Abstract
The US11 protein of herpes simplex virus type 1 (HSV-1) is a small, highly basic phosphoprotein expressed at late times during infection. US11 localizes to the nucleolus in infected cells, can associate with ribosomes, and has been shown to bind RNA. The RNA substrates of US11 identified thus far have no apparent role in the virus lytic cycle, so we set out to identify a novel, biologically relevant RNA substrate(s) for this protein in HSV-1-infected cells. We designed a reverse transcriptase PCR-based protocol that allowed specific selection of a 600-bp RNA binding partner for US11. This RNA sequence, designated 12/14, is present in the coterminal HSV-1 mRNAs UL12, UL13, and UL14. We show that the binding of US11 to 12/14 is sequence-specific and mediated by the C-terminal domain of the protein. To elucidate the role of US11 in the virus life cycle, we infected cells with wild-type virus, a cosmid-reconstructed US11 HSV-1 null mutant, and a cosmid-reconstructed wild-type virus and analyzed expression of UL12, -13, and -14 during a time course of infection. These experiments revealed that this interaction has biological activity; at early times of infection, US11 down-regulates UL13 protein kinase mRNA and protein.
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus of approximately 152 kb. It encodes over 76 polypeptides (22, 35) that are expressed as three temporal classes of genes, immediate early, early, and late, and are regulated in a coordinated cascade manner. The US11 gene of HSV-1 is termed nonessential, being dispensable for virus growth in tissue culture and in animal models (19, 23, 27). The US11 protein is a 21-kDa, highly basic phosphoprotein (40) expressed at late times during infection (15). US11 is present in the nucleus, particularly concentrated in the nucleolus, and the cytoplasm (20, 33, 36) and is present in the virion as a component of the tegument (approximately 600 to 1,000 molecules per virion). US11 can associate strongly with ribosomes and has been reported to copurify with rRNA and polysome fractions (10, 38). As this protein is delivered to the host cell upon infection, it has the potential to act at any stage during the lytic cycle (37).
The US11 protein has been shown to counteract host-mediated translational shutoff. When many viruses infect cells, cRNA is synthesized and triggers activation of the cellular double-stranded-RNA-dependent protein kinase R (PKR) (16, 44). This leads to phosphorylation of the α-subunit of the eukaryotic translation initiation factor (eIF2α) with complete inhibition of translation. Most viruses also express proteins or RNAs that can block this host cell response. For example, during HSV-1 infection, a large amount of viral cRNA is present, but the viral IPC34.5 protein (4) acts via its GADD34 homology domain to redirect the activity of protein phosphatase 1α to dephoshorylate eIF2α and therefore relieve the translational block. US11 was capable of suppressing the translational block exerted by an ICP34.5 null mutant when expressed ectopically as an early gene (3, 24, 25). Subsequent studies have shown that US11 can interact directly with PKR in vitro and may itself be a substrate for PKR phosphorylation, effectively competing with cellular substrates (2) in a mechanism similar to that of human immunodeficiency virus (HIV) Tat (1). This ability to interfere with PKR activation and eIF2α phosphorylation may also explain its ability to confer increased recovery after heat shock when expressed in HeLa cells (11).
The C-terminal domain of US11 possesses an RNA binding activity that is sequence and conformation specific (9, 36, 38). Although the protein does not contain any typical RNA recognition motifs, the C-terminal domain of the protein contains an RXP repeat (34) which forms a poly-l-proline type II helix that may be capable of recognizing nucleic acid motifs (40). To date, two unrelated HSV-1 RNA substrates for US11 binding have been identified, but these appear to have little potential role in the HSV-1 lytic cycle. The first is an antisense transcript of the 5′ untranslated region (UTR) of the US11 gene itself (36). This RNA, generated in vitro, has not been detected in infected cells. The second is a 3′-truncated, nonpolyadenylated transcript of the UL34 gene which does not appear to encode a protein product and is down-regulated six- to eightfold by US11 (38). In addition, US11 has been shown to bind two RNAs from retroviruses, the human T-cell lymphotropic virus type 1 Rex response element (XRE) and the HIV type 1 Rev response element (RRE) (9, 13, 40). These elements are both intronic nuclear export signals, which bind, respectively, the Rex and Rev proteins necessary for transporting incompletely spliced viral transcripts to the cytoplasm of infected cells (14, 21). US11 can substitute directly for the Rex and Rev proteins in promoting the cytoplasmic accumulation of unspliced env RNA and can bind the XRE and RRE elements directly. It is not clear how these substrates relate to the true in vivo role of the RNA binding properties of US11 in an HSV-1-infected cell.
The objective of this study was to discover whether US11 had the potential to bind any other transcripts from HSV-infected cells that would be relevant to its role in the virus lytic cycle. We designed a system to isolate and identify biologically relevant RNA species that can bind a known protein, in this case US11. As the RNA binding partners for US11 identified thus far have few features in common and indeed can be nonpolyadenylated, one essential feature of this selection protocol was that it should isolate polyadenylated and nonpolyadenylated RNAs equally effectively. Using this method, we isolated a novel RNA binding partner for US11: a 600-bp region within three coterminal HSV-1 RNAs, UL12, UL13, and UL14. These encode, respectively, an alkaline nuclease (43), a protein kinase (29, 41), and an essential protein of unknown function (8). We demonstrate using the electrophoretic mobility shift assay (EMSA) that US11 binds this RNA fragment directly in a sequence-specific manner via its C-terminal domain. Deletion studies indicate that the binding region overlaps the central portion of the UL14 3′ UTR, the 3′ end of the UL13 coding region, and the 5′ UTR of the UL12 gene. Using an HSV-1 US11 null mutant, we show that at early times of infection US11 regulates expression of the UL13 protein kinase.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Stocks of virus were grown as described previously (22). The wild-type virus was HSV-1 strain 17+. The US11 null mutant virus was a gift from C. Cunningham and A. Davidson, Institute of Virology, Glasgow, United Kingdom, who constructed this mutant. The virus was reconstructed from the cosmid-based system developed by Cunningham and Davidson (7). Cosmid 6, containing the US11 gene region (nucleotides [nt] 144761 ← 145246; accession number NC_001806) was linearized with SphI in the presence of ethidium bromide. The cosmid DNA was then blunt ended with T4 DNA polymerase, religated, and transfected into Escherichia coli DH5α. One cosmid lacking the SphI site at position 145165 (4 bp deleted at the cleavage site) was digested with PacI to release the insert, and this was cotransfected into rabbit skin cells with the other four wild-type cosmids, 28, 14, 56, and 48. Mutant virions were recovered and shown by Southern blotting to have lost the SphI site. This mutation results in a frame shift within the US11 coding region. Twenty-seven amino acids from the N terminus are synthesized, but as the RNA binding region of US11 resides in the C terminus (9, 40), this mutant is null for RNA binding. The mutation has no effect on the US10 or US12 genes that overlap US11 (A. Davidson, personal communication). A wild-type virus reconstructed from unaltered cosmids 6, 28, 14, 56, and 48 was used as a control for the null mutant virus.
Plasmids.
