Abstract
Adenovirus (Ad) DNA polymerase (pol) belongs to the distinct subclass of the polα family of DNA pols that employs the precursor terminal protein (pTP) as primer. Ad pol forms a stable heterodimer with this primer, and together, they bind specifically to the core origin in order to start replication. After initiation of Ad replication, the resulting pTP-trinucleotide intermediate jumps back and pTP starts to dissociate. Compared to free Ad pol, the pTP-pol complex shows reduced polymerase and exonuclease activities, but the reason for this is not understood. Furthermore, the interaction domains between these proteins have not been defined and the contribution of each protein to origin binding is unclear. To address these questions, we used oligonucleotides with a translocation block and show here that pTP binds at the entrance of the primer binding groove of Ad pol, thereby explaining the decreased synthetic activities of the pTP-pol complex and providing insight into how pTP primes Ad replication. Employing an exonuclease-deficient mutant polymerase, we further show that the polymerase and exonuclease active sites of Ad pol are spatially distinct and that the exonuclease activity of Ad pol is located at the N-terminal part of the protein. In addition, by probing the distances between both active sites and the surface of Ad pol, we show that Ad pol binds a DNA region of 14 to 15 nucleotides. Based on these results, a model for binding of the pTP-pol complex at the origin of replication is proposed.
Adenovirus (Ad) DNA replication requires three virally encoded proteins: Ad polymerase (pol), precursor terminal protein (pTP), and DNA binding protein (DBP). Replication initiates via a protein-priming mechanism involving Ad pol and the primer pTP, which form the tight heterodimer pTP-pol. The precise interaction surface(s) between pTP and Ad pol has yet to be determined, although it has been suggested that the contacts extend over a large surface (24). Initiation of replication is catalyzed by Ad pol and can be stimulated by the two cellular transcription factors nuclear factor I and Oct-1, which function to recruit and position the pTP-pol complex to the origin of replication. The Ad genome is a linear double-stranded DNA (dsDNA) of about 36 kb with two origins located in the inverted terminal repeats. A terminal protein (TP) is covalently coupled to each 5′ end of the genome. The first 20 bp of the linear genome has been defined as the minimal replication origin. This minimal origin is highly conserved in all Ad serotypes and contains the pTP-pol binding site (nucleotides 9 to 18). Since both pTP and Ad pol bind to origin DNA with some specificity (31), the contribution of each protein is unclear. Next to the core origin, an auxiliary region containing binding sites for the transcription factors is present (for a review, see references 9 and 13 and references therein).
After the formation of the preinitiation complex at the origin of replication, initiation starts opposite the fourth base of the template strand with the covalent coupling of the initiating nucleotide, dCTP, to a serine residue in the primer pTP (20, 21). When the third nucleotide is incorporated, the resulting trinucleotide intermediate (pTP-CAT) jumps back to base pair with template bases 1 to 3 (20). Concomitantly, pTP starts to dissociate from Ad pol (19). After dissociation, Ad pol replicates the Ad genome via a strand displacement mechanism that requires DBP and type I topoisomerase nuclear factor II (9). Late in infection, pTP is processed by a virus-encoded protease into the mature TP (30).
Ad pol is a 140-kDa protein that belongs to the polα family of DNA-dependent DNA pols, based on amino acid sequence comparison (15, 34, 36). Within this family, it is part of the subclass of protein-priming DNA pols (15). Mutational analysis of the polymerase domain has shown that, like other polα-like DNA pols, Ad pol is functionally conserved with the polymerase activity located at the C terminus (4, 5, 6, 17, 22, 26, 27). Biochemical analysis of Ad pol has shown that it replicates DNA in a processive manner but that it has a distributive 3′-5′ exonuclease activity on single-stranded DNA, although removal of a mismatched nucleotide and subsequent switching to polymerization proceeds processively (18). Both the polymerase and exonuclease activities are decreased when pTP is complexed with Ad pol, and dissociation likely increases processivity (18, 19). The lack of structural data for the pTP-pol complex or for any other protein-priming DNA pol has hampered the detailed characterization of Ad pol and its binding to pTP and DNA.
Here, we have further examined the interaction of the polymerase with pTP and DNA while it is in the polymerase or the exonuclease mode. By using a biotin-streptavidin translocation block developed by the Benkovic group (8, 12), we demonstrate that the exonuclease and polymerase active sites are spatially distinct and that, when bound to DNA, Ad pol covers a region of 14 to 15 nucleotides. Moreover, an exonuclease-deficient mutant was constructed by mutating a conserved residue located at the proposed exonuclease domain of Ad pol. Combined with mutational studies (22, 26, 27), these results suggest a molecular architecture for Ad pol similar to that of RB69 DNA pol, a model enzyme for the family B polymerases (11). Furthermore, we demonstrate that pTP binds at the primer binding groove of Ad pol. The decreased exonuclease and polymerase activity in the presence of pTP is therefore most likely the result of competition between pTP and the DNA, located at the primer binding groove. Based on these results, a model is proposed for the binding of the pTP-pol complex to the origin.
MATERIALS AND METHODS
DNA templates, nucleotides, and substrates.
