Abstract
Human cytomegalovirus (HCMV) glycoprotein US2 causes degradation of major histocompatibility complex (MHC) class I heavy-chain (HC), class II DR-α and DM-α proteins, and HFE, a nonclassical MHC protein. In US2-expressing cells, MHC proteins present in the endoplasmic reticulum (ER) are degraded by cytosolic proteasomes. It appears that US2 binding triggers a normal cellular pathway by which misfolded or aberrant proteins are translocated from the ER to cytoplasmic proteasomes. To better understand how US2 binds MHC proteins and causes their degradation, we constructed a panel of US2 mutants. Mutants truncated from the N terminus as far as residue 40 or from the C terminus to amino acid 140 could bind to class I and class II proteins. Nevertheless, mutants lacking just the cytosolic tail (residues 187 to 199) were unable to cause degradation of both class I and II proteins. Chimeric proteins were constructed in which US2 sequences were replaced with homologous sequences from US3, an HCMV glycoprotein that can also bind to class I and II proteins. One of these US2/US3 chimeras bound to class II but not to class I, and a second bound class I HC better than wild-type US2. Therefore, US2 residues involved in the binding to MHC class I differ subtly from those involved in binding to class II proteins. Moreover, our results demonstrate that the binding of US2 to class I and II proteins is not sufficient to cause degradation of MHC proteins. The cytosolic tail of US2 and certain US2 lumenal sequences, which are not involved in binding to MHC proteins, are required for degradation. Our results are consistent with the hypothesis that US2 couples MHC proteins to components of the ER degradation pathway, enormously increasing the rate of degradation of MHC proteins.
Human cytomegalovirus (HCMV) is a betaherpesvirus that can cause serious disease in children and patients who are immunosuppressed or immunodeficient. HCMV can infect diverse cell types, including epithelial, glial, and endothelial cells, fibroblasts, and monocytes/macrophages, and generally replicates slowly in most cells. The virus establishes a latent state in monocytes/macrophages (37). Periodic reactivation and replication occurs in the face of robust, fully primed host immunity. HCMV survives and spreads to other hosts, in part, by inhibiting recognition by T lymphocytes and natural killer cells (reviewed in references 22, 23, and 41).
The S component of the HCMV genome includes a region, US2-US11, that encodes eight membrane glycoproteins of similar size and showing limited homology one to another (20, 24; N. R. Hegde, R. A. Tomazin, T. W. Wisner, C. Dunn, J. M. Boname, D. M. Lewinsohn, and D. C. Johnson, unpublished data). These glycoproteins provide fascinating examples of diverse and, in some cases, redundant evasion of cellular immunity. The first notions about the functions of US2-US11 glycoproteins came from studies involving HCMV mutants that identified genes at either end of this region, US2 and US11, which downregulated major histocompatibility complex (MHC) class I proteins (24). Subsequent studies showed that four of the US2-US11 glycoproteins inhibit the MHC class I antigen presentation pathway (reviewed in references 22, 23, and 41). Expression of either US2, US3, US6, or US11 in cells by transfection or by using virus vectors reduces the cell surface MHC class I (2) and inhibits recognition by CD8+ T-cell clones (Hegde et al., unpublished), whereas the expression of US7, US8, US9, or US10 does not. US2 and US11 promote proteasome-mediated degradation of class I proteins (24, 25, 43, 44), and US2 causes degradation of HFE, a nonclassical MHC class I protein that regulates iron uptake (4). US3 causes retention of class I complexes in the endoplasmic reticulum (ER) (1, 26). US6 inhibits the transporter associated with antigen processing (TAP), reducing the access of antigenic peptides into the lumen of the ER (2, 19, 28). Recent studies indicate that all eight membrane glycoproteins expressed from this region—US2, US3, US6, US7, US8, US9, US10, and US11—are retained in the ER or Golgi and do not reach the cell surface (2, 20, 28).
Two of the glycoproteins from US2-US11 can also inhibit MHC class II antigen presentation to CD4+ T cells. When glycoproteins US2-US11 were independently expressed in class II-expressing cells, US2 and US3 inhibited recognition by CD4+ T-lymphocyte cells, whereas other glycoproteins from US2-US11 were not effective (40; Hegde et al., unpublished). US2 targets MHC class II HLA-DR-α and DM-α chains, as well as class I proteins, for destruction by the proteasome (40) and shows some limited preference for MHC class I when US2 is limiting (N. R. Hegde, unpublished data). US3 disrupts the class II pathway by inhibiting the association of invariant chains with class II DR-α/β dimers in the ER, so that class II proteins are mislocalized and not loaded with peptides in endosomal or lysosomal compartments (Hegde et al., unpublished). Thus, US2 and US3 act at different stages of the class II pathway, as is the case with the HCMV inhibitors of the class I pathway.
Normally, MHC class II proteins present exogenous or extracellular antigens that are taken up by phagocytosis or endocytosis into so-called “professional” antigen-presenting cells. However, given that US2 and US3 are expressed as membrane glycoproteins in the cytoplasm of HCMV-infected cells, we proposed that US2 and US3 act to inhibit class II-mediated presentation of endogenous, intracellular viral antigens (23, 40; Hegde et al., unpublished). Thus, during HCMV infection of class II-expressing cells, such as endothelial cells, glial cells, epithelial cells, or monocytes/macrophages, endogenous viral antigens would normally be presented to CD4+ T cells, and this can be blocked by US2 and US3 (C. Dunn, D. M. Lewinsohn, and D. Johnson, unpublished data). This may be especially important since the assembly of HCMV involves extensive targeting of viral proteins to endosomes, so that there is likely large-scale delivery of viral antigens to MHC class II antigen loading compartments (reviewed in reference 22).
The molecular mechanisms by which HCMV US2 and US11 cause degradation of the MHC class I and II proteins are not yet completely characterized. It appears that binding of MHC proteins by US2 and US11 triggers a normal cellular pathway, termed ER-associated degradation (ERAD), that removes aberrant or misfolded ER proteins (reviewed in references 6 and 21). There is retrotranslocation of class I heavy chains (HC) and class II DR-α into the cytoplasm through the Sec61 proteinaceous pores, followed by proteasome-mediated degradation (40, 43, 44). Compared to the normal rate of catabolism of MHC molecules, the US2-triggered process is very rapid. A deglycosylated form of class I HC accumulates in a soluble form in the cytoplasm of US2-expressing cells treated with inhibitors of the proteasome (43, 44). In the US11-induced pathway, there is a requirement for polyubiquitination of class I HC for degradation (27, 36). It appears that ubiquitination does not trigger retrotranslocation, since ubiquitination of the HC cytoplasmic domain is not required to initiate this process (35). However, there are some differences in the molecular mechanisms by which MHC proteins are degraded by US2. Class II DR-α chains remain associated with ER membranes in cells expressing US2 and treated with proteasome inhibitors (40) and do not accumulate in a deglycosylated form (40). We suggested that proteasomes are necessary for extraction of class II proteins from the ER membrane, supporting the view that proteasomes involved in this process are tethered onto the ER membrane.
In order to further study interactions between US2 and class I and II proteins, we constructed a panel of US2 mutants. Removal of just the cytoplasmic domain or the cytoplasmic and transmembrane domains of US2 led to a loss of degradation of both class I HC and class II DR-α proteins without affecting the binding to these proteins. Chimeric glycoproteins consisting of US2 and US3 sequences were also able to bind to MHC proteins but not mediate their degradation. Therefore, binding of US2 to both class I and II proteins is not sufficient to promote retrotranslocation and degradation. These data are consistent with the hypothesis that US2 bridges MHC proteins to other cellular factors that promote degradation through the ERAD pathway.
MATERIALS AND METHODS
Cells.
