Abstract
The early region 1A (E1A) gene is the first gene expressed after infection with adenovirus and has been most extensively characterized in human adenovirus type 5 (hAd5). The E1A proteins interact with numerous cellular regulatory proteins, influencing a variety of transcriptional and cell cycle events. For this reason, these multifunctional proteins have been useful as tools for dissecting pathways regulating cell growth and gene expression. Despite the large number of studies using hAd5 E1A, relatively little is known about the function of the E1A proteins of other adenoviruses. In 1985, a comparison of E1A sequences from three human and one simian adenovirus identified three regions with higher overall levels of sequence conservation designated conserved regions (CR) 1, 2, and 3. As expected, these regions are critical for a variety of E1A functions. Since that time, the sequences of several other human and simian adenovirus E1A proteins have been determined. Using these, and two additional sequences that we determined, we report here a detailed comparison of the sequences of 15 E1A proteins representing each of the six hAd subgroups and several simian adenoviruses. These analyses refine the positioning of CR1, 2, and 3; define a fourth CR located near the carboxyl terminus of E1A; and suggest several new functions for E1A.
The adenoviruses are a family of small nonenveloped viruses with double-stranded DNA genomes of approximately 35 kbp. These viruses infect a wide range of animal species, and 47 human adenovirus (hAd) serotypes have been isolated and grouped according to their abilities to agglutinate red blood cells (62). Forty years ago, hAd type 12 (hAd12) was shown to induce tumors when injected into newborn hamsters, providing the first evidence that a human virus could be oncogenic (71). Since then, adenoviruses have been tremendously useful for studying oncogenic transformation, cell cycle control, DNA replication, transcription, mRNA processing, immunological response, and apoptosis (62). In addition, adenoviruses are popular choices as vectors for gene therapy applications, given their ease of construction and growth and their ability to infect a diverse range of cell types (29).
The leftmost adenoviral gene, termed early region 1A (E1A), is the first gene expressed after infection and has been most extensively characterized in hAd5. In hAd5, the E1A gene encodes two major proteins of 289 and 243 residues that are expressed early after infection. These proteins arise from differential splicing of the same transcript and differ only by the presence of an internal sequence of 46 amino acids in the larger protein. The E1A proteins are localized in roughly equal amounts in both the cytoplasm and nucleus (58, 72). Three additional mRNA species are produced at later times that encode, or are predicted to encode, proteins of 217, 171, and 55 amino acids (67, 74). The E1A proteins are essential for a productive viral infection (35), as they activate expression of other viral early genes and reprogram cell growth to provide an optimal environment for viral replication (4).
hAd5 E1A interacts with a variety of cellular proteins, including transcriptional coactivators, such as the CREB binding protein (CBP) and p300 (2, 15, 42), the p300/CBP-associated factor (pCAF) (56), and the transcriptional repressor CtBP (61). E1A also interacts with various components of the general and specific transcriptional machinery, including the TATA-binding protein (TBP) (6, 22, 26, 64), several of the TBP-associated factors (21, 46), and a lengthy list of sequence-specific transcription factors (20). In addition, E1A also targets proteins that directly regulate the cell division cycle, such as the retinoblastoma tumor suppressor gene product (Rb) and the related family members p130 and p107 (16, 18, 25, 81) and the cyclin-dependent kinase inhibitors p21 and p27 (37, 45). Because of these many interactions with cellular regulatory proteins, the multifunctional E1A proteins influence a variety of transcriptional and cell cycle events (4, 14, 20, 49, 54, 63). Importantly, hAd5 E1A can function as an oncogene in rodent cells. Expression of hAd5 E1A alone is sufficient to immortalize primary rodent cells (30) and can fully transform them in cooperation with a second oncogene, such as adenovirus E1B (23) or activated ras (59). In human cells however, E1A can function as a tumor suppressor gene by inhibiting tumorigenesis and metastasis (50) and may have some utility in cancer therapy (73).
Despite the large numbers of studies using hAd5 E1A, relatively little is known about the function of the E1A proteins of other adenoviruses, raising the question of how representative hAd5 E1A is of the other E1A proteins. In 1985, comparison of E1A sequences from three hAds and one simian adenovirus (sAd) identified three regions with higher overall levels of sequence conservation designated conserved regions (CR) 1, 2, and 3 (38). Since that time, the sequences of a number of other human and sAd E1A genes have been determined. Using these, and two additional sequences that we determined, we report here a detailed comparison of 15 E1A proteins representing each of the six hAd subgroups.
MATERIALS AND METHODS
Provenances of virus and clones.
hAd21 was obtained from the College of American Pathologists (Northfield, Ill.) as a proficiency-testing isolate. The virus was initially propagated on monolayer cultures of A549 cells. The construction of plasmid pVM303, which contains the left end of hAd3, was described previously (39).
cDNA synthesis, cloning, and sequence determination.
Total RNA was extracted with Trizol (Sigma Aldrich, Oakville, Ontario, Canada) from human KB cells 6 h postinfection with hAd21 at an approximate multiplicity of infection of 20 PFU per cell. mRNA was subsequently isolated using Oligotex resin (Qiagen Inc., Mississauga, Ontario, Canada) and used as a template for Moloney murine leukemia virus reverse transcriptase (Invitrogen, Burlington, Ontario, Canada) to generate total cDNA. The E1A gene was amplified with primers JMO194 and JMO200 (GCGAATTCTTGAGTGCCAGCGAGTAGAGTTTTCTC and TAGTCGACCACAGCTGCAGGGCAC, respectively) designed to anneal in the highly conserved noncoding regions upstream and downstream of the gene in the subgroup B adenoviruses. The E1A gene was then subcloned as an EcoRI/SalI fragment into pAS1 (12) and sequenced using the flanking primers JMO26 (CATCATCGGAAGAGAGTAG) and JMO61 (CATAAATCATAAGAAATTCGC). Plasmid pVM303 was sequenced using primers JMO200 (described above), JMO188 (TACGAATTCATGAGACACCTGCGCTTC), and JMO201 (CTGCCACTTTATTTACAGTCCTGTGTCTGATGATG). JMO188 and JMO201 anneal to the N-terminal coding region and splice junction of hAd3, respectively. Sequencing was performed by the Robarts Research Institute DNA Sequencing Facility (London, Ontario, Canada).