All oligonucleotides, unlabeled nucleotides, [α-32P]deoxynucleoside triphosphates (dNTPs) (3,000 Ci/mmol), and [γ-32P]ATP (5,000Ci/mmol) were purchased from Amersham Pharmacia Biotech. Streptavidin was purchased from U.S. Biochemicals. T30 (5′-AATCCAAAATAAGGTATATTATTGATGATG-3′) represents the first 30 nucleotides of the template strand of the Ad type 5 genome, and D20 (5′-CATCATCAATAATATACCTT-3′) is the complementary (displaced) strand of T30. Three oligonucleotides were used with an incorporated biotin molecule: Tbio5′ (5′-bioATCCAAAATAAGGTATATTATTGATGATG-3′), which is identical to T30 except for the 5′dATP being replaced with a biotin group; D7bio (5′-CATCATbioCAATAATATACCT), which is identical to D20, only with an incorporated biotin group at position 7 and lacking the 3′-terminal nucleotide; and D7bio10 (5′-CATCATbioCAATAATATACCTTATTTTGGAT), which is identical to D7bio but with 10 extra nucleotides at the 3′ end. Labeling of the oligonucleotides was performed with T4 polynucleotide kinase (Amersham Pharmacia Biotech) and [γ-32P]ATP. The hybrid molecules D20/T30, D20/Tbio5′, and D7bio/T30 were obtained by boiling oligonucleotides in 60 mM Tris-HCl (pH 7.5)-200 mM NaCl, followed by slow cooling to room temperature. All oligonucleotides used were purified by 10% polyacrylamide-1× Tris-borate-EDTA gel electrophoresis.
Proteins and buffers.
All Ad proteins used were from serotype 2. Wild-type Ad DNA pol was expressed from a baculovirus expression system and purified to near homogeneity as previously described (4). The exonuclease-deficient mutant polymerase D422A was constructed by performing site-directed mutagenesis on full-length Ad pol cDNA as described previously (4). The oligonucleotides for the PCR mutagenesis were 5′-ATCACCGGCTTTGCCGAGATCGTGCTC-3′ and 5′-GAGCACGATCTCGGCAAAGCCGGTGAT-3′ (changes marked in bold). The presence of the desired mutation was confirmed by sequencing. Preparation of the recombinant baculoviruses, protein expression, and purification to near homogeneity were performed as described previously (4). The pTP-pol complex was purified as described previously (20). The buffer used for dilution of the polymerases and the pTP-pol complex contained 25 mM HEPES (pH 7.5), 100 mM NaCl, 1 mg of bovine serum albumin (BSA)/ml, and 20% glycerol.
Determination of the distance between the exonuclease active site and the entrance of the primer binding groove.
Exonucleolytic breakdown of 5′-labeled D7bio or D7bio10 was studied in the absence or presence of 7 nM streptavidin (preincubation for 5 min). The total reaction mixture (25 μl) contained 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol (DTT), 4% glycerol, 1 mg of BSA/ml, 10 mM MgCl2, and 0.05 ng of 5′-labeled D7bio. The reaction was started by adding Ad pol or the pTP-pol complex, respectively, to a final concentration of 28.5 nM. After incubation for the indicated times at 37°C, the reactions were stopped by the addition of formamide loading buffer (98% formamide, 0.5 M EDTA [pH 8.0], 0.025% bromphenol blue, 0.025% xylene cyanol). Samples were analyzed on 8 M urea-20% polyacrylamide electrophoresis gels, followed by autoradiography or analysis by phosphorimager. Exonucleolytic activity was detected as a decrease in size of the 5′-labeled D7bio primer.
Determination of the distance between the polymerase active site and the entrance of the template binding groove.
Partial duplex D20/Tbio5′ is a primer/template structure with a 9-nucleotide template overhang and a biotin at its 5′ terminus. The total reaction mixture (25 μl) contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% glycerol, 1 mg of BSA/ml, 10 mM MgCl2, 1 mM dNTPs, and 0.05 ng of 5′-labeled D20/Tbio5′. After incubation in the absence or presence of 7 nM of streptavidin for 5 min, the reaction was started by adding Ad pol or the pTP-pol complex to a final concentration of 28.5 nM. After incubation at 37°C for the indicated times, reactions were stopped by the addition of formamide loading buffer. Samples were analyzed on 8 M urea-20% polyacrylamide electrophoresis gels, followed by autoradiography or analysis by phosphorimager. Polymerization activity was detected as an increase in size of the 5′-labeled D20 primer.
Characterization of the exonuclease-deficient mutant polymerase D422A.
The 3′-5′ exonuclease assay and the DNA pol- and exonuclease-coupled assay used to characterize the exonuclease-deficient mutant polymerase D422A were performed as described previously (4) with the following changes. For the exonuclease assay, mutant or wild-type polymerase was used to a final concentration of 28.5 nM and degradation was studied at the times indicated in the figures. For the DNA pol- and exonuclease-coupled assay, mutant or wild-type polymerase at a final concentration of 28.5 nM was used in the presence of increasing amounts of dNTPs as indicated above. Incubation was carried out at 37°C for 10 min.
Determination of the distance between the polymerase active site and the entrance of the primer binding groove.
5′-Labeled D7bio was partially degraded by 1 μg of Ad pol in a reaction volume of 25 μl in the presence of 50 mM Tris-HCl (pH 7.5)-1 mM DTT-4% glycerol-1 mg of BSA/ml-10 mM MgCl2 in the absence of nucleotides. After incubation at 37°C for 2 min., the degraded products were boiled in order to inactivate Ad pol and hybridized to T30 in the presence of 60 mM Tris-HCl (pH 7.5) and 200 mM NaCl, followed by slow cooling to room temperature. The resulting primer/templates with various primer lengths were used in the polymerization assay and were incubated for 5 min, with or without 7 nM streptavidin. The total reaction mixture (25 μl) contained 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% glycerol, 1 mg of BSA/ml, 10 mM MgCl2, 1 mM dNTPs, and 0.05 ng of primer/template mixture. The reaction was started by the addition of mutant polymerase D422A to a final concentration of 28.5 nM and incubated at 30°C for the times indicated above. Reactions were stopped at the indicated times by the addition of formamide loading buffer. Samples were analyzed on 8 M urea-20% polyacrylamide gel electrophoresis, followed by autoradiography or analysis by phosphorimager.