U373-MG, a human astrocytoma cell line (ATCC, Rockville, Md.) was grown in Dulbecco modified Eagle medium (Mediatech, Herndon, Va.) supplemented with 10% fetal bovine serum (HyClone, Logan, Utah) and penicillin-streptomycin (BioWhittaker, Walkersville, Md.). U373-CIITA-HIS16 cells, denoted here as His16, were derived from U373-MG cells by stable transfection with the human class II transactivator (CIITA) gene (40) and were propagated in Dulbecco modified Eagle medium lacking histidine and supplemented with 10% fetal bovine serum and 0.5 mM histidinol (Sigma, St. Louis, Mo.).
Antibodies.
The mouse anti-class I monoclonal antibodies (MAbs) W6/32 (correctly assembled MHC I) (33) and HC10 (HC) (38) and the rabbit anti-class I heavy-chain serum (α-HC) (3) have been described previously. MAb specific for HLA-DR-α (DA6.147) (17), DR-β (HB10A) (8), DM-α (5C1) (9), or invariant chain (PIN.1) (34) were obtained from Peter Cresswell (Yale University, New Haven, Conn.). Rabbit antipeptide sera (US2N) specific for US2 have been described previously (40). A second polyclonal serum (α-US2) was prepared against the entire ectodomain of US2 by expressing US2 in insect cells. Briefly, the US2 signal sequence (residues 1 to 20) was replaced with the bee mellitin signal sequence and the transmembrane and cytoplasmic domains were removed so that US2 residues 21 to 155 were fused to a C-terminal epitope derived from herpes simplex virus glycoprotein D (gD; amino acids [aa] 263 to 284) that is recognized by MAb DL6 (11). This recombinant gene was introduced into baculovirus by using the BAC-to-BAC baculovirus expression system (Life Technologies, Rockville, Md.). The fusion protein was purified to homogeneity from Tn-5 insect cells infected with this baculovirus by using a DL6-affinity column as described previously (11) and then injected into rabbits as described elsewhere (40).
Construction of recombinant adenoviruses (Ads) expressing truncated forms of US2.
Wild-type, full-length US2 was expressed by using a replication-defective (E1−) Ad vector as described previously and denoted Adtet-US2 (40). Other recombinant Adtet vectors expressing truncated versions of US2 were constructed by using 293-cre4 cells that express the cre recombinase (7) and a shuttle plasmid, pAdtet7, that contains loxP sites (18). The US2 carboxy-terminal truncation mutants US2-186 (aa 1 to 186) (12), US2-160 (aa 1 to 160) (14), US2-150 (aa 1 to 150) (42), US2-140 (aa 1 to 140) (15), US2-130 (aa 1 to 130), US2-120 (aa 1 to 120), and US2-110 (aa 1 to 110) were constructed by inserting a stop codon into US2 sequences present in pCDNA 3.1(+) (Invitrogen, Carlsbad, Calif.). US2 mutants Kb21-199 (14), Kb28-199, Kb40-199, Kb50-199, and Kb60-199 are comprised of the murine MHC class I heavy chain H-2Kb (Kb) signal sequence (aa 1 to 20) fused to amino-terminal truncations of US2 from aa 21 to 199, aa 28 to 199, aa 40 to 199, aa 50 to 199, or aa 60 to 199. These truncations were then ligated to the Kb signal peptide (pSP72; Invitrogen). The chimeric cDNAs were then subcloned into plasmid pCDNA 3.1(+), followed by insertion into plasmid pAdtet7 downstream of an HCMV immediate-early promoter element that is regulated by a tetracycline transactivator element (16). 293-cre4 cells were cotransfected with these pAdtet7-derived shuttle plasmids and DNA derived from cells infected with psi5, an Ad vector that contains loxP sites flanking the DNA packaging sequences (18). We used cre-mediated recombination between pAdtet-US2 plasmids and psi5 DNA to produce Ad vectors, which were then passaged twice on 293-cre4 cells to remove psi5 helper viruses. For characterization or expression of US2, U373 or His16 cells were coinfected with the Adtet-US2 constructs (expressing truncated US2) and Adtet-trans, an Ad vector that expresses the tetracycline transactivator protein (18) by using an Adtet-US2/Adtet-trans ratio of 5:1.
Construction of Ad vectors expressing US2/US3 chimeric glycoproteins.
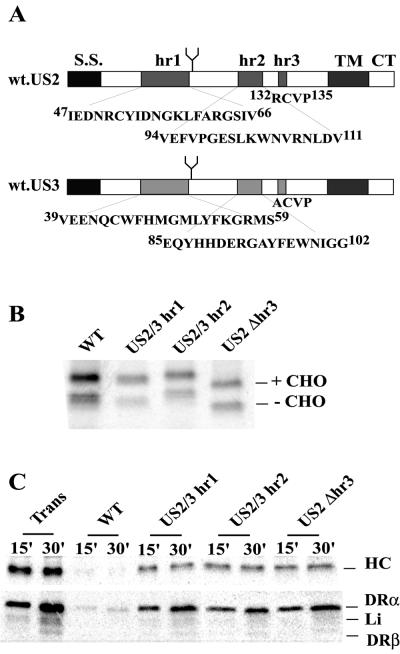
Two chimeric US2/US3 fusion proteins were engineered, inserting US3 sequences into US2, based on previous analyses suggesting that these glycoproteins contained three relatively homologous regions (hr) (1). US2/3-hr1 mutant contains US3 hr1 sequence (Fig. 6) inserted in place of US2 hr1 sequence (from I47 to V66). This chimera was engineered by using two rounds of PCR in a procedure adapted from the method of Morrison and Desrosiers (32). In the first PCR, the partial 5′ and 3′ fragments of US2/3-hr1 were amplified with four primers: 5′us2wt (5′-CGGGATCCATGAACAATCTCTGGAAAGCCTGG-3′; BamHI site underlined), 5′us2/3#1 (5′-GCTGGTTCCATATGGGCATGCTGTACTTCAAGGGGAGGATGTCGGGCAACATGAGTCGGTTCG-3′), 3′us2wt (5′-GCTCTAGATCAGCACACGAAAAACCGCATCC-3′; XbaI site underlined), and 3′us2/3#1 (5′-GCATGCCCATATGGAACCAGCATTGGTTCTCCTCCACTTGAAACCAGGGATGCTTGG-3′). The primers 5′us2/3#1 and 3′us2/3#1 overlap and encode the hr1 region of US3. The PCR products were gel purified and used as templates for the second round of PCR amplification, together with the outside primers 5′us2wt and 3′us2wt. The final product was digested with BamHI and XbaI and ligated into pUC19 (New England Biolabs, Boston, Mass.). The presence of the mutated region hr1 was confirmed by sequencing. The chimeric protein US2/3-hr2 has the hr2 domain of US2 (from V94 to V111) replaced with hr2 of US3 (Fig. 6). US2/3-hr2 was constructed with the ExSite PCR-based site-directed mutagenesis kit (Stratagene) with the primers 5′-TCCGCGCTCATCATGGTGGTACTGCTCGTCGTCGGCGTCTACGTACAG-3′ and 5′-GCCTATTTCGAGTGGAACATCGGTGGGATGCCGATCTTCGAGACGC-3′. The hr3 domain of US2 (132RCVP135) was deleted by ExSite PCR-based site-directed mutagenesis with the primers 5′-GTTCCCCTACACTAGACCGAC-3′ and 5′-GAACTGCGAGTGGATTACACG-3′. The US2/3-hr2 and US2Δhr3 sequences were subcloned into the pUC19 plasmid, verified by sequencing, and then inserted into pAdtet7 for recombination into Ad vectors as described above.
FIG. 6.
