Abstract
Treatment of patients with lamivudine (3TC) results in loss of detectable levels of hepatitis B virus (HBV) DNA from serum; however, the relapse rate, with regard to both reappearance of virus in the bloodstream and hepatic inflammation, is high when therapy is terminated. Although the rebound observed in patients has also been seen in animal hepadnavirus models, rebound has not been analyzed in an in vitro cell culture system. In this study, we used the HBV recombinant baculovirus/HepG2 system to measure the time course of antiviral agent-mediated loss of HBV replication as well as the time course and magnitude of HBV production after release from antiviral treatment. Because of the sensitivity of the system, it was possible to measure secreted virions, intracellular replicative intermediates, and nuclear non-protein-bound HBV DNA and separately analyze individual species of DNA, such as single-stranded HBV DNA compared to the double-stranded form and relaxed circular compared to covalently closed circular HBV DNA. We first determined that HBV replication in the HBV recombinant baculovirus/HepG2 system could proceed for at least 35 days, with a 30-day plateau level of replication, making it possible to study antiviral agent-mediated loss of HBV followed by rebound after cessation of drug treatment. All HBV DNA species decreased in a time-dependent fashion following antiviral treatment, but the magnitude of decline differed for each HBV DNA species, with the covalently closed circular form of HBV DNA being the most resistant to drug therapy. When drug treatment ceased, HBV DNA species reappeared with a pattern that recapitulated the initiation of replication, but with a different time course.
Hepadnaviruses are a family of small, enveloped DNA viruses that includes human hepatitis B virus (HBV) (12), woodchuck hepatitis virus (WHV) (43), ground squirrel hepatitis virus (30), and several avian hepatitis viruses, including duck hepatitis B virus (31). These viruses display profound liver tropism and cause acute and chronic liver infections in each of their hosts. During an infection, the 3.2-kb viral relaxed circular (RC) DNA is converted into covalently closed circular (CCC) DNA, which is found in the nuclei of the infected hepatocytes (46). These CCC DNA molecules act as the templates for transcription (by cellular RNA polymerase II) for the 3.5-, 2.4-, 2.1-, and 0.8-kb HBV viral RNAs (17). These transcripts encode the nucleocapsid polypeptides, the large surface antigen polypeptide, the middle and major surface antigen polypeptides, and the X-gene polypeptide, respectively (17). In addition, the 3.5-kb pregenomic RNA encodes the viral polymerase and is reverse transcribed by this polypeptide within the viral nucleocapsid to produce the 3.2-kb viral genomic DNA (35).
The mature nucleocapsids containing viral genomic DNA can be secreted from the cell in virus particles only by associating with surface antigen polypeptides within the endoplasmic reticulum membrane (7, 8, 18). Virus buds into the lumen of the endoplasmic reticulum and is transported out of the cell through the Golgi apparatus (23, 49). The ability of the mature nucleocapsid to form virus particles depends on the appropriate level of synthesis of the surface antigen polypeptides (7). Mature nucleocapsids can also transport the viral genome back into the nucleus, where the partially double-stranded viral genome is converted into a transcriptionally functional CCC DNA. Recycling of duck HBV DNA back to the nucleus is regulated by viral envelope proteins (28, 42). Presumably, the same mechanism may function in other hepadnavirus infections, including HBV.
HBV causes acute and chronic infections of the liver and is responsible for 1.2 million deaths annually (24). Approximately 0.5% of acute infections terminate with fatal, fulminant hepatitis. Chronic infections also have serious consequences, with nearly 25% terminating in untreatable liver cancer (4). Worldwide deaths from liver cancer caused by HBV infection probably exceed one million per year (36). Almost a third of the world's human population has been acutely infected with HBV, and about 5% are chronic carriers of the virus (24).
Until recently, the only approved treatment for chronic HBV infection was alpha interferon (20, 44, 48). Unfortunately, only 30 to 40% of chronically infected individuals respond to interferon with sustained elimination of HBV (34). This group of patients has low-level viremia and high alanine aminotransferase levels. Alpha interferon is not the ideal drug for HBV treatment because of its limited efficacy, parenteral administration, dose-limiting side effects, and high cost. Recently, a deoxycytidine analog, lamivudine (l-2′,3′-dideoxythiacytidine [3TC]), was approved for HBV treatment (16, 22, 41). Other nucleoside and nucleotide analogs such as famciclovir and adefovir are in phase III clinical trials (11). Others, such as clevudine (β-l-2′-deoxy-2′fluoro-5-methyl-arabinofuranosyl uracil [l-FMAU]), are still under preclinical investigation (9) and phase I/II evaluation (26).
