with modified vaccinia virus Ankara (MVA), assembly is blocked at a late stage of infection and immature virions (IVs) accumulate (G. Sutter and B. Moss, Proc. Natl. Acad. Sci. USA 89:10847-10851, 1992). In the present study the morphogenesis of MVA in HeLa cells was studied in more detail and compared to that under two conditions that permit the production of infectious particles: infection of HeLa cells with the WR strain of vaccinia virus (VV) and infection of BHK cells with MVA. Using several quantitative and qualitative assays, we show that early in infection, MVA in HeLa cells behaves in a manner identical to that under the permissive conditions. By immunofluorescence microscopy (IF) at late times of infection, the labelings for an abundant membrane protein of the intracellular mature virus, p16/A14L, and the viral DNA colocalize under permissive conditions, whereas in HeLa cells infected with MVA these two structures do not colocalize to the same extent. In both permissive and nonpermissive infection, p16-labeled IVs first appear at 5 h postinfection. In HeLa cells infected with MVA, IVs accumulated predominantly outside the DNA regions, whereas under permissive conditions they were associated with the viral DNA. At 4 h 30 min, the earliest time at which p16 is detected, the p16 labeling was found predominantly in a small number of distinct puncta by IF, which were distinct from the sites of DNA in both permissive and nonpermissive infection. By electron microscopy, no crescents or IVs were found at this time, and the p16-labeled structures were found to consist of membrane-rich vesicles that were in continuity with the cellular endoplasmic reticulum. Over the next 30 min of infection, a large number of p16-labeled crescents and IVs appeared abruptly under both permissive and nonpermissive conditions. Under permissive conditions, these IVs were in close association with the sites of DNA, and a significant amount of these IVs engulfed the viral DNA. In contrast, under nonpermissive conditions, the IVs and DNA were mostly in separate locations and relatively few IVs acquired DNA. Our data show that in HeLa cells MVA forms normal DNA replication sites and normal viral precursor membranes but the transport between these two structures is inhibited.
Modified vaccinia virus Ankara (MVA) is a highly attenuated vaccinia virus (VV) strain, derived from VV Ankara. The latter was passaged more than 500 times on chicken embryo fibroblasts (CEF), during the course of which the virus lost part of its genome. Compared to VV Copenhagen, the structural genes have mostly remained unchanged, but many genes on the left and right sides of the genome, which encode in particular host range and immune evasion factors, are deleted or fragmented (1). MVA grows permissively in CEF and BHK cells but fails to make substantial amounts of infectious progeny in all other mammalian cells tested, including those of human origin (4, 9). Nevertheless, in human trials in which MVA was tested as a putative vaccine against smallpox, the virus induced an efficient immunological response while having none of the side effects that can occur with nonattenuated VV strains, even in high-risk groups (27, 41). VV vaccination induces efficient T-cell immunity, including immunity to foreign proteins cloned into its double-stranded DNA genome, which become expressed upon infection. However, because of the potential side effects of VV vaccination, the use of attenuated VV strains has become mandatory. In this respect, MVA is emerging as an important alternative candidate, and an increasing number of studies indicate that recombinant MVA strains are promising candidates to be used as live vaccines against other infectious diseases and in cancer therapy (3, 5, 8, 14, 19, 36, 43).
VV infection starts with virus entry, which delivers the core into the cytoplasm (reviewed in reference 28). From this core, a number of early mRNAs are produced, the translation of which is required to initiate the subsequent process of DNA replication. This process occurs in distinct cytoplasmic sites that are enclosed by membranes of the rough endoplasmic reticulum (ER) (44) and sets off the production of late genes, the products of which are required for virion assembly. Virion assembly is complex; the first identifiable step is the formation of typical crescent-shaped membranes that are highly modified by viral membrane proteins. We have previously postulated that the virally modified membranes are part of the so-called intermediate compartment (IC) (39). The IC is now recognized as a specialized subdomain of the interconnected ER network that is involved in transport from the ER to the Golgi complex (15). For this reason, we now prefer to use the term specialized smooth ER domain for the membranes of the crescents (see also reference 18 and 38).
Although a recent study suggested that the crescent-shaped membranes contain only one lipid bilayer (20), we and others have accumulated compelling evidence that they are composed of two tightly apposed cisternal membranes derived from the cellular ER (31; see reference 38 for a review). The crescents mature into immature viruses (IVs), which are perfectly spherical particles composed of two membranes containing viral membrane proteins, which enclose the electron-dense core proteins (12, 49). Uptake of the viral DNA by the IVs (see references 17 and 18 for a possible model) is followed by maturation into the first infectious form of VV, the so-called intracellular mature virus (IMV). In contrast to the spherical IVs, the IMVs appear to be brick shaped and contain a morphologically and biochemically distinct (brick-shaped) core structure. A small percentage of the IMVs are wrapped by a double-membrane cisterna derived from the trans-Golgi network to form the intracellular enveloped virus. The intracellular enveloped virus has been shown to move on microtubules towards the plasma membrane, with which it fuses to release the extracellular enveloped virus (EEV) (reviewed in reference 29).
The gene product of A14L, a protein with a molecular mass of 15 to 16 kDa (p16), has previously been identified as a major membrane protein of the IMV (22, 32, 34). Subsequently, two independent studies have shown that p16/A14L is essential for IMV assembly. In its absence, crescent-shaped membrane are made, but they fail to interact with the viral core proteins and consequently no IMVs are formed (33, 45). By immunofluorescence microscopy (IF) of VV-infected cells, the protein was found to display a typical punctate pattern that precisely colocalized with the areas of DNA accumulation in the perinuclear area of the cell at late times of infection (34). By electron microscopy (EM), p16 localizes to the viral membranes of the crescents, IVs, and IMVs. Labeling was also detected on membranes of the ER-Golgi interphase, as assessed by the fact that these membranes could be double labeled with markers of that compartment, while low-level labeling was detected in the nuclear envelope and the rough ER (32, 34).
The stage of infection at which MVA is blocked depends on the cell type; in most cell lines of human origin, its life cycle appears to be inhibited at a late stage. HeLa cells infected with MVA synthesize late viral proteins, and IVs accumulate, but no IMVs are made (42). The reasons for this block in assembly have mostly remained elusive; attempts to rescue the phenotype in HeLa cells have suggested that several viral genes may be inactivated in this virus (50).
In the present study we have reinvestigated the assembly block of MVA in HeLa cells. Our data show that under nonpermissive conditions viral assembly is normal until 4 h 30 min to 5 h postinfection. At this time of infection, we show that two distinct domains of the cellular ER are modified, each with distinct functions in viral assembly. The first is the rough ER, which, starting at 2 h, temporally enwraps the replicating DNA. This ER is then disassembled at 5 to 5 h 30 min postinfection, concomitant with the appearance of crescents and IVs. The second domain is a membrane vesicle, enriched in p16/A14L, that first appears at 4 h 30 min and is part of the cellular smooth ER system. Under normal assembly conditions, these membrane domains are somehow transported to distinct sites on the surface of the viral DNA where IMV assembly occurs. This putative transport step does not occur in HeLa cells infected with MVA. Our data therefore suggest that MVA lacks the genes for proteins needed for the process of bringing together the IVs and the viral DNA in HeLa cells.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HeLa, BHK-21, and BSC40 cells were grown in Dulbecco's modified Eagle medium with 4.5 g of glucose per liter supplemented with 5% heat-inactivated fetal calf serum, 200 mM l-glutamine (GIBCO BRL, Life Technologies), and 100 U of penicillin per ml and 100 μg of streptomycin per ml (GIBCO BRL. Life Technologies) at 37°C with 5% CO2. The Western Reserve (WR) strain of VV (kindly provided by B. Moss) was purified from infected HeLa cells, and MVA (a kind gift of Gerd Sutter) was purified from infected BHK cells. To prepare virus stocks, cells were infected for 3 days at a multiplicity of infection (MOI) of 0.5 and IMV was isolated as described by Jensen et al. (22). Iodoxynol (Optiprep) gradient purification of MVA was done as described by Krijnse Locker et al. (24). To determine the titer, viruses were plaque titrated on BSC40 cells (WR) and BHK-21 cells (MVA) and fixed at 24 h postinfection in acetone-methanol (1:1). Viral foci were visualized by immunostaining with an anti-VV antibody as described by Earl et al. (11), using horseradish peroxidase and 0.66 mg of 3,3′-diaminobenzidine (Sigma) per ml in phosphate-buffered saline-0.024% H2O2, according to the instructions of the manufacturer. The titers obtained for both WR and MVA preparations were 1 × 108 to 4 × 108 PFU/ml. The PFU-per-particle ratios of purified MVA and WR preparations, determined as described by Joklik (23), were about 1 to 50 for both viral preparations.
The following antibodies were used throughout this study: anti-p16 (A14L) (34), anti-p35 (H5R) (44), anticore (30), rat monoclonal antibodies to anti-p42 (B5R) (35) (a kind gift of Gerhard Hiller), and anti-β-COP and anti-p27 (kindly provided by Rainer Pepperkok and Tommy Nilsson, respectively (10, 13). Human anti-DNA antibody was kindly supplied by Ivan Raska (University of Prague). Donkey anti-rabbit coupled to fluorescein isothiocyanate (FITC) or rhodamine was purchased from Jackson Immunoresearch Laboratories (Dianova, Hamburg, Germany).