Expression of US2/3 chimeric proteins and degradation of class I and II proteins. (A) Schematic representation of US2 and US3 including the homologous sequences: hr1, hr2, and hr3. US2/3-hr1 contains hr1 of US3 introduced into US2 in place of US2 hr1. US2/3-hr2 has hr2 of US3 introduced into US2 in place of US2 hr2. US2Δhr3 has a deletion of 4 aa (RCVP) in the hr3 region. (B) Expression of US2/3 chimeric proteins. His16 cells were infected with Ad vectors, the cells were radiolabeled for 5 min with 35S-labeled methionine and cysteine, and US2/3 proteins were immunoprecipitated with anti-US2 antibodies. (C) Degradation of class I and II proteins by US2/3 chimeric proteins. His16 cells were infected with Ad vectors and labeled with 35S-labeled methionine and cysteine for 1 min, and then the label chased for 15 or 30 min. MHC class I HC (upper panel or class II complexes (lower panel) were immunoprecipitated with MAb HC10 or DA6.147, respectively.
Radiolabeling of cells and immunoprecipitation of proteins.
His16 cells (6 × 106 to 7 × 106 cells) were infected with Ad vectors and then labeled with 35S-labeled methionine and cysteine (Promix; Amersham Pharmacia Biotech, Piscataway, N.J.) as described previously (40). Briefly, the cells were coinfected with Adtet-US2 or Ad vectors expressing mutant forms of US2 and in addition, Adtet-trans with, respectively, 100 and 20 PFU/cell (where plaques were determined on 293 cells). After 12 to 18 h, the cells were detached from the plates with trypsin-EDTA, washed twice with medium lacking methionine and cysteine, starved for 60 to 90 min in this medium, and then incubated in this medium supplemented with 35S-labeled methionine and cysteine (100 to 1,000 μCi/ml) for 1 to 30 min. The label was chased by incubating cells in medium containing 10-fold excess methionine and cysteine (2 mM). The cells were washed and lysed with NP-40-DOC lysis buffer (50 mM Tris-HCl, pH 7.5; 100 mM NaCl; 1% Nonidet P-40 [Calbiochem, San Diego, Calif.]; 0.5% sodium deoxycholate [Sigma]; 1 mg of bovine serum albumin/ml; 1 mM phenylmethylsulfonyl fluoride [Sigma]) and a cocktail of protease inhibitors (40). The cell extracts were then immunoprecipitated by mixing with the appropriate antibody for 2 h, followed by incubation with protein A-agarose beads (Amersham Pharmacia Biotech) for an additional 2 h. For endoglycosidase H (endo H) treatment, immunoprecipitated proteins were incubated with 0.5 mU of recombinant endo Hf (New England Biolabs) for 16 h at 37°C in 50 mM sodium acetate (pH 5.6) containing 0.3% sodium dodecyl sulfate (SDS) and 150 mM β-mercaptoethanol.
Sequential immunoprecipitations.
His16 cells (6 × 106 to 7 × 106 cells) were coinfected for 12 to 16 h with Adtet-US2 mutants and Adtet-trans by using 10 and 2 PFU/cell, respectively. The cells were then starved for methionine and cysteine for 60 to 90 min as described above in media supplemented with 35 μM proteasome inhibitor carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone (ZL3VS) (5). The cells were radiolabeled with 1 mCi of 35S-labeled methionine and cysteine/ml for 20 min in presence of ZL3VS. Cell extracts were prepared by using Tris saline (50 mM Tris-HCl, pH 7.5; 100 mM NaCl) containing 1% digitonin (Calbiochem). Samples were precleared by incubation with nonspecific rabbit polyclonal serum and protein A-agarose and then centrifuged. The extract was split and a fraction immunoprecipitated directly with anti-US2 antibodies. Other fractions were immunoprecipitated with anti-class I MAb W6/32 or anti-class II MAb DA6.147 for 2 h. The antigen-antibody complexes were captured by the addition of protein A-agarose beads for 2 h and washed with buffer containing 0.1% digitonin and 1 mM phenylmethylsulfonyl fluoride (Sigma). The proteins were next eluted by the addition of Tris saline-1% SDS, followed by boiling for 10 min. The eluted proteins were diluted 10-fold with Tris saline containing 0.5% NP-40, and the remaining primary antibody was captured by incubation with protein A-agarose for 2 h. A second immunoprecipitation was then performed by the addition of rabbit polyclonal anti-US2 sera for 2 h, followed by incubation with protein A-agarose for another 2 h.
Electrophoresis and autoradiography.
Immunoprecipitated proteins were eluted by the addition of Laemmli SDS sample buffer (4% SDS; 2% β-mercaptoethanol; 100 mM Tris-HCl, pH 6.8; 20% glycerol) and boiled for 5 min. Proteins were then subjected to electrophoresis in SDS-polyacrylamide gels, and then the gels were fixed in 30% acetic acid-10% methanol and incubated with Enlightning (New England Nuclear, Beverly, Mass.). The gels were dried and exposed to X-ray film (Eastman Kodak Company, Rochester, N.Y.) or analyzed by using a PhosphorImager BAS 2500 (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Expression of C- and N-terminal truncated mutants of HCMV US2.
A series of truncated forms of US2 were constructed by removing N- and C-terminal residues. These mutant US2 molecules were expressed by using recombinant Ad vectors, as in previous studies (29, 40, 45). Ad vectors are replication-defective (E1−) viruses and do not express detectable quantities of Ad proteins. In contrast to transfected cells, these Ad vectors allow variable protein expression in different cells with no pressure for cells to adapt to expression of foreign, often toxic, viral proteins.
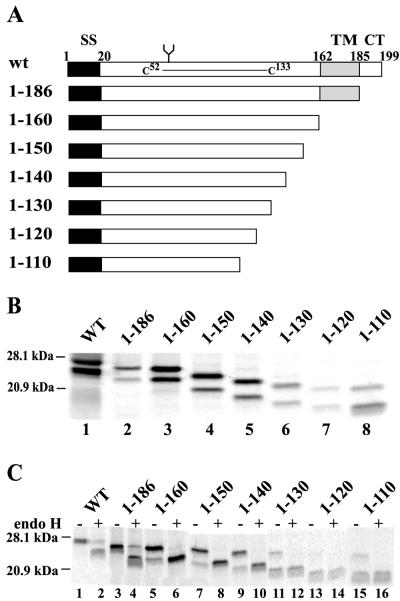
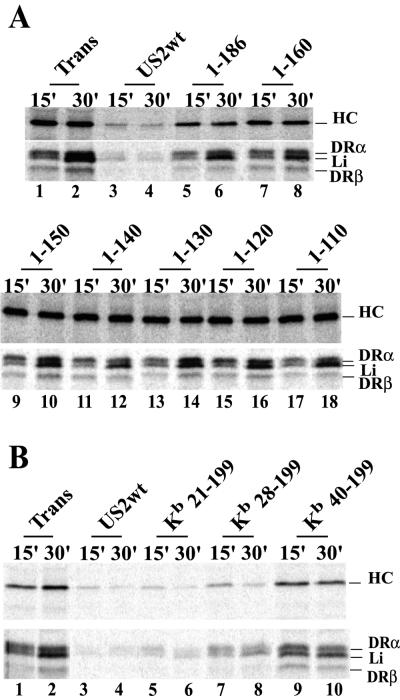
Figure 1A depicts a panel of US2 mutants truncated from the C terminus. US2 1-186 lacks 13 aa which constitute the cytoplasmic domain of the protein, US2 1-160 lacks the cytoplasmic domain and predicted transmembrane domain, and 1-150, 1-140, 1-130, 1-120, and 1-110 are deleted sequentially of 10 additional amino acids. The expression of these proteins was examined in His16 cells, U373 glial cells transfected with the CIITA transactivator so as to express MHC class II proteins (40). Cells were infected with Ad vectors, radiolabeled, and US2 immunoprecipitated with anti-US2 polyclonal antibodies. Glycoproteins smaller than wild-type US2 were detected in every case, although the expression of the smaller forms of US2 1-130, 1-120, and 1-110 was frequently lower than with wild-type US2 (Fig. 1B and C). When cells were radiolabeled with [35S]methionine in a short pulse, e.g., 5 min (Fig. 1B) or for longer periods in the presence of the proteasome inhibitor ZL3VS (Fig. 4A), there were two protein species detected with wild-type and mutant US2 proteins. The faster-migrating species were much less abundant, especially with wild-type US2 and 1-186, when cells were labeled for longer periods without proteasome inhibitor (Fig. 1C). Moreover, the more slowly migrating species was converted to the faster-migrating band by endo H (Fig. 1C). Therefore, the faster-migrating protein lacked N-linked oligosaccharides.