Treatment with 3TC is tolerable and effective in reducing or eliminating HBV DNA from the serum of chronically infected patients. Unfortunately, use of 3TC has raised specific problematic issues. The first concern is reappearance of HBV DNA in serum after cessation of drug therapy. The level and speed of rebound depend on the length of 3TC treatment (15, 27, 33). This rebound is assumed to be due to the persistence of CCC DNA in the nuclei of the infected hepatocytes (46). The rebound phenomenon has been studied experimentally in woodchucks infected with WHV and treated with 3TC (21). Upon cessation of 3TC therapy, WHV rebounded in a dose-dependent manner and resembled posttherapy virus “flares” reported in HBV-infected humans. The second concern is the emergence of drug-resistant mutants (2, 3, 19, 29, 38).
l-FMAU is a potent antihepadnaviral agent in vitro and in vivo and is a suitable candidate for antiviral therapy of chronic HBV infection (37). In woodchucks chronically infected with WHV, a single daily oral dose of l-FMAU for 4 weeks significantly reduced viremia, antigenemia, intrahepatic WHV replication, including CCC DNA, and intrahepatic expression of WHV core antigen in a dose-dependent manner. Withdrawal of l-FMAU resulted in a dose-related delay in rebound of the viremia. At the highest doses used, no evidence of drug-related toxicity was observed, and the viremia remained significantly suppressed in at least one-half of the treated animals for 10 to 12 weeks posttreatment (37).
We previously reported a novel transient in vitro system for studying HBV replication with HBV recombinant baculovirus to efficiently deliver the HBV genome to HepG2 cells (13). Baculoviruses are extremely efficient vectors for gene delivery of hepatic cells (5, 6, 40). In HBV baculovirus-infected HepG2 cells, HBV gene expression is driven exclusively from endogenous promoters, and HBV replication levels that are markedly higher than from conventional HBV-expressing cell lines can be obtained. HBV transcripts, intracellular and secreted HBV antigens, HBV DNA replicative intermediates (RI), and CCC HBV DNA are produced, and HBV virions are secreted. One of the advantages of the HBV baculovirus/HepG2 system is that the levels of HBV replication are sufficiently high that all forms of HBV DNA (RI, CCC, and extracellular) can be readily detected by Southern blot analysis. Indeed, the HBV baculovirus/HepG2 system is the only in vitro system in which it is possible to readily detect and quantify nuclear non-protein-bound HBV DNA. We also investigated the ability to use the HBV baculovirus system for antiviral agent testing (14) and showed that the HBV baculovirus-HepG2 system made it possible for the first time to systematically observe the effects of 3TC on CCC DNA in vitro.
In this study, we used the HBV recombinant baculovirus/HepG2 system to study the rebound phenomenon that has been seen in humans and woodchucks after cessation of 3TC therapy. We first determined whether a plateau level of HBV replication that lasts at least 30 days and hence long enough to measure rebound could be established with this system. Our goals were to quantify the relative time course and magnitude of disappearance of HBV DNA species involved in replication after drug therapy and quantify the time course and relative magnitude of reappearance of HBV DNA species after cessation of drug treatment.
MATERIALS AND METHODS
Cell culture.
The HepG2 cell line was maintained at 37°C in a humidified incubator at 5% CO2 (25). HepG2 cells were fed minimal essential medium (MEM; Gibco-BRL, Gaithersburg, Md.) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, Utah), designated MEM-FBS. Sf21 insect cells were maintained in Grace's insect medium supplemented with yeastolate and lactalbumin hydrolysate (Gibco-BRL, Gaithersburg, Md.) in a nonhumidified incubator at 28°C without CO2.
HBV baculovirus production and infection.
