Abstract
The simian virus 40 (SV40) large tumor (T) antigen is sufficient to transform cells in cultures and induce tumors in experimental animals. Transformation of primary cells in cultures requires both overcoming growth arrest by stimulating the cell cycle and blocking cell death activities presumably activated by oncogene-mediated hyperproliferation signals. The study presented here examined the ability of specific regions and activities of T antigen to modulate apoptosis in cells treated with the genotoxic agent 5-fluorouracil (5-FU). The results showed that the expression of full-length T antigen rendered rat embryo fibroblasts (REF) sensitive to 5-FU-induced apoptosis. Thus, neither the p53-binding region nor the Bcl-2 homology region of T antigen was sufficient to prevent cell death induced by the DNA-damaging agent. T-antigen-mediated sensitization occurred independently of retinoblastoma protein or p53 and p300 binding. An N-terminal segment containing the first 127 T-antigen amino acids (T1-127) was sufficient to sensitize cells. A C-terminal segment consisting of T-antigen amino acids 251 to 708 (T251-708) also sensitized cells to 5-FU-induced apoptosis. This sensitization did not occur when T251-708 was targeted to the nucleus by inclusion of the SV40 nuclear localization signal. The introduction of mutations into the T-antigen J domain resulted in mutation-specific and variable inhibition of apoptosis. This result suggested that either the structural or the functional integrity of the J domain is required to sensitize cells to apoptosis. Treatment of REF or REF expressing full-length T antigen, an N-terminal segment, or T251-708 resulted in increased expression of the p53-responsive MDM2 gene; apoptosis occurred through a p53-dependent pathway, as p53-null cells expressing these T antigens were resistant to 5-FU-induced apoptosis. Possible mechanisms involved in sensitizing cells to a p53-dependent apoptosis pathway in spite of the ability of T antigen to bind and inactivate the transcriptional transactivating activity of p53 are discussed.
The simian virus 40 (SV40) large tumor (T) antigen is sufficient to transform primary cells in cultures and induce tumors in transgenic animals (16, 31, 61). In contrast, most cellular and viral oncogenes do not individually transform primary cells. Rather, they require a cooperating oncogene to do so. A predominant characteristic of oncogenes that cooperate to transform primary cells is that at least one activity of the partners is involved in stimulating cell division (overcoming growth arrest) and at least one activity is involved in blocking apoptosis, activities presumably activated by oncogene-mediated hyperproliferation signals (4).
SV40 T antigen has both activities that stimulate cell cycle progression and activities that are expected to or that have been shown to inhibit apoptosis. T antigen contains four independent mitogenic activities (12). One activity of T antigen involved in stimulating cellular DNA synthesis is inhibited by the mutation D402N. This mutation limits the capacity of T antigen to bind the tumor suppressor p53 (28). The three remaining activities depend on the integrity of distinct regions within the N-terminal half of the protein. These activities are marked genetically by specific mutations in the region shared with small t antigen (the T/t common region) and the conserved LXCXE motif in the T-antigen-binding site for the tumor suppressor protein retinoblastoma protein (Rb) (and related family members p107 and p130) as well as the amino acid substitution S189N. Each of the alterations reduces but does not eliminate the ability of T antigen to stimulate cellular DNA synthesis; and the activities cooperate in maximal T-antigen-induced DNA synthesis in cultured cells (12, 13). Correspondingly, the expression of the N-terminal segment containing the first 121 amino acids of T antigen (T1-121) is sufficient to stimulate cellular DNA synthesis in specific tissues of transgenic mice, and this capability depends on Rb binding (41).
Rb is a multifunctional protein that arrests cells in the G1 phase of the cell division cycle by repressing the transcription of genes required for entry into the S phase (reviewed in reference 20). T-antigen-mediated inactivation of Rb functions involved in cell cycle arrest requires not only binding to the conserved LXCXE motif but also one or more activities contained within the T/t common region (49). This region shares sequence homology with the J domain of the DNA J family of proteins and functions as a J domain (6, 37). Correspondingly, the T-antigen J domain contains the histidine-proline-aspartate (HPD; amino acids 42 to 44) region that is conserved within papovavirus T-antigen homologues as well as other DnaJ homologues (37). Other Rb functions are inhibited independently of the J domain by virtue of binding to the LXCXE motif of T antigen (18, 39).
Inactivation of Rb in cells expressing a wild-type p53 protein can lead to apoptosis. The removal of functional Rb in specific tissues in mice either through gene knockout (34) or through expression of a T-antigen N-terminal segment (17) results in apoptosis. In a cell culture system, under specific conditions, the expression of the segment consisting of the first 138 amino acids of T-antigen (T1-138), which contains both the Rb-binding site and a functional J domain, was reported to cause morphological changes associated with apoptotic cells and cell death (11).
Three features of full-length T antigen suggest that it would act as an apoptosis inhibitor. First, T antigen binds and inactivates p53, providing a mechanism for derailing p53-dependent apoptosis pathways. Second, T antigen contains a region (amino acids 525 to 541) having approximately 60% amino acid homology with the BH-1 region of the antiapoptosis protein Bcl-2 (11). Previously, Conzen et al. (11) examined the involvement of the Bcl-2 homology region of T antigen in blocking apoptosis during serum withdrawal. They showed that cells expressing T antigens with mutations within the Bcl-2 homology region responded to serum withdrawal by undergoing apoptosis. Third, T antigen binds the p53 coactivator p300 (1, 14), although whether this binding is sufficient to prevent p300 activities is uncertain. Inactivation of p300 limits the ability of p53 to cause growth arrest through the transcriptional activation of growth arrest genes and apoptosis (2, 26). In transgenic mice, inactivation of p53 is required to block apoptosis in specific tissues in response to proliferation signals provided by an N-terminal T-antigen segment (52, 53).
Thus, T antigen renders cells functionally Rb and p53 deficient and potentially provides both apoptotic and antiapoptotic activities. The expectation that follows from this observation is that the expression of full-length T antigen or a T antigen capable of binding and inactivating p53 and containing the Bcl-2 similarity region would block p53-mediated apoptosis. In contrast, an N-terminal fragment not containing these regions either would not affect apoptosis or would enhance the process.
SV40 T antigen shares transformation-related activities with the adenovirus E1A protein 243R, the DNA viral oncogene product that has been examined most closely with respect to apoptosis induction (reviewed in reference 29). Cells expressing E1A either undergo apoptosis (54, 62) or are sensitized to apoptotic stimuli, including DNA-damaging agents (30, 33, 42). Additional adenovirus products, including the E1B proteins 55K, a p53-binding protein, and 19K, a Bcl-2 homologue, or an activated cellular ras oncogene blocks E1A-induced apoptosis. Nonetheless, cells expressing E1A proteins and activated Ras undergo apoptosis following treatment with genotoxic agents. This sensitization depends upon the ability of E1A to bind both the tumor suppressor protein Rb and the p53 coactivator p300 (42)
In order to define more closely the T-antigen activities involved in the induction or inhibition of apoptosis, we initiated an investigation into the response of T-antigen-expressing cells to the genotoxic agent 5-fluorouracil (5-FU). The results showed that the expression of full-length T antigen, rather than protecting cells from apoptosis, sensitized them to it. N-terminal T-antigen segments were sufficient for sensitization. Similarly, the C-terminal fragment consisting of T-antigen amino acids 251 to 708 (T251-708), which localizes to the cell cytoplasm, sensitized cells to 5-FU-induced apoptosis. However, T251-708NLS, which contains the SV40 nuclear localization signal (NLS), did not. Full-length T antigens bearing mutations that prevented Rb binding, p53 and p300 binding, or p53 function remained capable of sensitizing cells to apoptosis induction. The presence of certain mutations in the J domain blocked sensitization, suggesting that apoptosis following 5-FU treatment depends at least in part on the integrity of the J domain. The effect of mutations in the J domain was, however, allele specific. Additional results showed that apoptosis in response to the combined action of either a T-antigen fragment or full-length T antigen and the genotoxic insult occurs by a p53-dependent pathway.
MATERIALS AND METHODS
Plasmids.
Plasmids pPVU0 (23), pdl2005 (45), CAV83-708 (10), pdl501- 550 (23), pD44N (36), pH42Q (49), pT1-147Sac-dl (57), pdl2441 (59), pCAVT251-708 (9), pCAVT251-708NL (9), and WT-2 (10) have been described elsewhere. The T antigens they encode are listed in Table 1. Plasmid pSP72ras (9) contains an activated H-ras oncogene. E. White kindly provided plasmids pCMVE1A and pCMVE1B. They contain, respectively, the E1A and E1B coding regions of adenovirus and are expressed from the human cytomegalovirus major immediate-early enhancer-promoter. Plasmid LTRp53cG-ala contains wild-type murine p53 cDNA and was provided by A. Levine. The reporter p50-2luc contains the p53-responsive elements from the murine muscle-specific creatine kinase gene cloned 5′ of the luciferase gene (kindly provided by G. Zambetti).
TABLE 1.