Indirect immunofluorescence, entry assay, and relationship between intracellular cores and replication sites.
Indirect immunofluorescence was as described previously (6). Infected cells grown on coverslips were fixed at the indicated times with 3% paraformaldehyde in phosphate-buffered saline for 30 min at room temperature. After permeabilization with 1% Triton X-100, coverslips were incubated with the appropriate primary and secondary antibodies coupled to FITC or rhodamine and viewed using a Zeiss Axioskop 50 fluorescence microscope or a Zeiss Axioplan 2 confocal microscope. Viral DNA was visualized using DAPI (4′,6′-diamidino-2-phenylindole) at a concentration of 1 μg/ml. The entry kinetics was determined as described previously (24) using either sucrose-purified WR and MVA or iodoxynol-purified MVA to separate IMV from EEV. To visualize intracellular replication sites, infected cells were fixed at 4 h postinfection and labeled with antibodies to p35 (H5R). The amounts of intracellular cores and replication sites were counted in 30 cells.
EM.
Cryosections and Epon sections of infected and fixed cells were prepared as described previously (12, 46). To determine the amount of EEVs in different gradient-purified MVA preparations, fractions were absorbed to carbon-coated grids and labeled with anti-p42, rabbit anti-rat antibody, and 10-nm-diameter protein A-gold, followed by 10 min of floating on a mixture (8:2) of 2% methylcellulose and 3% uranyl acetate on ice. From this mixture the grid was looped out, blotted, and dried. Using this assay, it was found that MVA obtained from sucrose gradients contained 20% EEVs with intact p42-positive membranes and 10% with broken membranes. MVA purified on Optiprep gradients resulted in two light-scattering bands. The upper band contained 52% EEVs with intact membranes, with the remaining particles being IMVs. The lower Optiprep band contained IMVs only. Double labeling was carried out as described by Slot et al. (37). Images were taken with a Philips 400 EM.
RESULTS
MVA enters HeLa cells with the same kinetics as WR.
Although it had been shown that MVA infection in HeLa cells is blocked at a late stage of infection, it was not clear whether or not the earlier stages were also affected. It had been shown that the extent of DNA replication in these cells was similar to that for wild-type VV Ankara infection (42). We decided, therefore, to investigate a number of other parameters of the early phases of infection in more detail. In initial experiments we chose to compare MVA to the WR strain of VV because we have recently generated a variety of tools to study the early stages of the WR life cycle in these cells (24-26, 44). Throughout this study, infection of HeLa cells with MVA is referred to as nonpermissive, whereas infections of HeLa cells with WR and of BHK cells with MVA are called permissive.
Sucrose gradient-purified MVA stocks were prepared from infected BHK cells, while WR was isolated from HeLa cells. By negative-staining EM such preparations contained single particles only and lacked contamination (not shown). The PFU-per-particle ratios of such preparations were very comparable (see Materials and Methods); for both purified MVA and WR we calculated a ratio of 1 to 50, similar to what we and others have published before (24, 47).
Subsequently, the entry properties of both viruses were studied using an established assay. This assay relies on an antibody generated to VV cores that by IF recognizes intracellular cores but not extracellular virions. By fixing cells at different times after infection and subsequently labeling the cells with the anticore antibody, the average amount of cores per cell can be used as a quantitative entry assay (24).
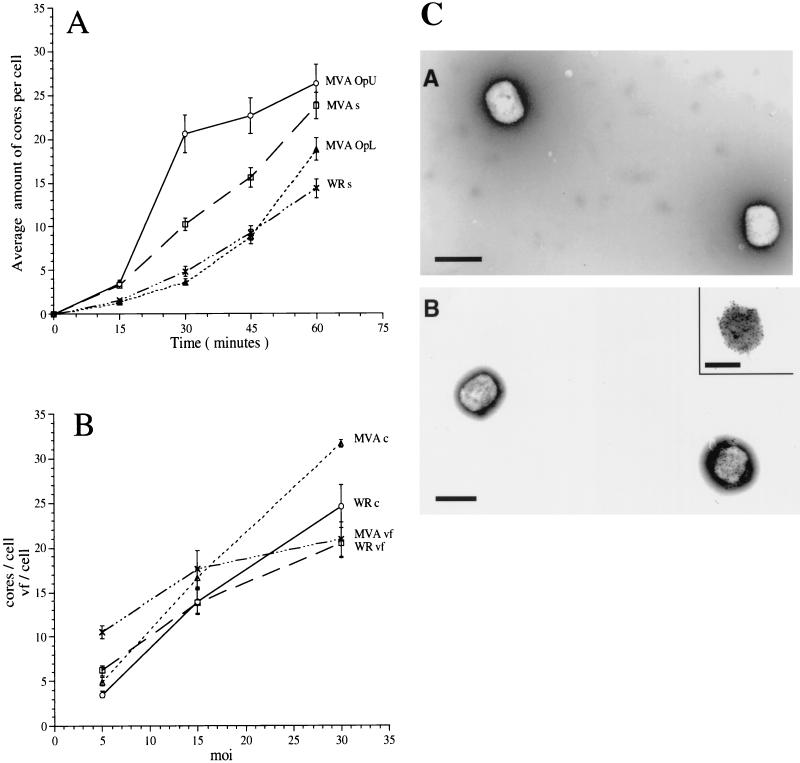
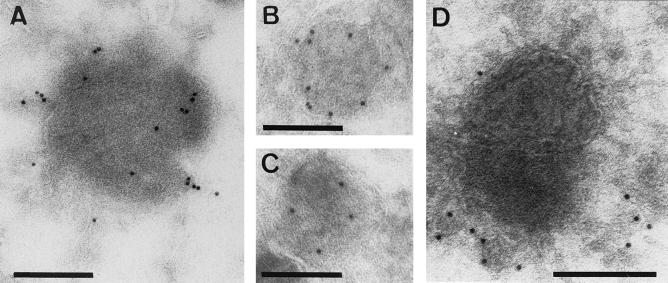
Upon comparing the entry kinetics of sucrose-purified MVA and WR by using this assay, we were initially surprised to see that the former virus apparently entered cells more efficiently than WR (Fig. 1A). At 60 min postinfection, MVA resulted in more intracellular cores than WR. We considered the possibility that our sucrose-purified MVA stock might contain substantial amounts of EEV, particles that are known to enter cells more efficiently than IMV (7, 24), MVA preparations were subjected to negative-staining EM after labeling with antibodies to the EEV-specific protein p42 (B5R gene). Since the entry assay was always performed after extensive sonication of the virus stocks, negative staining was also done on sonicated virus, a treatment thought to disrupt the EEV-specific membrane (21, 48). Despite sonication, up to 20% of the particles obtained from sucrose gradients appeared to be surrounded by an intact EEV-specific (p42-positive) membrane (Fig. 1C, panel B) (see Materials and Methods). In contrast, sucrose-purified WR contained only B5R-negative IMVs (Fig. 1C, panel A).
FIG. 1.
Entry of WR and MVA in HeLa cells (A) and relationship between MOI, intracellular cores, and DNA replication sites (B). (A) HeLa cells grown on coverslips were infected at an MOI of 30 in the presence of 300 μg of cycloheximide per ml with sucrose-purified WR (WR s) or MVA (MVA s), the upper band obtained from MVA purified on Optiprep gradients (MVA OpU) (enriched for EEV), or the lower band of the same gradient (MVA OpL) (containing IMV only). Cells were fixed at the indicated times postinfection and labeled with an anticore antibody followed by anti-rabbit-FITC to visualize intracellular cores. The graph represents the average amounts of cores per cell in 30 cells and the standard errors of the means. (B) HeLa cells grown on coverslips were infected with sucrose-purified WR or MVA at the indicated MOI. After 60 min at 37°C, one set of cells was fixed and labeled with the anticore antibody. A parallel set of cells was washed three times to remove unpenetrated virus and incubated for an additional 3 h before fixation. Intracellular cores and replication sites were visualized by IF (see Materials and Methods) and counted. The values represent the average amounts of cores per cell and standard errors of the means at 60 min postinfection with WR (WR c) or MVA (MVA c) or the average amounts of viral factories (vf) per cell and standard errors of the means at 4 h postinfection (WR vf or MVA vf) in 30 cells. (C) Negative-staining EM and immunolabeling with anti-p42 (B5R; 10-nm-diameter gold) of sucrose-purified WR (panel A) and MVA (panel B) isolated from infected HeLa or BHK cells, respectively. Bars, 300 μm.
Since we have recently shown that iodoxynol (Optiprep) gradients allowed a good separation of IMV from EEV (24), MVA was also purified on such gradients. In iodoxynol gradients two light-scattering bands were now clearly detected, and when analyzed by negative-staining EM after labeling with anti-p42, the upper band was enriched for EEVs while the lower band consisted of p42-negative IMVs (see Materials and Methods). When the entry assay was done with Optiprep-purified MVA IMV and EEV, the MVA IMV entry kinetics now resembled that of WR IMV WR and, at the MOI (of 30) used, resulted in similar amounts of intracellular cores (Fig. 1A). MVA EEV entered HeLa cells faster and more efficient than sucrose-purified MVA, consistent with the fact that the upper Optiprep band contained relatively more EEVs (Fig. 1A).
The combined results show that MVA IMV enters with the same kinetics and efficiency as WR IMV. When purified in sucrose gradients, the MVA stocks contained substantial amounts of EEV whose EEV-specific membrane apparently resisted rupture by sonication (see also reference 40).