The generation, amplification, and purification of the HBV baculovirus encoding a 1.3-unit-length replication-competent HBV genome have been described previously (13). The baculovirus infection procedure for HepG2 cells has also been described (13). Briefly, HepG2 cells were seeded at approximately 20 to 40% confluency and allowed to attach and grow for 16 to 24 h prior to infection. On the day of infection, duplicate plates of cells were trypsinized, and viable-cell number was determined with a hemacytometer with trypan blue exclusion. Average cell counts were calculated and used to determine the volume of high-titer virus stock necessary to infect cells at the indicated multiplicity of infection. Baculovirus was diluted in MEM-FBS to achieve the appropriate multiplicity of infection, with a volume of 1.0 or 0.5 ml used to infect 100- and 60-mm dishes, respectively. Baculovirus was adsorbed to HepG2 cells for 1 h at 37°C with gentle rocking every 15 min to ensure that the inoculum was evenly distributed. The inoculum was then aspirated, and the HepG2 cells were washed two times with phosphate-buffered saline (PBS) and refed MEM-FBS.
3TC and l-FMAU treatment.
3TC was kindly provided by BioChem Therapeutic Inc. (Laval, Quebec, Canada), and l-FMAU was a gift of Triangle Pharmaceuticals Inc. (Durham, N.C.). 3TC or l-FMAU was resuspended in sterile water, aliquoted, and frozen at −20°C to avoid repeated freezing and thawing of the drug. Medium containing drug was prepared daily as needed with fresh aliquots of drug. HepG2 cells were fed medium containing the indicated concentration of drug at the specified time postinfection (p.i.) with HBV baculovirus. Cells were fed fresh drug-containing medium daily until the time course for drug treatment was completed.
Analysis of intracellular RI.
Cytoplasmic preparations containing HBV core particles were isolated from HepG2 cells. Cells were lysed with 10% NP-40 for 20 min. The cytoplasmic fraction was separated from the nuclear fraction by centrifugation. Unprotected DNA was removed by adjusting cytoplasmic preparations to 10 mM MgCl2 and 500 μg of DNase I (Roche Diagnostic Corporation, Indianapolis, Ind.) per ml, followed by 1 h of incubation at 37°C. To extract replicative intermediates, EDTA, sodium dodecyl sulfate (SDS), NaCl, and proteinase K (Roche Diagnostic Corporation, Indianapolis, Ind.) were added separately and sequentially to final concentrations of 10 mM EDTA, 1% SDS, 100 mM NaCl, and 500 μg of proteinase K per ml. The sample was incubated for 2 h at 37°C and then subjected to sequential phenol and chloroform extractions and isopropanol precipitation. Precipitated nucleic acids were resuspended in a small volume of TE (10 mM Tris, 1 mM EDTA) and digested with 100 μg of RNase (Roche Diagnostic Corporation, Indianapolis, Ind.) per ml for 1 h at 37°C. RI were then analyzed by electrophoresis in 1% agarose gels, followed by Southern blotting as described before (39). Nucleic acid hybridization was performed. A full-length double-stranded HBV genome was used as a template to generate 32P-radiolabled probe with a Roche Diagnostic Corp. (Indianapolis, Ind.) random prime DNA labeling kit. HBV DNA bands were visualized and quantitated with a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
Detection of extracellular HBV DNA.
Conditioned medium was collected from HepG2 cells, centrifuged at 10,000 × g for 10 min, and transferred to clean tubes to remove cellular debris. HBV particles were precipitated from medium samples with polyethylene glycol 8000 (Sigma Chemical Co., St. Louis, Mo.) as described before (47). Virus pellets were resuspended in PBS, and DNA was extracted with lysis buffer (0.25% SDS, 0.25 M Tris, 0.25 M EDTA). The sample was then treated with proteinase K to achieve a final concentration of 500 μg/ml. Digestion was allowed to proceed for 2 h at 37°C, after which the sample was subjected to sequential phenol and chloroform extractions. DNA was precipitated with 0.1 volume of 3 M sodium acetate and 1 volume of isopropanol. Ten micrograms of tRNA (Roche Diagnostic Corporation, Indianapolis, Ind.) was added as a carrier during precipitation. Nucleic acid pellets were resuspended in a small volume of TE and digested with 100 μg of RNase/ml prior to analysis by electrophoresis and Southern blotting. HBV DNA bands were visualized and quantitated with a PhosphorImager and ImageQuant software.
Detection of non-protein-associated HBV DNA.