Cell lines used
| Cell linea | T antigen(s)b | Source or reference | No. of clonal cell lines tested | Plasmid(s) used to generate cell line |
|---|---|---|---|---|
| REF/T1-708+Ras | T1-708, t | This study; 9 | 7 | pPVU-0, pSP72-ras |
| REF/T1-708 | T1-708, t | This study; 9 | 4 | pPVU-0 |
| REF/T1-127+Ras | T1-127, t | This study | 3 | pCAVMT1-127, pSP72-ras |
| REF/T1-136+Ras | T1-136 | This study | 2 | pRSVBNeoN136, pSP72-ras |
| REF/T1-147+Ras | T1-147 | This study | 5 | pT147Sac-dl, pSP72-ras |
| REF/T251-708+Ras | T251-708 | This study; 9 | 5 | pCAV251-708, pSP72-ras |
| REF/T251-708NLS+Ras | T251-708NLS | This study; 9 | 5 | pCAV251-708NL, pSP72-ras |
| REF/T83-708+Ras | T83-708 | This study | 15 | pCAV83-708, pSP72-ras |
| REF/dl501-550+Ras | Tdl501-550, t | This study | 3 | pdl501-550, pSP72-ras |
| REF/T1-708E107K+Ras | T1-708E107K, t | This study | 5 | pSelectTE107K, pSP72-ras |
| REF/Tdl105-108+Ras | Tdl105-108, t | This study | 3 | pdl2441, pSP72-ras |
| REF/T1-708P43L/K45N+Ras | T1-708P43L/K45N, tP43L/K45N | This study; 5 | 3 | pTP43L/K45N, pSP72-ras |
| REF/T1-708D44E/G47R+Ras | T1-708D44E/G47R, tD44E/G47R | This study; 5 | 3 | pTD44E/G47R, pSP72-ras |
| REF/T1-708D44N+Ras | T1-708D44N, tD44N | This study | 4 | pD44N, pSP72-ras |
| REF/T1-708H42Q+Ras | T1-708H42Q, tH42Q | This study | 9 | pH42Q, pSP72-ras |
| REF/T1-708, t−+Ras | T1-708 | 9 | 4 | pSelectdl2005, pSP72-ras |
| REF/E1A+E1B | None | This study | 3 | pCMVE1A, pCMVE1B |
| REF/E1A+Ras | None | This study | 3 | pCMVE1A, pSP72-ras |
| B6MEF/E1A+Ras | None | This study; 9 | 4 | pCMVE1A, pSP72-ras |
| B6K/p53+/t/T1-708+Ras | T, t | This study | 2 | pPVU-0, pSP72-ras |
| B6Kp53−/−/T1-708+Ras | T, t | This study | 10 | pPVU-0, pSP72-ras |
| B6Kp53−/−/T1-127+Ras | T1-127, t | This study | 3 | pCAVMT1-127, pSP72-ras |
| B6Kp53−/−/T251-708+Ras | T251-708 | This study | 3 | pCAV251-708, pSP72-ras |
| B6Kp53−/−/E1A+Ras | None | This study | 1 | pCMVE1A, pSP72-ras |
| B6MEF/T1-708 | T1-708, t | This study | 2 | pWT-2 |
B6K, C57BL/6 kidney fibroblasts; B6Kp53−/−, C57BL/6 kidney fibroblasts isolated from a p53 homozygous mutant.
Amino acid substitutions are indicated by letters of the single amino acid code surrounding the amino acid altered; deletions are indicated by dl followed by the amino acids for the residues deleted.
Plasmid CAVMT1-127 was generated by releasing the SV40 sequences (nucleotides [nt] 5235 to 2533) from plasmid pE1T1-127 (55) with BglII and MluI, purifying this fragment by the GeneClean (Bio101) procedure, and inserting it into the corresponding sites of the CAVIL2M vector (9). The plasmid contains a large-T-antigen N-terminal segment encompassing codons 1 to 127 (T1-127) and small t antigen. Expression is under the control of the cytomegalovirus enhancer-promoter.
Plasmid pSelectESV was constructed by inserting the SV40 enhancer-promoter and early-region sequences from the KpnI to the BamHI sites (nt 294 to 2533) into the corresponding sites of the pSelect vector (Promega). The amino acid substitution mutation E107K was introduced into the large-T-antigen coding sequence by oligonucleotide-directed mutagenesis of plasmid pSelectESV with an Altered Sites mutagenesis kit (Promega) to produce plasmid pSelectTE107K. The presence of the mutation was confirmed by DNA sequence analysis.
Plasmids pw2tP43L/K45N and pw2tD44E/G47R were kindly provided by K. Rundell and have been described elsewhere (38). They express small t antigens with the two amino acid substitutions indicated. Plasmids (pTP43L/K45N and pTD44E/G47R) expressing large T antigens containing these amino acid substitutions were generated by replacing the SV40 sequences from the BstXI site (nt 4759) to the BamHI site (nt 2533) of pw2tP43L/K45N and pw2tD44E/G47R with the corresponding wild-type fragment from pPVU0.
Cell lines.
The rodent cell lines used in this study are listed in Table 1. Each cell line was derived from a single colony of cells following calcium phosphate-mediated transfection of primary cell cultures with the plasmids indicated and as described previously (9, 23). Primary Fischer 344 rat embryo fibroblasts (REF) and primary C57BL/6 mouse embryo fibroblasts (B6MEF) were generated as described previously in detail (58). Primary kidney fibroblast cultures from adult C57BL/6 and p53-null (−/−) mice were generated in a similar fashion.
Cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS) and supplemented with 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, 100 U of penicillin per ml, 0.03% glutamine, 25 mM HEPES, and 0.15% NaHCO3 (DMEM5×2+HEPES) and were incubated at 37°C. Primary cells were maintained similarly by using DMEM containing 10% FBS and supplemented with 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, 100 U of penicillin per ml, 0.03% glutamine, and 0.07% NaHCO3 (DMEM10×1).
Treatment with 5-FU.
For each concentration of 5-FU used, two 100-mm-diameter dishes were each seeded with 2 × 106 viable cells in 10 ml of DMEM5×2+HEPES for cell lines or DMEM containing 10% FBS and 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, 100 U of penicillin per ml, 0.03% glutamine, and 0.14% NaHCO3 for primary REF. Cells were allowed to adhere for approximately 3 h at 37°C in a 5% CO2 atmosphere. Following this incubation period, 5 ml of DMEM5×2+HEPES or 5 ml of DMEM5×2+HEPES containing three times the final concentration of 5-FU per ml indicated in the figures was added to each dish. The dishes were incubated overnight, and the cells were collected and subjected to apoptosis assays.
Apoptosis assays.
For the detection of nucleosomal DNA ladders, adherent cells were scraped from culture dishes into medium. The cells were pelleted by centrifugation at 912.8 × g for 5 min at room temperature. The pellets were suspended in 5 ml of Tris-buffered saline (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 0.9 mM CaCl2, 0.5 mM MgCl2, 0.1% dextrose). The cells were pelleted again, and the pellets were resuspended in 0.1 ml of Tris-buffered saline. Then, 0.6 ml of lysis buffer A (10 mM Tris [pH 8], 5 mM EDTA, 100 mM NaCl, 0.5% sodium dodecyl sulfate [SDS], 1 μg of proteinase K/μl) was added, and the cell suspensions were incubated at 37°C for approximately 2 h. Low-molecular-weight DNA was isolated by using a modified Hirt procedure (22). Specifically, following the incubation period, 0.2 ml of 4 M NaCl was added to the cell lysates, and the lysates were mixed gently by inversion and incubated overnight at 4°C. The lysates then were centrifuged at 20,000 × g for 30 min at 4°C, and the viscous pellets were removed. The low-molecular-weight DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. The resulting DNA pellets were dissolved in 20 μl of 10 mM Tris (pH 7.5)-1 mM EDTA- 20 μg of RNase A/ml and incubated for 1 h at 37°C. DNA fragments were resolved by electrophoresis for 1.5 h at 100 V in 1% agarose gels in Tris-borate buffer containing 0.5 μg of ethidium bromide per ml.
Cell-mixing experiments were performed to determine the number of cells that were capable of undergoing 5-FU-induced apoptosis and that were needed to visualize nucleosomal DNA ladders in low-molecular-weight DNA extracts of treated cells. REF lines transformed by adenovirus E1A and E1B are resistant to 5-FU-induced apoptosis, whereas REF transformed by full-length T antigen are sensitive (Fig. 1). REF expressing E1A and E1B were mixed with REF expressing T antigen in decreasing proportions from 100 to 0%, seeded at a combined density of 2 × 106 cells per 100-mm dish, and treated with 5-FU as indicated above. Examination of low-molecular-weight DNA extracts of treated cells for DNA ladders indicated that 1.6 × 106 cells expressing T antigen were sufficient to consistently detect DNA ladders. In subsequent experiments, a minimum of 2 × 106 cells were used for the detection of DNA ladders (data not shown). A time course analysis of DNA ladder detection following 5-FU treatment of cells expressing T-antigen amino acids 1 to 708 (T1-708) showed that ladders were not detected at 4 or 8 h after 5-FU treatment; however, they were readily detected at 16 h after treatment (data not shown). In subsequent experiments, cells were treated with 5-FU for 16 h prior to extraction of low-molecular-weight DNA.
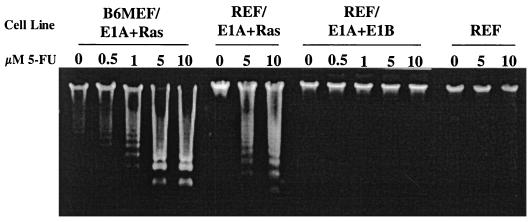
FIG. 1.