RESULTS
The distance between the exonuclease active site and the entrance of the primer binding groove is 5 nucleotides.
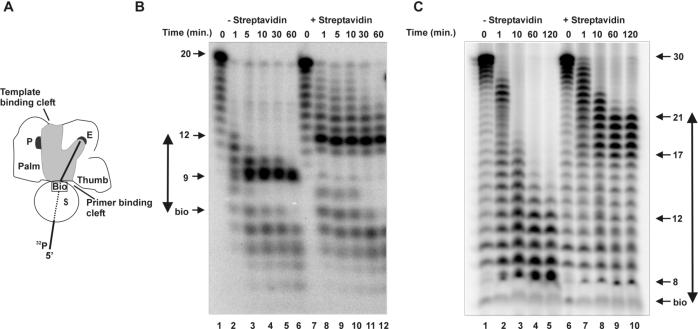
To measure the distance between the exonuclease active site and the entrance of the primer binding groove, we used a technique developed for T4 DNA pol based on a bulky biotin-streptavidin block located at a specific position within an oligonucleotide (8, 12). The 20-mer D7bio contains a biotin at the seventh position from the 5′ end to which streptavidin strongly binds (Kd ≅ 10−15 M) (35). D7bio can be degraded by the exonuclease activity of Ad pol, resulting in different product lengths. When streptavidin contacts the enzyme, it blocks further entry of the oligonucleotide at the primer binding groove due to steric hindrance (8, 12). This approach allowed us to determine the distance between the exonuclease active site and the entrance of the primer binding groove as schematically depicted in Fig. 1A.
FIG. 1.
Distance between the exonuclease active site and the entrance of the primer binding groove. (A) Schematic representation of Ad pol based on the crystal structure of the replicating RB69 DNA pol complex (11). The lower part of the polymerase shows the palm domain containing the polymerase active center (P) and the thumb. The upper part of the polymerase shows the exonuclease domain with its active center (E). The N-terminal domain and the fingers have been left out for clarity. The gray area indicates the grooves where DNA can bind. The entrances of the primer and template binding grooves are depicted. 5′-Labeled D7bio is schematically presented as a solid line or as a broken line when is covered by streptavidin (S). The biotin group (Bio) is depicted as a box. (B) For Ad pol, exonucleolytic degradation was studied on 5′-labeled 20-mer D7bio, which contains a biotin group at position 7. Degradation was studied for the indicated times in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of streptavidin. Arrows indicate accumulated degradation products and the position of the biotin group (bio). The measured distance is presented as a double-headed arrow. (C) For the pTP-pol complex, exonucleolytic degradation of the 5′-labeled 30-mer D7bio10 was studied in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of streptavidin for the indicated times. Arrows indicate accumulated degradation products and the position of the biotin group. The measured distance is presented as a double-headed arrow.
When D7bio was degraded by Ad pol for the indicated times (Fig. 1B), accumulation of a product of 9 nucleotides was observed starting after 5 min of incubation in the absence of streptavidin (lanes 3 to 6). D7bio could thus be degraded up to 2 bases 3′ of the biotin group. Under these conditions, degradation of oligonucleotides without the biotin group resulted in products of 3 nucleotides (data not shown [18]). This indicates that the biotin group probably sterically impaired translocation before it reached the exonuclease active site, preventing further degradation. When D7bio was preincubated with streptavidin and subsequently degraded, a drastic change in the degradation pattern was observed (Fig. 1B), showing accumulation of a 12-nucleotide-long product (lanes 8 to 12). Streptavidin therefore blocked the entry and further degradation of D7bio once it reached this length. This indicates that the distance between the enzyme surface at the primer binding groove and the exonuclease active site (Fig. 1B) was 5 nucleotides. To ascertain the entry of D7bio at the primer binding groove, degradation was also performed on the dsDNA primer/template D7bio/T30. The same accumulation product of 12 nucleotides was observed (data not shown).
pTP binds at the primer binding groove of Ad pol.
A previous study showed that the pTP-pol complex has a decreased rate of replication and exonuclease activities compared to free Ad pol (18). Furthermore, a difference in product lengths for the exonuclease activity of the pTP-pol complex was found (18). By using the experimental setup described for the previous experiment, we probed the pTP-pol interaction.