Non-protein-bound circular HBV DNA was extracted from HepG2 cells with a modification of a previously described procedure (42). For total HBV CCC, cells were lysed with TE (50:1) and 4% SDS for 20 min. The extract was precipitated with 2.5 M KCl and centrifuged at 10,000 rpm for 10 min. For the nuclear extraction, nuclei were separated from the whole-cell lysate as described above for isolation of RI. The nuclear pellets were suspended in TE (50:1), and the sample was then lysed with 4% SDS, followed by precipitation with 2.5 M KCl. For either isolation procedure, the supernatants were phenol and chloroform extracted, and nucleic acid was recovered by isopropanol precipitation. Ten micrograms of tRNA was added as a carrier during precipitation. Extracted nucleic acid was resuspended in water and digested with 100 μg of RNase/ml and 30 U of Plasmid-Safe ATP-dependent DNase (Epicenter Technologies, Madison, Wis.) for 3 h at 37°C. Samples were analyzed by electrophoresis and Southern blotting. HBV DNA bands were visualized and quantitated with a PhosphorImager and ImageQuant software.
Analysis of secreted HBV antigens.
Detection of HBV surface and e antigens (HBsAg and HBeAg, respectively) was carried out with a Sorin Biomedica (Saluggia, Italy) radioimmunoassay kit according to the manufacturer's instructions. Medium samples collected from HepG2 cells were centrifuged at 10,000 × g to remove cellular debris, transferred to clean tubes, and stored at −20°C until analyzed.
Generation of baculovirus probe.
To generate a baculovirus probe, pBlueBac4.5 (Invitrogen, Carlsbad, Calif.) was digested with Bsp1407I (MBI Fermentas, Hanover, Mass.) and SnaBI (New England Biolabs, Beverly, Mass.) restriction enzymes for 3 h and then fractionated by electrophoresis in a 1.0% agarose gel. The smaller 332-bp band was cut out of the gel and purified with a Geneclean III kit (Bio 101, Carlsbad, Calif.). The 332-bp fragment was used as a template to generate a radiolabeled probe with a Roche Diagnostic Corp. (Indianapolis, Ind.) random prime DNA labeling kit.
Total DNA extraction for detection of baculovirus DNA.
At the indicated times p.i. with HBV recombinant baculovirus, HepG2 cells were trypsinized and resuspended in MEM-FBS. The cells were subjected to centrifugation at 6,000 × g for 10 min, washed in PBS, and repelleted at 6,000 × g for 10 min. The resulting pellet was resuspended in 1 ml of PBS. Then 3 ml of lysis buffer (50 mM Tris [pH 8.0], 50 mM NaCl, 5 mM EDTA [pH 8.0], 0.5% SDS, 500 μg of proteinase K per ml) was added, and incubation was carried out overnight at 37°C. Phenol extraction was carried out. The sample was treated with RNase to remove contaminating RNA. Following phenol-chloroform extraction, total DNA was precipitated with 3 M sodium acetate (0.1:1, vol/vol) and isopropanol (2:1, vol/vol). Total DNA was resuspended in TE, normalized by optical density at 260 nm, and analyzed by electrophoresis and Southern blotting. The baculovirus DNA band was visualized and quantitated with a PhosphorImager and ImageQuant software.
RESULTS
Extension of the HBV baculovirus/HepG2 system to 30 days.
In our initial description of the use of HBV recombinant baculovirus to initiate HBV replication in HepG2 cells, the time course analysis of HBV gene expression and replication was terminated at 11 days p.i. (13). In the previous studies, HepG2 cells were infected 1 day postseeding at a multiplicity of 50 PFU/cell. Cells were fed daily but were not subcultured. The 3.5-, 2.4-, and 2.1-kb HBV transcripts were readily detectable by Northern blot analysis by day 1 p.i. and remained highly expressed through 11 days p.i. Production of HBV RI and extracellular HBV DNA became maximal at approximately 3 days p.i. and remained at a plateau level until 11 days p.i. Culture medium was collected daily, and the extracellular DNA detected represented HBV virions released into the medium over each 24-h period before collection. These data suggested that HBV replication was ongoing at a reasonably constant rate from days 3 to 11 p.i. and that there was reason to believe that replication should continue for more than 11 days.
In the present study, we wanted to determine whether HBV replication could be extended beyond 11 days with this system. A period of 30 to 35 days was selected because we knew that HepG2 cells would remain viable without passage for this length of time. A 30- to 35-day period should provide sufficient time to measure antiviral agent-mediated loss of HBV production as well as the relative time course and magnitude of reappearance of HBV RI, extracellular DNA, and CCC DNA after release from antiviral treatment.