B6MEF and REF expressing E1A and Ras undergo apoptosis following 5-FU treatment. A B6MEF line expressing adenovirus E1A and activated Ras (B6MEF/E1A+Ras), REF expressing E1A and Ras (REF/E1A+Ras) or E1A, E1B, and Ras (REF/E1A+E1B), and primary REF were treated overnight with the amounts of 5-FU indicated. As described in Materials and Methods, low-molecular-weight DNA was isolated and subjected to electrophoresis through 1% agarose gels. Nucleosomal DNA fragments were visualized by ethidium bromide staining.
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed by using an in situ cell death detection kit (Boehringer Mannheim). Specifically, 2 × 106 viable cells were added to 100-mm dishes containing 18-mm coverslips and treated with 5-FU as described above. Following overnight incubation, TUNEL assays were performed by using the conditions specified by the manufacturer. Coverslips were mounted on glass slides with glycerol-phosphate-buffered saline (PBS) (1:9) containing propidium iodide as a counterstain, and labeled cells were visualized by using a Nikon Microfot-FXA fluorescence microscope.
Immunoprecipitation.
To assess the levels of expression of T antigens in cell lines, cells were passaged 1:2 in 75-cm2 cell culture flasks containing DMEM5×2+HEPES and incubated overnight at 37°C. The medium was removed, and the cell monolayers were washed twice with 5 ml of cold PBS. The second PBS wash was aspirated, and 1 ml of cold lysis buffer B (50 mM Tris, 5 mM EDTA, 150 mM NaCl, 0.5% IGEPAL CA-630 [Sigma I3021], 1 mM phenylmethylsulfonyl fluoride, 0.01 mg of aprotinin per ml) was added. Flasks were incubated for 30 min to 1 h at 4°C on a rocking platform. Cell lysates were transferred to 1.5-ml microcentrifuge tubes, and cellular debris was pelleted by centrifugation at 20,000 × g for 5 min at 4°C. The protein concentration of each cell lysate was quantified by using a Bio-Rad protein assay.
Immunoprecipitations were performed with samples that had been equalized for protein content as described previously (23). T antigens were immunoprecipitated by using either 2 μl of polyclonal antibody (Pab) 901, which recognizes an epitope between T-antigen amino acids 684 and 698 (23), or 100 μl of PAb 416 or 2 μl of PAb 902, which recognizes an epitope between amino acids 82 and 127 (43) or amino acids 1 and 82 (21), respectively. For detection of p53, the protein was immunoprecipitated from 2 mg of total cell lysate by using 180 μl of PAb 421 (21). MDM2 was immunoprecipitated from 300 μg of total cell lysate by using 10 μl of Ab-2 from Oncogene Science.
Western blot analysis.
Immunoprecipitates were subjected to SDS-10% polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. MDM2, T antigen, and p53 were detected by using antibody Ab-1 (Santa Cruz), PAb 901, and PAb 421, respectively. Reacting antibodies were detected by using either a horseradish peroxidase-conjugated goat anti-mouse antibody or protein A-horseradish peroxidase and were visualized by using a Renaissance Western blot chemiluminescence system (NEN).
Transactivation of a p53-responsive promoter.
Confluent cultures of SAOS-2 cells were passaged 1:2 in DMEM10×1 and incubated overnight at 37°C. On the following day, for each DNA combination tested, each of four 60-mm culture dishes was seeded with 3 × 105 cells in 5 ml of DMEM10×1; the cultures were incubated overnight. Each culture was transfected by using the calcium phosphate method with 5 μg of salmon sperm DNA, 2.5 μg of Bcl-2-expressing plasmid, 2.5 μg of the p50-2luc reporter construct, 1 μg of wild-type mouse p53-expressing plasmid, and various amounts of T-antigen-expressing plasmids. Specifically, 0.5 ml of the DNA-calcium phosphate precipitate was added directly to the culture medium, and the cultures were incubated at 37°C for 6 h. The culture medium was removed, and the cell monolayers were washed twice with DMEM supplemented with 100 μg of streptomycin per ml, 100 μg of kanamycin per ml, 100 U of penicillin per ml, 0.03% glutamine, and 0.15% NaHCO3. Fresh DMEM10×1 was added to the cultures, and incubation was continued for an additional 24 h. Samples then were harvested, and luciferase activity was assayed by using a Promega luciferase assay system according to the manufacturer's instructions. Relative light units were monitored by using a Femtomaster FB12 luminometer (Zylux Corp.).
RESULTS
Influence of T antigen on apoptosis following 5-FU treatment.
The purpose of the investigations reported here was to define activities of T antigen that modulate apoptosis. As the experimental system, cell lines were individually derived from clones of primary REF transformed by full-length or mutant T antigens and Ras and then were examined for their response to treatment with the DNA-damaging agent 5-FU. The ras oncogene was included to permit analysis of cell lines expressing mutant T antigens that have lost the capacity to immortalize primary cells (56) yet have retained the ability to cooperate with a ras oncogene in transforming primary cells (5, 9). Initially, cell lines expressing full-length T antigen and Ras and either untreated or treated with various concentrations of 5-FU were examined for evidence of apoptosis. The nucleosomal DNA fragment ladder assay was selected to monitor apoptosis. Apoptosis is divided into three general stages, early, middle, and late. In the early stage, death signals are received and death precursor initiation caspases (procaspases) are cleaved into active caspases. If unchecked by survival signals, apoptosis will occur. Survival signals can be of two general types, those that interfere with the activity of proapoptotic proteins and those that inactivate caspases. When the pathway to apoptosis has not been defined and additional proteins may have an impact on the level of survival activities, it is prudent to use an assay that addresses the late stage of apoptosis. The presence of DNA ladders indicates that apoptosis has progressed to a late stage.
Previously, Lowe et al. (30) showed that mouse embryo fibroblast lines expressing the adenovirus E1A gene and Ras, while protected from E1A-induced apoptosis, were sensitized to apoptosis following exposure to DNA-damaging agents. The results presented in Fig. 1 compare the response of B6MEF expressing E1A and Ras to that of REF expressing the same oncoproteins. As controls, primary REF cultures and REF expressing E1A and the adenovirus E1B gene products 55K, a p53-binding protein, and 19K, a homologue of the antiapoptosis protein Bcl-2, were included. The results showed that, at the concentrations used, 5-FU did not induce apoptosis in REF. In contrast, 5-FU induced apoptosis in B6MEF or REF expressing E1A and Ras in a dose-dependent manner. REF expressing E1A and E1B proteins were protected from apoptosis following 5-FU treatment. In this and subsequent analyses of T-antigen-mediated sensitization to apoptosis, multiple clonally derived cell lines of each type responded similarly to treatment. The numbers of clonally derived cell lines examined are shown in Table 1. These results validated the use of REF for investigating the response of cells expressing T antigen to 5-FU treatment.
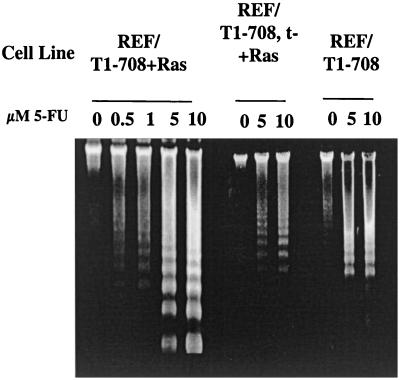
The response of REF expressing full-length T antigen and Ras (REF/T1-708+Ras) to 5-FU treatment is shown in Fig. 2. These cells were sensitized to 5-FU-induced apoptosis. The sensitization occurred both in cells expressing T1-708 and small t antigen (REF/T1-708+Ras) and in cells expressing T1-708 alone (REF/T1-708,t−+Ras). The expression of Ras was not required for sensitization. REF immortalized by T1-708 in the absence of Ras (REF/T1-708) underwent apoptosis following 5-FU treatment. Two related conclusions follow from these data. The presence of the p53-binding and Bcl-2 homology regions is not sufficient to protect cells expressing T antigen from apoptosis following treatment with 5-FU. In contrast, the expression of T antigen is sufficient to sensitize cells to 5-FU-induced apoptosis.
FIG. 2.
Responses of cells expressing SV40 T antigens in the presence and absence of Ras to 5-FU treatment. REF expressing large T and small t antigens and Ras (REF/T1-708+Ras), large T antigen only and Ras (REF/T1-708,t−+Ras), or large T and small t antigens but no Ras (REF/T1-708) were treated overnight with the concentrations of 5-FU indicated prior to the isolation and visualization of nucleosomal DNA as described in the legend to Fig. 1
Influence of Rb or p53 and p300 binding on sensitization.
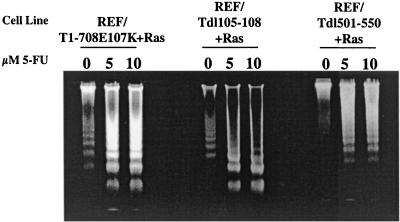
The influence of binding of the tumor suppressor protein Rb (and related family members) or p53 and the p53 coactivator p300 on sensitization was examined by using REF lines cotransformed by Ras and T antigens with amino acid substitutions or deletions in the binding regions for these proteins. Rb and its family members p107 and p130 bind to T antigen through the T-antigen LXCXE motif (amino acids 103 to 107). As shown in Fig. 3, REF expressing a T antigen with the amino acid substitution E107K (REF/T1-708E107K+Ras) or with a deletion of amino acids 105 to 108 (REF/Tdl105- 108+Ras) remained sensitive to apoptosis following 5-FU treatment. It was shown previously that a mutant T antigen missing amino acids 501 to 550 (Tdl501- 550) could not bind p53 (23) or its coactivator p300 (27). The results presented in Fig. 3 show that REF transformed by Tdl501- 550 and Ras remained sensitive to 5-FU-induced apoptosis. The results suggested that an activity other than binding of the Rb family of proteins, p53, and p300 is required for T-antigen-mediated sensitization to apoptosis. The possibility that multiple T-antigen activities individually sensitize cells cannot be excluded, however.