When the 20-mer D7bio primer was incubated with streptavidin and the pTP-pol complex, no exonucleolytic degradation was observed (data not shown). This suggests that either the pTP-pol complex could not bind to the oligonucleotide in the presence of streptavidin or the 3′ end of D7bio could not reach the exonuclease active site because it is too short. To distinguish between these possibilities, a larger oligonucleotide (D7bio10) with 10 additional nucleotides at its 3′ end was designed, keeping the internal biotin molecule at position 7. In the absence of streptavidin, degradation of this oligonucleotide to 8 nucleotides was observed (Fig. 1C, lane 5). Comparison of the degradation patterns in Fig. 1B and C showed that the exonucleolytic activity of the pTP-pol complex was slower than that of free Ad pol, in agreement with previous results (18). Furthermore, degradation of D7bio10 could now proceed up to 1 nucleotide from the biotin group rather than to 2 nucleotides (Fig. 1C), suggesting a more open exonuclease active site when pTP is complexed to Ad pol. When the experiment was performed in the presence of streptavidin, degradation led to the accumulation of products between 17 and 21 bp (Fig. 1C, lanes 9 and 10). This result showed that the pTP-pol complex was indeed able to bind to oligonucleotide D7bio10 in the presence of streptavidin and that pTP was located at the primer binding groove of Ad pol. The absence of an accumulated product of 12 bp further indicated that pTP was complexed to Ad pol throughout the experiment. The distance from the biotin to the exonuclease active site is therefore estimated at 10 to 14 nucleotides. Since the distance between the entrance of the primer binding groove and the exonuclease active site was 5 nucleotides (Fig. 1B), pTP may occupy a region between 5 and 9 nucleotides at the primer binding groove of Ad pol. The presence of several products of almost equal intensity might be explained by a flexible structure of pTP. When D7bio10 is degraded, streptavidin might approach pTP under various angles at the primer binding groove, leading to a range of product lengths dependent on the geometry of the pTP surface. In addition, the flexibility of the biotin group could play a role.
The distance between the polymerase active site and entrance of the template binding groove is 5 nucleotides.
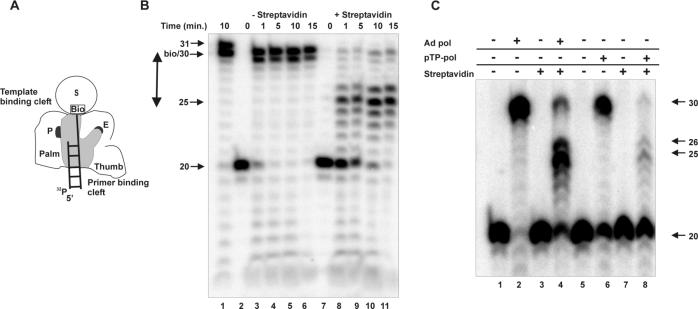
Next, we wanted to determine the distance between the entrance of the template binding groove and the polymerase active site. For this, primer D20 was hybridized to the 30-mer Tbio5′, creating a primer/template with a 9-nucleotide overhang at the 5′ end and a terminal biotin group. In the presence of streptavidin, the D20/Tbio5′ was elongated by Ad pol until streptavidin blocked further the entrance of the template strand at the template binding groove, as indicated in the experimental scheme (Fig. 2A).
FIG. 2.
Distance between polymerase active site and entrance of the template binding groove. (A) Schematic representation of the experiment. See the legend to Fig. 1A for details. (B) Elongation of Ad pol was studied on primer/template D20/Tbio5′ in the absence (lanes 2 to 6) or presence (lanes 7 to 11) of streptavidin for the indicated times. Lane 1 is a control elongation reaction performed on primer/template D20/T30. Arrows indicate accumulated products. The double-headed arrow depicts the measured distance between the entrance of the template binding groove and the polymerase active site. (C) Elongation of D20/Tbio5′ was studied on Ad pol and the pTP-pol complex in the absence (−) or presence (+) of streptavidin for 15 min at 37°C.
When D20/Tbio5′ was incubated with all four dNTPs and Ad pol for the indicated times, D20 was elongated to a main product of 30 nucleotides (Fig. 2B, lanes 3 to 6). Since the template is 29 nucleotides long, this result shows that 1 base was added opposite the 5′ biotin molecule of Tbio5′. To test if this was due to the presence of the biotin group or due to a nontemplated nucleotide addition, the same experiment was performed with primer/template D20/T30, which has a base instead of biotin at its 5′ end. As shown in Fig. 2B, both a 30-mer and a 31-mer were found (lane 1). This indicates that Ad pol under these conditions could add a nontemplated nucleotide to a blunt-ended DNA substrate, as has been described previously for several DNA pols (7).
When the primer/template D20/Tbio5′ was preincubated with streptavidin and subsequently elongated by Ad pol and dNTPs, a product of 25 nucleotides accumulated (lanes 10 and 11). Also, some longer read-through products were formed, possibly caused by the flexibility of the translocation block. Since the main product was 25 nucleotides long, our results suggested that the distance between the polymerase active site and the entrance of the template binding groove (Fig. 2B) was 5 nucleotides.
pTP does not block the entrance of the template binding groove of Ad pol.
The same experimental setup as described above (Fig. 2A) was used to determine if pTP could contact Ad pol at the entrance of the template binding groove in addition to the entrance of the primer binding groove. As can be seen in Fig. 2C, both Ad pol and the pTP-pol complex were able to fully elongate primer/template D20/Tbio5′, albeit with a lower activity for the pTP-pol complex (compare lanes 2 and 6) in agreement with previous results (18). In the presence of streptavidin, elongation stalled for both pTP-pol (lane 8) and free Ad pol (lane 4) at 25 nucleotides with the formation of some read-through product. Longer incubation for pTP-pol resulted in further accumulation of the 25-nucleotide product (data not shown). Therefore, these results suggest that, in contrast to the primer binding groove, pTP does not block the entrance of the template binding groove and thus any contacts in that region, if they exist, do not disturb passage of the template strand.
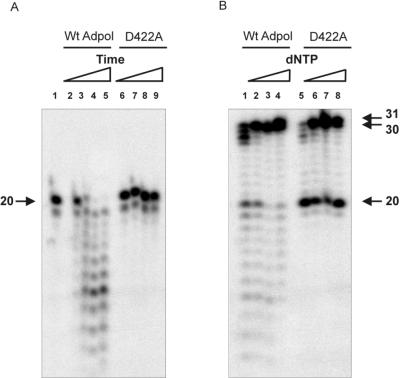
Mutant polymerase D422A is exonuclease deficient.