Sequence manipulation.
Stretches of overlapping and complementary strand sequences from each viral DNA fragment were manually assembled into a coherent sequence. Splice junctions for the largest E1A products of hAd3 were predicted based on BLAST alignment (1) of the nucleotide sequence with the closely related hAd7 nucleotide sequence. The sequences of hAd3 and hAd21 E1As have been deposited in GenBank; additional E1A sequences were obtained from GenBank, and all of the accession numbers are listed in Table 1. The sequence of hAd17 was predicted from the published sequence (10) following BLAST alignment with the related hAd9 nucleotide sequence. Isoelectric points (pI) and amino acid compositions were determined using the ProtParam Tool at the Expert Protein Analysis System website (http://ca.expasy.org/). Alignments of the largest predicted E1A products were performed with CLUSTAL W (69) at the European Molecular Biology Laboratory European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/) using default parameters except that the gap open cost was set to 2. The evolutionary tree (Fig. 1) was displayed using the program TreeView (53). The aligned sequence file produced by CLUSTAL W was imported into GeneDoc (52), edited manually, and shaded to four levels of conservation (Fig. 2). Overall sequence identities and similarities were also calculated using GeneDoc. Sequence identities at each position in the alignment were calculated manually and plotted using Microsoft Excel (Fig. 3). Sequence identities and similarities for various subregions of E1A were calculated with respect to hAd5 using GeneDoc, averaged, and plotted using Microsoft Excel (Fig. 4). Nuclear import sequences (Table 2) were predicted using the program PSORT (51). Secondary-structure predictions for each E1A protein (see Fig. 6) were performed using the program PSIPRED (47).
TABLE 1.
GenBank accession numbers and properties of the largest adenovirus E1A protein products
| Virus | Human subgroup | Accession no. | E1A size (aa)a | pI | mol% Proline | mol% Acidic | mol% Basic |
|---|---|---|---|---|---|---|---|
| hAd2 | C | P03254 | 289 | 4.56 | 15.6 | 17.3 | 7.3 |
| hAd3 | B | AF492352 | 261 | 4.33 | 12.3 | 19.2 | 7.7 |
| hAd4 | E | P10407 | 257 | 4.41 | 9.7 | 20.2 | 8.2 |
| hAd5 | C | P03255 | 289 | 4.56 | 15.9 | 17.3 | 7.3 |
| hAd7-I | B | P03256 | 261 | 4.38 | 12.3 | 18.8 | 8.0 |
| hAd7-II | B | AAA42455 | 261 | 4.37 | 11.9 | 19.2 | 8.0 |
| hAd9 | D | AAD16301 | 251 | 4.09 | 10.0 | 23.1 | 6.8 |
| hAd12 | A | P03259 | 266 | 4.18 | 9.4 | 20.7 | 6.4 |
| hAd17 | D | AF108105 | 253 | 4.10 | 10.7 | 22.1 | 6.3 |
| hAd21 | B | AF492353 | 261 | 4.35 | 11.9 | 19.5 | 8.0 |
| hAd40 | F | P10541 | 249 | 3.99 | 9.2 | 24.1 | 6.0 |
| hAd41 | F | P10542 | 251 | 4.03 | 8.0 | 22.7 | 6.0 |
| sAd7-I | P06499 | 266 | 4.35 | 9.4 | 19.5 | 7.1 | |
| sAd7-II | CAA25511 | 266 | 4.33 | 9.0 | 19.2 | 6.4 | |
| sAd25 | AAL35510 | 257 | 4.38 | 10.5 | 19.8 | 8.6 |
aa, amino acids.
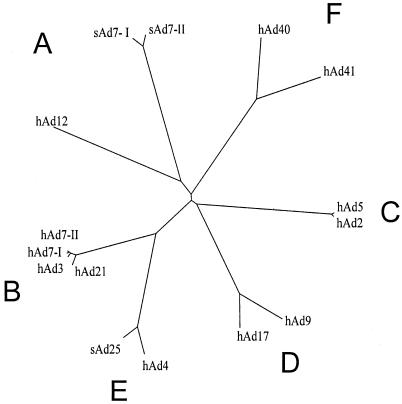
FIG. 1.
Phylogenetic tree for the adenovirus E1A proteins. An unrooted tree was generated for the E1A proteins (Table 1) with the program TreeView. Each species of E1A is labeled at the tip of its representative branch. The adenoviral subgroups are labeled A to F according to convention.
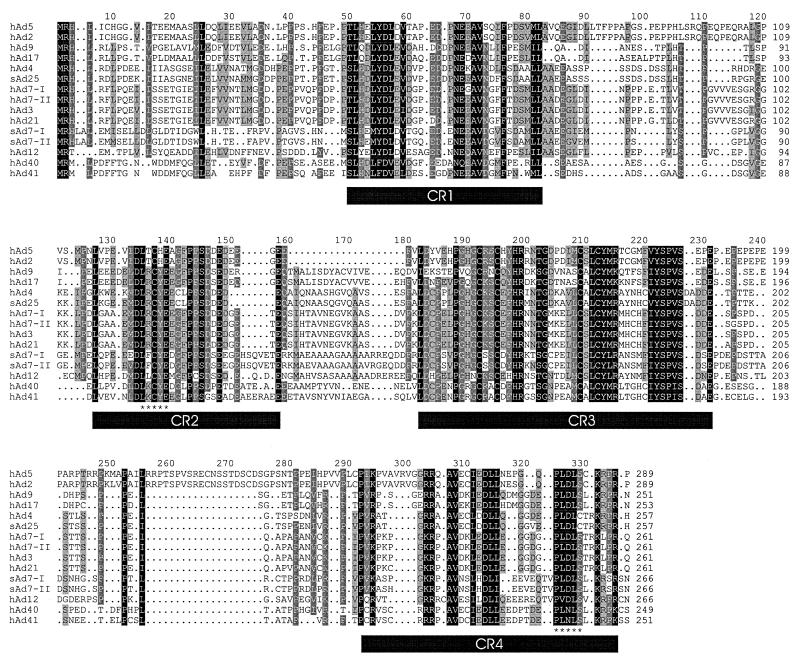
FIG. 2.