FIG. 3.
Effect of mutationally preventing Rb or p53 and p300 from binding to T antigen on sensitization to 5-FU-induced apoptosis. REF lines expressing Ras and T antigens with alterations in the Rb-binding region (REF/T107E107K+Ras and REF/T105- 108+Ras) or a T antigen with a deletion in the p53-binding site (REF/Tdl501- 110+Ras) were treated overnight with the concentrations of 5-FU indicated prior to the isolation and visualization of nucleosomal DNA as described in the legend to Fig. 1
Independent regions of T antigen sensitize cells to 5-FU-induced apoptosis.
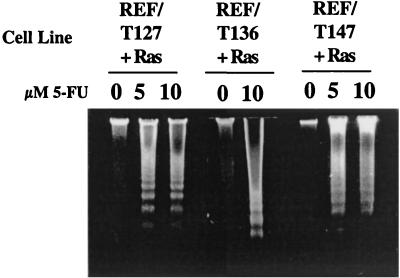
In order to identify a region of T antigen that was sufficient to sensitize cells, REF cotransformed by N- or C-terminal T-antigen fragments and Ras were tested for the generation of nucleosomal DNA ladders following 5-FU treatment. The results presented in Fig. 4 show that the expression of a T-antigen fragment consisting of amino acids 1 to 147 (T1-147), 1 to 136 (T1-136), or 1 to 127 (T1-127) was sufficient to sensitize cells. Each of these N-terminal T-antigen fragments contains two known functional regions, the Rb-binding region and the J domain. All of the proteins are expected to accumulate in the cell nucleus. T1-147 and T1-136 contain an intact NLS (T-antigen amino acids 126 to 132). Although T1-127 does not contain an NLS, it is expected to accumulate in both the nucleus and the cytoplasm, presumably due to its small size, as shown previously for T antigens of similar lengths (41).
FIG. 4.
T1-127 is sufficient to sensitize cells to 5-FU-induced apoptosis. REF lines expressing Ras and N-terminal fragment T1-127 (REF/T127+Ras), T1-136 (REF/T136+Ras), or T1-147 (REF/T147+Ras) were treated overnight with the concentrations of 5-FU indicated prior to the isolation and analysis of low-molecular-weight DNA as described in the legend to Fig. 1.
Role of the J domain in sensitization.
The first 82 amino acids of T antigen share homology with J domains of the DnaJ family of molecular chaperones (6, 37). Within this region, the hexapeptide motif (HPDKGG), at T-antigen amino acids 42 to 47, is highly conserved among papovaviruses. The T-antigen J domain interacts with the ATPase domain of the DnaK protein Hsc70 through the J domain HDP loop and stimulates Hsc70 ATPase activity (6, 37). Hydrolysis of ATP can disrupt multiprotein complexes or alter the conformation of bound substrates. Two classes of activities are associated with the T-antigen J domain. The first class consists of activities that appear to depend solely on the integrity of this domain. These J domain-dependent, Rb-binding-independent functions include the activation of Hsc70 ATPase (48) and transactivation of the cyclin A promoter (5). The second class includes J domain activities required to inactivate a specific subset of Rb family member functions. Thus, several T-antigen activities depend not only on binding of the Rb family of proteins but also on an intact J domain. Among these J domain-dependent, Rb-binding-dependent activities are the relief of Rb/p130-dependent E2F repression (64), hypophosphorylation of p107/p130, degradation of p130 (50), growth of cells in low-serum medium (50), override of Rb/p107/p130-mediated growth arrest (64), and the ability of N-terminal T-antigen segments to cooperate with Ras transformation assays (5, 9). The finding (Fig. 3) that T antigens rendered incapable of binding Rb (T1-708E107K and Tdl105- 108) still sensitized cells to apoptosis following 5-FU treatment suggested that binding and inactivating Rb were not required.
The possibility that a J domain activity was involved was investigated by examining a series of T antigens with amino acid substitutions or a deletion within that region. Four amino acid substitution mutants were examined. The substitutions D44N and H42Q reside within the J domain HDP loop and, among J domain mutants available, these mutants have been examined most thoroughly for loss of function (18, 47-50, 64). The double amino acid substitution mutants P43L/K45N andD44E/G47R have replacements of amino acids within the hexapeptide but, with the exception of D44E, outside of the HDP loop. In the J domain mutant T83-708 (consisting of T-antigen amino acids 83 to 708), the first 82 amino acids of T antigen are replaced by the first 7 amino acids of β-galactosidase followed by 31 amino acids of the cloning vector pBluescript SK (Stratagene) and 2 amino acids encoded by the linker used to generate the T83-708-expressing plasmid (9).
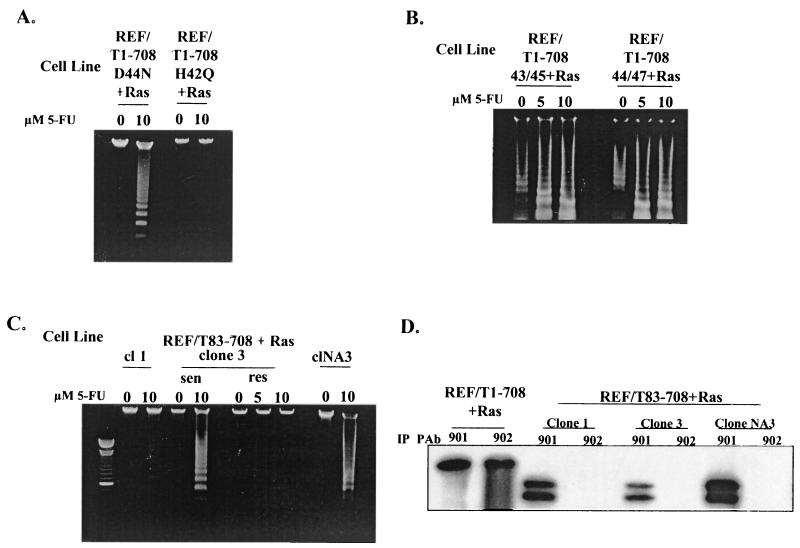
REF lines expressing each of these T antigens were generated and examined for evidence of apoptosis following 5-FU treatment. The results are presented in Fig. 5. All seven clonally derived cell lines expressing T1-708D44N remained sensitive to apoptosis. A representative result is shown in Fig. 5A. The three lines expressing T1-708P43L/K45N and the three lines expressing T1-708D44E/G47R all remained sensitive. Representative results are shown in Fig. 5B. These results suggested that an intact J domain may not be needed for cells to be rendered sensitive to apoptosis. However, examination of other J domain mutants provided an additional perspective. The majority (seven of nine) of the individual clonally derived cell lines expressing T1-708H42Q were resistant to 5-FU-induced apoptosis. For cells expressing T83-708, considerable clonal variation in responses was noted (Fig. 5C). Approximately half (7 of 15) of the cell lines expressing this T antigen were sensitized to apoptosis, and half were not. Each clonal line was tested multiple times and, in general, each time the response was consistent. Occasionally, however, a specific clonal line expressing T1-708H42Q (1 of 9 lines) or T83-708 (1 of 15 lines) showed variation (intraclonal) in responses during repeated assays. An example of such intraclonal variation is shown in Fig. 5C for clone 3.
FIG. 5.
Effects of specific mutations in the J domain on sensitization to apoptosis. Nucleosomal DNA ladder assays were performed with cell lines expressing T antigen containing the amino acid substitution D44N (REF/T1-708D44N+Ras) or H42Q (REF/T1-708H42Q+Ras) (A) or P43L/K45N (left) or D44E/G47R (right) (B) or T antigen missing the J domain (REF/T83-708+Ras) (C) after treatment with the concentrations of 5-FU indicated or after no treatment. (D) Steady-state level of wild-type T antigen (REF/T1-708+Ras) or T83-708 protein (REF/T83-708+Ras) in individual clonally derived cell lines, as determined by immunoprecipitation (IP) and Western blot analysis. cl, clone; sen, sensitive; res, resistant.
The possibility that resistant cell lines expressed a lower steady-state level of T antigen than did sensitive cell lines was examined by immunoprecipitation and Western blot analyses. Representative results for cell lines expressing T83-708 are shown in Fig. 5D. As expected, T antigen in the REF/T1-708+Ras cell line was immunoprecipitated by a monoclonal antibody (PAb 902) which recognizes an epitope in the N terminus and a monoclonal antibody (PAb 901) which recognizes an epitope in the C terminus of the protein. The T antigen in REF/T83-708+Ras lines was immunoprecipitated by the C-terminus-specific antibody only. The three REF/T83-708+Ras cell lines shown in Fig. 5 each accumulated amounts of T83-708 approximately equal to or greater than the amounts immunoprecipitated from extracts of REF/T1-708+Ras. However, one of the lines (clone 1) was resistant, one (clone NA3) was sensitive, and one (clone 3) was variable in its response to 5-FU. Similarly, the difference in sensitization between cell lines expressing T1-708D44N and T1-708H42Q did not correlate with the level of the T antigen in the cells (data not shown). The finding that specific mutations in the J domain can prevent sensitization of the mutant T-antigen-expressing cells to apoptosis suggests that an activity within or dependent on the integrity of this region is involved in the sensitization process. Possible reasons for the clonal variation are addressed in the Discussion.