To complete the measurements of the various DNA binding grooves within Ad pol, we wanted to determine the distance between the polymerase active site and the entrance of the primer binding groove. However, since Ad pol possesses a distributive 3′-5′ exonuclease activity (Fig. 1B) (21, 25), discrimination between the elongation and degradation of wild-type Ad pol is difficult. Therefore, the exonuclease-deficient mutant polymerase D422A was constructed by changing the catalytic aspartate residue present in the highly conserved Exo II motif (1) into an alanine residue (D422A). Mutant polymerase D422A was characterized by a 3′-5′ exonuclease assay and a DNA pol- and exonuclease-coupled assay as shown in Fig. 3. In contrast to wild-type Ad pol (Fig. 3A, lanes 2 to 5), no exonucleolytic breakdown for mutant polymerase D422A on 5′-labeled oligonucleotide D20 was observed (Fig. 3A, lanes 6 to 9), confirming the exonuclease-deficient phenotype. The polymerase- and exonuclease-coupled assay showed that at low nucleotide concentrations, wild-type Ad pol could both polymerize and degrade the primer/template (Fig. 3B). The polymerase activity of mutant polymerase D422A was only mildly affected (Fig. 3B), but as expected, no degradation was observed, explaining at least in part the lower elongation activity. At higher nucleotide concentrations (e.g., 1 mM), the polymerase activity of D422A was wild type-like (data not shown). Both enzymes could also elongate primer D20 up to 31 nucleotides, indicating that a nontemplated nucleotide was added, as was shown previously in Fig. 2B.
FIG. 3.
Characterization of the exonuclease-deficient mutant polymerase D422A. Mutant polymerase D422A was tested for exonuclease activity on 5′-labeled D20 and for polymerase activity on primer/template D20/T30 as described in Materials and Methods. (A) Exonucleolytic degradation was studied on D20 for wild-type (Wt) Ad pol and mutant polymerase D422A. The incubation times were as follows: 1 min (lanes 2 and 6), 5 min (lanes 3 and 7), 10 min (lanes 4 and 8), and 15 min (lanes 5 and 9). (B) To study the elongation of Wt Ad pol and mutant polymerase D422A, a DNA pol- and exonuclease-coupled assay on D20/T30 was performed in the presence of the following increasing concentrations of dNTPs: 25 nM (lanes 1 and 5), 125 nM (lanes 2 and 6), 625 nM (lanes 3 and 7), and 2,500 nM (lanes 4 and 8). Incubation was at 37°C for 10 min.
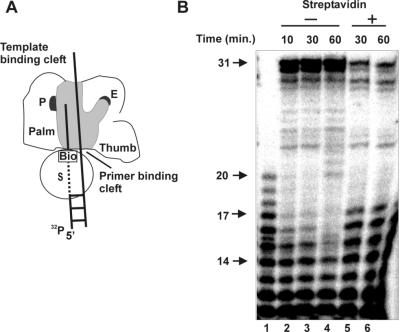
The distance between the polymerase active site and the entrance of the primer binding groove is 9 to 10 nucleotides.
With the exonuclease-deficient mutant polymerase D422A, the distance between the polymerase active site and the entrance of the primer binding groove could be measured. The 20-mer D7bio was partially degraded at the 3′ end as described in Materials and Methods and hybridized to T30, generating a primer/template mixture with various primer lengths. When these primer/templates were elongated in the presence of streptavidin, only primers that were long enough to reach the polymerase active site (measured from the streptavidin-biotin block) could support elongation. This assay therefore allowed us to determine the distance (in nucleotides) between the polymerase active site and the entrance of the primer binding groove (Fig. 4A). Figure 4B shows the result of the elongation of the partially degraded D7bio/T30 mixture (lane 1). In the absence of streptavidin, all primers longer than 14 nucleotides could be elongated by mutant polymerase D422A after 60 min (Fig. 4B, lane 4) with products up to 31 nucleotides. Quantitation of the products of 13 and 14 nucleotides showed a decreased intensity after 60 min (lane 4), indicating that small amounts of these products were also elongated. Longer incubation did not change this pattern (data not shown). When the mixture was preincubated with streptavidin and elongated, a change in the elongation pattern was observed (lanes 5 and 6). Only primers longer than 17 nucleotides could be efficiently elongated, even after 60 min. Since the 17-mer product also showed some decreased intensity, we estimated a distance of 9 to 10 nucleotides from the entrance of the primer binding groove to the polymerase active site.
FIG. 4.
Distance between the polymerase active site and the entrance of the primer binding groove. (A) Schematic representation of the experiment. See the legend to Fig. 1A for details. (B) Partially degraded D20 was hybridized to template T30. The resulting primer/template mixture (lane 1) was elongated at 30°C for the indicated times in the absence (lanes 2 to 4) or presence (lanes 5 and 6) of streptavidin by the exonuclease-deficient mutant polymerase D422A. Arrows indicate the length of the elongation products.
Combining the data for exonucleolytic degradation and elongation, we propose that there is a difference of 4 to 5 nucleotides in the length of the primer in contacting the polymerase active site (9 to 10 nucleotides) or the exonuclease active site (5 nucleotides). Therefore, these results support the notion that the exonuclease active site and the polymerase active site in Ad pol are spatially distinct. Furthermore, since we already showed a distance of 5 nucleotides between the polymerase active site and the entrance of the template binding groove (Fig. 2B), Ad pol can bind a DNA region of approximately 14 to 15 nucleotides in length.