The glutathione S-transferase (GST)-US11 fusion constructs and GST-US11 N-terminal (GST-US11 m6) and GST-US11 C-terminal (GST-US11 m11) deletion mutants were gifts from J.-J. Diaz, Universite Claude Bernard, Lyons, France (40). The sequence containing the Δ34 sequence was amplified from the KpnC′ fragment of HSV-1 using the primers 5′-GTGTGCACGGCGAGCTGCTC-3′ and 5′-AGCACCGACACACGCCGGTG-3′ and ligated directly into the pGEM-T Easy vector (Promega). The 600-bp 12/14 DNA and its deletions were made by PCR amplification from the HSV-1 EcoRI D fragment and incorporate a T7 promoter sequence at the 5′ end to allow in vitro transcription. The 12/14 DNA was also cloned into the pGEM T Easy vector and used for synthesis of both sense and antisense transcripts. The primer sequences are given in Table 1. The plasmid pGAPDH contained the human GAPDH coding region. All clones were checked by sequencing.
TABLE 1.
Primers used in this study
| Name of 12/14 template | Forward primer | Reverse primer |
|---|---|---|
| 12/14-SEN | CAGAGATGTAATACTCACTATAGGGCCTGGAGT TGGTGGTTAGCGTAC | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ1/6 | CAGAGATGTAATACGACTCACTATAGGGCCACC AGCTGTCGCCGGACTTC | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ2/6 | CAGAGATGTAATACGACTCACTATAGGGCGAG ACCAACACCCACGGCCTG | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ3/6 | CAGAGATGTAATACTCACTATAGGGCCGGATCG GTGTATAAATTAC | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ4/6 | CAGAGATGTAATACTCACTATAGGGCCGCGCTGT CGTGAGAATCAG | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ5/6 | CAGAGATGTAATACTCACTATAGGGCCAGCATGT CCGCCGGGACGC | GTGGTTGTCAGCGGAAGACTGTTAG |
| Δ2-4 | CAGAGATGTAATACGACTCACTATAGGGCGAGAC CAACACCCACGGCCTG | CTGATTCTCACGACAGCGCG |
| Δ2-5 | CAGAGATGTAATACGACTCACTATAGGGCGAGACC AACACCCACGGCCTG | GCGTCCCGGCGGACATGCTG |
Cell infection and preparation of extracts.
Eighty percent confluent HeLa cell monolayers (4 × 107 cells) were infected with wild-type, cosmid-reconstructed wild-type, or US11 null mutant HSV-1 at a multiplicity of infection of 10 or with no virus (mock infected). Adsorption was for 1 h at 37°C. For the preparation of cell extracts, the monolayers were washed with phosphate-buffered saline (PBS), and the cells were lysed by suspension in 1 ml of cell extract buffer (50 mM HEPES, pH 7.5, 50 mM NaCl, 0.1% NP-40 containing 1 μl of protease inhibitor cocktail [Roche Molecular Biochemicals]). Extracts were sonicated on ice, cell debris was pelleted, and the protein concentration was determined by Bradford assay (Bio-Rad).
Purification of fusion proteins.
GST-US11, GST-US11-m6, and GST-US11-m11 were expressed and purified as described by Schaerer-Uthurralt et al. (40) and dialyzed against RNA binding buffer (10 mM HEPES, pH 7.6, 60 mM KCl, 3 mM MgCl2, 1 mM DTT, 5% glycerol).
Western blotting and immunoprecipitation.
Proteins were fractionated on sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gels and electroblotted onto nitrocellulose membranes. The blots were blocked using PBS with 5% (wt/vol) dried milk powder and washed in PBS. Mouse monoclonal antibodies against US11 (a gift of B. Roizmann, University of Chicago) were used at a dilution of 1 in 5,000. The Q1 monoclonal antibody against UL12 (N. Stow, University of Glasgow) was used at a dilution of 1 in 3,000. The monoclonal antibody against ICP27 (Advanced Biotechnologies Ltd.) was used at a dilution of 1 in 3,000. Polyclonal antibodies against UL13 (Y. Nishiyama, Nagoya University School of Medicine) and UL14 (J. Baines, Cornell University) were used at dilutions of 1 in 50 and 1 in 500, respectively, in PBST (PBS with 0.05% Tween). After the primary antibody was washed off in PBST, secondary antibody (either anti-mouse-horseradish peroxidase conjugate or protein A-horseradish peroxidase) was added at a dilution of 1 in 1,000 in PBST for 1 h at room temperature. The membranes were washed as before and visualized using enhanced-chemiluminescence reagents (Amersham) according to the manufacturer's protocol.
RNA preparation, riboprobes, and RNA EMSAs.
Total cellular RNA was prepared using Trizol (Life Technologies) extraction according to the manufacturer's protocol. Riboprobes for EMSA were transcribed, using the Promega in vitro transcription kit, from PCR-amplified DNA, including a T7 promoter at the 5′ end, or from inserts cloned into pGEM-T Easy using T7 or SP6 polymerase in the presence of [α-32P]UTP. For the EMSAs, 250 ng of GST or GST fusion protein (unless otherwise indicated) was incubated on ice in 10 mM HEPES (pH 7.6)-60 mM KCl-3 mM MgCl2-1 mM DTT-5% glycerol in the presence of 10 μg of tRNA and 40 U of RNasin for 10 min; 20,000 cpm of probe was denatured for 1 min at 80°C and renatured for 2 min on ice before being added to the binding reaction mixture, which was then incubated at 4°C for 10 min. The complexes were resolved on native 4% polyacrylamide gels electrophoresed at 14 V/cm.
Northern and Southern blotting.
RNA was prepared using Trizol (Life Technologies) extraction according to the manufacturer's protocol, phenol-chloroform extracted twice, chloroform extracted once, and ethanol precipitated. Polyadenylated RNA was prepared by affinity chromatography on oligo(dT) cellulose columns as described previously (39). One microgram of poly(A)+ RNA was fractionated on a denaturing 1.2% agarose-2.2 M formaldehyde gel and Northern blotted exactly as described previously (39). Southern blots were prepared as described previously (Hybond protocol; Amersham). Northern and Southern blots were hybridized with 32P-labeled in vitro-transcribed Δ34 riboprobe in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-50% formamide at 55°C for 16 h. The blots were washed to 0.1× SSC at 65°C and subjected to autoradiography.
GST-US11 RNA pull downs.