FIG. 1.
Expression of HCMV US2 C-terminal truncated proteins. (A) Schematic representation of C-terminal mutants of HCMV US2. Wild-type US2 (residues 1 to 199) contains an N-terminal signal sequence (SS) of 20 aa, followed by an ER-lumenal domain that includes a glycosylation site (Asn68) and a disulfide bridge between Cys52 and Cys133. The lumenal domain is followed by a predicted transmembrane domain (from residues 162 to 185) and a 14-residue cytosolic tail (residues 186 to 199). (B) Wild-type (WT) US2 and US2 mutants were expressed in His16 cells by infecting cells with Ad vectors for 12 to 16 h; the cells were radiolabeled with 35S-labeled methionine and cysteine for 5 min, and US2 proteins were immunoprecipitated with rabbit polyclonal anti-US2 sera. Molecular sizes of marker proteins of 28.1 and 20.9 kDa are indicated on the left. (C) His16 cells infected with Ad vectors expressing various US2 were radiolabeled for 30 min, and then US2 proteins were immunoprecipitated and either treated with endo H (+) or not treated with endo H (−).
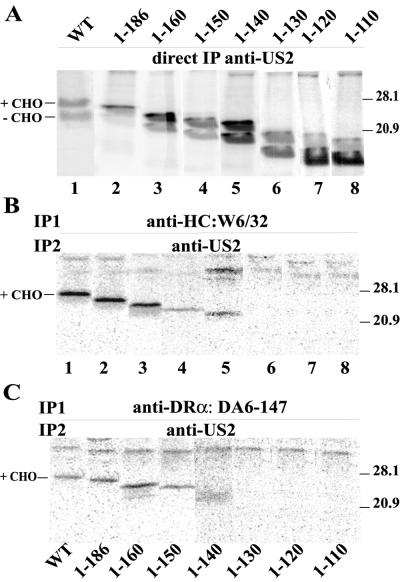
FIG. 4.
Sequential immunoprecipitation of US2 C-terminal truncation proteins with MHC class I and II proteins. His16 cells were infected with Ad vectors expressing US2 proteins for 12 to 16 h, incubated with 35 μM proteasome inhibitor ZL3VS for 60 to 90 min, and then radiolabeled with 35S-labeled methionine and cysteine for 20 min in the presence of ZL3VS. Cell extracts were made with 1% digitonin buffer, and US2 was immunoprecipitated directly with polyclonal anti-US2 antibodies (A), class I HC was precipitated with MAb W6/32 (B), or class II complexes were precipitated with MAb DA6-147. In panels B and C, the class I or II complexes were denatured in 1% SDS and reprecipitated with anti-US2 antibodies.
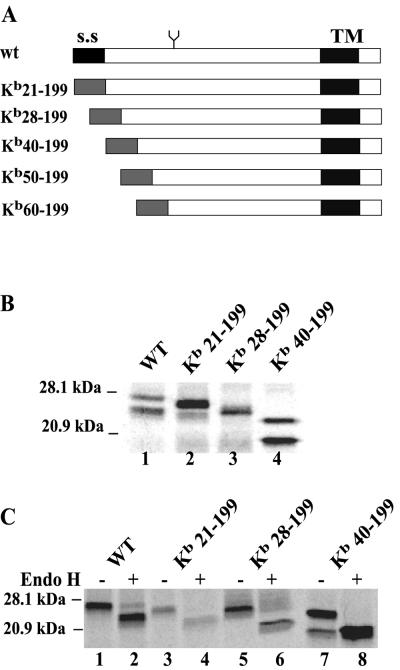
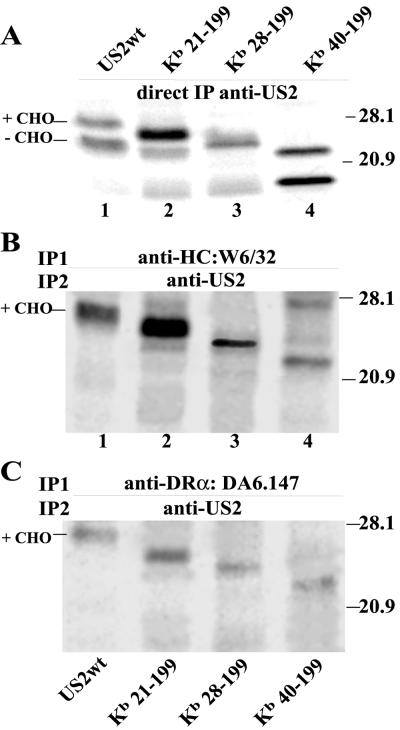
A second panel of mutants truncated from the N terminus was constructed by replacing the US2 signal sequence (residues 1 to 20) with the signal sequence of the mouse MHC class I HC molecule Kb. US2 sequences 21-199, 28-199, 40-199, 50-199, and 60-199 were fused onto the Kb signal sequence (Fig. 2A). Although there is evidence that the US2 signal sequence is not cleaved (14), the Kb signal sequences in these N-terminal truncation mutants appeared to be cleaved, since Kb21-199 was significantly smaller than wild-type US2 (Fig. 2B and C). The deletion mutants Kb50-199 and Kb60-199 were not stable in cells and were not characterized further. Kb21-199, which contains all US2 coding sequences except the native signal sequence, appeared as two protein species when cells were labeled in a short pulse, as observed with wild-type US2 (Fig. 2B). There were also two protein bands with Kb40-199, and less so with Kb28-199. Again, the upper band in each case was modified with N-linked oligosaccharides and was sensitive to endo H (Fig. 2C).
FIG. 2.
Expression of HCMV US2 N-terminal truncated proteins. (A) Schematic representation of a panel of US2 mutants truncated from the N terminus. The native signal sequence of US2 was replaced with the murine MHC class I Kb signal sequence. (B) Expression of N-terminal truncated mutants of US2 in His16 cells infected with Ad vectors and radiolabeled for 5 min with 35S-labeled methionine and cysteine. US2 proteins were immunoprecipitated with rabbit polyclonal anti-US2 sera. Note that Kb50-199 and Kb60-199 were not stable, and the expression was not shown. (C) Cells infected with Ad vectors expressing mutant US2 proteins were radiolabeled for 30 min, US2 immunoprecipitated, and then either treated (+) or not treated (−) with endo H.
The cytosolic domain of US2 is necessary for degradation of MHC class I HC and class II DR-α proteins.
Wild-type and C-terminal truncated forms of US2 were expressed in His16 cells by infection with Ad vectors, and the effects on MHC class I HC and class II DR-α were assessed. Cells were radiolabeled for 1 min, the label was chased for 15 or 30 min, and MHC class I HC or class II DR-α proteins were immunoprecipitated from cell lysates. Note that there was some acquisition of DR-α immunoreactivity for the MAb DA6.147 during the chase periods (Fig. 3A, lanes 2 and 10, and B, lane 2), as described previously (40). To control for the effects of Ad proteins, cells were infected with similar amounts of the Ad vector expressing the transactivator protein (Adtet-trans). In most experiments, infection of these cells with Adtet-trans or other Ad vectors increased the expression of class I and II proteins compared to uninfected cells, probably through interferon induction of MHC expression. Expression of wild-type US2 caused marked loss of both HC and DR molecules (Fig. 3). Previous results showed that the other components of DR complex, DR-β and the invariant chain Ii, are not degraded in cells expressing US2 (40) and that losses of DR-β and Ii bands are caused by loss of DR-α because the class II complex was immunoprecipitated with DA6.147, a DR-α-specific MAb. Mutant 1-186 showed little or no effect on class I or II proteins in this experiment (Fig. 3A, lanes 5 and 6), although in other experiments in which high levels of 1-186 were expressed there was some limited degradation of both class I and II proteins. Mutants 1-160, 1-150, 1-140, 1-130, 1-120, and 1-110 produced no obvious degradation of class I HC and class II DR even at the highest levels of expression (Fig. 3A). We concluded that US2 proteins lacking just the 13 aa comprising the cytoplasmic domain or the cytoplasmic and transmembrane domains are largely inactive in promoting degradation of DR-α and class I HC (see Table 1 for summary).