Relationship between intracellular cores and DNA replication sites.
By conventional IF, the gene product of H5R, a 35-kDa protein, colocalizes precisely with the sites of viral DNA synthesis at 3 h postinfection (2, 44), allowing us to estimate the number of cytoplasmic DNA sites at a given MOI. As shown before, the combination of anti-p35 and the anticore antibodies allows one to estimate the relationship between intracellular cores and DNA replication sites (25). In HeLa cells infected with WR, at up to a certain MOI an approximately one-to-one ratio exists between the number of cores and replication sites, suggesting that each incoming core is infectious (25). Above this MOI the number of replication sites does not further increase, suggesting that only a limited number of cytoplasmic DNA sites can be made in HeLa cells. In the present study we wanted to test whether a similar one-to-one ratio also existed in HeLa cells infected with MVA.
HeLa cells were infected with MVA and WR at MOIs of 5, 15, and 30, and the intracellular cores were counted at 1 h postinfection. A parallel set of cells was incubated for an additional 3 h before fixation and labeling with anti-p35 (H5R). The DNA replication sites were counted at 4 h postinfection, because this appeared to be the earliest time postinfection at which the DNA factories could be detected by IF at MOIs of 5 and 15. For both MVA and WR the amount of intracellular cores increased linearly with increasing MOI (Fig. 1B). Since sucrose-purified MVA containing up to 20% EEVs, was used in this experiment, MVA infection resulted in more intracellular cores than WR infection at the same MOI. Nevertheless, the relationship between cores and replication sites followed a similar pattern for both viruses; at and MOI of 5, there appeared to be more DNA sites than cores at 4 h postinfection (Fig. 1B). This is most likely because at low MOIs it is more difficult to accurately count intracellular cores, because not all cells are infected. At an MOI of 15, however, the ratio between cores and replication sites was approximately one to one for both viruses (Fig. 1B). For both MVA and WR at an MOI of 30, fewer DNA sites than cores were counted (Fig. 1B). Compared to an MOI of 15, however, the number of replication sites obtained at an MOI of 30 was slightly higher, suggesting that the MOI which results in the maximum amount of replication sites that can be made in HeLa cells (see above) was somewhere between 15 and 30 under the present experimental conditions.
These data show that upon infection of HeLa cells with MVA and WR, the relationship between cores and replication sites appeared to be comparable, with a one-to-one ratio between these two structures at an MOI of 15.
Upon infection of HeLa cells with MVA, p16/A14L does not colocalize with the DNA replication sites.
Having established that MVA behaved similarly to WR in two parameters of the early phase of infection in HeLa cells, we next investigated later stages of infection. The nonpermissive infection of MVA in HeLa cells was compared to the permissive conditions of WR in HeLa cells and MVA in BHK cells.
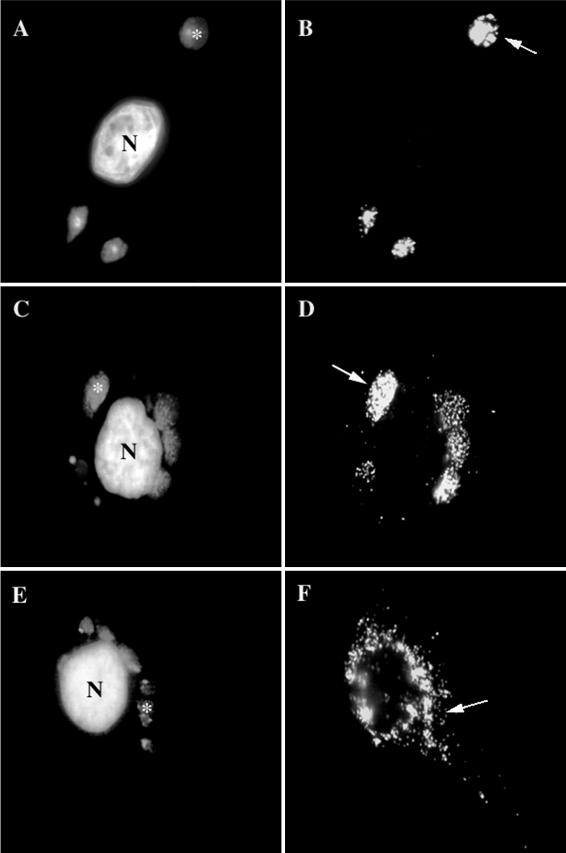
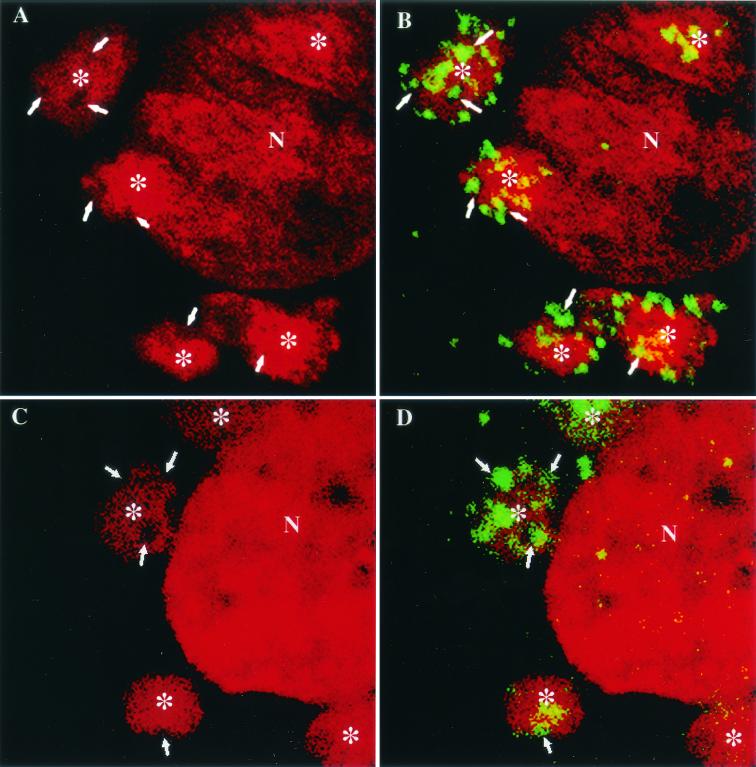
As mentioned above, as determined by conventional light microscopy in HeLa cells late in infection, p16 (A14L) and the viral DNA precisely colocalize (34). When HeLa cells infected with WR or BHK cells infected with MVA were fixed at 8 h postinfection, the regions of p16 and DAPI labeling indeed appeared to completely overlap (Fig. 2A to D). However, when we analyzed these structures in more detail, the respective labeling patterns were quite different; whereas anti-p16 (A14L) showed a striking punctate pattern, the DAPI-positive areas of viral DNA appeared as a uniformly labeled structure (Fig. 2A to D). Confocal microscopy analyses of these same preparations were then carried out. HeLa cells infected with WR or BHK cells infected with MVA were fixed at 6 h postinfection and double labeled for DAPI and anti-p16. By confocal sections it now appeared that when the p16 labeling was artificially removed from the image by shutting off the FITC channel, there was no DNA labeling at the sites where the p16 labeling was concentrated. Instead, the uniformly labeled DNA structures enclosed DNA-free gaps on their surfaces that were filled by the p16-positive structures. It thus appeared that the p16 and DNA labeling represented two different structures that were closely associated but distinct. The fact that these results were obtained for both BHK cells infected with MVA (Fig. 3A and B) and HeLa cells infected with WR (Fig. 3C and D) suggested that this observation was general for a permissive VV infection and was not cell type dependent.
FIG. 2.
As determined by IF, p16 and the viral DNA do not colocalize to the same extent in MVA-infected HeLa cells as with permissive infections of MVA in BHK cells or WR in HeLa cells. BHK cells (A and B) or HeLa cells (E and F) infected with MVA or HeLa cells infected with WR (C and D), all at an MOI of 10, were fixed at 8 h postinfection. Fixed cells were double labeled with DAPI (A, C, and E) and anti-p16 (A14L) (B, D, and F). In panels A, C, and E, asterisks indicate DNA factories, while in panels B, D, and F, arrows point to the same DNA factories. In panels B and D, p16 overlaps completely with the DNA factories in panels A and C, while in panel F, the p16 labeling is scattered in the cytoplasm, not obviously colocalizing with the DNA in panel E. N, nucleus.
FIG. 3.
Confocal sections of VV-infected cells double labeled for DNA (DAPI) and p16 (A14L). BHK cells infected with MVA (A and B) and HeLa cells infected with WR (C and D) were fixed at 6 h postinfection and double labeled with DAPI (red) and anti-p16 (green). In panels A and C the DAPI labeling is shown alone, and in panels B and D the merge of DAPI and p16 labeling is shown. The arrows indicate clusters of p16 labeling in panels B and D and the corresponding area in panels A and C. Asterisks indicate a DNA factory region. N, nucleus.
In HeLa cells infected with MVA, however, the punctate p16 pattern appeared to overlap much less with the DNA structure and instead was more dispersed in the cytoplasm (Fig. 2E and F). Confocal microscopy analysis confirmed that the two sets of structure were mostly separate under these conditions (not shown).
Thus, under nonpermissive conditions at late times of infection, an abundant membrane protein failed to reach its normal sites on the surface of the viral DNA.