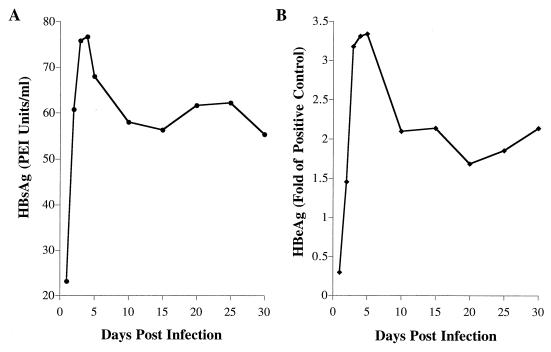
We first measured the expression of HBV secreted antigens through 30 days p.i. HepG2 cells were infected at 100 PFU/cell, and conditioned medium was collected for analysis of HBsAg and HBeAg by radioimmunoassay. Samples were collected from cells that had been fed fresh medium 24 h prior to sampling and, as such, contained the HBV antigens secreted over the previous 24-h period. HBsAg levels became maximal at 3 to 4 days p.i. and remained at a high plateau level from days 10 to 30 p.i. (Fig. 1A). HBeAg levels became maximal at 3 to 5 days p.i. (Fig. 1B). A high plateau level of secreted HBeAg was detected from 10 to 30 days p.i., as had been observed for HBsAg.
FIG. 1.
Time course analysis of HBV antigens secreted by HBV baculovirus-infected HepG2 cells. HepG2 cells were seeded in 100-mm dishes at approximately 20 to 40% confluency, grown for 16 to 24 h, and then infected with 100 PFU of HBV baculovirus/cell. Cells were fed daily. Conditioned medium exposed to the infected cells for 24 h was collected at the indicated times p.i. and analyzed by radioimmunoassay for the presence of (A) HBsAg and (B) HBeAg. The amount of HBsAg was plotted against days postinfection. For HBeAg, the ratio of counts per minute for the samples to that for the positive control was plotted against days postinfection.
Having determined that HepG2 cells infected with HBV recombinant baculovirus continued to secrete high levels of HBV antigens for at least 30 days p.i., we decided to next assess the length of time that the cells could continue to synthesize HBV RI and secrete HBV virions. HepG2 cells were infected with HBV recombinant baculovirus at a multiplicity of 100 PFU/cell, and cytoplasmic cores were isolated at various times p.i. When HBV replication is initiated by HBV recombinant baculovirus, the magnitude of HBV expression is sufficiently high that it is possible to follow the temporal appearance first of nucleocapsids containing only single-stranded (SS) HBV DNA and subsequently the formation of nucleocapsids containing more complex HBV DNA species.
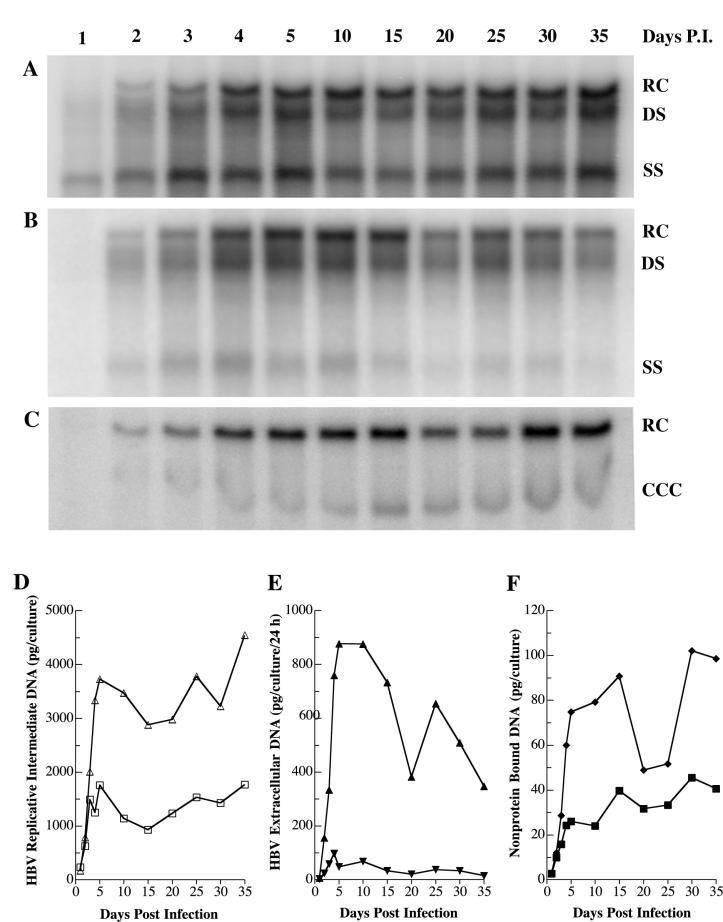
Nucleocapsids containing SS HBV DNA species were apparent by 1 day p.i. (Fig. 2A). Nucleocapsids containing SS and RC/double-stranded (DS) DNA became readily detectable by 2 days p.i. Production of all forms of HBV RI became maximal by 4 days p.i. and remained at that level through 35 days p.i. Examination of HBV DNA from the medium of HepG2 cells revealed that virions were not present at 1 day p.i., were readily detectable by 2 days p.i., and became maximal by 4 days p.i. (Fig. 2B). Failure to observe HBV virions in the medium until 2 days p.i. was in agreement with our previous study (13) and followed directly from the finding that nucleocapsids containing partially DS HBV DNA were not detected until 2 days p.i. The level of HBV DNA in the medium remained high through 35 days p.i. It is important to note that, as was the case for detection of secreted HBsAg and HBeAg, medium used for analysis of virion production was collected from cells fed fresh medium 24 h prior to sampling and, as such, contained the virions freshly secreted over the previous 24-h period.