FIG. 3.
Degradation of class I and II proteins by US2 C- and N-terminal truncation mutants. His16 cells were infected for 12 to 16 h with Adtet-trans alone or coinfected with Adtet-trans (Trans) and AdtetUS2 (wild-type US2 [US2wt]) or Ad vectors expressing US2 mutants: 1-186, 1-160, 1-150, 1-140, 1-130, 1-120, or 1-110 (A) or Kb21-199, Kb28-199, or Kb40-199 (B). The cells were radiolabeled with 35S-labeled methionine and cysteine for 1 min, and then the label was chased for 15 or 30 min. The cell extracts were immunoprecipitated with MAb HC10 (anti-class I HC, upper panels) or DA6.147 (anti-class II DR-α, lower panels).
TABLE 1.
Class I and class II binding and degradation properties of US2 mutantsa
| US2 or US2 mutant (residues) | Binding
|
Degradation
|
||
|---|---|---|---|---|
| Class I | Class II | Class I | Class II | |
| US2 wild type | ++++ | +++ | ++++ | ++++ |
| 1-186 | ++++ | +++ | − | − |
| 1-160 | ++++ | +++ | − | − |
| 1-150 | ++ | ++ | − | − |
| 1-140 | ++ | + | − | − |
| 1-130 | − | − | − | − |
| 1-120 | − | − | − | − |
| 1-110 | − | − | − | − |
| Kb21-199 | ++++ | +++ | ++++ | ++++ |
| Kb28-199 | ++++ | +++ | +++ | +++ |
| Kb40-199 | +++ | + | + | + |
| US2/3-hr1 (47-66) | ++++ | +++ | − | − |
| US2/3-hr2 (94-111) | − | +++ | − | − |
| US2 Δhr3 (132-135) | − | + | − | − |
The signal sequence and N-terminal residues of US2 are not required for MHC protein degradation.
US2 N-terminal truncation mutants Kb21-199, Kb28-199, and Kb40-199 were expressed in cells by using Ad vectors, and the degradation of class I HC and class II DR-α was examined. Kb21-199, in which only the native US2 signal sequence was replaced, behaved like wild-type US2 (Fig. 3B, lanes 5 and 6). Similarly, Kb28-199 caused degradation of both HC and DR-α (Fig. 3B, lanes 7 and 8), although in most experiments there was reduced activity compared with Kb21-199. The mutant Kb40-199 was much less active, producing limited degradation of class I and II proteins (Fig. 3B, lanes 9 and 10). We concluded that the extreme N-terminal residues of US2, certainly aa 1 to 28, including the native signal sequence (residues 1 to 20) are not required for the degradation of HC and DR-α. The wild-type US2 signal sequence is apparently not cleaved, at least in some cells (14), but our results show that the US2 signal sequence can be replaced by a heterologous signal sequence which is cleaved without affecting its function. We have no direct evidence that there was cleavage of Kb28-199 or Kb40-199 but, based on the cleavage of Kb21-199, this is likely the case.
Binding of US2 truncation mutants to class I and II proteins.
To examine whether US2 mutant proteins could bind to MHC class I and II proteins, we performed sequential immunoprecipitation experiments. Cells were infected with Ad vectors expressing mutant US2 molecules; cell extracts were then immunoprecipitated with antibodies specific for MHC class I HC (W6/32) or class II DR-α (DA6.147). Precipitated proteins were denatured and immunoprecipitated in a second round with polyclonal antibodies specific for US2. In parallel, US2 was precipitated directly from cell lysates with anti-US2 antibodies to control for levels of US2 expression in the cells. Wild-type US2 was found in MHC class I and class II complexes precipitated from cells, although at lower levels in class II complexes (Fig. 4B and C). This finding may relate to the affinities or steric effects of the primary antibodies. Alternatively, since the cells were labeled in a 20-min pulse, the class II complexes that assemble slowly may acquire US2 in a delayed fashion.
C-terminal truncations of US2, 1-186, and 1-160 were immunoprecipitated with anti-class I HC and anti-DR-α antibodies at levels similar or identical to those observed with wild-type US2 (Fig. 4B and C). Mutants 1-150 and 1-140 bound detectable but lower levels of class I HC and class II DR complexes (Fig. 4B and C). There was no detectable binding of US2 1-130, 1-120, and 1-110 to either class I or II proteins (Fig. 4B and C). The glycosylated forms of wild-type and mutant US2 proteins, the slower-migrating bands, were preferentially bound to class I and class II proteins (Fig. 4B and C). The strong binding of 1-186 and 1-160 to class I and class II complexes was especially interesting given that these mutants were unable to affect the degradation of class I or II proteins.
Similar experiments were performed to test binding of US2 mutants truncated from the N terminus. Kb21-199, Kb28-199, and Kb40-199 all bound to class I HC and class II DR complexes (Fig. 5B and C). Note that degradation of class I and II proteins by Kb40-199 was markedly reduced (Fig. 3B). As observed above, only the glycosylated forms of the N-terminal US2 mutants were able to bind HC and DR-α. In summary, certain mutant forms of US2, e.g., 1-186, 1-160 and Kb40-199, can bind well to class I and class II proteins but are unable to cause their degradation (see Table 1 for summary).
FIG. 5.
Sequential immunoprecipitation of US2 N-terminal truncations with MHC class I and II proteins. His16 cells were infected with Ad vectors for 12 to 16 h and incubated with ZL3VS for 60 to 90 min, and the cells were labeled for 20 min and then lysed in 1% digitonin buffer. (A) An aliquot of the samples was immunoprecipitated directly with polyclonal anti-US2 antibodies. (B) MHC class I was precipitated with MAb W6/32, and precipitated proteins were denatured and reprecipitated with polyclonal anti-US2 antibodies. (C) Class II complexes were immunoprecipitated with MAb DA6.147, and the precipitated proteins were denatured and reprecipitated with anti-US2 antibodies.
Analysis of chimeric US2/US3 proteins.
To characterize further the lumenal domains of US2 which contact MHC proteins directly, we constructed chimeras composed largely of US2 with short sequences replaced by homologous sequences from HCMV US3. US2 and US3 can bind to both class I and class II proteins, US2 causes degradation of both MHC proteins, whereas US3 causes ER retention of class I and mislocalizes class II (1, 26; Hegde et al., unpublished). Fruh and coworkers noted that US2 and US3 display more extensive homology in three regions, termed hr1 (residues 47 to 66 in US2), hr2 (residues 94 to 111 in US2) and hr3 (residues 132 to 135 in US2) (1). Thus, proteins with these regions exchanged are more likely to fold correctly. By characterizing these chimeric mutants we sought to further characterize regions of US2 that can couple class I and II proteins to cellular molecules that cause ER degradation.