For the GST fusion protein pull downs, 50 μg of purified recombinant GST-US11 or GST alone was pretreated with 50 U of micrococcal nuclease at 30°C for 15 min. The reaction was stopped by the addition of EDTA to a final concentration of 5 mM, and the reaction mixture was incubated for 10 min on ice. Glutathione-agarose beads were preswollen and washed in binding buffer (150 mM NaCl, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris-Cl [pH 8.0], 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride) and resuspended to form a 50% slurry. Seventy-five microliters of the 50% slurry was incubated with 50 μg of protein on a rotating wheel for 5 min at room temperature. The beads were then pelleted, washed five times with binding buffer, and resuspended in 100 μl of binding buffer. Cellular extracts were precleared with GST-glutathione-agarose by adding 100 μl of GST-glutathione-agarose (prepared as described above) to 100 μl total (pooled nuclear and cytoplasmic fractions prepared by the method of Dignam et al. [12]) HeLa cell extract (diluted with binding buffer to 500 μg/ml) and incubated for 30 min at 4°C on a rotating wheel. The beads were then pelleted, and the precleared extract was used in the pull-down experiment. Heparin was added to the extract at 1 μg/μl along with 100 μl of either GST-glutathione-agarose or GST-US11-glutathione-agarose, and the mixture was incubated at 4°C for 1 h. The beads were pelleted and washed three times in binding buffer containing 5 mg of bovine serum albumin/ml, once with binding buffer containing 200 mM NaCl, and three times with binding buffer alone. The beads were resuspended in 500 μl of Tris-EDTA-0.5% sodium dodecyl sulfate and boiled for 2 min to elute the protein-RNA complexes. The RNA was extracted twice with phenol-chloroform and once with chloroform and ethanol precipitated.
Reverse transcriptase (RT) PCR.
For first-strand synthesis of cDNA, 5 μl of the pulled-down RNA was reverse transcribed using the 5′ rapid amplification of cDNA ends (RACE) system (Life Technologies) as specified in the manufacturer's protocol except that 2.5 pmol of a random 6-mer or 9-mer primer (Life Technologies) was used instead of the first gene-specific primer. The cDNA was isolated using GlassMAX spin columns and terminal deoxynucleotidyl transferase homopolymer tailing with CTP. For the first round of PCR, the reaction mixture contained 5 μl of dC-tailed cDNA; 20 pmol of random primer; 20 pmol of abridged anchor primer (Life Technologies); 1 mM (each) dATP, dCTP, dTTP, and dGTP; 45 mM Tris-HCl, pH 8.8; 11 mM (NH4)2SO4; 4.5 mM MgCl2; 6.7 mM 2-mercaptoethanol; 4.4 μM EDTA, pH 8.0; 113 μg of bovine serum albumin/ml; and 4 U of Taq DNA polymerase (Applied Biosystems). The PCR cycle consisted of a hot start followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 4 min with a final extension at 72°C for 15 min. For the second round of PCR, 5 μl of a 1-in-50 dilution of the first PCR mixture was used under the same reaction conditions as detailed above, except that 20 pmol of the abridged universal amplification primer (Life Technologies) was used instead of the abridged anchor primer. The PCR products were fractionated on 6% native acrylamide gels. The PCR products were cloned into pGEM-T Easy vector and sequenced, using the recommended primers for pGEM, by ABI-PRISM automated sequencing.
RESULTS
Isolation of RNA substrates for US11.
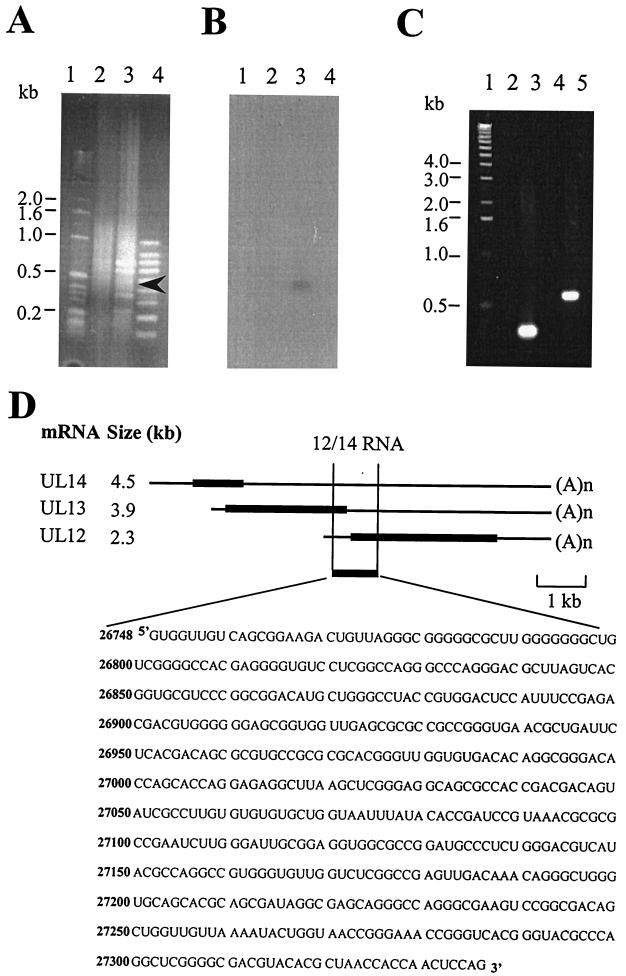
Figure 1 summarizes the method we devised to isolate RNA substrates for the US11 protein. In brief, GST or GST-US11 protein was incubated with total HSV-1-infected HeLa extract (16 h postinfection) and subjected to a pull-down using glutathione-agarose. A time of 16 h postinfection was chosen because US11 is expressed as a true late protein (15). After being washed extensively, the RNA was eluted, phenol-chloroform extracted, and ethanol precipitated. RNA was reverse transcribed using 6- or 9-mer random primers. The single-stranded cDNA fragments were C-tailed and subjected to two rounds of PCR with either 6- or 9-mer random primers and 5′ RACE primer sets (Life Technologies). Figure 2A shows fractionation on an ethidium bromide-stained gel of the PCR products from a typical reaction carried out using 6-mer primers. While a smear of products with few distinct bands was seen with GST alone (Fig. 2A, lane 2), the GST-US11 pull-down yielded prominent bands of between 200 and 600 bp (Fig. 2A, lane 3). Since one of these products was expected to represent Δ34 RNA, the gel was Southern blotted and hybridized with a radiolabeled in vitro-transcribed Δ34 probe. Figure 2B, lane 3, shows that a single band of <500 bp hybridized with this probe, indicating that this procedure was capable of isolating a known RNA bound by US11. In contrast to the result with the 6-mer primers, only a single 600-bp fragment was obtained in a similar experiment using the 9-mer primers (Fig. 2C, lane 5), and again, no products were seen with GST alone (Fig. 2C, lane 4). As a positive control, gene-specific primers were used to amplify the Δ34 RNA from the pull-down RNA populations. As before, no products were seen with GST alone (Fig. 2C, lane 2), but the expected 300-bp fragment was obtained with GST-US11 (Fig. 2C, lane 3). The identity of this species was confirmed as Δ34 when it was cloned into pGEM-T Easy and sequenced.
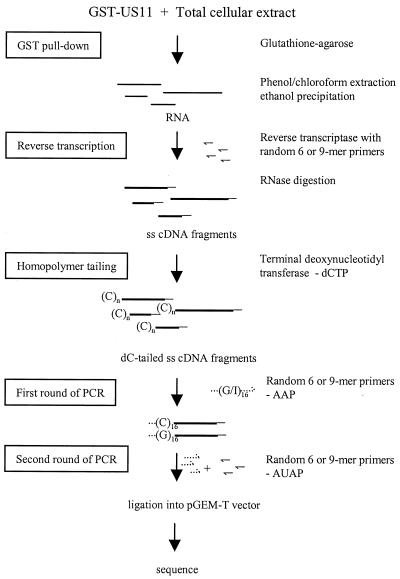
FIG. 1.