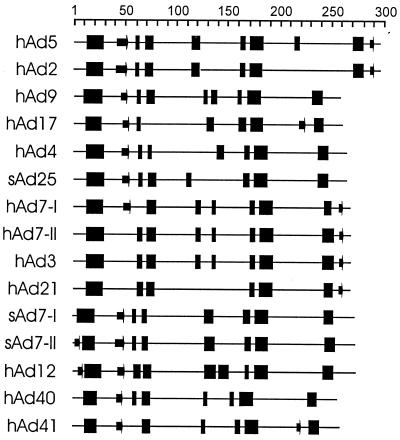
Sequence alignment of the adenovirus E1A proteins. The sequences of the indicated adenovirus E1A proteins were aligned and shaded for conservation. Darker shading corresponds to higher levels of conservation. Gaps are indicated as dots. The positions of the CR are indicated as solid bars. The binding sites for Rb and CtBP in CR2 and CR4, respectively, are indicated with asterisks.
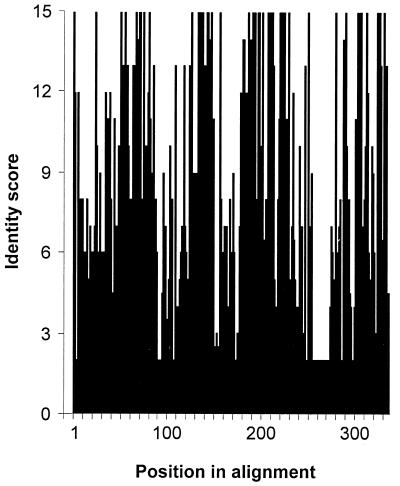
FIG. 3.
Variations in the level of sequence identity along E1A. Protein sequences were aligned (Fig. 2), and the plurality of the consensus sequence was calculated as described in Materials and Methods and plotted as a function of the amino acid position. Note that gaps introduced during the alignment are reflected in the numbering of the consensus sequence, which therefore does not correspond to any of the individual sequences. The axis on the left indicates the number of sequences out of 15 that were identical. A nonintegral value indicates that two or more different amino acids were equally prevalent at a given position.
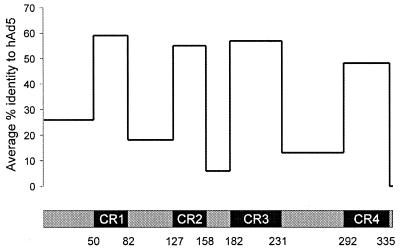
FIG. 4.
Graph of average sequence identities along E1A. Four CR within the E1A sequences were identified as described in Materials and Methods. The percent identity of each adenoviral E1A sequence to that of hAd5 was calculated for each of the indicated regions and averaged. Note that hAd2 E1A was excluded from these calculations as it is virtually identical to that of hAd5.
TABLE 2.
Sequences and locations of predicted NLSs within E1A proteins
| Virus | Predicted NLS(s) | Location(s) (aa)a |
|---|---|---|
| hAd2, hAd5 | PTRRPKM | 203-209 |
| RRPK | 205-208 | |
| KRPRb | 285-288 | |
| hAd3, hAd7-I, hAd7-II, hAd21, sAd7-I, sAd7-II | None | NA |
| hAd4, sAd25 | RKRP | 252-255 |
| KRPR | 253-256 | |
| hAd9 | KRPR | 247-250 |
| hAd12 | KRPR | 261-264 |
| hAd17 | KRPR | 249-252 |
| hAd40 | RRRP | 218-221 |
| KRPK | 244-247 | |
| hAd41 | RRRP | 220-223 |
| KRPK | 246-249 |
aa, amino acids. NA, not applicable.
Function confirmed experimentally (42).
FIG. 6.
Secondary-structure predictions for the E1A proteins. Prediction of secondary structure for each of the indicated E1A proteins was performed using the PSIPRED program as described in Materials and Methods. Predicted α-helices and β-strands four or more residues in length are shown as blocks or arrows, respectively. The scale at the top indicates the amino acid positions within each E1A protein.
Nucleotide sequence accession numbers.
The sequences of the hAd3 and hAd21 E1As have been deposited in GenBank under accession numbers AF492352 and AF492353, respectively.
RESULTS AND DISCUSSION
Sequence relationships among larger E1A proteins of hAds and sAds.
The last detailed analysis of E1A sequences compared three hAd E1A proteins, representing each of the subgroups A, B, and C, as well as a sAd (38). With the two new sequences reported here, a total of 15 E1A sequences are available in GenBank (Table 1), allowing a more thorough comparison of their similarities and differences. We chose to include the two available independent sequences for hAd7 and sAd7, although they are 99 and 95% identical, respectively. This decision was made because similar levels of identity exist between the E1A sequences of hAd2 and hAd5 (98%) and between hAd3 and hAd7 (98%), which are treated as distinct virus types. Of these 15 proteins, there is a single representative (each) from subgroups A and E, four from subgroup B, two from subgroup C, two from subgroup D, and two from subgroup F, and three are sAd sequences. A variety of physical and chemical properties of the E1A proteins are reported in Table 1. These proteins range in size from 249 to 289 residues, with the subgroup C proteins being the largest and the subgroup D and F proteins being the smallest. All the E1A proteins are acidic, with calculated pI values ranging from 3.99 to 4.56. Subgroup C proteins are the least acidic, whereas subgroup F proteins have the highest overall acidity. The E1A proteins contain 17.3 to 24.1 mol% acidic residues and 6.0 to 8.8 mol% basic residues. The proline content of the E1A proteins ranged from 8.0 to 15.9 mol%, with subgroup C proteins having the highest percentages.