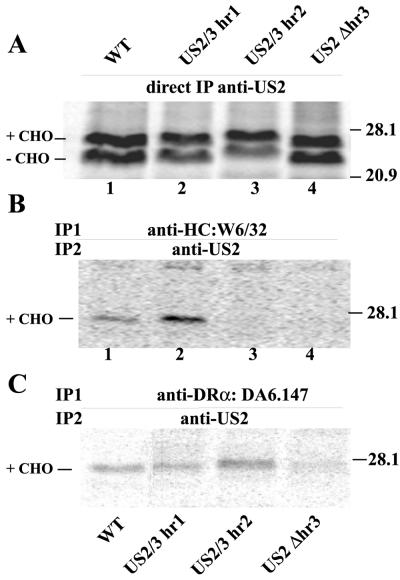
Using PCR, chimeric US2/US3 proteins were constructed: (i) US2/3-hr1 is largely US2 except that US3 hr1 sequences (aa 39 to 59) replace those of US2 (aa 47 to 66); (ii) US2/3-hr2 has US3 hr2 sequences (aa 85 to 102) replacing US2 hr2 sequences (aa 94 to 111) (Fig. 6A). For hr3, US2 and US3 sequences are short (4 aa) and largely identical and, therefore, we constructed a US2 mutant deleted of residues 132 to 135, including cysteine 133. These glycoproteins were expressed by using recombinant Ad vectors that expressed proteins of the expected size; again, doublets were observed (Fig. 6B). The slower-migrating species was converted to a species comigrating with the lower band by endo H treatment (not shown).
Degradation of class I HC and DR-α was not observed with the chimeric proteins US2/3-hr1 and US2/3-hr2 and with the deletion mutant US2Δhr3 (Fig. 6C). The binding of the chimeric US2/3 glycoproteins was studied as described above by sequential immunoprecipitation. Binding of mutant US2/3-hr1 to class I HC was observed, and in all experiments the amount of US2 present in class I immunoprecipitates was greater than that observed with wild-type US2 (Fig. 7B). This apparently does not relate to wild-type US2-mediated degradation of class I and concomitant loss of US2 because the labeling was performed in the presence of a proteasome inhibitor, ZL3VS, that abolishes proteolysis of both class I and US2 (40). Moreover, expression of US2/3-hr1 was not higher than and the binding of US2/3-hr1 to class II was not different from that of wild-type US2 (Fig. 7C). Thus, US2/3-hr1 apparently binds with higher affinity to class I compared to wild-type US2. Mutant US2/3-hr2 was not able to bind to class I but bound to class II complexes as well as wild-type US2 (Fig. 7B and C). Mutant US2Δhr3 bound to class I HC and class II complexes poorly or not at all. Note that US2Δhr3 lacks residues 132 to 135, including Cys133, which is involved in a disulfide bridge with Cys52 (13), and may be folded aberrantly. The results with US2/3-hr1 and -hr2 were quite striking because these chimeric proteins displayed preferences for class I versus class II proteins, something not previously observed.
FIG. 7.
Binding of chimeric US2/3 chimeras to MHC I and II proteins. His16 cells were infected for 12 to 16 h with Ad vectors expressing chimeric proteins, and cells were incubated with ZL3VS for 60 to 90 min then radiolabeled for 20 min. (A) Digitonin cell extracts were immunoprecipitated directly with anti-US2 antibodies. (B) Cell extracts were immunoprecipitated with anti-class I HC (W6/32), denatured, and then immunoprecipitated with anti-US2 antibodies. (C) Cell extracts were immunoprecipitated with anti-class II MAb DA6.147 and then immunoprecipitated with anti-US2 antibodies.
DISCUSSION
US2 causes rapid proteasome-mediated degradation of MHC class I HC, HFE (a nonclassical MHC class I protein), and class II DR-α and DM-α proteins (4, 40, 43, 44). Binding of US2 to class I triggers association of class I HC with the Sec61 translocon so that there is retrotranslocation into the cytoplasm and proteolysis by the proteasome (44). US2 similarly causes MHC II DR-α to be rapidly degraded, but in this case there is evidence that proteasomes are required for the retrotranslocation process (40). The requirement for active proteasomes is similar to observations involving the free DR-β chain, destabilized in the absence of its partner DR-α, which is degraded by proteasomes, but remains present in the membrane when proteasomes are blocked (10). It appears that proteasomes anchored onto the cytosolic surface of the ER play an active role in extracting MHC proteins from the ER membrane. However, little is known about how retrotranslocation is triggered, apart from the binding of US2 to MHC proteins and the association of HC with Sec61 (44). It appears likely that US2 bridges MHC proteins to other cellular proteins that are regulatory or structural components of the ERAD pathway.
We have defined here US2 sequences important for binding to and degradation of class I HC and class II DR-α. These studies have provided new insights into the mechanism by which US2 functions. A mutant in which the 20-aa US2 signal sequence was replaced by another signal sequence was not compromised for degradation of class I or II proteins. There is evidence that the US2 signal sequence is not cleaved, at least in some cells (14). However, the Kb signal sequence was cleaved. Thus, removal of this signal sequence and exposure of residue 21 does not reduce functional properties of US2. Kb28-199 which is missing 8 additional amino acids also bound class I and II proteins and caused their degradation, but with reduced activity. There was a more extensive loss of function in Kb40-199. We conclude that the extreme N terminus of US2, at least as far as residue 27, is not required for binding and degradation of both MHC proteins. A β-sheet structure, beginning at residue 45, has been described for US2 (13). Deletion of residues 1 to 27 or even of residues 1 to 39 should not affect this structure. However, Kb40-199 caused little degradation of MHC proteins, although the protein bound to both class I and II. Therefore, the N terminus of US2 between 27 and 39 is apparently required for a process other than binding MHC proteins or, alternatively, this region strengthens binding to both MHC class I and II proteins in cells.
Removal of the small cytoplasmic domain of US2 in 1-186 reduced the degradation of class I and II proteins by >90%. The mutant 1-160, lacking both the transmembrane and cytoplasmic domains lost all degradation activity. In other studies, US2 1-186 produced by in vitro translation did not degrade class I HC (12). Thus, US2 must be anchored in the ER membrane and retain cytosolic sequences in order to cause degradation. US2 1-186 and 1-160 mutants were especially interesting because both bind well to class I and II proteins. We concluded that these membrane-bound and soluble US2 molecules can bind efficiently and tightly to class I and II proteins but cannot cause their degradation. Therefore, binding of US2 to MHC substrates is not sufficient for degradation. A similar conclusion was drawn from studies of a class I HC protein lacking all but four of the residues making up the cytoplasmic domain (39). US2 mutants of this type should be useful in ongoing efforts to identify cellular components of the ERAD pathway.
Our studies indicate that the majority of the lumenal domain of US2, including residues 28 to 160, is required for efficient or tight binding to both class I and II proteins, but smaller fragments, including aa 40 to 140, may be able to bind less tightly. These observations compare well with the recently described structure of a fragment of US2 (residues 15 to 140) bound to soluble class I HLA-A2 (13). US2 sequences, including residues 43 to 137, formed an immunoglobulin G-like fold and interacted along a face of US2 including contact residues 71, 72, 73, 75, 76, 77, 86, 87, 88, 120, 125, and 130. Mutant 1-130 should contain all of these contact residues, but the truncation probably affects the correct folding of the protein through the loss of Cys133 involved in a disulfide bond with Cys52 (13). Mutant US2Δhr3 carries a similar deletion of Cys133 and cannot bind class I or II proteins, further supporting an essential role for the disulfide bond.
Analysis of US2/US3 chimeras provided additional information on binding of US2 to class I and class II proteins. Class I HC, class II DR-α, and DM-α share only 25 to 30% amino acid identity, yet US2 binds to all three of these proteins and causes their degradation (40). Other proteins with similar homology, DR-β and DM-β, are not affected. US2/3-hr2, which contains US3 sequences in place of US2 domain hr2 (residues 94 to 111), was able to bind class II DR-α normally, yet could not bind class I HC. Chimera US2/3-hr1 in which US2 residues (i.e., residues 47 to 66) were replaced with homologous US3 sequences bound class I HC better than wild-type US2, but there was no change in the binding of class II. Neither of these chimeras could cause degradation of either class I or II proteins. This is the first evidence that US2 sequences involved in binding to class I HC are subtly different from those involved in binding to class II. We note that most other US2 mutations affected binding to class I and II proteins equally.