DISCUSSION
Spatial relationship between polymerase and exonuclease active sites.
We observed that the distance between the enzyme surface and the exonuclease active site is approximately 5 nucleotides. This is very similar to the distances observed for T4 DNA pol (4 to 5 nucleotides [12]) and for φ29 DNA pol (5 nucleotides [10]). The distance between the entrance of the primer binding groove and the polymerase active site (9 to 10 nucleotides) was shown to be 2 to 4 nucleotides longer than that which was measured for T4 DNA pol (7 nucleotides) and φ29 DNA pol (6 nucleotides). This difference may, as in the case of φ29 DNA pol, simply reflect its smaller size (66 kDa for φ29 DNA pol versus 140 kDa for Ad pol). Ad pol, T4 DNA pol, and φ29 DNA pol all belong to a family of polα-like DNA pols. Recently, the structures of the replicating and editing complexes of RB69 DNA pol that can be used as a model for DNA pols belonging to this family have been resolved (11, 29). The structure of RB69 DNA pol shows that it contains a polymerase domain that, like other DNA pols, resembles the shape of a right hand consisting of a palm, fingers, and a thumb. In addition, an exonuclease domain is present, with its catalytic site located away from the polymerase active site (29). When the primer/template is bound, it is stabilized by numerous interactions between residues in the thumb domain and the minor groove of the DNA (11). Based on the structure of this replicating complex, it can be estimated that the distance between the polymerase active site and the entrance of the primer binding groove is approximately 10 nucleotides (11). The switch from the polymerase to the exonuclease active site is accompanied by a conformational change, as observed for RB69 DNA pol (11). The thumb domain confers a closed conformation when the polymerase is in the polymerizing mode but is in a more open conformation when the polymerase is in the editing mode, having fewer contacts with the DNA (29). The distance between the exonuclease active site and the entrance of the primer binding groove, measured in the editing mode, is estimated at approximately 6 nucleotides (29). These distances are close to that measured in this study for Ad pol (9 to 10 nucleotides and 5 nucleotides, respectively).
The similar spatial relationship for the exonuclease and polymerase active sites for RB69 DNA pol and for Ad pol and the fact that they all belong to the same family of α-like DNA pols support the proposal that they all have a similar structural organization. This conclusion is further supported by mutational analysis of a set of conserved residues in the C-terminal part of Ad pol that suggests an arrangement of conserved motifs in Ad pol similar to that in RB69 DNA pol (22). Moreover, we confirmed that the exonuclease activity resides in the N-terminal part of Ad pol since mutant polymerase D422A lost its exonuclease activity while the polymerase activity remained almost wild type-like (Fig. 3), similar to what was found for other characterized α-like DNA pols.
pTP interacts at the primer binding groove of Ad pol.
Here, we present data suggesting that pTP binds at the entrance of the primer binding groove of Ad pol (Fig. 1C). This finding is in agreement with the proposed role of pTP to present its priming serine residue at the polymerase active site. Mutations in Ad pol, including amino acids Y1080, E1057, and Y673, resulted in a strong reduction in pTP interaction, initiation activity, and DNA binding (22). Accordingly, extensive mutational analysis of the protein-priming φ29 DNA pol (reviewed in reference 2) has indicated that several amino acids proposed to interact with the DNA primer/template cause defects in TP interaction (3, 32), suggesting that both primers are bound by the enzyme in a similar fashion (11). Furthermore, a partial proteolysis study on φ29 DNA pol revealed that the protection and digestion pattern of TP was similar to that obtained with DNA, suggesting that both primers DNA and TP fit in the same dsDNA-binding channel and protect the same regions of φ29 DNA pol (33). All these data are in agreement with the location of pTP at the entrance of the primer binding groove.
When the pTP-pol complex was probed with primer/template and a terminal biotin, it was demonstrated that pTP did not block the entrance of the template binding groove (Fig. 2C). This result indicates that pTP does not bind this side of the polymerase, although it cannot be excluded that pTP dissociates first before the primer/template is elongated. Two observations, however, argue against this option. First, the rate of polymerization is much lower for the pTP-pol complex (Fig. 1 and 2), suggesting that pTP remains bound to Ad pol when it is in the polymerase mode, and second, it was shown that dissociation of pTP is not a prerequisite for DNA-primed polymerization (20).
It was shown that, in the presence of pTP, Ad pol is able to perform both exonuclease activity and polymerase activity (18, 19). For this, Ad pol needs to accommodate both DNA and pTP at the primer binding groove. Both the exonuclease and polymerase activities are decreased in the presence of pTP (18, 19). This is not caused by an altered DNA binding affinity (31). Rather, we assume that catalysis or the translocation of DNA after each catalytic event is hampered. This could be caused by competition for DNA and pTP binding at the primer binding groove of Ad pol. At least 3 nucleotides need to be incorporated before pTP starts to dissociate (19), suggesting that some flexibility in the priming part of pTP exists. The crystal structure of the pTP-pol complex or of any other protein-priming polymerase is required to determine the exact space constraints of both proteins.
Origin binding of the pTP-pol complex.