The 15 E1A sequences were aligned using CLUSTAL W. As expected, the highest levels of identity and similarity occurred between viruses within the same subgroup. The two sAd7 sequences were most like hAd12, while the sAd25 sequence most resembled hAd4, suggesting that they should be placed in human subgroups A and E, respectively. The organization of the E1A sequences into six subgroups is also displayed as an unrooted phylogenetic tree (Fig. 1). This tree also supports the placement of sAd25 in subgroup E, while the degree of divergence of the sAd7 proteins from hAd12 suggests that they could be considered a new subgroup rather than members of subgroup A. Subgroup C members are most closely related to subgroup D, whereas subgroup B is closest to E, and A is closest to F. The sequences within subgroups B and C are the most highly related, whereas those within subgroup A have the greatest degree of divergence. The high degree of relatedness between hAd4 and sAd25 suggests that the distinction between at least some sAds and hAds may be relatively artificial.
Redefinition of the conserved regions of E1A.
Previous work showed that three separate regions of E1A had a higher overall degree of homology than the rest of E1A (38, 75). With the expanded number of sequences now available, we decided to revisit the definition of these CR. An inspection of the overall sequence alignment (Fig. 2) shows that there are indeed a number of regions conserved among all types. We determined the number of times the most common residue occurred at each position in the alignment and plotted this measure of sequence identity graphically (Fig. 3). This type of analysis suggests that there may be four regions of higher homology, separated by regions with distinctly lower overall identity. Three of the highly conserved regions correspond approximately to those identified previously, but a fourth conserved region has become apparent near the C terminus of E1A. To define the boundaries of these regions precisely, we decided to set a stringent cutoff for each edge of the CR based on 100% identity or similarity, with no more than nine consecutive less conserved residues occurring between absolutely conserved residues. This arbitrary cutoff does not preclude the possibility that residues adjacent to the absolutely conserved core regions may be critical. However, the fact that those adjacent residues are not absolutely conserved in all E1A proteins suggests that they may be of secondary importance. Using this definition, CR1 spans residues 50 to 82, CR2 spans residues 127 to 158, CR3 spans residues 182 to 231, and CR4 spans residues 292 to 335 of the alignment. These values correspond to residues 42 to 72, 115 to 140, 144 to 191, and 251 to 288, respectively, in hAd5. The generally accepted prior boundaries of CR1, -2, and -3 for hAd2 and -5 are residues 41 to 80, 121 to 139, and 140 to 189 (62), and these compare favorably with the regions our analysis delimits. The stringent cutoffs we chose resulted in a somewhat smaller CR1 and -3 but an expanded CR2.
The relationship between the region we have defined as CR3 and its ability to function as a transcriptional activator can be evaluated because of the tremendous amount of study that this region has received. Interestingly, a comprehensive mutational analysis of a region containing CR3 in hAd5 E1A demonstrated that mutation of residues 137 to 144 had no effect, whereas mutation of residue 145 impaired transcriptional activation (77). Another study showed that deletion of residues 188 to 204 also blocked transcriptional activation (33). Taken together, these results support our definition of a smaller CR3 that is shifted leftward with respect to the original to encompass several extra amino acid residues on its C-terminal edge. Within CR3, the four cysteine residues that form the zinc finger (11) are absolutely conserved (alignment positions 192, 195, 209, and 212).
We determined the relative sequence identity for each CR and the less conserved regions with respect to the prototype hAd5. The average sequence identities for all sequences, except the closely related hAd2, are plotted in Fig. 4. The average identity ranged from 48 to 59% among the CR and from 5 to 25% among the other regions. CR1 had the highest level of sequence conservation, whereas CR4 had the lowest level, perhaps explaining its oversight in prior analyses. The extreme N-terminal portion had the highest average identity of the less conserved regions, suggesting that it could also be involved in activities common to different E1A proteins. This is supported by the observation that mutations in this region abolish the ability of both hAd5 and hAd12 E1A proteins to transform primary rodent cells and repress transcription (5, 34, 60, 66, 76, 82). The region linking CR2 and CR3 had the lowest average identity, as it is almost completely absent in the subgroup C E1A proteins. This region is exceptionally rich in alanine residues in the subgroup A E1A proteins. Studies using subgroup A and C chimeras have shown that this region influences tumorigenicity (32, 68), perhaps by repressing major histocompatibility complex class I antigens or by conferring resistance to lysis by cytotoxic T lymphocytes or natural killer cells (31, 55). The targets of this region have not yet been identified.
Conservation of protein interaction sites among E1A types.
We next examined whether the well-defined binding sites for several of the known E1A-interacting proteins were conserved in all species of E1A. The Rb protein is known to interact with a consensus site composed of the core LXCXE, where X can be any amino acid (13). This sequence resides within CR2 and is absolutely conserved in all E1A sequences examined here (Fig. 2, positions 135, 137, and 139), indicating the importance of this interaction for viral activation of the cell cycle and transcription (4). All the E1A sequences possess an invariant aspartic acid residue amino terminal to this consensus site. Interestingly, previous studies with the human papillomavirus E7 proteins, which also bind Rb via an LXCXE motif, have shown that high-affinity interaction with Rb requires the presence of an aspartic acid residue at the same position (27). This suggests that the different E1A types presented here all associate with Rb with high affinity.
The interaction of the transcriptional corepressor CtBP with E1A requires the sequence PLDLS near the C termini of hAd5 and hAd12 E1As (48, 61). This motif, or homologous variants, are present in all E1A types with the exception of the proteins of subgroup D viruses, which contain the variant PLDLC. It remains to be determined if these proteins retain interaction with CtBP, but it appears that most if not all of the E1A proteins target this transcriptional regulator. Interestingly, the lysine residues at position 332 of the alignment are absolutely identical in all E1A proteins (Fig. 2). In hAd5 E1A, this lysine is acetylated by p300 and pCAF, and this modification blocks the interaction of E1A with CtBP (83), suggesting that this method of regulating CtBP binding may exist in common in all E1A proteins.