During the examination of cells expressing various T antigens for apoptosis, the responses of cultures treated with 5-FU were compared to those of untreated cultures. As expected, no nucleosomal DNA ladders were observed in low-molecular-weight extracts of untreated cells, except in two instances. Nucleosomal DNA ladders were observed in extracts of untreated cells expressing T antigens with an amino acid substitution (T1-708E107K) or a deletion (Tdl105- 108) in the Rb-binding region (Fig. 3) or the double amino acid substitution P43L/K45N or D44E/G47R in the J domain (Fig. 5). This basal level of apoptosis was observed consistently in repeated assays and for all clonal lines expressing these T antigens (Table 1 shows the numbers of lines tested). The observation that apoptosis could be detected in untreated cultures suggested that specific alterations in the N terminus of T antigen promote low levels of apoptosis in the absence of 5-FU treatment. Ladders formed by low-molecular-weight DNA extracts from untreated cultures were less intense and composed of higher-molecular-weight DNA fragments than ladders formed by extracts from their treated counterparts. The increase in the intensity of lower-molecular-weight DNA bands following treatment with 5-FU indicated that cells expressing the mutant T antigens, like cells expressing wild-type T antigen, were sensitized to apoptosis.
Sensitization of cells expressing T251-708 to apoptosis following 5-FU treatment.
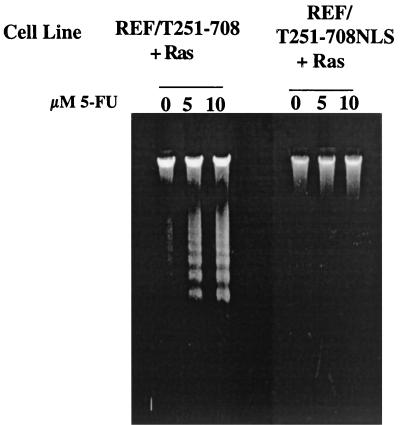
The results presented in Fig. 4 indicated that N-terminal segment T1-127 was sufficient to sensitize cells to apoptosis following 5-FU treatment. The expectation derived from that observation was that a C-terminal T-antigen segment devoid of that region would have no effect on sensitization. As shown in Fig. 6, however, cells expressing the C-terminal segment T251-708 (REF/T251-708+Ras) also were sensitized. This result suggested that the C-terminal portion of T antigen contains an independent activity responsible for sensitization.
FIG. 6.
Effect of subcellular localization of T antigen on sensitization to apoptosis. Nucleosomal ladder assays were performed as described in the legend to Fig. 1 with REF lines expressing Ras and C-terminal fragment T251-708 (REF/T251-708+Ras) or T251-708 containing the SV40 NLS between amino acids 650 and 651 (REFT251-708NLS+Ras) after treatment with the amounts of 5-FU indicated.
The T251-708 protein accumulates in the cell cytoplasm (56), whereas full-length T antigen and the N-terminal T-antigen segments accumulate in the nucleus. In order to determine whether the subcellular localization of the C-terminal T-antigen segment would influence its ability to sensitize cells, REF were cotransformed with Ras and T251-708 into which the SV40 NLS had been inserted between residues 650 and 651 (T251-708NLS) (9, 56), and the resulting cell lines were examined for their responses to 5-FU treatment. The results presented in Fig. 6 show that cells expressing T251-708NLS did not undergo apoptosis following treatment with 5-FU. Thus, the function or functions in T251-708 that sensitize cells to apoptosis were only active when the protein was expressed in an abnormal cellular location. The finding that T251-708NLS did not sensitize cells to apoptosis is of key importance in interpreting the results of the genetic analyses assessing the role of N-terminal T-antigen activities in sensitization. T antigens with mutations in the Rb-binding motif or the J domain accumulate in the nucleus. Since the C-terminal sensitization activity depends on the abnormal localization of T251-708 in the cytoplasm, the responses of treated cell lines expressing the full-length nuclear mutant T antigens to 5-FU directly reflect the involvement of the N-terminal activity. Immunoprecipitation and Western blot analyses (data not shown) confirmed that the REF/T251-708+Ras and REF/T251-708NLS+Ras lines each expressed steady-state levels of the truncated T antigen equivalent to or higher than the level expressed by the REF/T1-708+Ras lines.
5-FU-induced apoptosis in T-antigen-expressing cells occurs through a p53-dependent pathway.
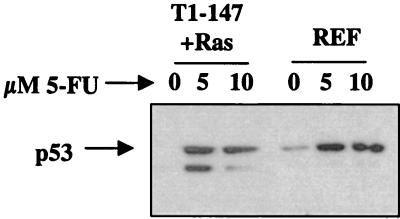
Treatment with genotoxic agents is known to increase intracellular p53 levels and thereby initiate the p53-dependent apoptosis pathway. Figure 7 shows that the steady-state levels of p53 increase in primary REF cultures and REF lines expressing T1-147 following treatment with 5-FU. The demonstration of increased levels of p53 in extracts from treated cells expressing T1-708 was complicated by the stabilization of p53 through binding to T antigen. Binding of p53 to wild-type T antigen interferes with the ability of p53 to activate transcription (3, 15). Presumably, an increase in the level of unbound p53 would be required to transactivate p53-responsive genes. An increase in the level of unbound p53 following treatment could not be demonstrated consistently by immunoprecipitation and Western blot analyses, even after multiple sequential immunoprecipitations with an anti-T-antigen monoclonal antibody to deplete the lysate of T-antigen-p53 complexes prior to immunoprecipitation with anti-p53 antibody (data not shown).
FIG. 7.
Steady-state levels of p53 following treatment of REF or REF lines expressing N-terminal segment T1-147 and Ras (T1-147+Ras) with 5-FU. Cells were treated overnight with the indicated amounts of 5-FU. Protein extracts were prepared, and p53 was immunoprecipitated from 2 mg of total cell lysate by using PAb 421. Immunoprecipitates were subjected to SDS- 10% PAGE and transferred to PVDF membranes, and p53 was detected by using PAb 421. The migration position of p53 is indicated.
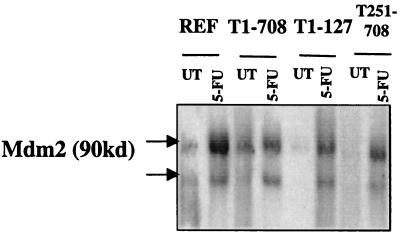
Therefore, transactivation of a gene expressed from a p53-responsive promoter was used as a more sensitive assay for detecting transcriptionally active p53 in T-antigen-expressing cells following 5-FU treatment. The results presented in Fig. 8 show that in REF and in cells expressing wild-type T antigen, T1-127, or T251-708, the steady-state levels of the p53-responsive gene product MDM2 increased following treatment. The increased levels of MDM2 indicated that active p53 was present in the cell lines and suggested that apoptosis was occurring through the p53-dependent pathway.
FIG. 8.
Levels of the p53-responsive gene product MDM2 following treatment of REF and REF lines expressing wild-type T antigen (T1-708), N-terminal fragment T1-127, or C-terminal fragment T251-708. REF or REF lines expressing the T antigens indicated were either treated overnight with 10 μM 5-FU (5-FU) or left untreated (UT). Protein extracts were prepared, and MDM2 was immunoprecipitated from 300 μg of lysate by using Ab-2. Immunoprecipitates were subjected to SDS- 10% PAGE, proteins were transferred to PVDF membranes, and MDM2 was detected by using Ab-1. The migration positions of the MDM2 proteins are indicated.
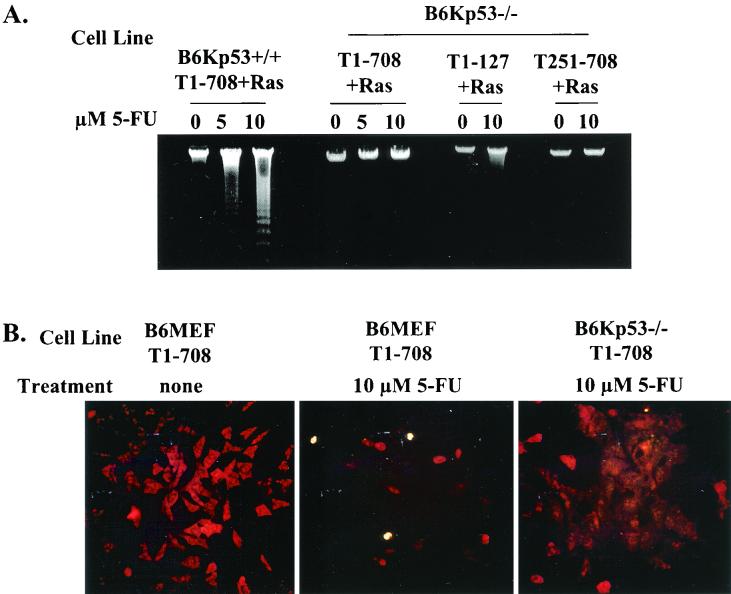
In order to explore the expectation that 5-FU induced apoptosis in T-antigen-expressing cells through the p53-dependent apoptosis pathway, primary kidney fibroblasts were generated from p53-null mice and normal mice. These primary cultures were cotransformed with T1-708 and Ras, T1-127 and Ras, or T251-708 and Ras. Each of the cell lines generated expressed the expected T antigens (data not shown). The cell lines were treated with 5-FU and examined for evidence of apoptosis by two methods. The results presented in Fig. 9A show that low-molecular-weight DNA extracts of p53-null cells expressing T1-708 (B6Kp53−/−/T1-708+Ras), T1-127 (B6Kp53−/−/T1-127+Ras), or T251-708 (B6Kp53−/−/T251-708+Ras) did not contain nucleosomal DNA fragments, whereas B6Kp53+/+ cells expressing T1-708 and Ras did. The results presented in Fig. 9B confirm the absence of apoptosis in p53-null cells expressing wild-type T antigen in the in situ TUNEL assay. Untreated and treated B6MEF expressing wild-type T antigen were used as negative and positive controls, respectively, for the TUNEL assay.