Scheme for the isolation and identification of RNAs that bind US11. The thin straight lines represent RNA molecules. The thick straight lines represent single-stranded (ss) cDNA molecules. The doubly thick straight lines represent double-stranded cDNA molecules. The solid hooked lines represent single-stranded random primers. The dotted hooked lines represent 5′ RACE primers (Life Technologies). Abbreviations: AAP, abridged anchor primer; AUAP, abridged universal amplification primer; (C)n, dC tail.
FIG. 2.
Analysis of the products of the 6-mer and 9-mer RT-PCRs of the US11 RNA pull down. (A) Fractionation of the products of the 6-mer RT-PCR on an ethidium bromide-stained agarose gel. Lanes: 1, 1-kb ladder marker; 2, RT-PCR with RNA isolated using GST alone in the pull down; 3, RT-PCR with RNA isolated using GST-US11 fusion protein in the pull down (the arrowhead indicates the band that hybridizes to the Δ34 probe in the blot in panel B); 4, 100-bp marker. (B) Southern blot of the gel shown in panel A hybridized with an in vitro-transcribed 32P-labeled Δ34 probe. (C) Fractionation of the products of the 9-mer RT-PCR on an ethidium bromide-stained agarose gel. Lanes: 1, 1-kb ladder marker; 2, control RT-PCR using Δ34-specific primers and RNA isolated using GST alone in the pull down; 3, control RT-PCR using Δ34-specific primers and RNA isolated using GST-US11 fusion protein in the pull down; 4, RT-PCR with random and anchor primers and RNA isolated using GST alone in the pull down; 5, RT-PCR with random and anchor primers and RNA isolated using GST-US11 fusion protein in the pull down. (D) Diagram of the three coterminal transcripts UL12, UL13, and UL14 showing the position of the US11-pulled-out 12/14 RNA and the sequence of the 585-nt 12/14 RNA. Thick lines, coding regions; thin lines, 5′ and 3′ UTRs; (A)n, end of the message.
Since the 9-mer RT-PCR protocol yielded a single specific product, of a size very similar to that of one of the products of the 6-mer reaction, that was not homologous to Δ34 RNA, we analyzed this cDNA further. The 600-bp fragment was cloned into pGEM-T Easy and sequenced. The RNA was found to map to the unique long (UL) region of the HSV-1 genome and corresponded to the 5′ end of the UL12 gene, the 3′ end of the UL13 coding region, and a portion of the 3′ UTR and the central portion of the UL14 3′ UTR (Fig. 2D). This RNA was designated 12/14.
US11 binds 12/14 RNA directly in a sequence-specific manner.
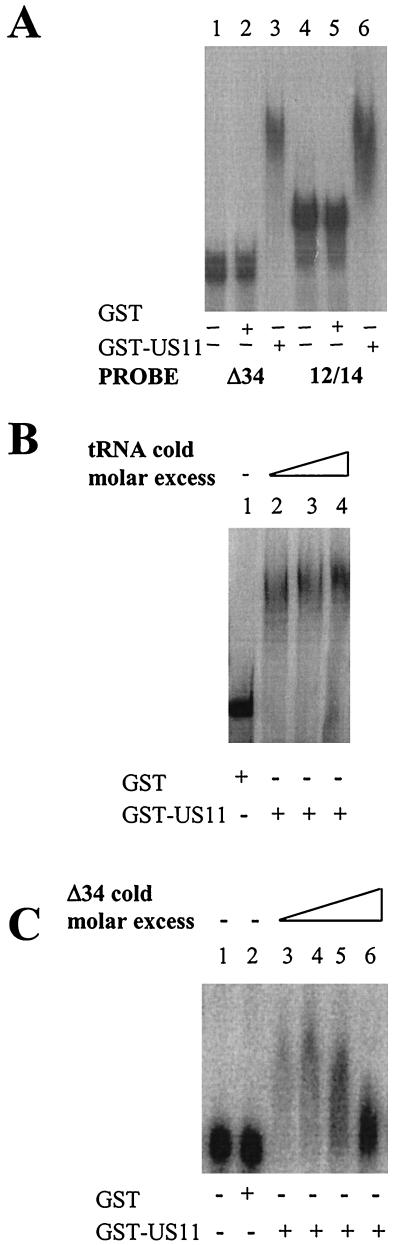
In order to assay the interaction of these putative partners, radiolabeled 12/14 RNA was tested for binding to US11 in EMSAs. While GST alone generated no shift in the mobility of the probe (Fig. 3A, lane 5), GST-US11 caused significant retardation of the probe (Fig. 3A, lane 6). In the same experiment, an in vitro-transcribed radiolabeled Δ34 probe was also significantly retarded by the GST-US11 protein as expected (Fig. 3A, lane 3). EMSA was also carried out in the presence of a nonspecific competitor, yeast tRNA, and no decrease in 12/14 probe binding was observed up to a 1,000-fold molar excess of cold competitor tRNA (Fig. 3B, lanes 2 to 4). EMSA was also carried out with a nonspecific 310-nt RNA. This RNA, in vitro transcribed from a PCR product homologous to the late 3′ UTR of HPV-31, was not bound by US11 (data not shown). Finally, a specific competition experiment with unlabeled Δ34 RNA was carried out. Figure 3C shows that only at over fourfold molar excess of Δ34 RNA and above is there reduced binding of US11 to 12/14 RNA. This result demonstrates that US11 protein can bind the 12/14 sequence directly and specifically in vitro in a manner similar to its binding of the Δ34 probe.
FIG. 3.
EMSA analysis of binding of radiolabeled 12/14 RNA to US11 protein. (A) EMSA of the binding of US11 to 32P-labeled RNAin vitro transcribed from PCR-amplified Δ34 or 12/14 DNA. Protein (0.25 μg) was added (+) in each reaction. Lanes 1 and 4, no added protein; lanes 2 and 5, GST alone; lanes 3 and 6, GST-US11 fusion protein. (B) EMSA of US11/-12/-14 binding in the presence of nonspecific competitor tRNA. Lanes: 1, 12/14 probe incubated with GST protein; 2 to 4, 12/14 probe and GST-US11 protein bound in the presence of increasing concentrations of tRNA. (C) EMSA of binding of US11 to 32P-labeled 12/14 RNA in the presence of cold specific competitor Δ34 RNA. Lanes: 1, no protein; 2, GST protein; 3 to 6, 12/14 probe (1.5 pmol) and GST-US11 protein bound in the presence of increasing concentrations (one- to eightfold molar excess) of unlabeled competitor Δ34 RNA.
The C-terminal domain of US11 binds 12/14 RNA.