By EM at late times of infection, IVs do not localize to the DNA sites in HeLa cells infected with MVA.
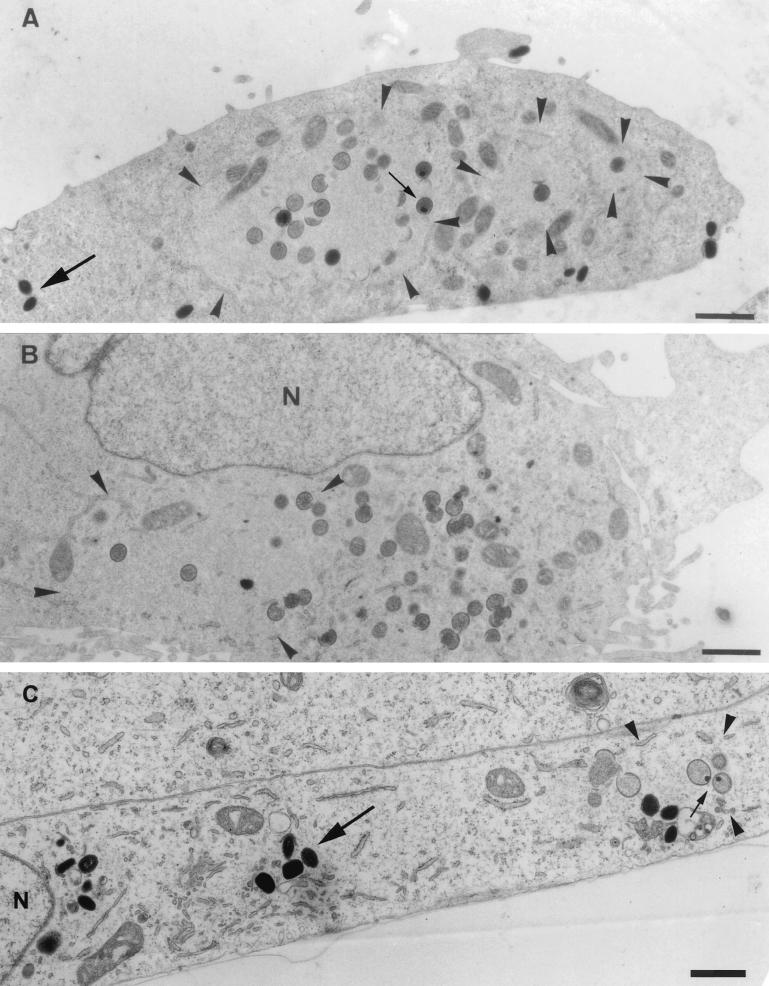
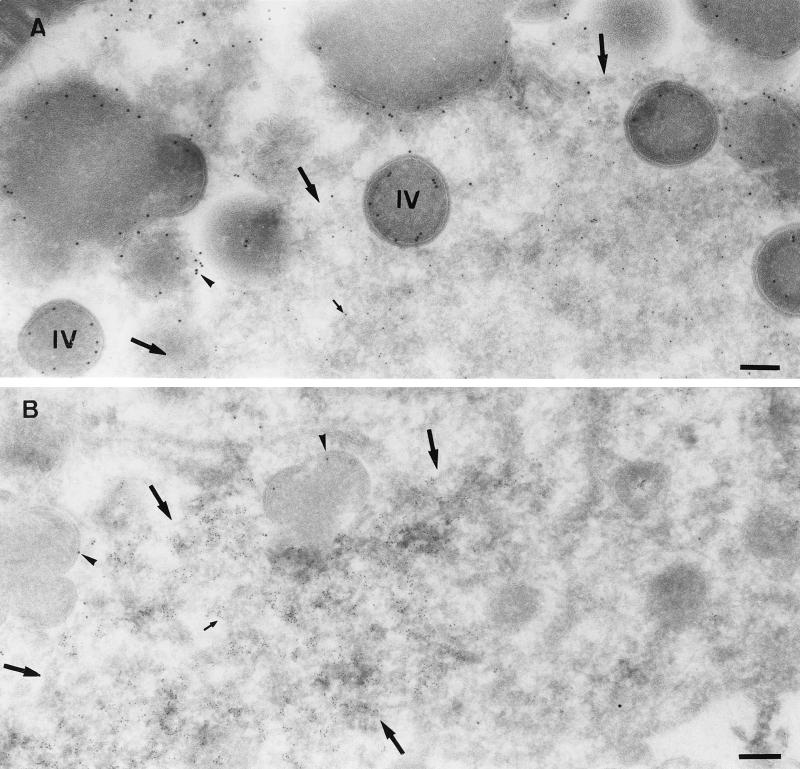
We next used EM to visualize the viral membrane and DNA structures at a higher resolution under permissive and nonpermissive conditions at 8 h postinfection. For this, sections of Epon-embedded samples were analyzed. The regions of DNA accumulation could easily be identified in such sections, because they have a different electron density than the surrounding cytoplasm (see Fig. 5 and reference 44). At early times of infection (4 h 30 min), under both permissive and nonpermissive conditions, the DNA replication sites were almost entirely surrounded by membranes of the rough ER, in agreement with our previous study (44), as seen in both Epon sections and cryosections labeled with anti-DNA (Fig. 4). These data argue that the process of ER wrapping around the DNA replication sites occurs normally under nonpermissive conditions. Moreover, at late times of infection, under both permissive and nonpermissive conditions the ER had mostly disassembled from the DNA regions, consistent with our recent data (44). Under permissive conditions (WR-infected HeLa cells [Fig. 5A ] or MVA-infected BHK cells [Fig. 5C]), IVs and crescents that localized exclusively within the DNA regions were seen. No IMVs accumulated at the DNA regions; instead, virions were always found in different regions of the cell, suggesting that once assembled, IMVs moved away from the sites where the DNA accumulated (Fig. 5A and C).
FIG. 5.
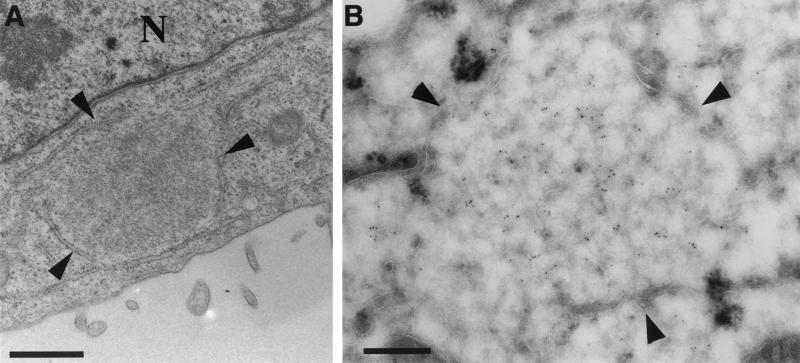
Epon sections of HeLa cells infected with WR (A) or MVA (B) or BHK cells infected with MVA (C) at 8 h postinfection. In all cases cells were infected at an MOI of 10. The region of viral DNA accumulation in the cytoplasm of infected cells is indicated with arrowheads. This region is covered with IVs in HeLa cells infected with WR (A) and BHK cells infected with MVA (C), while no IVs are observed outside this region. Small arrows point to IVs containing a nucleoid. In HeLa cells infected with MVA (B), the region of DNA accumulation (arrowheads) has a few IVs, but more IVs can be found in the rest of the cytoplasm, not localizing to the DNA site. IMVs in panels A and C are indicated with large arrows and do not appear to be present in the DNA region. The latter viral form cannot be detected in HeLa cells infected with MVA (B). N, nucleus. Bars, 1 μm.
FIG. 4.
Viral DNA replication site in HeLa cells infected with MVA at 4 h 30 min postinfection. HeLa cells were infected with MVA at a MOI of 10 and fixed at 4 h 30 min postinfection. (A) Epon section in which one DNA replication site that is almost entirely surrounded by membranes of the ER can be seen. (B) Cryosections were double labeled with anti-p16 (10-nm-diameter gold) and anti-DNA (5-nm-diameter gold). The image shows a replication site that is almost completely ER enwrapped. The central part of this region is labeled abundantly with anti-DNA. The ER membrane around the replication site is not significantly labeled with anti-p16. Bars, 200 nm. N, nucleus.
In agreement with the results reported by Sutter and Moss (42), in HeLa cells infected with MVA no IMVs could be detected, and many IVs appeared to accumulate instead. In contrast to the case under permissive conditions, only a few of those IVs were seen at the DNA sites, and the bulk of these particles were found scattered in the cytoplasm (Fig. 5B). Like the IVs, and in contrast to permissive conditions, the crescents were also found predominantly outside the DNA regions (Fig. 5B). These data supported the evidence obtained by light microscopy that the viral membranes were not properly reaching the DNA sites.
To test whether the failure of IV-DNA colocalization might also be reflected in a reduced uptake of DNA by these particles, the number of IVs that contained DNA was counted and compared to those under three permissive conditions. The viral DNA is known to enter particles in distinct, concentrated structures that can easily be seen by EM of Epon-embedded samples (see for, instance, Fig. 5A and C). HeLa and BHK cells were infected with WR and MVA and fixed at 8 h postinfection, and the fixed cells were embedded in Epon. The total number of IVs in 15 sections of infected cells was counted (this number ranged from 76 to 151 IVs in 15 cell profiles). Under conditions of permissive infection, the percentage of IVs containing a nucleoid was between 28 and 35% (34.7, 32.5, and 28.6% for WR-infected BHK cells, WR-infected HeLa cells, and MVA-infected BHK cells, respectively). However, in HeLa cells infected with MVA, this percentage was on average only 5.6%, showing that significantly fewer IVs could acquire DNA at this time of infection.