In a recent publication, Rehm et al. (33a) were unable to find evidence that US2 could cause degradation of class II proteins in dendritic cells (DCs) infected with an Ad vector expressing US2 (33a). In general, Ad vectors express transgenes poorly in DCs, and our efforts to deliver functional quantities of US2 or other US2-US11 proteins into DCs and other blood-derived adherent cells, including monocytes/macrophages, have not met with substantial success (R. Tomazin, J. A. Nelson and D. C. Johnson, unpublished results). DCs express relatively high levels of MHC class I and II proteins. Rehm et al. observed no obvious reductions in cell surface class I or class II proteins in DCs infected with AdUS2 by using 1,200 PFU/cell. In order to make conclusions about the effects of US2 on class II proteins, it was important to determine that US2 affected class I proteins. There was an extensive loss of class I HC in uninfected DCs during chase periods, and it appeared that US2 did not add significantly to the loss of HC. Similarly, US2 had little or no effect on class II DR-α in DCs. Although the authors concluded that US2 caused degradation of HC and was not effective with class II proteins, we concluded that US2 was expressed at insufficient levels in these cells to cause degradation of either class I or II proteins. These authors did observe deglycosylated forms of class II-α in US2-expressing DCs and suggested that this was further evidence that US2 did not affect class II proteins. However, this view fails to take into account our observations that deglycosylated intermediates of class II-α do not accumulate in US2-expressing cells treated with proteasome inhibitors (40). Therefore, general conclusions about US2's effects on II proteins based on these studies of DCs are not warranted.
HCMV infects a broad assortment of cells in vivo. It appears unlikely to us that US2 acts to reduce class I or II antigen presentation in professional antigen-presenting cells or in monocytes/macrophages or DCs. Rather, we believe that other cells, e.g., endothelial, epithelial, or even glial cells, that express lower levels of both class I and II proteins may be more plausible targets of these proteins. Once infected by HCMV, these cells would present endogenous viral antigens and be subject to the antiviral effects of CD4+ T cells. Obviously, cells will differ, not only in the quantities of MHC proteins but also in other cellular factors. The recent suggestion that U373 cells are somehow not appropriate for studies of HCMV's effects on class II proteins (33a) fails to recognize several observations: (i) that HCMV infects glial cells in vivo (44) and causes frequent neurologic damage in children and (ii) that glial cells naturally express low levels of class II proteins, can present antigens to CD4+ T cells (40), and can be readily induced to express higher levels of class II. Moreover, it must also be kept in mind that US2 does not act in isolation; HCMV infection leads to the expression of other viral proteins that can inhibit the class II pathway, including US3 that causes mislocalization of class II complexes preventing peptide loading (Hegde et al., unpublished), and unidentified inhibitors of class II transcription (30, 31).
Considerable care must also be taken in interpreting negative results involving the binding of US2 to class II proteins. A soluble US2 (aa 15 to 140) produced in bacteria did not bind to truncated forms of class II DR or DM proteins in gel shift assays, whereas this US2 did bind to a soluble form of HLA-A2 (15). Soluble US2 was also unable to bind to HLA-B7 and -B27, yet these class I alleles are degraded in mammalian cells (15). This US2 was extracted from inclusion bodies of bacterial cells and refolded in vitro and could be abnormally folded or else lacking important posttranslational modifications. The binding of our US2 1-140 mutant to both class I and II proteins in cells was reduced compared to the binding of 1-160, suggesting that sequences outside the region from residues 15 to 140 stabilize interactions with both class I and II proteins. Moreover, we presented evidence here that binding to class I and class II can involve different US2 sequences.
There is now extensive evidence that US2 effectively initiates degradation MHC class II proteins in cells, as well as class I proteins. In human His16 cells that express similar quantities of class I and II proteins, there was some preference, i.e., 1.5- to 2.5-fold, for degradation of class I versus class II when US2 was limiting, but both class I HC and class II DR-α were rapidly and extensively (>90%) degraded when US2 was expressed at higher levels (N. Hegde and D. C. Johnson, unpublished data). Similarly, DR-α and DM-α were efficiently degraded in HeLa cells transfected with just the DR-α or DM-α DNAs, whereas DR-β, DM-β, and Ii remained unharmed (Hegde and Johnson, unpublished). The expression of US3, US8, US9, and US10 in a variety of cell types has no effect on the stability of class I or class II proteins and other ER-resident proteins, whereas US2 leads to the degradation of DR-α in every case (Hegde et al., unpublished; Hegde and Johnson, unpublished). However, the best evidence for US2's effects on MHC class II proteins involves the inhibition of class II-mediated presentation to CD4+ T lymphocytes. The original results involved anti-tuberculosis antigen CD4+ T cells (40) and have been extended to include other US2-US11 glycoproteins, including US3, US8, US9, US10, and US11 (Hegde et al., unpublished). Only US2 and US3 diminished presentation to anti-tuberculosis antigen CD4+ T cells. Moreover, anti-HCMV gB CD4+ T cells have recently been isolated, and these clones are unable to recognize gB in His16 or Neo6 cells that also express US2 or US3 (Dunn et al., unpublished).
Not only are MHC proteins degraded after US2 binding, but US2 itself is also degraded by proteasomes (44). It is not clear whether retrotranslocation and degradation of US2 occurs simultaneously with that of MHC proteins or in a separate step. Cells treated with proteasome inhibitors display two forms of US2: glycosylated and unglycosylated molecules, with the latter lacking N-linked oligosaccharides (40, 42, 44). The unglycosylated form of wild-type US2 is unstable and is not observed when cells are labeled for longer periods or in chases in the absence of proteasome inhibitor. Here, we observed that mutant forms of US2 that did not cause degradation of MHC proteins (e.g., 1-186, 1-160, 1-150, or Kb40-199) accumulated in significant quantities as unglycosylated species in the absence of proteasome inhibitors. In contrast, US2 molecules that mediated degradation displayed rapid and preferential loss of the unglycosylated US2. Thus, the retrotranslocation of MHC molecules and proteasome-mediated degradation of these substrates must occur in order that unglycosylated US2 is degraded by the proteasome. One possibility is that unglycosylated US2 is an intermediate in the degradation pathway, there is deglycosylation of US2 as part of this process, followed by rapid proteolysis of US2 and MHC proteins. When degradation of MHC substrates does not occur, in the case of mutants, deglycosylated US2 accumulates and is more stable. Therefore, the degradation of US2 apparently occurs simultaneously with that of MHC proteins or the two processes are coupled.
In summary, our observations provide several important new insights into how US2 functions. First, certain mutations affected binding to class II, and not class I, or showed enhanced binding of class I. Therefore, the surfaces of US2 involved in contacting class I and II differ to some extent. Second, unglycosylated US2 appears to be an intermediate in the degradation pathway and is apparently degraded simultaneously or synchronously with MHC proteins. Third, binding of US2 to MHC substrates is not sufficient for degradation; other processes or proteins must be involved. It is likely that degradation occurs by the triggering of a normal cellular process, with MHC proteins being perceived as misfolded or aberrant by ER proteins that monitor quality. Thus, US2 likely bridges MHC proteins to these ERAD proteins and US2 sequences outside those required to bind class I and II appear to be involved in these connections. Consistent with this hypothesis, expression of US2 in cells with 30- to 40-fold differences in the amount of class II DR-α leads to similar levels of degradation when US2 is constant (Hegde and Johnson, unpublished). This view is consistent with the notion that proteins other than Sec61 and class II are limiting for degradation of MHC proteins in cells. US2 and US2 mutants will be valuable tools for the identification and characterization of cellular proteins involved in ERAD.
Acknowledgments
We thank Domenico Tortorella and Hidde Ploegh who provided truncated forms of US2 in plasmids. We are grateful to Kim Goldsmith for constructing and purifying baculovirus-derived US2. We appreciate the advice of Peter Cresswell, Claire Dunn, and Nag Hegde.
This work was supported by NIH grants from the National Eye Institute (EY11245) and the National Cancer Institute (CA73996).