FIG. 9.
Sensitization of cells to 5-FU-induced apoptosis occurs through a p53-dependent process. (A) Nucleosomal ladder assays for apoptosis in p53 wild-type cells expressing T1-708 (B6Kp53+/+/T1-708+Ras) and p53-null cells expressing T1-708 (B6Kp53−/−/T1-708+Ras), T1-127 (B6Kp53−/−/T1-127+Ras), or T251-708 (B6Kp53−/−/T251-708+Ras) after overnight treatment with the indicated amounts of 5-FU. (B) TUNEL assays for apoptosis in B6MEF expressing wild-type T antigen (B6MEF/T1-708) and either left untreated (none) or treated overnight with the indicated amount of 5-FU and in p53-null cells expressing wild-type T antigen (B6Kp53−/−/T1-708+Ras) and treated overnight with the indicated amount of 5-FU.
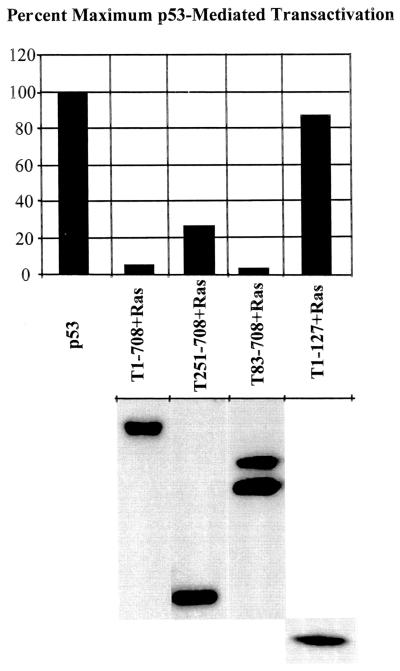
C-terminal T-antigen segments retain the ability to inactivate p53.
The observation that T251-708-mediated sensitization to 5-FU-induced apoptosis involved the p53 pathway raised the question as to whether this C-terminal T-antigen fragment is capable of inhibiting the transcriptional transactivating activity of p53. It was shown previously that T251-708 binds p53 (9). However, the possibility that binding did not eliminate p53-mediated transactivation could not be ruled out. Therefore, SAOS-2 (p53-null) cells were transiently transfected with the p53-responsive reporter p50-2luc, a p53-expressing vector, and various T-antigen-expressing plasmids. Transactivation was monitored as relative light units generated and was expressed as a percentage of the maximum value (relative light units from extracts of cells transfected with the reporter and the p53-expressing vector). The results presented in Fig. 10 (upper panel) show that full-length T antigen (T1-708), a C-terminal T-antigen segment that accumulates in the nucleus (T83-708), and T251-708 suppressed p53-mediated transactivation. The N-terminal segment T1-127, which is incapable of binding p53, did not suppress transactivation. The results presented in Fig. 10 (lower panel) show that the steady-state levels of the T antigens in extracts of parallel cultures were similar.
FIG. 10.
Segment T251-708 retains the ability to inhibit p53-mediated transactivation in transient transfection assays. The p53-null cell line SAOS-2 was transfected with wild-type p53- and Bcl-2-expressing plasmids for the T antigens indicated and the p53-responsive reporter p50-2luc. Forty-eight hours later, samples were assayed for luciferase activity. The results (upper panel) are presented as the percentage of activity obtained in cells transfected with p53- and Bcl-2-expressing plasmids and the p53-responsive reporter. The Bcl-2-expressing plasmid was included in all transfections to eliminate the possible loss of cells through p53-mediated apoptosis (5). The lower panel shows the steady-state levels of T antigens in parallel transfected cultures as determined by immunoprecipitation and Western blot analysis.
DISCUSSION
In investigating the ability of T antigen to modulate apoptosis, we found that the expression of full-length T antigen (T1-708), the N-terminal segment T1-127, and the C-terminal segment T251-708 each sensitized REF to apoptosis following treatment with the chemotherapeutic agent 5-FU. These finding indicated that one or more activities in the N terminus of T antigen are sufficient to sensitize cells and that, in cells expressing wild-type T antigen and the C-terminal segment, the presence of the region with limited homology to Bcl-2 (amino acids 525 to 541) (11) and the p53- and p300-binding regions (amino acids 351 to 450 and 533 to 626) (23) is not sufficient to prevent the chemosensitization.
The finding that cells expressing T1-708 and T251-708 responded to 5-FU treatment by undergoing apoptosis was not anticipated. In normal human cells in cultures, the induction of apoptosis following 5-FU treatment occurs through a p53-dependent pathway. Targeted disruption of the p53 gene in these cells prevents 5-FU-induced apoptosis (7). Thus, the expectation underlying the experiments reported here was that T1-708, by virtue of binding and inactivating the transcriptional transactivating property of p53, would prevent 5-FU-induced apoptosis. We considered three possibilities regarding the ability of a T antigen with an intact p53-binding region to sensitize cells to 5-FU-induced apoptosis. First, apoptosis may occur through a p53-independent process. Studies have shown that cells expressing T antigen undergo apoptosis following growth factor withdrawal (11, 46) or asbestos-mediated DNA damage (25) through a p53-independent process. Second, transiently elevated levels of unbound p53 may be sufficient to initiate the p53-dependent apoptosis pathway and allow its progression to a point at which apoptosis cannot be reversed. Third, p53 bound to T antigen may retain an activity involved in apoptosis induction.
We showed that 5-FU-induced apoptosis in cells expressing T1-708, T1-127, or T251-708 occurred through a p53-dependent pathway. Whereas normal C57BL/6 kidney cells expressing T1-708 were sensitive to 5-FU-induced apoptosis, p53-null C57BL/6 kidney cells expressing T antigens were resistant. These results suggested that activated p53 is present in 5-FU-treated REF expressing T antigens in spite of the ability of T1-708 and T251-708 to bind the tumor suppressor protein. In order to explore this possibility, we examined the level of endogenous MDM2 protein in T-antigen-expressing cells before and after 5-FU treatment. Increased levels of MDM2 were observed, as expected, in REF and cells expressing T1-127, which does not contain the p53-binding region. MDM2 levels were also increased in cells expressing T1-708, T1-127, or T251-708. This result indicated that transcriptionally active p53 was at least transiently available to transactivate responsive endogenous genes following treatment.
Our finding that cells expressing the C-terminal fragment T251-708 also responded to 5-FU treatment by undergoing apoptosis was unexpected in light of the finding that the N-terminal fragment T1-127 was sufficient to sensitize cells. As was the case with full-length T antigen, sensitization occurred through a p53-dependent pathway, and T251-708 inactivated the transcriptional transactivating property of p53 in transient transfection assays; however, in cells expressing T251-708, treatment with 5-FU increased the level of p53-dependent endogenous MDM2. These results indicated that either an independent apoptosis function is located in the C-terminal fragment or its abnormal accumulation in the cell cytoplasm is involved in sensitization or both. To investigate the modulating effect of the subcellular localization of the C-terminal fragment on apoptosis, we generated cell lines expressing T251-708NLS. This T-antigen fragment contains an SV40 NLS inserted between amino acids 650 and 651 and accumulates in the nucleus (56). Cells expressing T251-708NLS were resistant to 5-FU-induced apoptosis.
The results raised interrelated questions as to how endogenous genes such as the MDM2 gene and presumably proapoptosis genes can be transactivated even though T1-708 and T251-708 effectively limit the transcriptional transactivating capability of p53 in transient transfection assays and why T251-708, when localized to the nucleus, does not sensitize cells. It is known that newly synthesized p53 does not associate efficiently with T antigen (8, 23, 24). Coupled with the rapid transport of p53 to the nucleus following 5-FU treatment, sufficient p53 may reach target genes to signal the onset of apoptosis before T antigen-p53 binding occurs. It is likely that T251-708, by virtue of its cytoplasmic location, cannot inactivate nuclear p53, whereas T251-708NLS can and in that way acts as an apoptosis survival factor. Importantly, the finding that T251-708NLS does not sensitize cells to apoptosis validates the conclusion that activities in the N terminus of T antigen are involved in the sensitization of cells expressing the full-length protein.
The finding that an N-terminal T-antigen segment (T1-127) was sufficient to sensitize cells to 5-FU-induced apoptosis implicated activities in this region of the protein in the sensitization process. We examined the responses of cells expressing full-length T antigens with mutations in the J domain (amino acids 1 to 82) or in the LXCXE motif (amino acids 103 to 107) of the Rb-binding region to 5-FU treatment. Mutant T antigens that could not bind Rb still sensitized cells to apoptosis, indicating that binding Rb in the context of full-length T antigen was not required. This result is in contrast to results obtained by Samuelson and Lowe (42) for the E1A protein. Sensitization to apoptosis induced by genotoxic agents in cells expressing E1A depends on the ability of E1A to bind both Rb and the p53 coactivator p300 (42). Sensitization by full-length T antigen, however, was independent of Rb binding. Similarly, mutant T antigens that were defective in p53 and p300 binding still sensitized cells to 5-FU-induced apoptosis. This observation adds to the body of evidence indicating that, although E1A and T antigen contain similar activities, the consequences of the activities in the contexts of the two proteins differ.