Conservation of phosphorylation sites.
hAd5 E1A is phosphorylated at serine residues 89, 96, 132, 185, 188, and 219 (see reference 79 and references therein). Mutations at these sites affect a variety of E1A activities, suggesting that phosphorylation may regulate E1A function (44, 78, 79). These sites correspond to positions 99, 107, 145, 223, 226, and 260 in the aligned sequence. Only serines 145, 223, and 226 are invariant, and they are flanked by residues that are highly conserved in virtually every sequence, suggesting that they quite likely are phosphorylated similarly to hAd5 E1A. In each of the adenovirus E1A sequences, serines 145 and 226 fit the consensus sequence for substrates of casein kinase II (S/TXXD/E), although in hAd5 E1A, only the first of these sites is phosphorylated in vitro by this kinase (78).
NLSs.
hAd5 has been shown to contain a functional nuclear localization signal (NLS), comprised of the sequence KRPRP, at its extreme C terminus (43). Several of the residues within this sequence are not well conserved, suggesting that a functional NLS may not be present in some of the other E1A proteins. We used the program PSORT (51) to predict NLS sequences in each of the E1A proteins (Table 2). While most E1A proteins contain putative NLSs near their carboxyl termini, the subgroup B and sAd7 proteins do not. Additionally, no other NLS sequences are predicted to exist in these proteins. In contrast, E1A proteins from subgroups C, D, and F are predicted to contain a second NLS in addition to the one located at the carboxyl terminus. Interestingly, hAd5 E1A contains additional functional NLS sequences within residues 23 to 120 and CR3 that do not match standard consensus NLS motifs (57, 65). It seems likely that these signals could function in the absence of a carboxyl-terminal signal to mediate import into the nucleus. It remains unclear why only about half of either hAd5 or hAd12 E1A is localized to the nucleus (19, 58, 72) despite such a variety of nuclear import signals and the absence of any predicted nuclear export signal sequences.
Potential interactions with proteins containing SH3 domains.
Proteins containing src homology-3 (SH3) domains recognize and bind proline-rich ligands, generally those possessing a motif containing the core sequence PXXP (36). We noticed that hAd5 E1A contains 11 PXXP sequences, suggesting the possibility that E1A may target cellular proteins with SH3 domains. Inspection of the alignment indicates that none of these putative motifs are highly conserved, with the possible exception of the PIKP sequence starting at alignment position 292, which is present only in the E1A proteins of subgroups B, C, and D. Whether this sequence actually interacts with any cellular SH3 domain-containing protein remains to be determined.
Sequence unique to subgroup C.
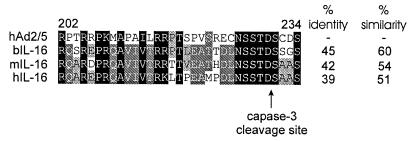
Inspection of the alignment indicates that subgroup C E1A proteins contain a lengthy insertion shortly after CR3 that is not present in any of the other proteins. We performed a BLAST analysis of this protein sequence and determined that it possesses significant homology to a short portion of interleukin-16 (IL-16) from a variety of species (Fig. 5). However, no significant homology could be observed at the nucleotide level (data not shown), suggesting that this sequence is not likely to represent an integration of a cellular sequence into E1A. This is further supported by the observation that overall homology with bovine or mouse IL-16 is higher than with the human sequence. Interestingly, this portion of IL-16 represents the site at which secreted IL-16 is cleaved from the much larger precursor protein by capase-3 (84). A comparison of the cleavage site indicates that the aspartic acid residue and adjacent sequences are identical in subgroup C E1A proteins, suggesting that under some circumstances they too might be substrates for caspase-3. In this way, E1A could compete for cleavage by caspase-3, possibly reducing its effectiveness at cleaving cellular substrates, such as the precursor of IL-16. Alternatively, cleavage of E1A could potentially release the amino-terminal and carboxyl-terminal portions of E1A to perform separate functions. Given the role of caspase-3 in apoptosis (8) and the known ability of E1A to induce apoptosis (80), it is intriguing to speculate that hAd2 and -5 E1As may have evolved to be specifically cleaved during the apoptotic process.
FIG. 5.
Sequence alignment of the region unique to hAd2 and hAd5 E1As with precursor IL-16. The sequences of hAd2 and -5 E1As (residues 202 to 234) and precursor IL-16 proteins of bovine (b), mouse (m), and human (h) were aligned and shaded for conservation. Darker shading corresponds to higher levels of sequence conservation. The percentages of identical and similar amino acid residues are shown to the right. Cleavage of precursor IL-16 by caspase-3 occurs following the aspartic acid and is indicated with an arrow.
This region unique to subgroup C E1A proteins has previously been implicated in the ability of E1A to bind DNA in vitro (3, 85) and contains a number of basic amino acid residues. However, our in vivo studies with the yeast Saccharomyces cerevisiae suggest that this region does not actually bind DNA and that the observed in vitro binding is an artifact resulting from an ionic interaction between the basic amino acids in this region and the negatively charged phosphates in DNA (3).
Protein secondary-structure analysis.
Secondary-structure predictions of the E1A proteins (Fig. 6) were generated using the PSIPRED program (47). Predicted α-helices and β-strands of four or more residues in length are shown. Interestingly, despite the limited degree of sequence homology, many of the predicted secondary structures are common to virtually all of the E1A proteins. Specifically, all E1A proteins are predicted to contain α-helices near their N termini, one or two in CR1, two within CR3, and an additional helix within CR4. The presence of a putative α-helix near the N-terminal portion of each of the E1A proteins is interesting, as the sequence in this region is not highly conserved. In E1A from hAd2 and -5, this region extends from residues 13 to 38, which is required for interaction with a variety of proteins, including p300/CBP (17), the S4 and Sug1 components of the proteasome (24, 72), and the cyclin-dependent kinase inhibitor p21 (9). In hAd12 E1A, this predicted region is considerably shorter, extending from amino acids 12 to 27, and is preceded by a short β-strand. Interestingly, the amino-terminal portion of hAd12 differs from those of hAd2 and -5 in that it functions as a transcriptional activation domain (41) and is sufficient to interact with p300 (40). The α-helix in CR3 corresponds closely to the zinc finger and extends typically 5 amino acids past the final zinc-coordinating cysteines. In hAd5, the zinc finger is required for interaction with the TBP (22) and the Sur-2 component of the transcription mediator complex (7). The α-helix in CR4 contains the sequence LXXLL in the E1A proteins of subgroups B, E, and F or homologous variants in all other E1A sequences (positions 311, 314, and 315). This motif is present in a variety of transcriptional coactivators, including p300 and CBP, which interact with liganded nuclear receptors (28, 70). Whether this region of E1A can indeed bind to nuclear receptors remains to be determined.