REFERENCES
- 1.Ahn, K., A. Angulo, P. Ghazal, P. A. Peterson, Y. Yang, and K. Fruh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 3.Beersma, M. F., M. J. Bijlmakers, and H. L. Ploegh. 1993. Human cytomegalovirus downregulates HLA class I expression by reducing the stability of class I H chains. J. Immunol. 151:4455-4464. [PubMed] [Google Scholar]
- 4.Ben-Arieh, S. V., B. Zimerman, N. I. Smorodinsky, M. Yaacubovicz, C. Schechter, I. Bacik, J. Gibbs, J. R. Bennink, J. W. Yewdell, J. E. Coligan, H. Firat, F. Lemonnier, and R. Ehrlich. 2001. Human cytomegalovirus protein US2 interferes with the expression of human HFE, a nonclassical class I major histocompatibility complex molecule that regulates iron homeostasis. J. Virol. 75:10557-10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogyo, M., J. S. McMaster, M. Gaczynska, D. Tortorella, A. L. Goldberg, and H. L. Ploegh. 1997. Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA 94:6629-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky, J. L., and A. A. McCracken. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell. Dev. Biol. 10:507-513. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., M. Anton, and F. L. Graham. 1996. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat. Cell. Mol. Genet. 22:477-488. [DOI] [PubMed] [Google Scholar]
- 8.Clark, E. A., and R. Yakoshi. 1984. Leucocyte typing. Springer-Verlag, Berlin, Germany.
- 9.Denzin, L. K., N. F. Robbins, C. Carboy-Newcomb, and P. Cresswell. 1994. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity 1:595-606. [DOI] [PubMed] [Google Scholar]
- 10.Dusseljee, S., R. Wubbolts, D. Verwoerd, A. Tulp, H. Janssen, J. Calafat, and J. Neefjes. 1998. Removal and degradation of the free MHC class II beta chain in the endoplasmic reticulum requires proteasomes and is accelerated by BFA. J. Cell Sci. 111:2217-2226. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg, R. J., M. Ponce de Leon, D. Pereira, L. Long, and G. B. Cohen. 1982. Purification of glycoprotein gD of herpes virus simplex type 1 and 2 by use of a monoclonal antibody. J. Virol. 41:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furman, M. H., H. L. Ploegh, and D. Tortorella. 2002. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J. Biol. Chem. 277:3258-3267. [DOI] [PubMed] [Google Scholar]
- 13.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, H. L. Ploegh, and D. C. Wiley. 2001. Antigen presentation subverted: structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA 98:6794-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gewurz, B. E., H. L. Ploegh, and T. D. Tortorella. US2, an HCMV-encoded type I membrane glycoprotein, contains a non-cleavable amino-terminal signal peptide. J. Biol. Chem., in press. [DOI] [PubMed]
- 15.Gewurz, B. E., E. W. Wang, D. Tortorella, D. J. Schust, and H. L. Ploegh. 2001. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 75:5197-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy, K., V. Van Heyningen, B. B. Cohen, D. L. Deane, and C. M. Steel. 1982. Differential expression and serologically distinct subpopulations of human Ia antigens detected with monoclonal antibodies to Ia alpha and beta chains. Eur. J. Immunol. 12:942-948. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, S., M. Kitamura, T. Harris-Stansil, Y. Dai, and M. L. Phipps. 1997. Construction of adenovirus vectors through Cre-lox recombination. J. Virol. 71:1842-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 20.Huber, M., R. Tomazin, T. W. Wisner, J. Boname, and D. C. Johnson. 2002.. HCMV US7, US8, US9, and US10 are cytoplasmic glycoproteins, not found at cell surfaces, and US9 does not mediate cell-to-cell spread. J. Virol. 76:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, A. E., and N. G. Haigh. 2000. The ER translocon and retrotranslocation: is the shift into reverse manual or automatic? Cell 102:709-712. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. C., and N. Hegde. Inhibition of the MHC class II antigen presentation pathway by human cytomegalovirus. Curr. Top. Microbiol. Immunol., in press. [DOI] [PubMed]
- 23.Johnson, D. C., and G. McFadden. 2001. Viral immune evasion, p. 357-378. In S. H. E. Kaufman, A. Sher, and R. Ahmed (ed.), Immunology of infectious diseases. ASM Press, Washington, D.C.
- 24.Jones, T. R., L. K. Hanson, L. Sun, J. S. Slater, R. M. Stenberg, and A. E. Campbell. 1995. Multiple independent loci within the human cytomegalovirus unique short region downregulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69:4830-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, T. R., and L. Sun. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 71:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikkert, M., G. Hassink, M. Barel, C. Hirsch, F. J. Van Der Wal, and E. Wiertz. 2001. Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 358:369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehner, P. J., J. T. Karttunen, G. W. Wilkinson, and P. Cresswell. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. USA 94:6904-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, D. M., C. M. Cebulla, B. M. Rahill, and D. D. Sedmak. 2001. Cytomegalovirus and transcriptional downregulation of major histocompatibility complex class II expression. Semin. Immunol. 13:11-18. [DOI] [PubMed] [Google Scholar]
- 31.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Human cytomegalovirus inhibits IFN-α-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-α signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 32.Morrison, H. G., and R. C. Desrosiers. 1993. A PCR-based strategy for extensive mutagenesis of a target DNA sequence. BioTechniques 14:454-457. [PubMed] [Google Scholar]
- 33.Parham, P., and H. L. Ploegh. 1980. Molecular characterization of HLA-A, B homologues in owl monkeys and other nonhuman primates. Immunogenetics 11:131-143. [DOI] [PubMed] [Google Scholar]
- 33a.Rehm, A., A. Engelsber, D. Tortorella, I. J. Korner, I. Lehman, H. L. Ploegh, and U. E. Hopken. 2002. Human cytomegalovirus gene products US2 and US11 differ in their ability to attack major histocompatibility class I heavy chains in dendritic cells. J. Virol. 76:5043-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche, P. A., M. S. Marks, and P. Cresswell. 1991. Formation of a nine-subunit complex by HLA class II glycoproteins and the invariant chain. Nature 354:392-394. [DOI] [PubMed] [Google Scholar]
- 35.Shamu, C. E., D. Flierman, H. L. Ploegh, T. A. Rapoport, and V. Chau. 2001. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol. Biol. Cell 12:2546-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shamu, C. E., C. M. Story, T. A. Rapoport, and H. L. Ploegh. 1999. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 147:45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soderberg-Naucler, C., and J. A. Nelson. 2000. Immunomodulation of cytomegalovirus, p. 399-418. In M. W. Cunningham and R. S. Fujinami (ed.), Effects of microbes on the immune system. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 38.Stam, N. J., T. M. Vroom, P. J. Peters, E. B. Pastoors, and H. L. Ploegh. 1990. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in Western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immunoelectron microscopy. Int. Immunol. 2:113-125. [DOI] [PubMed] [Google Scholar]
- 39.Story, C. M., M. H. Furman, and H. L. Ploegh. 1999. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc. Natl. Acad. Sci. USA 96:8516-8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomazin, R., J. Boname, N. R. Hegde, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1043. [DOI] [PubMed] [Google Scholar]
- 41.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 42.Tortorella, D., C. M. Story, J. B. Huppa, E. J. Wiertz, T. R. Jones, I. Bacik, J. R. Bennink, J. W. Yewdell, and H. L. Ploegh. 1998. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142:365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 44.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 44a.Wiley, C. A., R. D. Schrier, F. J. Denaro, J. A. Nelson, P. W. Lampert, and M. B. Oldstone. 1986. Localization of cytomegalovirus proteins and genome during fulminant central nervous system infection in an AIDS patient. J. Neuropathol. Exp. Neurol. 45:127-139. [DOI] [PubMed] [Google Scholar]
- 45.York, I. A., C. Roop, D. W. Andrews, S. R. Riddell, F. L. Graham, and D. C. Johnson. 1994. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77:5225-5235. [DOI] [PubMed] [Google Scholar]