Genetic analysis of the T-antigen J domain revealed a complex, mutant-specific response. The J domain has an impact on multiple T-antigen functions. Each of the mutants used has been tested for multiple J domain-dependent activities. For instance, in the context of full-length T antigen, the amino acid substitutions H42Q and D44N inactivate multiple J domain functions, including the promotion of increased cell saturation density and growth in low-serum medium (50), override of Rb/p107/p130-mediated growth arrest (64), relief of Rb/p130-dependent E2F repression (18, 64), and disruption of p130-E2F DNA-binding complexes (64). In addition, the D44N mutation prevents T-antigen- Hsc70 complex formation (51). The double amino acid substitutions at positions 43 and 45 or 44 and 47 in the context of full-length T antigen inhibit the formation of dense foci on human cells unless complemented by small t antigen (38). Finally, it was shown recently that full-length T antigens containing the mutations H42Q, D44N, P43L/K45N, and D44E/G47R lose the J domain-dependent function required to transactivate the cyclin A promoter (5, 35). However, the P43L/K45N and D44E/G47R mutants retain the ability to override Rb-mediated growth arrest (5), while T antigens containing either the D44N or the H42Q substitution lose both activities (5, 64). Thus, the J domain may contain mutationally separable activities or, alternatively, activities that overlap, and each of the J domain mutations used in this investigation inactivated at least one of them.
Clonally derived cell lines expressing T1-708D44N, T1-708P43L/K45N, or T1-708D44E/G47R uniformly were sensitized to apoptosis following 5-FU treatment. However, most clonally derived cell lines expressing T1-708H42Q were resistant. Cell lines expressing T83-708 showed variation in their responses to 5-FU treatment. Approximately half of the lines examined were sensitive, and half were resistant. Such responses would suggest that sensitization was partially dependent on the J domain. The partial dependence of T-antigen activities on an intact J domain was demonstrated previously for both SV40 and polyomavirus large T antigens (18, 44). Nonetheless, since resistant cell lines were consistently observed when specific mutations in the J domain were present, we concluded that this region was involved either structurally or functionally in conferring sensitivity to apoptosis following 5-FU treatment. The finding that specific mutations in the J domain inactivated an activity dependent on the integrity of the J domain while other mutations did not was expected. It is well recognized that the penetrance displayed by J domain mutants is variable (6).
The variation between clonally derived cell lines expressing the same mutant T antigen was less easily explained. The variation did not relate to the levels of T antigens expressed in the individual lines. It is possible that the key function involved in sensitizing cells to apoptosis lies outside of the J domain and is differentially affected by specific mutational distortions of the J domain. Since the cell lines showing variable responses were derived from individual cell clones, it is also possible that an interaction of T antigen with a cellular protein is involved and that the level of the cellular protein or the ability of the mutant T antigen to interact with it dictates the response to genotoxic damage. One cellular protein that may influence the ability of T-antigen to sensitize cells to apoptosis is the recently described proapoptotic protein p193. Overexpression of p193 leads to apoptosis (60). T antigen binds p193 and prevents p193-mediated apoptosis. Using in vitro transcribed and translated products, Tsai et al. (60) showed that a fragment containing T-antigen amino acids 1 to 92 (T1- 92) did not bind p193, whereas T1-147 did. These results tentatively mapped the p193-binding site to between T-antigen amino acids 92 and 147. Whether the N-terminal 92 T-antigen amino acids are required for binding was not determined. Clearly, binding of p193 to full-length T antigen does not prevent T-antigen-mediated sensitization to 5-FU-induced apoptosis. It remains possible, however, that the levels of p193 differ among clonally derived cell lines or that specific mutations within the J domain render T antigen more or less effective in binding the cellular apoptosis-promoting protein. For instance, alterations in the J domain may inhibit T-antigen-mediated sensitization to apoptosis only if binding to p193 remains undisturbed. Additional investigations will be needed to explore the relationship among the J domain, sensitization to apoptosis, and p193 binding.
The possibility also remains that the p53-dependent apoptosis observed following exposure of T-antigen-expressing cells to 5-FU occurs independently of p53-mediated transactivation. Recently, Matas et al. (32) showed that p53-mediated apoptosis following DNA damage can occur via a transcription-independent pathway. The pathway is presumably silent under normal growing conditions. An additional explanation can be formulated. It is possible that p53, in its T-antigen-bound form, although unable to transactivate p53-responsive reporters, retains other p53 activities. Several investigations into the relationship between p53 activities and apoptosis have indicated that p53-mediated repression may be more closely tied to apoptosis than is p53-mediated transactivation (63 and references therein). Inhibition of p53-mediated transrepression by T antigen has not been investigated in detail.
It should be pointed out that although both T1-127 and cytoplasmic T251-708 independently sensitized cells to apoptosis, the roles of the J domain, Rb-binding motif, and p53- and p300-binding regions were examined only in the context of full-length T antigen. The experimental strategy used in the experiments reported here depended on generating cell lines by way of cotransformation with Ras. It was shown previously that N-terminal T-antigen segments with mutations in either the J domain or the Rb-binding motif will not cooperate with Ras (5). Similarly, C-terminal T-antigen segments with mutations that have been reported to decrease p53 binding do not cooperate with Ras (5). Thus, alternative strategies will be needed to investigate the role of T-antigen activities in the context of truncated T antigens.
Recently, Gjoerup et al. (19) presented evidence that small t antigen induces apoptosis in specific cell types. The various biological activities of small t antigen correlate with either its J domain or its second major activity, binding protein phosphatase 2A (reviewed in reference 40). The J domains of large T and small t antigens are encoded in the same reading frame of the SV40 genome and are identical in amino acid sequence. J domain mutations introduced into the SV40 genome will result in identical amino acid substitutions or deletions in both large T and small t antigens. However, small t antigen does not play a major role in sensitizing REF to 5-FU-induced apoptosis, as cells expressing only large T antigen were sensitized. In addition, induction of apoptosis by small t antigen correlates with protein phosphatase 2A binding (19), an activity not located in the J domain of large T antigen. Also, small-t-antigen-induced apoptosis occurs through a p53-independent process, whereas the large-T-antigen-mediated sensitization described here is p53 dependent.
Acknowledgments
This work was supported by grant CA 24964 from the National Cancer Institute of the National Institutes of Health.
The excellent technical assistance of Holly K. Lacko is gratefully acknowledged.