Predicted β-strands are generally shorter and not as conserved as the α-helices. However, most E1A proteins are predicted to contain a short β-strand at the start of CR1, with the exception of three of the subgroup B proteins. In hAd5 E1A, this region spans residues 42 to 50 and is necessary for interaction with p300/CBP (17). In addition, point mutations at residue 47 impair binding to Rb and p130 (76), suggesting that this putative β-strand may form an interaction surface with these and perhaps other cellular regulatory proteins. Subgroup B and C protein are also predicted to contain a short β-strand that overlaps the CtBP binding site.
Conclusions.
The analyses presented here refine the positioning of CR1, -2, and -3 and define a fourth CR near the carboxyl termini of the E1A proteins. Despite the differences among the E1A sequences, numerous protein interaction motifs and predicted regions of secondary structure remain recognizably present in all E1A species. These observations suggest a strong selective pressure to maintain specific protein-protein interactions, such as those between E1A and Rb or CtBP. The alignment presented here should aid in defining the surfaces of E1A required for interaction with other cellular targets. In addition, this sequence analysis suggests the possibility that at least some of the E1A proteins may interact with SH3 domain-containing proteins or liganded nuclear hormone receptors.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and The London Health Sciences Centre awarded to J.S.M. N.A. held a McLauchlin Foundation studentship and is currently supported by a Premier's Research Excellence Award. J.S.M. holds a Canadian Institutes of Health Research Scholarship.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 3.Avvakumov, N., M. Sahbegovic, Z. Zhang, M. Shuen, and J. S. Mymryk. 2002. Analysis of DNA binding by the adenovirus type 5 E1A oncoprotein. J. Gen. Virol. 83:517-524. [DOI] [PubMed] [Google Scholar]
- 4.Bayley, S. T., and J. S. Mymryk. 1994. Adenovirus E1A proteins and transformation. Int. J. Oncol. 5:425-444. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, J. M., P. M. Loewenstein, Q. Q. Tang, L. Yu, and M. Green. 2002. Adenovirus E1A N-terminal amino acid sequence requirements for repression of transcription in vitro and in vivo correlate with those required for E1A interference with TBP-TATA complex formation. J. Virol. 76:1461-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, T. G., and A. J. Berk. 1993. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 7:1810-1823. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 8.Budihardjo, I., H. Oliver, M. Lutter, X. Luo, and X. Wang. 1999. Biochemical pathways of caspase activation during apoptosis. Annu. Rev. Cell Dev. Biol. 15:269-290. [DOI] [PubMed] [Google Scholar]
- 9.Chattopadhyay, D., M. K. Ghosh, A. Mal, and M. L. Harter. 2001. Inactivation of p21 by E1A leads to the induction of apoptosis in DNA-damaged cells. J. Virol. 75:9844-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chillon, M., A. Bosch, J. Zabner, L. Law, D. Armentano, M. J. Welsh, and B. L. Davidson. 1999. Group D adenoviruses infect primary central nervous system cells more efficiently than those from group C. J. Virol. 73:2537-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culp, J. S., L. C. Webster, D. J. Friedman, C. L. Smith, W.-J. Huang, F. Y.-H. Wu, M. Rosenberg, and R. P. Ricciardi. 1988. The 289-amino acid E1A protein of adenovirus binds zinc in a region that is important for trans-activation. Proc. Natl. Acad. Sci. USA 85:6450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durfee, T., K. Becherer, P.-L. Chen, S.-H. Yeh, Y. Yang, A. E. Kilburn, W.-H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, N., P. Guida, K. Munger, and E. Harlow. 1992. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J. Virol. 66:6893-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, N., and E. Harlow. 1992. Adenovirus E1A targets key regulators of cell proliferation. Cancer Surv. 12:161-195. [PubMed] [Google Scholar]
- 15.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 16.Egan, C., S. T. Bayley, and P. E. Branton. 1989. Binding of the Rb1 protein to E1A products is required for adenovirus transformation. Oncogene 4:383-388. [PubMed] [Google Scholar]
- 17.Egan, C., T. N. Jelsma, J. A. Howe, S. T. Bayley, B. Ferguson, and P. E. Branton. 1988. Mapping of cellular protein-binding sites on the products of early region 1A of human adenovirus type 5. Mol. Cell. Biol. 8:3955-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewen, M. E., Y. Xing, J. B. Lawrence, and D. M. Livingston. 1991. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell 66:1155-1164. [DOI] [PubMed] [Google Scholar]
- 19.Fax, P., C. R. Carlson, P. Collas, K. Tasken, H. Esche, and D. Brockmann. 2001. Binding of PKA-RIIα to the adenovirus E1A(12S) oncoprotein correlates with its nuclear translocation and an increase in PKA-dependent promoter activity. Virology 285:30-41. [DOI] [PubMed] [Google Scholar]
- 20.Gallimore, P. H., and A. S. Turnell. 2001. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20:7824-7835. [DOI] [PubMed] [Google Scholar]
- 21.Geisberg, J. V., J. L. Chen, and R. P. Ricciardi. 1995. Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol. Cell. Biol. 15:6283-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisberg, J. V., W. S. Lee, A. J. Berk, and R. P. Ricciardi. 1994. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 91:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham, F. L., A. J. van der Eb, and H. L. Heijneker. 1974. Size and location of the transforming region in human adenovirus DNA. Nature 251:687-691. [DOI] [PubMed] [Google Scholar]