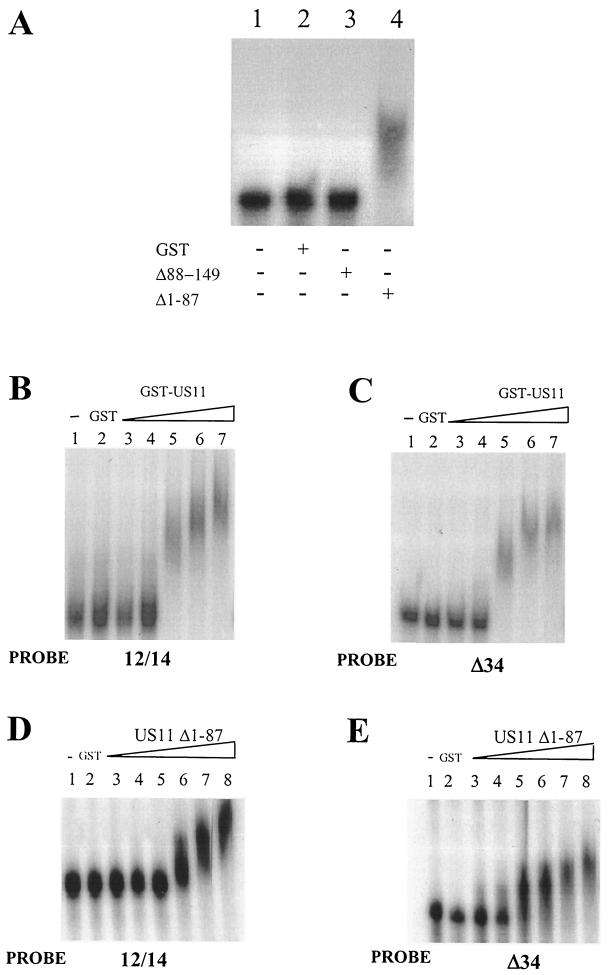
Previously, US11 had been shown to bind Δ34 and RRE/XRE RNA through its C-terminal domain (9, 40). We determined whether 12/14 RNA also interacted with US11 through its C-terminal domain. Figure 4A, lane 3, shows that the N-terminal domain of the protein (US11 Δ88-149) does not retard the 12/14 RNA but the C-terminal domain (US11 Δ1-87) retards the RNA significantly (Fig. 4A, lane 4). Using the full-length GST-US11 protein, we investigated the effects of increasing concentrations of protein on formation of the US11/-12/-14 RNA complex. At low protein concentrations, no complex formed (Fig. 4B, lanes 3 and 4). However, when the protein concentration was increased further, a retarded complex began to form (Fig. 4B, lane 5). We found that the US11/-12/-14 complex became more retarded with increasing protein concentrations (Fig. 4B, lanes 6 and 7). A very similar effect was observed with a Δ34 probe (Fig. 4C). This effect was specific for the RNA binding C-terminal protein (Fig. 4D). Higher-order complexes could be seen clearly with the 12/14 probe (Fig. 4D, lanes 6 to 8) and also with the Δ34 probe (Fig. 4E, lanes 5 to 8). It has been suggested that US11 may multimerize on Δ34 RNA (9, 40), although this has not yet been formally proven and other explanations for reduced mobility of US11/RNA complexes in EMSA may exist.
FIG. 4.
EMSA of binding of wild-type and truncated US11 proteins to radiolabeled 12/14 RNA. (A) Binding of N- and C-terminal truncations of US11 to 12/14 RNA using 32P-labeled 12/14 probe. Lane 1, no protein; lane 2, GST protein; lane 3, C-terminal truncation; lane 4, N-terminal truncation. +, present; −, absent. (B) Multimerization of US11 on 12/14 RNA. Lane 1, no added protein; lane 2, GST protein; lanes 3 to 7, increasing concentrations (110 to 550 nM) of wild-type GST-US11 fusion protein. (C) Multimerization of US11 on Δ34 RNA. Lane 1, no added protein; lane 2, GST protein; lanes 3 to 7, increasing concentrations (110 to 550 nM) of wild-type GST-US11 fusion protein. (D) Multimerization of the US11 N-terminal truncation protein on 12/14 RNA. Lane 1, no added protein; lane 2, GST protein; lanes 3 to 8, increasing concentrations (150 to 1,500 nM) of GST-US11Δ1-87 fusion protein. (E) Multimerization of the US11 N-terminal truncation protein on Δ34 RNA. Lane 1, no added protein; lane 2, GST protein; lanes 3 to 8, increasing concentrations (150 to 1,500 nM) of GST-US11Δ1-64 fusion protein.
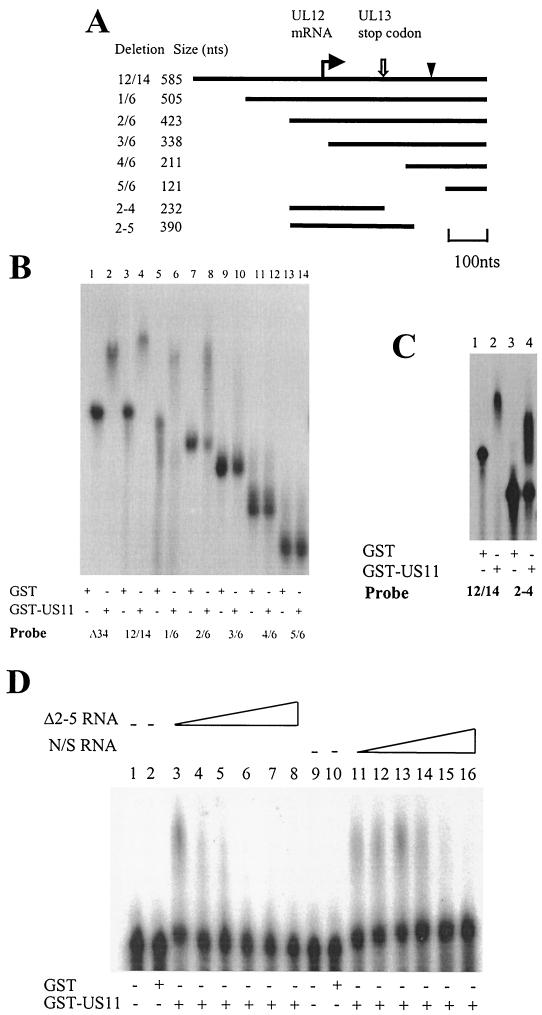
Mapping of the US11 binding site on 12/14 RNA.