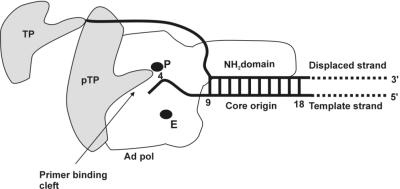
Based on the results discussed above, a model for origin binding of the pTP-pol complex preceding replication initiation can be proposed (Fig. 5). pTP is located in the model as binding to the entrance of the primer binding groove, with its priming part located at the polymerase active site close to the fourth nucleotide of the template strand. Since pTP is a DBP (31), it could be located near or even in contact with the displaced parental TP-containing DNA strand. Furthermore, the location of pTP at the entrance of the primer binding groove of Ad pol positions it close to the parental TP. Parental TP has been shown to be involved in stabilizing the binding of the pTP-pol complex at the origin, possibly via a direct interaction with pTP (25) (R. N. de Jong, unpublished data). A direct interaction between parental TP and pTP (p2 in φ29) has been previously described for φ29 (14, 28).
FIG. 5.
Model for origin binding of the pTP-pol complex preceding replication initiation. The Ad origin DNA (thick lines) containing the core origin (nucleotides 9 to 18) are bound by Ad pol. pTP is complexed to Ad pol with its priming part depicted at the primer binding cleft (arrow) close to the polymerase active site (P) and the templating nucleotide for initiation (nucleotide 4 of the template strand). The exonuclease active site (E) has been indicated for clarity. The NH2 domain represents the putative N-terminal domain of Ad pol that could bind to the core origin. See text for more details.
The distance from the polymerase active site to the entrance of the template binding groove is 5 nucleotides (Fig. 2) and 9 to 10 nucleotides to the entrance of the primer binding groove (Fig. 4), indicating that Ad pol covers 14 to 15 nucleotides of DNA when it is in the polymerase mode. Although these data are approximate due to the flexibility of the biotin group, they correspond well to what has been observed in RB69 DNA pol. Here, a minimum of 12 nucleotides was covered, although the exact number was difficult to estimate since the template strand of the replicating complex was unstructured at its 5′ end (11). The template strand enters RB69 DNA pol in a groove formed between the NH2 and exonuclease domains (11). A DNase I footprint of Ad pol on the Ad type 2 origin showed protection of the first 20 nucleotides (31). The pTP-pol complex showed increased specificity on origin-containing DNA and protected bases 9 to 18 (31), but the contribution of each protein is unclear (23). Since the distance between the polymerase active site and entrance of the template binding groove is 5 nucleotides, our results indicate that at preinitiation complex formation, nucleotides 1 to 9 are covered by Ad pol. These data then do not explain the DNase I footprinting results that show that nucleotides 9 to 18 are covered. In order to explain this discrepancy, we propose that the N terminus of Ad pol folds into a domain that binds to the core origin sequence (Fig. 5). Although this N-terminal domain is not conserved between DNA pols, it may play a role in origin recognition in the case of Ad pol. Indeed, there are about 250 amino acids located N terminally that contain a putative Zn finger motif. Point and linker insertion mutants in this motif retained DNA elongation but had severely reduced initiation activity and lost the ability to bind the Ad core origin DNA, which is essential for Ad DNA replication (16). Furthermore, one of the four fragments of Ad pol obtained after partial digestion with endolys C contained these 250 amino acids, suggesting that it could fold as an independent domain (24). Alternatively, pTP could contribute to binding of the core origin. However, this is unlikely, as no blocking of the entrance of the template binding cleft by pTP was observed, which would be expected when pTP is binding the core origin. Interestingly, the N-terminal domain of Ad pol was the only domain that could not bind to pTP, as was demonstrated in the partial proteolysis study (24), further indicating that it is unlikely that pTP binds at this side of the polymerase.
In summary, our results and those reported previously support the proposal that Ad pol has a molecular architecture similar to that of RB69 DNA pol. Furthermore, the location of pTP was directly probed, binding at the primer binding groove of Ad pol, providing an explanation for the observed decrease in polymerase and exonuclease activity in the presence of pTP and allowing insight into the use of a protein to prime replication. These results have led to a model for pTP-pol binding on the origin of replication. Since no structural information exists on any protein-priming polymerase or any priming protein, these results are an important contribution to the understanding of Ad DNA replication and protein-primed replication in general.
Acknowledgments
We thank Marinus R. Heideman for purification of the wild-type Ad pol and Rob de Jong for stimulating discussions and critical reading of the manuscript.
This work was supported by The Netherlands Organization for Scientific Research (NWO).