These results strongly suggest that the localization of the IVs to the regions of DNA accumulation is required for efficient DNA uptake into the particle.
Targeting of p16 to the viral DNA.
Since the above-described results showed that under nonpermissive conditions MVA assembly at late stages was abnormal, we decided to make a more detailed time course to see at what point the p16-labeled structures failed to assemble properly. One possibility was that under nonpermissive conditions p16-labeled IVs initially colocalized with the DNA. As more IVs that failed to mature into IMVs started to accumulate, they were being displaced from the DNA, resulting over time in a dispersed localization pattern of p16 as shown in Fig. 2E and F. Alternatively, p16 simply failed to reach the DNA at all times of infection.
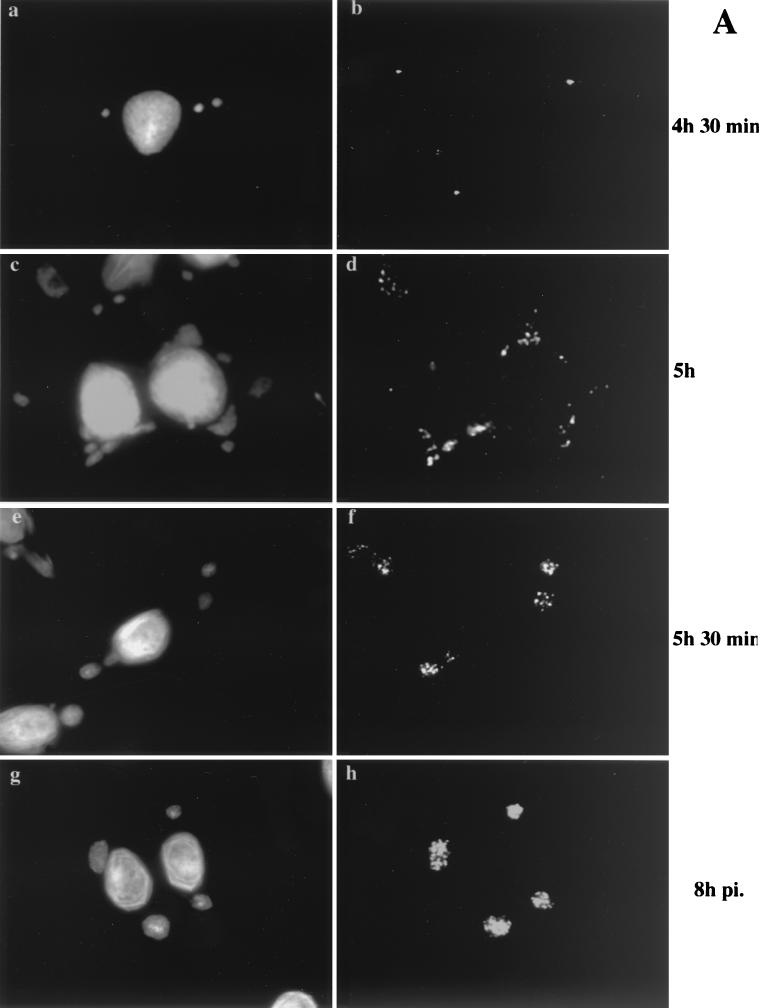
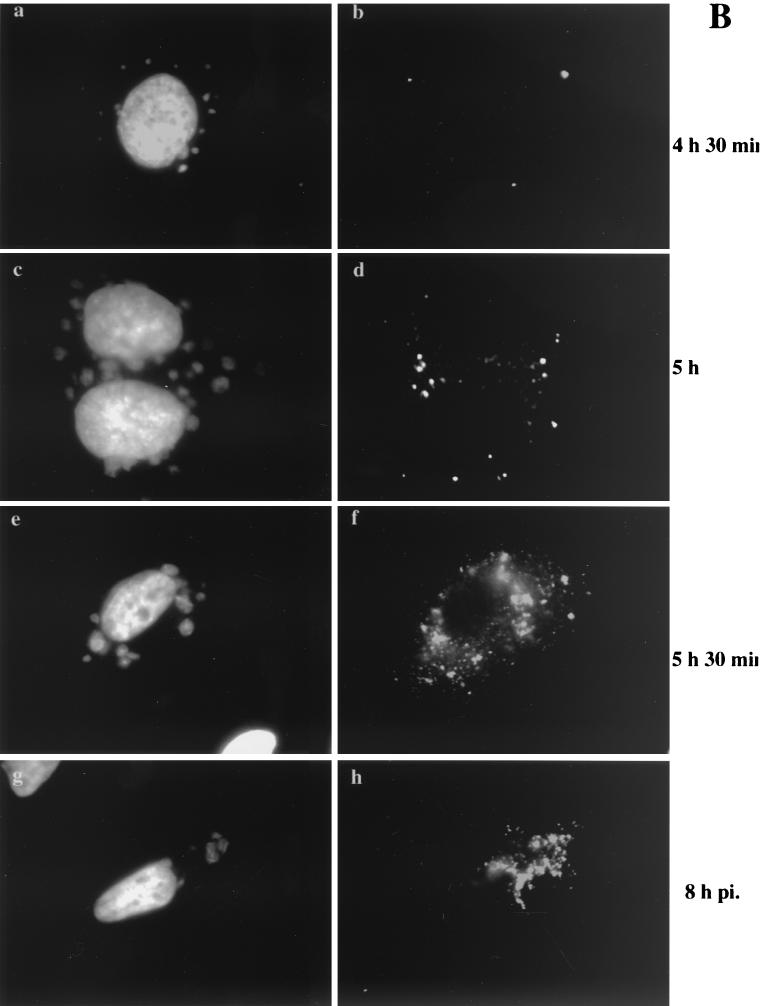
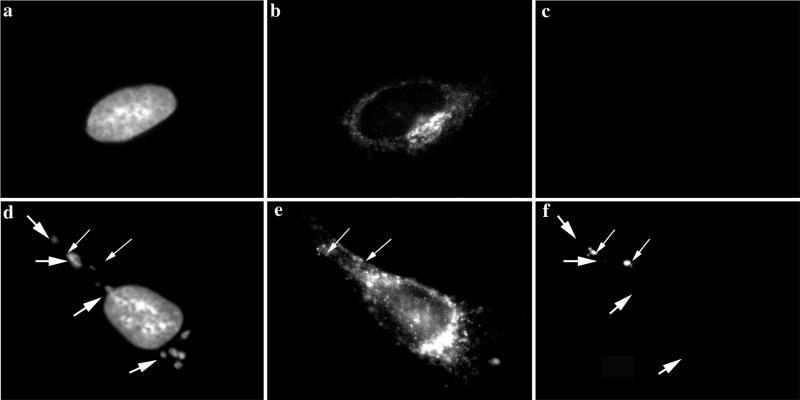
As MVA in BHK cells behaved identically to WR in HeLa cells, we decided to compare MVA infection in BHK cells (permissive conditions) (Fig. 6A ) to MVA infection in HeLa cells (nonpermissive conditions) (Fig. 6B). Cells were infected and fixed starting at 4 h 30 min postinfection, since this appeared to be the earliest time point at which we were able to detect p16 labeling in both HeLa and BHK cells at the MOI (of 10) used. They were also fixed at 5 h, 5 h 30 min, and 8 h postinfection, and the fixed cells were double labeled with DAPI and anti-p16.
FIG. 6.
Localization of p16 at different times of infection. BHK cells (A) or HeLa cells (B) were grown on coverslips and infected with MVA at an MOI of 10. The cells were fixed at the indicated times postinfection (pi.) and double labeled with DAPI (a, c, e, and g) and anti-p16 (A14L) followed by anti-rabbit antibody coupled to rhodamine (b, d, f, and h). Note that p16 perfectly overlaps with the regions labeled with DAPI in BHK cells infected with MVA from 5 h 30 min postinfection onwards. p16 does not overlap to the same extent in HeLa cells infected with MVA. At 4 h 30 min postinfection under both permissive and nonpermissive conditions, p16-positive spots that do not colocalize with the viral DNA are detected.
At 4 h 30 min postinfection under permissive and nonpermissive conditions the labeling patterns were identical. In both cases the labeled DNA and p16 structures were distinct and, in contrast to late times of permissive infection, did not colocalize. Whereas the DNA was found in three to five discrete sites, the p16 labeling was prominently seen in a small number of (three to six) of distinct puncta per cell (Fig. 6 A, panels a and b, and B, panels a and b). A faint cytoplasmic labeling was also observed, but no obvious pattern could be recognized. At 5 h postinfection under permissive conditions, clear signs of colocalization between p16-labeled puncta and the DNA were seen. Punctate p16 labeling was found on the DNA structure, as before, but did not cover the entire surface to the extent seen at 6 h, while a variable amount of the p16 puncta was still located outside the DNA site (Fig. 6A, panels c and d). At 5 h 30 min and 8 h postinfection, p16 was found to precisely colocalize with the DNA sites under permissive conditions, consistent with the above-described results (Fig. 6A, panels e to h). In contrast, under nonpermissive conditions, at all time points tested, p16 and DNA did not colocalize to the same extent. Although some of the p16 labeling was adjacent to the DNA, the bulk of this label appeared to be randomly scattered in the cytoplasm (Fig. 6B, panels c to h).