Deletion mutants of the 12/14 RNA were synthesized by PCR using the primers shown in Table 1. Figure 5A shows the extents of these deletions on the RNA. EMSA with radiolabeled RNA, in vitro transcribed from PCR-amplified templates corresponding to each of the 12/14 deletions, showed that deletions of 80 (deletion 1/6 [Fig. 5B, lane 6]) and 162 nt (deletion 2/6 [Fig. 5B, lane 8]) from the 5′ end retained binding activity. However, the next deletion of 247 nt (deletion 3/6) showed no retardation of the probe (Fig. 5B, lane 10), and further deletions from the 5′ end failed to bind US11. Thus, the central portion of 12/14 appeared to be necessary for binding the US11 protein. To determine whether this region was sufficient for binding, a template corresponding to the 5′ end of deletion 2/6 through to the UL13 stop codon was constructed (Fig. 5A). Figure 5C, lane 4, shows that the 2-4 deletion was efficiently retarded by the US11 protein, confirming that the central portion of the 12/14 RNA contains the GST-US11 protein binding site. We were unable to detect binding of US11 to any smaller 12/14 deletion probes, indicating that the US11 binding site is quite large at around 230 nt. The binding site lies within the 5′ UTR of UL12 RNA, the 3′ end of the coding region of UL13 RNA, and the 3′ UTR of UL14 RNA (Fig. 5A). We PCR amplified the 290-nt region within 12/14 RNA that included the US11-interacting region down to just before the start codon of the UL12 gene. This Δ2-5 deletion is bound efficiently by the US11 protein (Fig. 5D, lanes 3 and 11). We carried out a specific and nonspecific EMSA competition experiment with this probe to confirm that binding of US11 was sequence specific. With increasing concentrations of cold Δ2-5 RNA, less US11/RNA complex is formed (Fig. 5D, lanes 4 to 8). However, when the competitor is an unrelated RNA of a similar size (HPV-31 late 3′ UTR; 310 nt), competition does not occur until >8-fold molar excess of transcript is present in the reaction (Fig. 5D, lanes 11 to 16).
FIG. 5.
Deletion analysis of the US11 binding site on 12/14 RNA. (A) Diagram of the 12/14 RNA showing locations of the deletion probes used. The bent arrow indicates the start of the UL12 RNA. The open arrow indicates the position of the UL13 stop codon. The arrowhead represents the start codon of the UL12 RNA. (B) EMSA of GST and GST-US11 protein binding to sequential 5′-3′ deletions of the 12/14 RNA. Protein (0.25 μg) was added in each reaction. The RNA probes used are indicated beneath the panel. (C) EMSA comparing GST-US11 binding to α-32P-labeled RNA in vitro transcribed from PCR-amplified 12/14 and 2-4 deletion DNAs. Lanes 1 and 3, GST protein; lanes 2 and 4, GST-US11 protein. +, present; −, absent. (D) EMSA of binding of US11 protein to α-32P-labeled Δ2-5 RNA in the presence of specific (Δ2-5 RNA, lanes 3 to 8) and nonspecific (N/S) (HPV-31 late 3′ UTR; lanes 11 to 16) cold competitor RNA. Protein (0.5 μg) was added in each reaction. Lanes 1 and 9, no protein; lanes 2 and 10, GST protein; lanes 3 to 8 and 11 to 16, GST-US11 protein.
US11 regulates expression of UL13.
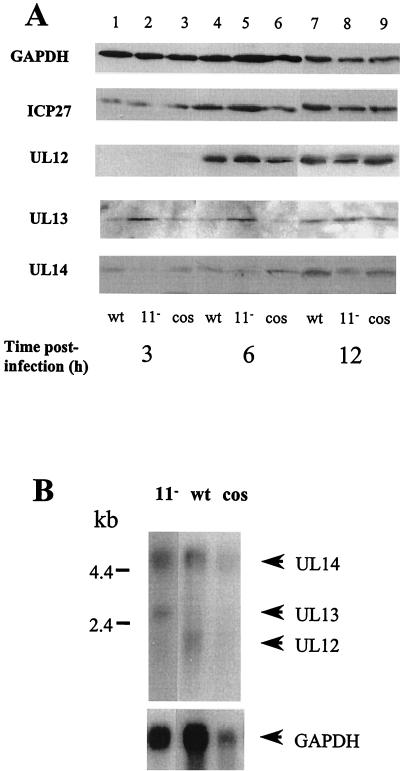
As the US11 protein binds RNA corresponding to the coterminal region of the UL12, -13, and -14 genes, we tested whether US11 could regulate expression of these proteins in HSV-1-infected cells. HeLa cells were infected with wild-type HSV-1, the cosmid-reconstructed wild-type virus, and the cosmid-reconstructed HSV-1 US11 null mutant for 3, 6, and 12 h. Cellular protein was isolated and used in Western blotting to determine whether there was any change in expression of the three proteins. Only half the amount of protein was electrophoresed in each of the 12-h sample lanes in order to avoid overexposure of the autoradiograph following enhanced-chemiluminescence development. Figure 6A shows that steady-state levels of all three proteins increased with time postinfection as expected. There was very little difference in the levels of UL12 protein in any of the infected cell extracts at each of the time points examined. For UL13 at early times of infection, there was a consistent increase in the amount of UL13 protein present in cells infected with the US11 null mutant virus compared with cells infected with the wild-type viruses. In contrast, there was a small decrease in levels of UL14 protein at all time points in the US11 null mutant-infected cell extracts compared to the wild-type and cosmid-reconstructed virus-infected cell extracts. IPC27 protein is expressed from a region of the genome outside the 12/14 region. This protein increased in abundance during infection as expected, but there were no significant differences in the levels of this protein in lysates from the wild-type and mutant viruses. The cellular GAPDH protein was equally abundant in lysates of cells infected with the wild-type and mutant viruses at each time postinfection. These results indicate that US11 down-regulates expression of UL13 protein at early times of infection and may up-regulate expression of UL14 protein throughout an infection.
FIG. 6.
US11 specifically regulates expression of UL13 RNA and protein. (A) Western blot analysis of levels of ICP27; UL12, -13, and -14 proteins; and GAPDH (to assess protein loading) during a time course of infection of HeLa cells with wild-type (wt), cosmid-reconstructed wild-type (cos), and US11 null mutant (11−) viruses. Only half the amount of protein was electrophoresed in each lane for the 12-h samples. Mock-infected cell extracts did not show binding to any HSV-1 protein antibody (data not shown). Each blot was repeated three times with very similar results. (B) Northern blot analysis of abundances of UL12, -13, and -14 polyadenylated transcripts in HeLa cells at 6 h postinfection with wild-type, cosmid-reconstructed wild-type, and US11 deletion mutant viruses. The probe was α-32P-radiolabeled RNA in vitro transcribed from the cloned portion of the HSV-1 genome containing the UL12/-13/-14 coterminal RNAs. The bottom blot shows the result of stripping and rehybridizing the blot with a probe for GAPDH RNA. The blot was hybridized in 50% formamide-5× SSC at 42°C for 16 h and was washed to 0.1× SSC at 65°C.
US11 protein regulates expression of UL13 at the level of the RNA.
To check whether regulation of expression of the UL12, UL13, and UL14 proteins was reflected in RNA levels, poly(A)+ RNA was isolated from HeLa cells infected for 6 h with wild-type virus, cosmid-reconstructed virus, and US11− mutant virus. Northern blot analysis of these RNAs fractionated on a denaturing formaldehyde gel and probed with an antisense 12/14 RNA probe showed that while UL14 RNA was expressed at similar steady-state levels in cells infected with the wild-type and null mutant virus, the level of UL13 RNA was significantly increased in cells infected with the US11 null mutant virus. In contrast, there was less UL12 RNA in cells infected with this virus. The level of hybridization to the lane containing cosmid-reconstructed wild-type virus was much weaker than the levels of hybridization to the other lanes due to less RNA loaded (cf. the GAPDH control lane), but very little UL13 RNA could be detected. This result provides further evidence that US11 regulates expression of UL13 RNA in virus-infected cells but may also affect the steady-state levels of UL12 RNA.