- 24.Grand, R. J., A. S. Turnell, G. G. Mason, W. Wang, A. E. Milner, J. S. Mymryk, S. M. Rookes, A. J. Rivett, and P. H. Gallimore. 1999. Adenovirus early region 1A protein binds to mammalian SUG1—a regulatory component of the proteasome. Oncogene 18:449-458. [DOI] [PubMed] [Google Scholar]
- 25.Hannon, G. J., D. Demetrick, and D. Beach. 1993. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 7:2378-2391. [DOI] [PubMed] [Google Scholar]
- 26.Hateboer, G., H. T. M. Timmers, A. K. Rustgi, M. Billaud, L. J. van't Veer, and R. Bernards. 1993. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 90:8489-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heck, D. V., C. L. Yee, P. M. Howley, and K. Munger. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 89:4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 29.Hitt, M. M., and F. L. Graham. 2000. Adenovirus vectors for human gene therapy. Adv. Virus Res. 55:479-505. [DOI] [PubMed] [Google Scholar]
- 30.Houweling, A., P. J. van den Elsen, and A. J. van der Eb. 1980. Partial transformation of primary rat cells by the left most 4.5% fragment of adenovirus 5 DNA. Virology 105:537-550. [DOI] [PubMed] [Google Scholar]
- 31.Huvent, I., C. Cousin, A. Kiss, C. Bernard, and J. C. D'Halluin. 1996. Susceptibility to natural killer cells and down regulation of MHC class I expression in adenovirus 12 transformed cells are regulated by different E1A domains. Virus Res. 45:123-134. [DOI] [PubMed] [Google Scholar]
- 32.Jelinek, T., D. S. Pereira, and F. L. Graham. 1994. Tumorigenicity of adenovirus-transformed rodent cells is influenced by at least two regions of adenovirus type 12 early region 1A. J. Virol. 68:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelsma, T. N., J. A. Howe, C. M. Evelegh, N. F. Cunniff, M. H. Skiadopoulos, M. R. Floroff, J. E. Denman, and S. T. Bayley. 1988. Use of deletion and point mutants spanning the coding region of the adenovirus 5 E1A gene to define a domain that is essential for transcriptional activation. Virology 163:494-502. [DOI] [PubMed] [Google Scholar]
- 34.Jelsma, T. N., J. A. Howe, J. S. Mymryk, C. M. Evelegh, N. F. A. Cunniff, and S. T. Bayley. 1989. Sequences in E1A proteins of human adenovirus 5 required for cell transformation, repression of a transcriptional enhancer, and induction of proliferating cell nuclear antigen. Virology 171:120-130. [DOI] [PubMed] [Google Scholar]
- 35.Jones, N., and T. Shenk. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 76:3665-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 37.Keblusek, P., J. C. Dorsman, A. F. Teunisse, H. Teunissen, A. J. van der Eb, and A. Zantema. 1999. The adenoviral E1A oncoproteins interfere with the growth-inhibiting effect of the cdk-inhibitor p21(CIP1/WAF1). J. Gen. Virol. 80:381-390. [DOI] [PubMed] [Google Scholar]
- 38.Kimelman, D., J. S. Miller, D. Porter, and B. E. Roberts. 1985. E1a regions of the human adenoviruses and of the highly oncogenic simian adenovirus 7 are closely related. J. Virol. 53:399-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leite, J. P., C. Niel, and J. C. D'Halluin. 1986. Expression of the chloramphenicol acetyl transferase gene in human cells under the control of early adenovirus subgroup C promoters: effect of E1A gene products from other subgroups on gene expression. Gene 41:207-215. [DOI] [PubMed] [Google Scholar]
- 40.Lipinski, K. S., P. Fax, B. Wilker, H. Hennemann, D. Brockmann, and H. Esche. 1999. Differences in the interactions of oncogenic adenovirus 12 early region 1A and nononcogenic adenovirus 2 early region 1A with the cellular coactivators p300 and CBP. Virology 255:94-105. [DOI] [PubMed] [Google Scholar]
- 41.Lipinski, K. S., G. Kronerlux, H. Esche, and D. Brockmann. 1997. The e1a n terminus (aa 1-29) of the highly oncogenic adenovirus type 12 harbours a trans-activation function not detectable in the non-oncogenic serotype 2. J. Gen. Virol. 78:413-421. [DOI] [PubMed] [Google Scholar]
- 42.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. L. Harter, and R. H. Goodman. 1995. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 374:85-88. [DOI] [PubMed] [Google Scholar]
- 43.Lyons, R. H., B. Q. Ferguson, and M. Rosenberg. 1987. Pentapeptide nuclear localization signal in adenovirus E1a. Mol. Cell. Biol. 7:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mal, A., A. Piotrkowski, and M. L. Harter. 1996. Cyclin-dependent kinases phosphorylate the adenovirus e1a protein, enhancing its ability to bind prb and disrupt prb-e2f complexes. J. Virol. 70:2911-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mal, A., R. Y. C. Poon, P. H. Howe, H. Toyoshima, T. Hunter, and M. L. Harter. 1996. Inactivation of p27(kip1) by the viral e1a oncoprotein in TGF-beta-treated cells. Nature 380:262-265. [DOI] [PubMed] [Google Scholar]
- 46.Mazzarelli, J. M., G. B. Atkins, J. V. Geisberg, and R. P. Ricciardi. 1995. The viral oncoproteins Ad5 E1A, HPV16 E7 and SV40 TAg bind a common region of the TBP-associated factor-110. Oncogene 11:1859-1864. [PubMed] [Google Scholar]
- 47.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 48.Molloy, D. P., A. E. Milner, I. K. Yakub, G. Chinnadurai, P. H. Gallimore, and R. J. Grand. 1998. Structural determinants present in the C-terminal binding protein binding site of adenovirus early region 1A proteins. J. Biol. Chem. 273:20867-20876. [DOI] [PubMed] [Google Scholar]
- 49.Moran, E. 1994. Cell growth control mechanisms reflected through protein interactions with the adenovirus E1A gene products. Semin. Virol. 5:327-340. [Google Scholar]
- 50.Mymryk, J. S. 1996. Tumour suppressive properties of the adenovirus 5 E1A oncogene. Oncogene 13:1581-1589. [PubMed] [Google Scholar]
- 51.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 52.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. NEWS 4:14. [Google Scholar]