It thus seemed that under both permissive and nonpermissive conditions, p16 initially accumulated in structures that were distinct and separate from the DNA. Whereas under permissive conditions p16 and the DNA were subsequently found in close association, this step appeared to be blocked under nonpermissive conditions.
Analysis of the p16-labeled early membrane structures by EM.
The IF analyses showed that distinct p16-enriched puncta assembled at 4 h 30 min postinfection and in permissive assembly could move towards the DNA (or vice versa). We therefore used immuno-EM in order to see these structures directly. It was possible that these structures represented the first signs of crescents. In any case, since they were enriched in p16, we expected them to be membrane structures that would be either in continuity with or derived from the cellular ER.
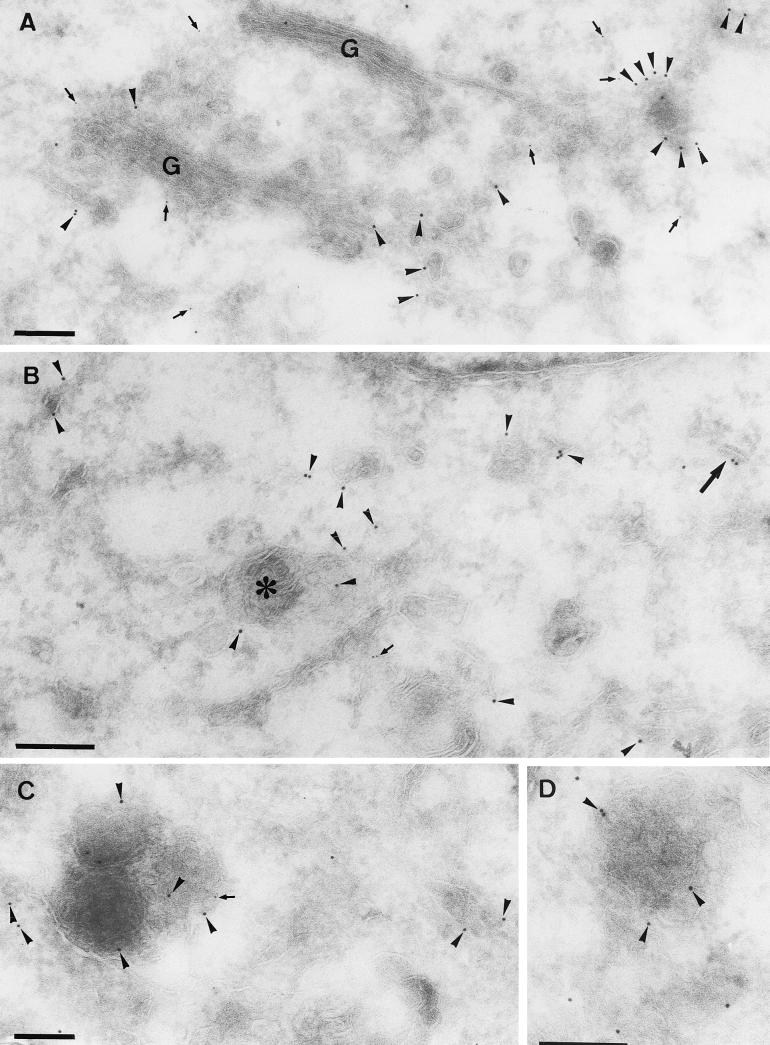
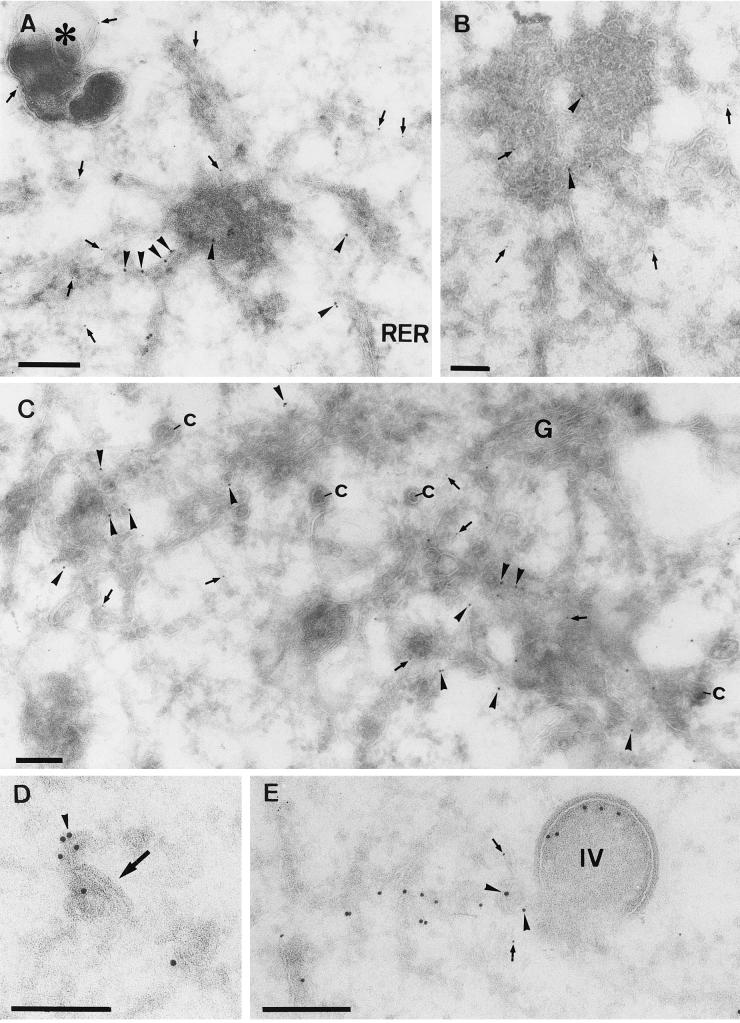
Cryosections were prepared at 4 h 30 min and 5 h postinfection of BHK or HeLa cells with MVA. Under both permissive and nonpermissive conditions, at 4 h 30 min postinfection no crescents or IVs could be observed (Fig. 4), which implies that at this time of infection, these structures cannot be responsible for the labeling of p16 seen by IF. No difference between the permissive and nonpermissive conditions was seen at this time point. The replication sites were still almost completely enwrapped by membranes of the rough ER (Fig. 4). Consistent with the light microscopy results, these ER membranes were not detectably labeled with anti-p16 both under permissive (not shown) and nonpermissive (Fig. 4B) conditions. Instead, and in contrast to the IF results, consistent labeling was found on smooth ER membranes, including tubular vesicular membranes that were often located on one side of the Golgi complex (Fig. 7A). When sections were double labeled for p16 and a marker of the ER-Golgi boundary region, gp27 (13), the two antigens were found to be closely intermingled in these areas, demonstrating that p16 accumulated in these membranes at the earliest time point of infection at which it becomes significantly expressed (Fig. 7A and B). In addition, clear sites of p16 segregation into distinct subdomains, in which the labeling was more concentrated, were observed (Fig. 7A, C, and D; see Fig. 9). At this time we also observed one example of what we interpret to be the first hint of a mini-crescent domain at 4 h 30 min (Fig. 7B).
FIG. 7.
Double labeling of p16 and gp27 at 4 h 30 min postinfection. Cryosections were labeled with anti-p16 (10-nm-diameter gold) (arrowheads) and anti-gp27 (5-nm-diameter gold) (arrows). (A, C, and D) MVA in HeLa cells; (B) MVA in BHK cells. Panel A shows scattered labeling for both antigens in the vicinity of the Golgi stack (G). Note the concentration of p16 in structures that are in continuity with the smooth ER membranes. These domains are mostly devoid of gp27. In panel B, in addition to scattered ER labeling of p16, some is found on a multivesicular body-like structure (asterisk). The large arrow in panel B indicates the first hint, at 4 h 30 min, of a small crescent domain labeled for p16. In panels C and D examples of the membrane-rich vesicles described in the text are shown. Bars, 200 nm.
FIG. 9.
Examples of p16-labeled membrane-rich vesicles in HeLa cells infected with MVA. Cells were fixed at 4 h 30 min (A to C) or 8 h (D) postinfection. Bars, 200 nm.
In contrast to the labeling observed over the ER-Golgi region, which was rather prominent, we had to search many sections in order to find the structures (three to five per cell) that were labeled abundantly for p16 by IF. These structures emerged as a striking kind of vesicular aggregate of membranes that labeled variably, but often strongly, for p16 (Fig. 7C and D, 8A and B, and 9). We consider it likely that these tightly knit structures may be quite inaccessible to p16 labeling and that the EM labeling may be an underestimate. These structures were mostly devoid of label for gp27, although gp27-labeled membranes were often in their close vicinity (Fig. 7C and 8A and B).
FIG. 8.