DISCUSSION
Previous studies of the RNA binding properties of the HSV-1 US11 protein identified three RNA substrates: an in vitro-generated antisense transcript of the 5′ UTR of the US11 transcript (36); a truncated, nonpolyadenylated transcript of the HSV-1 UL34 gene (38); and the RREs and XREs of HIV type 1 and human T-cell lymphotropic virus type 1, respectively (9). However, as the two HSV-1 RNAs most likely do not encode a protein product and the XREs and RREs are from a heterologous viral system, it is unclear how they might be relevant to the HSV-1 life cycle. We set out to determine if US11 could bind any biologically relevant RNAs in HSV-1-infected cells. To accomplish this, we developed a novel method for isolating RNA molecules that bind a specific protein and used this technique to isolate an RNA that bound the US11 protein. Our data show that this RNA is derived from a sequence present in the HSV-1 3′-coterminal mRNAs encoding UL12, UL13, and UL14. In agreement with what has been found for other US11/RNA complexes, US11 binding to this RNA is sequence specific and is mediated by the C-terminal domain. Moreover, we demonstrate that this interaction has biological activity, as US11 can down-regulate expression of UL13 at the RNA and protein levels and up-regulate UL12 RNA and UL14 protein in HSV-1-infected cells.
Designing an RT-PCR protocol to isolate RNA binding partners of US11 presented an interesting challenge for several reasons. First, the RNA substrates of US11 identified thus far have no real sequence similarities and the Δ34 RNA and the antisense transcript of the 5′ UTR of US11 are nonpolyadenylated, while the XREs and RREs are polyadenylated. There are very few methods detailed for the isolation of RNA binding partners to proteins; those that exist have been developed largely using hnRNPs and are dependent on the presence of a poly(A) tail as a priming site for RT-PCR (1a, 42). However, US11 appears to bind a small number of RNAs (38), and so the protocol we devised had to combine sensitivity with the capacity to isolate nonpolyadenylated RNAs as efficiently as polyadenylated species. Thus, random primers were used to ensure that no sequence constraints were imposed upon RT-PCR of the US11 binding RNA, and use of 9-mer primers, rather than the shorter random primers, conferred increased specificity on the reaction. With the 6-mer primers, we isolated at least five cDNA species, one of which is the known binder Δ34 RNA, while with the 9-mer primers, we isolated a single RNA species, the 12/14 RNA that bound US11. The remaining cDNA species pulled out in the 6-mer RT-PCR are being identified and checked for US11 binding. We expect that the method we have devised will be applicable to other known proteins that bind unknown RNA substrates.
Our RNA binding studies demonstrate that the 12/14 RNA binds US11, specifically the C terminus of the protein, as previously reported for US11 binding of Δ34 RNA. Although similar concentrations of full-length GST-US11 fusion protein were required to bind the Δ34 and 12/14 RNAs, a twofold-higher concentration of the N-terminally truncated GST-US11 protein was required to bind 12/14 RNA than was needed for binding Δ34 RNA. This suggests that efficient binding of 12/14 RNA may require other regions of the US11 protein in cooperation with the RNA binding domain, perhaps to provide stability of binding of this much larger transcript.
It has been reported that the US11 protein can multimerize on Δ34 RNA via its C-terminal domain (9, 40). Although other explanations for this phenomenon may exist, e.g., novel protein-RNA conformations at higher concentrations of US11 protein, our EMSA data with the Δ34 probe confirm the previously observed effect and show further that this also occurs with the 12/14 RNA. There is a significant increase in retardation of the probe with increasing concentrations of either wild-type or N-terminally truncated GST-US11 protein. Deletion analyses indicate that the US11 binding site on the 12/14 RNA is at 230 nt, around the size of the binding site for Δ34 RNA (38). This binding site maps to a region in the 5′ UTR of UL12 RNA, the 3′ end of the coding region of UL13 RNA, and the 3′ UTR of UL14 RNA.
One problem with our studies comparing UL12, -13, and -14 expression in HeLa cells infected with wild-type and US11 null mutant viruses is that the virus we have used as a control is a wild-type cosmid-reconstructed virus. Following isolation, restriction enzyme digest patterns of this cosmid were as expected, but the entire mutant cosmid was not sequenced. The four cosmids not containing the US11 locus are the same in the cosmid-reconstructed and US11 null mutant viruses, ruling out the possibility that the phenotypes we report are the result of any secondary mutation in these cosmids. However, without isolating a virus in which the mutation has been repaired, we cannot exclude the possibility that the US11 locus-containing cosmid contains a secondary mutation or that a mutation arose during isolation of the US11 null mutant virus that might contribute to the effects we have observed.
US11 can bind to and down-regulate the expression of the truncated product of the UL34 gene, Δ34 RNA. The mechanism of the control of expression is unknown, but it has been suggested that US11 may be acting as an antiterminator allowing increased read-through of the UL34 gene or that US11 may act by inducing the rapid nucleocytoplasmic export of full-length UL34 mRNA, hence decreasing the chance of nuclear endonucleolytic cleavage to yield the Δ34 RNA (6, 38). Clearly, US11 also negatively regulates expression of UL13. Although our experiments do not rule out the possibility that US11 controls the transcription of the UL13 gene, US11 binds UL13 transcripts, making it most likely that US11 also regulates UL13 RNA posttranscriptionally. Unlike the Rev and Rex proteins, US11 is associated with polysomes as well as nucleolar ribosomes at early times of infection (37). RNA degradation is a polysome-associated process, and it is possible that US11 acts to direct UL13 and Δ34 RNAs to the cytoplasm and to these sites for degradation. The situation is very different for UL14. Although levels of UL14 RNA are very similar in cells infected with wild-type and US11 null mutant viruses, UL14 protein levels are up-regulated by US11 protein, indicating some form of translational control. In contrast, steady-state levels of UL12 transcripts are reduced in cells infected with the US11 null mutant, but this is not reflected in the protein levels. However, we cannot conclude that there exists a direct correlation between the various RNA and protein levels, as we have not assayed the rate of de novo synthesis of each protein. It remains to be determined how US11 binds and regulates the expression of UL12 RNA and the UL13 and UL14 proteins.
UL13 is a protein kinase that is nonessential for virus growth (5). It is expressed with late gene kinetics (29) but is also a component of the tegument and is thus present in cells from the start of infection (5, 29). UL13 has been implicated in the phosphorylation of ICP22 in particular but also of VP22, US1.5 (29, 32), ICP0 (28), gE, gI (26), eF-1δ (17), and RNA polymerase II (18). Purves et al., (31) demonstrated that in the absence of UL13 there is a decrease in accumulation of a subset of late mRNAs, including US11 itself and ICP35; the phosphorylation of ICP22 by UL13 late in infection is likely required to allow this accumulation. One possibility that fits with the timing of the regulatory interaction of US11 with UL13 RNA is that US11 may suppress expression of UL13 at early times of infection to keep it below a certain threshold level to stall the accumulation of these late RNAs. UL13 protein kinase counterparts are found in all members of the family Herpesviridae, whereas US11 is found only in HSV-1 and HSV-2. This implies that US11 is not required to modulate the UL13 counterparts in the other members of the virus family. It would be reasonable to assume that if inhibition of UL13 activity at early times of infection is functionally important in the infected cell, another regulation pathway must be employed in these related viruses.