- 53.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 54.Peeper, D. S., and A. Zantema. 1993. Adenovirus-E1A proteins transform cells by sequestering regulatory proteins. Mol. Biol. Rep. 17:197-207. [DOI] [PubMed] [Google Scholar]
- 55.Pereira, D. S., K. L. Rosenthal, and F. L. Graham. 1995. Identification of adenovirus E1A regions which affect MHC class I expression and susceptibility to cytotoxic T lymphocytes. Virology 211:268-277. [DOI] [PubMed] [Google Scholar]
- 56.Reid, J. L., A. J. Bannister, P. Zegerman, M. A. Martinez-Balbas, and T. Kouzarides. 1998. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 17:4469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richter, J. D., P. Young, N. C. Jones, B. Krippl, M. Rosenberg, and B. Ferguson. 1985. A first exon-encoded domain of E1A sufficient for posttranslational modification, nuclear localization, and induction of adenovirus E3 promoter expression in Xenopus oocytes. Proc. Natl. Acad. Sci. USA 82:8434-8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowe, D. T., F. L. Graham, and P. E. Branton. 1983. Intracellular localization of adenovirus type 5 tumor antigens in productively infected cells. Virology 129:456-468. [DOI] [PubMed] [Google Scholar]
- 59.Ruley, H. E. 1983. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 304:602-606. [DOI] [PubMed] [Google Scholar]
- 60.Sawada, Y., M. Ishino, K. Miura, E. Ohtsuka, and K. Fujinaga. 1997. Identification of specific amino acid residues of adenovirus 12 E1A involved in transformation and p300 binding. Virus Genes 15:161-170. [DOI] [PubMed] [Google Scholar]
- 61.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 979-1016. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 63.Shenk, T., and J. Flint. 1991. Transcriptional and transforming activities of the adenovirus E1A proteins. Adv. Cancer Res. 57:47-85. [DOI] [PubMed] [Google Scholar]
- 64.Song, C. Z., P. M. Loewenstein, K. Toth, and M. Green. 1995. Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc. Natl. Acad. Sci. USA 92:10330-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Standiford, D. M., and J. D. Richter. 1992. Analysis of a developmentally regulated nuclear localization signal in Xenopus. J. Cell Biol. 118:991-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stephens, C., and E. Harlow. 1987. Differential splicing yields novel adenovirus 5 E1A mRNAs that encode 30 kd and 35 kd proteins. EMBO J. 6:2027-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Telling, G. C., and J. Williams. 1994. Constructing chimeric type 12/type 5 adenovirus E1A genes and using them to identify an oncogenic determinant of adenovirus type 12. J. Virol. 68:877-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 71.Trentin, J. L., Y. Yabe, and G. Taylor. 1962. The quest for human cancer viruses. Science 137:835-841. [DOI] [PubMed] [Google Scholar]
- 72.Turnell, A. S., R. J. Grand, C. Gorbea, X. Zhang, W. Wang, J. S. Mymryk, and P. H. Gallimore. 2000. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 19:4759-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ueno, N. T., D. Yu, and M. C. Hung. 2001. E1A: tumor suppressor or oncogene? Preclinical and clinical investigations of E1A gene therapy. Breast Cancer 8:285-293. [DOI] [PubMed] [Google Scholar]
- 74.Ulfendahl, P. J., S. Linder, J.-P. Kreivi, K. Nordqvist, C. Sevensson, H. Hultberg, and G. Akusjarvi. 1987. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 6:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Ormondt, H., J. Maat, and R. Dijkema. 1980. Comparison of nucleotide sequences of the early E1a regions for subgroups A, B, and C of human adenoviruses. Gene 12:63-76. [DOI] [PubMed] [Google Scholar]
- 76.Wang, H.-G., Y. Rikitake, M. C. Carter, P. Yaciuk, S. E. Abraham, B. Zerler, and E. Moran. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol. 67:476-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Webster, L. C., and R. P. Ricciardi. 1991. trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol. Cell. Biol. 11:4287-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whalen, S. G., R. C. Marcellus, D. Barbeau, and P. E. Branton. 1996. Importance of the ser-132 phosphorylation site in cell transformation and apoptosis induced by the adenovirus type 5 e1a protein. J. Virol. 70:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whalen, S. G., R. C. Marcellus, A. Whalen, N. G. Ahn, R. P. Ricciardi, and P. E. Branton. 1997. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J. Virol. 71:3545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.White, E. 2001. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene 20:7836-7846. [DOI] [PubMed] [Google Scholar]
- 81.Whyte, P., K. J. Buchkovich, J. M. Horowitz, S. H. Friend, M. Raybuck, R. A. Weinberg, and E. Harlow. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124-129. [DOI] [PubMed] [Google Scholar]
- 82.Whyte, P., H. E. Ruley, and E. Harlow. 1988. Two regions of the adenovirus early region 1A proteins are required for transformation. J. Virol. 62:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, Q., H. Yao, N. Vo, and R. H. Goodman. 2000. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl. Acad. Sci. USA 97:14323-14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang, Y., D. M. Center, D. M. Wu, W. W. Cruikshank, J. Yuan, D. W. Andrews, and H. Kornfeld. 1998. Processing and activation of pro-interleukin-16 by caspase-3. J. Biol. Chem. 273:1144-1149. [DOI] [PubMed] [Google Scholar]
- 85.Zu, Y.-L., Y. Takamatsu, M.-J. Zhao, T. Maekawa, H. Handa, and S. Ishii. 1992. Transcriptional regulation by a point mutant of adenovirus-2 E1a product lacking DNA binding activity. J. Biol. Chem. 267:20181-20187. [PubMed] [Google Scholar]