Double labeling of p16 and gp27 at 5 h postinfection. (A, B, and C) HeLa cells (A and B) or BHK cells (C) infected with MVA. All sections were double labeled with anti-p16 (10-nm-diameter gold) (arrowheads) and anti-gp27 (5-nm-diameter gold) (arrows). (A and B) Variable p16 labeling of the multimembrane vesicle that is connected to the rough ER (RER). These structures contain little to no labeling for gp27, while the membranes in continuity with them are clearly labeled for the cellular protein. In panel A gp27 but not p16 also labels a distinct dense vesicular structure (asterisk). (C) Relatively large amounts of label for both antigens in membranes adjacent to the Golgi complex are seen. The significant number of cop buds-vesicles (c) in continuity with these membranes indicates that they are on the cis side of the Golgi stack (G). (D) What we believe is the beginning of crescent formation (arrow) that is labeled with anti-p16 (arrowhead) is shown. (E) Double labeling with p16 (10-nm-diameter gold) (arrowheads) and gp27 (5-nm-diameter gold) (arrows), showing an IV, the inner membranes of which are labeled with anti-p16 and that is in continuity with tubular membranes that are labeled with both p16 and gp27. Bars, 200 nm.
Collectively, these data argue that the p16-enriched smooth ER domains segregated from the cellular marker gp27 of the smooth ER/IC/cis-Golgi already at an early stage. To extend these observations we also tested by IF a second marker of this region, β-COP, a component of COP-1 vesicles that also localizes to the ER-Golgi boundary region (10, 16). Infected and mock-infected cells were fixed at 4 h 30 min and double labeled with the anti-p16 and anti-β-COP. As shown in Fig. 10, the labeling for COP was more extended in infected cells than in uninfected cells, where it is mostly in the perinuclear area of the cell, close to the Golgi region. In MVA-infected cells, the COP label was closely apposed to the p16-labeled vesicles, but the two sets of structures were clearly distinct. This supports our proposal that the p16-labeled multimembrane vesicles have segregated from cellular markers of the smooth ER network.
FIG. 10.
IF localization of β-COP in HeLa cells infected with MVA at 4 h 30 min postinfection. HeLa cells grown on coverslips were infected (d to f) or mock infected (a to c) with MVA at an MOI of 10 and fixed at 4 h 30 min after infection. Fixed cells were triple labeled with DAPI (a and d), anti-β-COP (b and e), and anti-p16 (c and f). The large arrows indicate viral DNA replication sites, and the small arrows indicate p16-labeled structures. In panel e the small arrows indicate p16 structures that are adjacent to COP-labeled structures.
Between 4 h 30 min and 5 h, IVs form but fail to associate with the viral DNA under nonpermissive conditions.
When we examined at 5 h infected cells that had been double labeled for p16 and gp27, we also saw significant label for both in the vicinity of the Golgi complex (Fig. 8C). More striking at this time was the appearance of crescents and IVs that, as expected, labeled strongly for p16 but excluded gp27 (Fig. 8D and E). As shown before for later times of infection, these crescents and IVs were in continuity with tubular membranes that were also abundantly labeled with anti-p16 (34) and weakly labeled with gp27 (Fig. 8E). Some images showed rare intermediate stages in which p16-labeled membranes appeared to be transforming into crescents (Fig. 8D). There was no obvious difference in the pattern of labeling between permissive and nonpermissive conditions, in which similar amounts of IVs were seen (not shown).
We next double labeled these 5-h infected cells for p16 and anti-DNA or anti-p35 (H5R). We have previously shown that p35 associates precisely with the viral replication sites from 3 h until at least 6 h postinfection by IF and EM (44). This antibody gave a labeling pattern identical to that for anti-DNA in this study also.
As shown in Fig. 11, under normal assembly conditions, the IVs and crescents were intimately associated with the boundary between the DNA (anti-p35 or anti-DNA labeled) region and the surrounding cytoplasm (see also reference 39). In agreement with the analysis of the Epon sections (at 8 h) shown in Fig. 5, in HeLa cells infected with MVA for 5 h, we failed to see any consistent associations of IVs with these regions (not shown). Between 4 h 30 min and 5 h postinfection, under both permissive and nonpermissive conditions, the ER around the DNA site disassembled, exposing free DNA to the surrounding cytoplasm, in agreement with the EM analysis at 8 h (above) (Fig. 11; see also reference 44). The disassembly of the ER around the factory coincided precisely with the appearance of crescents and IVs.
FIG. 11.
BHK cells infected with MVA and fixed at 5 h postinfection. (A) Sections were double labeled with anti-p16 (10-nm-diameter gold) (arrowheads) and anti-p35 (H5R) (5-nm-diameter gold) (small arrow). The image shows an extended p35-positive area, the boundary of which is indicated with large arrows, that contains several IVs. (B) Sections were double labeled with anti-DNA (5-nm-diameter gold) (small arrow) and p16 (10-nm-diameter gold) (arrowheads). Also in this example there is an abrupt boundary (large arrows) where the DNA labeling stops. p16-labeled IVs are found within this region. Bars, 200 nm.
DISCUSSION
In the present study we have reinvestigated the morphogenesis of MVA in HeLa cells. We show that MVA behaved similarly to WR in several parameters of the early stages of infection; both entry and the formation of DNA replication sites were comparable. At late times of infection, however, MVA assembly in HeLa cells was inhibited. We identify the step in which the block occurs as being due to the mistargeting of IVs to the regions of DNA. The time course of p16 expression enabled us to identify a novel p16-enriched membrane structure that we propose to be the first viral membrane domain that segregates from the smooth ER immediately prior to the formation of the crescent.
Viral entry and production of DNA replication are normal under nonpermissive conditions.
Although it was known that the infection of MVA in HeLa cells was blocked at a late stage of infection, we considered it important to test whether the virus behaved similarly to a VV strain that grows permissively in these cells. Since MVA is an important candidate to be used for live vaccination against other pathogens and in human cancer therapy, it seemed worthwhile to undertake a thorough analysis of its assembly in cultured human cells. It is important to know, for instance, whether MVA infection in such cells requires substantially more or less virus to obtain a similar number of infected cells compared to, for instance, the WR strain of VV. While it is clear that MVA has been adapted to grow very efficiently on CEF cells, it is conceivable that during the course of this adaptation the virus has lost some of its ability to initiate efficient infection in other cells. We show, however, using assays that we had established previously to study the initiation of the early stages of infection in HeLa cells (24, 25), that MVA behaved identically to WR. We show for the first time that MVA purified from BHK cells has a PFU/particle ratio similar to that for purified WR stocks. Moreover, MVA entered HeLa cells with similar kinetics, resulting in a similar amount of intracellular cores at the same MOI. MVA also appeared to be as efficient as WR in producing replication sites, with a one-to-one ratio between incoming cores and sites of DNA synthesis at an MOI of 15.
Early p16 structures as precursors of the viral crescent.
At 4 h 30 min postinfection, the earliest time we could detect p16 by IF, we found that this abundant late membrane protein became enriched in a small number of distinct puncta. By EM these structures were identified as distinct membrane-rich structures. They vaguely resembled multivesicular bodies but were clearly distinct from typical endocytic organelles. Although first seen early in infection, these structures are also visible at later times of infection by EM (Fig. 9D). This suggests that from 4 h 30 min postinfection the viral protein accumulated in such membrane structures before being converted to crescents. Of course, we cannot formally exclude the possibility that the p16 vesicles might have no functional relationship with the assembly sites and may represent dead-end products of p16 synthesis. However, the same structures labeled with p16 were observed under all three conditions of infection used throughout this study. This argues that rather than being artifacts of infection, they are more likely to have a specific function in VV morphogenesis. We propose that the p16-rich structures represent smooth ER-derived membranes that are the precursors of the viral crescents. The lack of colocalization of abundant cellular marker proteins of the SER-IC region with the p16 convoluted membranes is consistent with this idea. It suggests that the membranes may serve to segregate p16 (and likely other viral membrane proteins) from host proteins and is consistent with the fact that the virally modified membranes of crescents and IVs generally exclude host proteins (38).
By IF the bulk of p16 was seen to concentrate in these vesicular structures. However, by EM only a relatively small fraction of the total label was found on them. There was consistent labeling throughout the rough ER and nuclear envelope, with a concentration in a variety of smooth ER structures that were continuous with the ER-Golgi boundary. It is difficult to quantify the fraction of p16 that is in the vesicular membrane aggregates because of the likelihood that the highly packed membranes may be more inaccessible to anti-p16 than the more open smooth ER membranes. However, even without quantitation, the fact that the EM labeling on rough and smooth ER could be seen in practically every section, whereas the (three to five) vesicular structures were seldom seen, argues that only a small fraction of the total p16 is in these vesicles.
VV morphogenesis uses two distinct ER domains.
In the present study we show that all of the processes occurring early in infection are normal in HeLa cells infected with MVA, including the process of segregating the ER into two distinct viral domains. In a recent study we showed that early in infection the viral replication sites becomes almost completely surrounded by membranes of the rough ER by 3 h postinfection. We also described how these ER membranes around the DNA dissociate from this site at about the time that IVs and crescents appear, late in infection (44). Extending that study, we now show that also in MVA-infected BHK and HeLa cells, the replication site was almost completely enclosed by the rough ER. In addition, when crescents and IVs were first observed, from 5 h postinfection onwards, we observed that the ER around the replication site had disassembled, under both permissive and nonpermissive conditions. Thus, assembly and disassembly of this ER domain involved in DNA replications occurs normally under nonpermissive conditions.