REFERENCES
- 1.Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, L., and M. Salas. 1995. Mutational analysis of bacteriophage phi 29 DNA polymerase. Methods Enzymol. 262:283-294. [DOI] [PubMed] [Google Scholar]
- 3.Blasco, M. A., J. Mendez, J. M. Lazaro, L. Blanco, and M. Salas. 1995. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J. Biol. Chem. 270:2735-2740. [DOI] [PubMed] [Google Scholar]
- 4.Brenkman, A. B., M. R. Heideman, V. Truniger, M. Salas, and P. C. Der Vliet. 2001. The (I/Y)XGG motif of adenovirus DNA polymerase affects template DNA binding and the transition from initiation to elongation. J. Biol. Chem. 276:29846-29853. [DOI] [PubMed] [Google Scholar]
- 5.Chen, M., and M. S. Horwitz. 1989. Dissection of functional domains of adenovirus DNA polymerase by linker-insertion mutagenesis. Proc. Natl. Acad. Sci. USA 86:6116-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, M., N. Mermod, and M. S. Horwitz. 1990. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J. Biol. Chem. 265:18634-18642. [PubMed] [Google Scholar]
- 7.Clark, J. M. 1988. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16:9677-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowart, M., K. J. Gibson, D. J. Allen, and S. J. Benkovic. 1989. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry 28:1975-1983. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, R. N., and P. C. van der Vliet. 1999. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236:1-12. [DOI] [PubMed] [Google Scholar]
- 9a.de Jong, R. N., P. C. van der Vliet, and A. B. Brenkman. Curr. Top. Microbiol. Immunol., in press. [DOI] [PubMed]
- 10.de Vega, M., L. Blanco, and M. Salas. 1999. Processive proofreading and the spatial relationship between polymerase and exonuclease active sites of bacteriophage phi29 DNA polymerase. J. Mol. Biol. 292:39-51. [DOI] [PubMed] [Google Scholar]
- 11.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105:657-667. [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan, V., and S. J. Benkovic. 1994. Spatial relationship between polymerase and exonuclease active sites of phage T4 DNA polymerase enzyme. J. Biol. Chem. 269:21123-21126. [PubMed] [Google Scholar]
- 13.Hay, R. T. 1996. Adenovirus DNA replication, p. 699-719. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Illana, B., J. M. Lazaro, C. Gutierrez, W. J. Meijer, L. Blanco, and M. Salas. 1999. Phage phi29 terminal protein residues Asn80 and Tyr82 are recognition elements of the replication origins. J. Biol. Chem. 274:15073-15079. [DOI] [PubMed] [Google Scholar]
- 15.Ito, J., and D. K. Braithwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joung, I., and J. A. Engler. 1992. Mutations in two cysteine-histidine-rich clusters in adenovirus type 2 DNA polymerase affect DNA binding. J. Virol. 66:5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joung, I., M. S. Horwitz, and J. A. Engler. 1991. Mutagenesis of conserved region I in the DNA polymerase from human adenovirus serotype 2. Virology 184:235-241. [DOI] [PubMed] [Google Scholar]
- 18.King, A. J., W. R. Teertstra, L. Blanco, M. Salas, and P. C. van der Vliet. 1997. Processive proofreading by the adenovirus DNA polymerase. Association with the priming protein reduces exonucleolytic degradation. Nucleic Acids Res. 25:1745-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, A. J., W. R. Teertstra, and P. C. van der Vliet. 1997. Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J. Biol. Chem. 272:24617-24623. [DOI] [PubMed] [Google Scholar]
- 20.King, A. J., and P. C. van der Vliet. 1994. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 13:5786-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichy, J. H., M. S. Horwitz, and J. Hurwitz. 1981. Formation of a covalent complex between the 80,000-dalton adenovirus terminal protein and 5′-dCMP in vitro. Proc. Natl. Acad. Sci. USA 78:2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, H., J. H. Naismith, and R. T. Hay. 2000. Identification of conserved residues contributing to the activities of adenovirus DNA polymerase. J. Virol. 74:11681-11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mul, Y. M., and P. C. van der Vliet. 1992. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 11:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker, E. J., C. H. Botting, A. Webster, and R. T. Hay. 1998. Adenovirus DNA polymerase: domain organisation and interaction with preterminal protein. Nucleic Acids Res. 26:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pronk, R., and P. C. van der Vliet. 1993. The adenovirus terminal protein influences binding of replication proteins and changes the origin structure. Nucleic Acids Res. 21:2293-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roovers, D. J., P. F. Overman, X. Q. Chen, and J. S. Sussenbach. 1991. Linker mutation scanning of the genes encoding the adenovirus type 5 terminal protein precursor and DNA polymerase. Virology 180:273-284. [DOI] [PubMed] [Google Scholar]
- 27.Roovers, D. J., F. M. van der Lee, J. van der Wees, and J. S. Sussenbach. 1993. Analysis of the adenovirus type 5 terminal protein precursor and DNA polymerase by linker insertion mutagenesis. J. Virol. 67:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serna-Rico, A., B. Illana, M. Salas, and W. J. Meijer. 2000. The putative coiled coil domain of the phi 29 terminal protein is a major determinant involved in recognition of the origin of replication. J. Biol. Chem. 275:40529-40538. [DOI] [PubMed] [Google Scholar]
- 29.Shamoo, Y., and T. A. Steitz. 1999. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99:155-166. [DOI] [PubMed] [Google Scholar]
- 30.Smart, J. E., and B. W. Stillman. 1982. Adenovirus terminal protein precursor. Partial amino acid sequence and the site of covalent linkage to virus DNA. J. Biol. Chem. 257:13499-13506. [PubMed] [Google Scholar]
- 31.Temperley, S. M., and R. T. Hay. 1992. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO J. 11:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truniger, V., L. Blanco, and M. Salas. 1999. Role of the “YxGG/A” motif of Phi29 DNA polymerase in protein-primed replication. J. Mol. Biol. 286:57-69. [DOI] [PubMed] [Google Scholar]
- 33.Truniger, V., L. Blanco, and M. Salas. 2000. Analysis of phi 29 DNA polymerase by partial proteolysis: binding of terminal protein in the double-stranded DNA channel. J. Mol. Biol. 295:441-453. [DOI] [PubMed] [Google Scholar]
- 34.Wang, T. S., S. W. Wong, and D. Korn. 1989. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 3:14-21. [DOI] [PubMed] [Google Scholar]
- 35.Weber, P. C., D. H. Ohlendorf, J. J. Wendoloski, and F. R. Salemme. 1989. Structural origins of high-affinity biotin binding to streptavidin. Science 243:85-88. [DOI] [PubMed] [Google Scholar]
- 36.Wong, S. W., A. F. Wahl, P. M. Yuan, N. Arai, B. E. Pearson, K. Arai, D. Korn, M. W. Hunkapiller, and T. S. Wang. 1988. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 7:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]