Acknowledgments
We thank Andrew Davidson and Charles Cunningham for providing the US11 null mutant virus, Jean-Jacques Madjar and Jean-Jacques Diaz for generously providing the GST-US11 constructs, Y. Nishiyama for providing the anti-UL13 antibody, Joel Baines for the anti-UL14 antibody, and Nigel Stow for the anti-UL12 antibody. We are grateful to Alasdair MacLean for critical reading of the manuscript.
REFERENCES
- 1.Brand, S. R., R. Kobayashi, and M. B. Matthews. 1997. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J. Biol. Chem. 272:8388-8395. [DOI] [PubMed] [Google Scholar]
- 1a.Brooks, S. A., and W. F. Rigby. 2000. Characterisation of the mRNA ligands bound by the RNA binding protein hnRNP A2 utilizing a novel in vivo technique. Nucleic Acids Res. 28:E49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus Us11 protein effectively compensates for the gamma134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassady, K. A., M. Gross, and B. Roizman. 1998. The second-site mutation in the herpes simplex virus recombinants lacking the gamma1 34.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J. Virol. 72:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, J., and B. Roizman. 1992. The gamma134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, B. R. 1996. Virology. New tricks from an old foe. Nature 379:208-209. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, C., and A. J. Davidson. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:115-124. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham, C., A. J. Davidson, A. R. MacLean, N. S. Taus, and J. D. Baines. 1995. Herpes simplex virus type1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 74:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, J.-J., M. D. Dodon, N. Schaerer-Uthurralt, D. Simonin, K. Kindbeiter, L. Gazzolo, and J.-J. Madjar. 1996. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 379:273-277. [DOI] [PubMed] [Google Scholar]
- 10.Diaz, J.-J., D. Simonin, T. Masse, P. Deviller, K. Kindbeiter, L. Denoroy, and J.-J. Madjar. 1993. The herpes simplex virus type 1 US11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J. Gen. Virol. 74:397-406. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Latoud, C., J.-J. Diaz, N. Fabre-Jonca, K. Kindbeiter, J.-J. Madjar, and A. P. Arrigo. 1997. Herpes simplex virus Us11 protein enhances recovery of protein synthesis and survival in heat shock-treated HeLa cells. Cell Stress Chaperones 2:119-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. LeBovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duc-Dodon, M., I. Mikaelian, A. Sergeant, and L. Gazzolo. 2000. The herpes simplex virus 1 Us11 protein cooperates with suboptimal amounts of human immunodeficiency virus type 1 (HIV-1) Rev protein to rescue HIV-1 production. Virology 270:43-53. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, M., J. Inoue, H. Toyoshima, T. Akizawa, M. Higashi, and M. Yoshida. 1989. HTLV-1 rex and HIV-1 rev act through similar mechanisms to relieve suppression of unspliced RNA expression. Oncogene 4:1275-1279. [PubMed] [Google Scholar]
- 15.Johnson, P. A., C. MacLean, H. S. Marsden, R. G. Dalziel, and R. D. Everett. 1986. The product of gene US11 of herpes simplex virus type 1 is expressed as a true late gene. J. Gen. Virol. 67:871-883. [DOI] [PubMed] [Google Scholar]
- 16.Katze, M. G. 1995. Regulation of the interferon-inducible PKR: can viruses cope? Trends Microbiol. 3:75-78. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1998. Eukaryotic elongation factor 1 delta is hyperphosphorylated by the protein kinase encoded by the U(L)13 gene of herpes simplex virus 1. J. Virol. 72:1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longnecker, R., and B. Roizman. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573-576. [DOI] [PubMed] [Google Scholar]
- 20.MacLean, C. A., F. J. Rixon, and H. S. Marsden. 1987. The products of gene US11 of herpes simplex virus type 1 are DNA-binding and localised to the nucleoli of infected cells. J. Gen. Virol. 68:1929-1937. [DOI] [PubMed] [Google Scholar]
- 21.Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev transactivator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch, D. J., M. A. Dalrymple, A. J. Davidson, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 23.Meignier, B., R. Longnecker, P. Mavromara-Nazos, A. G. Sears, and B. Roizman. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251-254. [DOI] [PubMed] [Google Scholar]
- 24.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey, M., J. Poppers, A. Ladd, and I. Mohr. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage on mutants deficient in a GADD34-related function. J. Virol. 73:3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with gylcoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama, Y., R. Kurachi, T. Daikoku, and K. Umene. 1993. The US 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419-423. [DOI] [PubMed] [Google Scholar]
- 28.Ogle, W. O., T. I. Ng, K. L. Carter, and B. Roizman. 1997. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology 2:413.. [DOI] [PubMed] [Google Scholar]
- 29.Overton, H., D. McMillan, L. S. Klavinshis, L. Hope, A. J. Ritchie, and P. Wong-Kai-In. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 30.Overton, H., D. McMillan, L. Hope, and P. Wong-Kai-In. 1994. Production of host shutoff-defective mutants of herpes simplex virus type 1 by inactivation of the UL13 gene. Virology 202:97-106. [DOI] [PubMed] [Google Scholar]
- 31.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puvion-Dutilleul, F. 1987. Localisation of viral-specific 21 kDa protein in nucleoli of herpes simplex infected cells. Eur. J. Cell Biol. 343:487-498. [PubMed] [Google Scholar]
- 34.Rixon, F. J., and D. J. McGeoch. 1984. A 3′ co-terminal family of mRNAs from the herpes simplex virus type 1 short region: two overlapping reading frames encode unrelated polypeptides one of which has a highly reiterated amino acid sequence. Nucleic Acids Res. 12:2473-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman, B., and A. E. Sears. 1990. Herpes simplex viruses and their replication, p. 1795-1894. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. R. Roizman (ed.), Fields virology. Raven Press, New York, N.Y.
- 36.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein Us11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roller, R. J., and B. Roizman. 1991. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J. Virol. 65:5873-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schaerer-Uthurralt, N., M. Erard, K. Kindbeiter, J.-J. Madjar, and J.-J. Diaz. 1998. Distinct domains in herpes simplex virus type 1 US11 protein mediate post-transcriptional transactivation of human T-lymphotropic virus type 1 envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593-1602. [DOI] [PubMed] [Google Scholar]
- 41.Smith, R. F., and T. F. Smith. 1989. Identification of new protein kinase-related genes in three herpes viruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 63:450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trifillis, P., N. Day, and M. Kiledjian. 1999. Finding the right RNA: identification of cellular mRNA substrates for RNA-binding proteins. RNA 5:1071-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weller, S. K., M. R. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterisation of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]
- 44.Williams, B. R. G. 1999. PKR, a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]