REFERENCES
- 1.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Bargonetti, J., I. Reynisdottir, P. N. Friedman, and C. Prives. 1992. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53 Genes Dev. 6:1886-1898. [DOI] [PubMed]
- 4.Bates, S., and K. H. Vousden. 1999. Mechanisms of p53-mediated apoptosis. Cell. Mol. Life Sci. 55:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beachy, T. M., S. L. Cole, J. F. Cavender, and M. J. Tevethia. 2002. Regions and activities of simian virus 40 T antigen that cooperate with an activated ras oncogene in transforming primary rat embryo fibroblasts. J. Virol. 76:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky, J. L., and J. M. Pipas. 1998. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol. 72:5329-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunz, F., P. M. Hwang, C. Torrance, T. Waldman, Y. Zhang, L. Dillehay, J. Williams, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Investig. 104:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, R. B., and E. G. Gurney. 1982. Time-dependent maturation of the simian virus 40 large T antigen-p53 complex studied by using monoclonal antibodies. J. Virol. 44:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavender, J. F., A. Conn, M. Epler, H. Lacko, and M. J. Tevethia. 1995. Simian virus 40 large T antigen contains two independent activities that cooperate with a ras oncogene to transform rat embryo fibroblasts. J. Virol. 69:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavender, J. F., C. Mummert, and M. J. Tevethia. 1999. Transactivation of a ribosomal gene by simian virus 40 large-T antigen requires at least three activities of the protein. J. Virol. 73:214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conzen, S. D., C. A. Snay, and C. N. Cole. 1997. Identification of a novel antiapoptotic functional domain in simian virus 40 large T antigen. J. Virol. 71:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickmanns, A., A. Zeitvogel, F. Simmersbach, R. Weber, A. K. Arthur, S. Dehde, A. G. Wildeman, and E. Fanning. 1994. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J. Virol. 68:5496-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobbelstein, M., A. K. Arthur, S. Dehde, K. van Zee, A. Dickmanns, and E. Fanning. 1992. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene 7:837-847. [PubMed] [Google Scholar]
- 14.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farmer, G., J. Bargonetti, H. Zhu, P. Friedman, R. Prywes, and C. Prives. 1992. Wild-type p53 activates transcription in vitro. Nature 358:83-86. [DOI] [PubMed] [Google Scholar]
- 16.Fowlis, D. J., and A. Balmain. 1993. Oncogenes and tumour suppressor genes in transgenic mouse models of neoplasia. Eur. J. Cancer 4:638-645. [DOI] [PubMed] [Google Scholar]
- 17.Fromm, L., W. Shawlot, K. Gunning, J. S. Butel, and P. A. Overbeek. 1994. The retinoblastoma protein-binding region of simian virus 40 large T antigen alters cell cycle regulation in lenses of transgenic mice. Mol. Cell. Biol. 14:6743-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjoerup, O., H. Chao, J. A. DeCaprio, and T. M. Roberts. 2000. pRB-dependent, J domain-independent function of simian virus 40 large T antigen in override of p53 growth suppression. J. Virol. 74:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gjoerup, O., D. Zaveri, and T. M. Roberts. 2001. Induction of p53-independent apoptosis by simian virus 40 small t antigen. J. Virol. 75:9142-9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour, J. W., and D. C. Dean. 2000. Rb function in cell-cycle regulation and apoptosis. Nat. Cell Biol. 2:E65-E67. [DOI] [PubMed]
- 21.Harlow, E., L. V. Crawford, D. C. Pim, and N. M. Williamson. 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39:861-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Kierstead, T. D., and M. J. Tevethia. 1993. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J. Virol. 67:1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraiss, S., A. Quaiser, M. Oren, and M. Montenarh. 1988. Oligomerization of oncoprotein p53. J. Virol. 62:4737-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levresse, V., S. Moritz, A. Renier, L. Kheuang, F. Galateau-Salle, J. P. Mege, P. Piedbois, B. Salmons, W. Guenzburg, and M. C. Jaurand. 1998. Effect of simian virus large T antigen expression on cell cycle control and apoptosis in rat pleural mesothelial cells exposed to DNA damaging agents. Oncogene 16:1041-1053. [DOI] [PubMed] [Google Scholar]
- 26.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 27.Lill, N. L., M. J. Tevethia, R. Eckner, D. M. Livingston, and N. Modjtahedi. 1997. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J. Virol. 71:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J. Y., and D. T. Simmons. 1991. Stable T-p53 complexes are not required for replication of simian virus 40 in culture or for enhanced phosphorylation of T antigen and p53. J. Virol. 65:2066-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe, S. W. 1999. Activation of p53 by oncogenes. Endocr. Relat. Cancer 6:45-48. [DOI] [PubMed] [Google Scholar]
- 30.Lowe, S. W., H. E. Ruley, T. Jacks, and D. E. Housman. 1993. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74:957-967. [DOI] [PubMed] [Google Scholar]
- 31.Manfredi, J. J., and C. Prives. 1994. The transforming activity of simian virus 40 large tumor antigen. Biochim. Biophys. Acta 1198:65-83. [DOI] [PubMed] [Google Scholar]
- 32.Matas, D., A. Sigal, P. Stambolsky, M. Milyavsky, L. Weisz, D. Schwartz, N. Goldfinger, and V. Rotter. 2001. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCurrach, M. E., T. M. Connor, C. M. Knudson, S. J. Korsmeyer, and S. W. Lowe. 1997. bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc. Natl. Acad. Sci. USA 94:2345-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgenbesser, S. D., B. O. Williams, T. Jacks, and R. A. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72-74. [DOI] [PubMed] [Google Scholar]
- 35.Oshima, J., K. E. Steinmann, J. Campisi, and R. Schlegel. 1993. Modulation of cell growth, p34cdc2 and cyclin A levels by SV-40 large T antigen. Oncogene 8:2987-2993. [PubMed] [Google Scholar]
- 36.Peden, K. W., and J. M. Pipas. 1992. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes 6:107-118. [DOI] [PubMed] [Google Scholar]
- 37.Pipas, J. M. 1998. Molecular chaperone function of the SV40 large T antigen. Dev. Biol. Stand. 94:313-319. [PubMed] [Google Scholar]
- 38.Porras, A., J. Bennett, A. Howe, K. Tokos, N. Bouck, B. Henglein, S. Sathyamangalam, B. Thimmapaya, and K. Rundell. 1996. A novel simian virus 40 early-region domain mediates transactivation of the cyclin A promoter by small-t antigen and is required for transformation in small-t-antigen-dependent assays. J. Virol. 70:6902-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quartin, R. S., C. N. Cole, J. M. Pipas, and A. J. Levine. 1994. The amino-terminal functions of the simian virus 40 large T antigen are required to overcome wild-type p53-mediated growth arrest of cells. J. Virol. 68:1334-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rundell, K., and R. Parakati. 2001. The role of the SV40 ST antigen in cell growth promotion and transformation. Semin. Cancer Biol. 11:5-13. [DOI] [PubMed] [Google Scholar]
- 41.Saenz Robles, M. T., H. Symonds, J. Chen, and T. Van Dyke. 1994. Induction versus progression of brain tumor development: differential functions for the pRB- and p53-targeting domains of simian virus 40 T antigen. Mol. Cell. Biol. 14:2686-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samuelson, A. V., and S. W. Lowe. 1997. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl. Acad. Sci. USA 94:12094-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawai, E. T., G. Rasmussen, and J. S. Butel. 1994. Construction of SV40 deletion mutants and delimitation of the binding domain for heat shock protein to the amino terminus of large T-antigen. Virus Res. 31:367-378. [DOI] [PubMed] [Google Scholar]
- 44.Sheng, Q., T. M. Love, and B. Schaffhausen. 2000. J domain-independent regulation of the Rb family by polyomavirus large T antigen. J. Virol. 74:5280-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleigh, M. J., W. C. Topp, R. Hanich, and J. F. Sambrook. 1978. Mutants of SV40 with an altered small t protein are reduced in their ability to transform cells. Cell 14:79-88. [DOI] [PubMed] [Google Scholar]
- 46.Slinskey, A., D. Barnes, and J. M. Pipas. 1999. Simian virus 40 large T antigen J domain and Rb-binding motif are sufficient to block apoptosis induced by growth factor withdrawal in a neural stem cell line. J. Virol. 73:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sock, E., J. Enderich, and M. Wegner. 1999. The J domain of papovaviral large tumor antigen is required for synergistic interaction with the POU-domain protein Tst-1/Oct6/SCIP. Mol. Cell. Biol. 19:2455-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stubdal, H., J. Zalvide, and J. A. DeCaprio. 1996. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J. Virol. 70:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan, C. S., S. P. Gilbert, and J. M. Pipas. 2001. ATP-dependent simian virus 40 T-antigen- Hsc70 complex formation. J. Virol. 75:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Symonds, H., L. Krall, L. Remington, M. Saenz Robles, T. Jacks, and T. Van Dyke. 1994. p53-dependent apoptosis in vivo: impact of p53 inactivation on tumorigenesis. Cold Spring Harbor Symp. Quant. Biol. 59:247-257. [DOI] [PubMed] [Google Scholar]
- 53.Symonds, H., L. Krall, L. Remington, M. Saenz-Robles, S. Lowe, T. Jacks, and T. Van Dyke. 1994. p53-dependent apoptosis suppresses tumor growth and progression in vivo. Cell 78:703-711. [DOI] [PubMed] [Google Scholar]
- 54.Teodoro, J. G., G. C. Shore, and P. E. Branton. 1995. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene 11:467-474. [PubMed] [Google Scholar]
- 55.Tevethia, M. J., R. H. Bonneau, J. W. Griffith, and L. Mylin. 1997. A simian virus 40 large-T-antigen segment containing amino acids 1 to 127 and expressed under the control of the rat elastase 1 promoter produces pancreatic acinar carcinomas in transgenic mice. J. Virol. 71:8157-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tevethia, M. J., H. A. Lacko, and A. Conn. 1998. Two regions of simian virus 40 large T-antigen independently extend the life span of primary C57BL/6 mouse embryo fibroblasts and cooperate in immortalization. Virology 243:303-312. [DOI] [PubMed] [Google Scholar]
- 57.Tevethia, M. J., H. A. Lacko, T. D. Kierstead, and D. L. Thompson. 1997. Adding an Rb-binding site to an N-terminally truncated simian virus 40 T antigen restores growth to high cell density, and the T common region in trans provides anchorage-independent growth and rapid growth in low serum concentrations. J. Virol. 71:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tevethia, M. J., and H. L. Ozer. 2001. SV40-mediated immortalization. Methods Mol. Biol. 165:185-199. [DOI] [PubMed] [Google Scholar]
- 59.Tevethia, M. J., J. M. Pipas, T. Kierstead, and C. Cole. 1988. Requirements for immortalization of primary mouse embryo fibroblasts probed with mutants bearing deletions in the 3′ end of SV40 gene A. Virology 162:76-89. [DOI] [PubMed] [Google Scholar]
- 60.Tsai, S. C., K. B. Pasumarthi, L. Pajak, M. Franklin, B. Patton, H. Wang, W. J. Henzel, J. T. Stults, and L. J. Field. 2000. Simian virus 40 large T antigen binds a novel Bcl-2 homology domain 3-containing proapoptosis protein in the cytoplasm. J. Biol. Chem. 275:3239-3246. [DOI] [PubMed] [Google Scholar]
- 61.Viney, J. L. 1995. Transgenic and gene knockout mice in cancer research. Cancer Metastasis Rev. 14:77-90. [DOI] [PubMed] [Google Scholar]
- 62.White, E., and B. Stillman. 1987. Expression of adenovirus E1B mutant phenotypes is dependent on the host cell and on synthesis of E1A proteins. J. Virol. 61:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yap, D. B., J. K. Hsieh, F. S. Chan, and X. Lu. 1999. mdm2: a bridge over the two tumour suppressors, p53 and Rb. Oncogene 18:7681-7689. [DOI] [PubMed] [Google Scholar]
- 64.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]