These data illuminate an important aspect of viral membrane biogenesis as it occurs in normal and in MVA-infected cells. During the viral life cycle, two distinct functional ER subdomains are assembled. The first is the rough ER, which assembles around the DNA replication site at between 2 and 3 h postinfection, a process that may be mediated by early viral membrane proteins (44). These rough ER domains, which contain normal ER markers, serve as a transient scaffold for DNA replication and need to be dismantled later in infection. Before dismantling occurs, however, a second, smooth ER subdomain emerges, which is enriched in viral late membrane proteins. In contrast to the DNA replication domains, these domains exclude host proteins and are therefore a means for segregating and concentrating the viral proteins which subsequently self-assemble into viral crescents. Our results suggest that the membranes and the DNA need to be transported towards each other for normal assembly. This might, at first glance, be considered to be the movement of one distinct membrane structure along, for example, microtubules, towards a second distinct structure, the DNA factory. However, our recent assembly model (18) provides another, more likely explanation. This model postulates that the crescent-IV domain cisternae are directly connected to separate ER domains that bind to a distinct piece of DNA (likely one genome; see also reference 12). Upon assembly, the ER-bound genome is pushed inside the IV membranes. The resulting IMV is composed of one continuous S-shaped cisternal membrane, in which the genome is surrounded by at least two cisternae. We suggest that MVA in HeLa cells is blocked at a stage that normally allows the ER-bound genome to wrap inside the IV membranes. We therefore predict that the inability of MVA to assemble in HeLa cells is due to a defect in viral self-assembly rather than in a defect in transport per se. This process can be envisioned to be exceedingly complex, and it is therefore not surprising that attempts to rescue the aborted phenotype of MVA in HeLa cells suggested that the failure of MVA to produce infectious progeny in these cells is indeed complex and most likely related to more than one viral gene product (50). The colocalization by IF of IVs and DNA upon permissive infection is, however, so striking that it could be used as a convenient assay for attempts to rescue the aborted transport of viral precursor membranes to the viral DNA replication sites. Such experiments could perhaps shed more light on a possible interaction between the DNA and assembly factories that is apparently essential for efficient assembly of the IMV.
Acknowledgments
We thank Jens Rietdorf for assistance with the confocal microscopy.
M.C.S. was supported by a fellowship from the Ministerio de Educacion, Cultura y Deporte, Spain.
REFERENCES
- 1.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 2.Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. [DOI] [PubMed] [Google Scholar]
- 3.Belyakov, I. M., L. S. Wyatt, J. D. Ahlers, P. Earl, D. Pendleton, B. L. Kelsall, W. Strober, B. Moss, and J. A. Berzofsky. 1998. Induction of a mucosal cytotoxic T-lymphocyte response by intrarectal immunization with a replication-deficient recombinant vaccinia virus expressing human immunodeficiency virus 89.6 envelope protein. J. Virol. 72:8264-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, M. W., W. W. Overwijk, R. S. Charmberlain, S. A. Rosenberg, B. Moss, and N. P. Restifo. 1997. Highly attenuated modified vaccinia virus Ankara (MVA) as an effective recombinant vector: a murine tumor model. Vaccine 15:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Den Boon, J. A., E. J. Snijder, J. Krijnse Locker, M. C. Horzinek, and P. J. M. Rottier. 1991. Another triple-spanning envelope protein among intracellular budding RNA viruses: the torovirus E protein. Virology 182:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doms, R. W., R. Blumenthal, and B. Moss. 1990. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 64:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drexler, I., E. Antunes, M. Schmitz, T. Woelfel, C. Huber, V. Erfle, P. Rieber, M. Theobald, and G. Sutter. 1999. Modified virus Ankara for delivery of human tyrosinase as melanoma-associated antigen: induction of tyrosinase- and melanoma-specific human leukocyte antigen A*0201-restricted cytotoxic T cells in vitro and in vivo. Cancer Res. 59:4955-4963. [PubMed] [Google Scholar]
- 9.Drexler, I., K. Heller, B. Wahren, V. Erfle, and G. Sutter. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79:347-352. [DOI] [PubMed] [Google Scholar]
- 10.Duden, R., G. Griffiths, R. Frank, P. Argos, and T. E. Kreis. 1991. Beta-COP, a 110kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to beta-adaptin. Cell 64:649-665. [DOI] [PubMed] [Google Scholar]
- 11.Earl, P. L., N. Cooper, L. S. Wyatt, B. Moss, and M. W. Carroll. 1998. Titration of MVA stocks by immunostaining, p. 16.9-16.11. In F. M. Ausubel, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene/Wiley Interscience, New York, N.Y.
- 12.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. Krijnse Locker. 1995. Characterization of ts 16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fullekrug, J., T. Suganuma, B. L. Tang, W. Hong, B. Storrie, and T. Nilsson. 1999. Localization and recycling of gp27 (hp24gamma3): complex formation with other p24 family members. Mol. Biol. Cell 10:1939-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, S. C., J. Schneider, M. Plebanski, C. M. Hannan, T. J. Blanchard, G. L. Smith, and A. V. Hill. 1999. Ty virus-like particles, DNA vaccines and modified vaccinia virus Ankara: comparison and combinations. Biol. Chem. 380:299-303. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths, G. 1996. On vesicle and membrane compartments. Protoplasma 195:37-58. [Google Scholar]
- 16.Griffiths, G., R. Pepperkok, J. Krijnse Locker, and T. E. Kreis. 1995. Immunocytochemical localization of beta-COP to the ER-Golgi boundary and the TGN. J. Cell Sci. 108:2839-2856. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths, G., N. Roos, S. Schleich, and J. Krijnse Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analyses. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths, G., R. Wepf, T. Wendt, J. Krijnse Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., T. R. Fuerst, G. Sutter, M. W. Carroll, L. C. Yang, S. Goldstein, M. Piatak, Jr., W. R. Elkins, W. G. Alvord, D. C. Montefiori, B. Moss, and J. D. Lifson. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichihashi, Y. 1996. Extracellular enveloped virus escapes neutralization. Virology 217:478-485. [DOI] [PubMed] [Google Scholar]
- 22.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 24.Krijnse Locker, J., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 11:2497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallardo, M., E. Leithe, S. Schleich, N. Roos, L. Doglio, and J. Krijnse Locker. 2002. Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites. J. Virol. 76:5167-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallardo, M., S. Schleich, and J. Krijnse Locker. 2001. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs to distinct cytoplasmic structures. Mol. Biol. Cell 12:3875-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayr, A., H. Stickl, H. K. Müller, K. Danner, and H. Singer. 1978. Der pockenimpfstamm MVA: Marker, genetische Struktur, Erfahrungen mit der parenteralen Schutzimpfung und Verhalten im abwehrgeschwächten Organismus. Zentbl. Bakteriol. Hyg. Abt. 1 Orig. B 167:375-390. [PubMed] [Google Scholar]
- 28.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 29.Moss, B., and B. M. Ward. 2001. High-speed mass transit for poxviruses on microtubules. Nat. Cell Biol. 3:E245-246. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. Krijnse Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implication of the 21-kilodalton and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. Krijnse Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmelz, M., B. Sodeik, M. Ericsson, E. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, J., S. C. Gilbert, T. J. Blanchard, T. Hanke, K. J. Robson, C. M. Hannan, M. Becker, R. Sinden, G. L. Smith, and A. V. S. Hill. 1998. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat. Med. 4:397-402. [DOI] [PubMed] [Google Scholar]
- 37.Slot, J. W., H. J. Geuze, S. Gigengack, G. E. Lienhard, and D. E. James. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik, B., and J. Krijnse Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 39.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spehner, D., R. Drillien, F. Proamer, C. Houssais-Pêcheur, M.-A. Zanta, M. Geist, K. Dott, and J.-M. Balloul. 2000. Enveloped virus is the major virus form produced during productive infection with modified vaccinia virus Ankara strain. Virology 273:9-15. [DOI] [PubMed] [Google Scholar]
- 41.Stickl, H., V. Hochstein-Mintzel, A. Mayr, H. Huber, H. Schaefer, and A. Holzner. 1974. MVA-stufenimpfung gegen Pocken. Dtsch. Med. Wochenschr. 99:2386-2392. [DOI] [PubMed] [Google Scholar]
- 42.Sutter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 89:10847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutter, G., L. S. Wyatt, P. L. Foley, J. R. Bennink, and B. Moss. 1994. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine 12:1032-1040. [DOI] [PubMed] [Google Scholar]
- 44.Tolonen, N., L. Doglio, S. Schleich, and J. Krijnse Locker. 2001. Vaccinia virus DNA replication occurs in ER-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12:2031-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. Krijnse Locker. 1999. The localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanderplasschen, A., and G. L. Smith. 1997. A novel binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 71:4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanderplasschen, A., M. Hollinshead, and G. L. Smith. 1998. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol. 79:877-887. [DOI] [PubMed] [Google Scholar]
- 49.Van Slyke, J. K., and D. E. Hruby. 1994. Immunolocalization of vaccinia virus structural proteins during virion formation. Virology 198:624-635. [DOI] [PubMed] [Google Scholar]
- 50.Wyatt, L. S., M. W. Carroll, C.-P. Czerny, M. Merchlinsky, J. R. Sisler, and B. Moss. 1998. Marker rescue of the host range restriction defects of modified vaccinia virus Ankara. Virology 251:334-342. [DOI] [PubMed] [Google Scholar]