FIG. 2.
Time course analysis of HBV DNA species produced by HBV baculovirus-infected HepG2 cells. HepG2 cells were seeded in 100-mm dishes at approximately 20 to 40% confluency, grown for 16 to 24 h, and then infected with 100 PFU of HBV baculovirus/cell. On the indicated days p.i., cells were harvested and analyzed for HBV RI DNA by Southern blotting (A and D). The HBV DNA bands were visualized with a PhosphorImager, and the digital image is shown in panel A. The medium was collected and analyzed for extracellular HBV DNA by Southern blotting (B and E). The HBV DNA bands were visualized with a PhosphorImager, and the digital images are shown in panel B. Cultures were fed daily. The HBV DNA bands indicate the amount of extracellular HBV DNA released into the medium over the 24-h period before collection. On the indicated days p.i., cells were harvested and analyzed for non-protein-bound HBV DNA (C and F). Both the RC and CCC forms of HBV DNA were detected by Southern blotting analysis. The HBV DNA bands were visualized with a PhosphorImager, and the digital image is shown in panel C. The amount of HBV DNA was also plotted in terms of picograms of HBV DNA. (D) Replicative intermediates: RC/DS, Δ; SS, □. (E) Extracellular HBV DNA: RC/DS, ▴; SS, ▾. (F) Non-protein-bound DNA: RC, ♦; CCC, ▪.
One unique aspect of the HBV baculovirus/HepG2 system is that nuclear HBV DNA in both the RC and CCC DNA forms is readily detectable by Southern blot analysis. We previously reported that nuclear HBV DNA is strongly expressed through at least 11 days p.i. of HepG2 cells with HBV recombinant baculovirus (13). In the present study, HepG2 cells were infected with HBV recombinant baculovirus at a multiplicity of 100 PFU/cell, and non-protein-bound low-molecular-weight DNA was extracted from HepG2 cells at various times through 35 days p.i. Time course analysis revealed that CCC HBV DNA was detectable from days 2 through 35 p.i. (Fig. 2C).
The Southern blots were quantitated with a PhosphorImager. The levels of the various HBV DNA species produced during the 35-day period of HBV replication were plotted in picograms per culture or picograms per culture per 24 h (Fig. 2D, 2E, and 2F). HepG2 cells divide over the first 5 to 6 days after plating (4 to 5 days p.i.), at which time the cells reach confluence. From that time until day 35, some cell division can be observed, but the rate is markedly reduced. It is interesting that from days 5 to 35, the level of intracellular HBV RI remained reasonably constant (Fig. 2D) and the level of nuclear CCC DNA continued to increase steadily (Fig. 2F). In contrast, as the cell cultures aged, the capacity to produce extracellular HBV declined by approximately 50% (Fig. 2E).
Decline in baculovirus DNA levels with time after baculovirus infection.
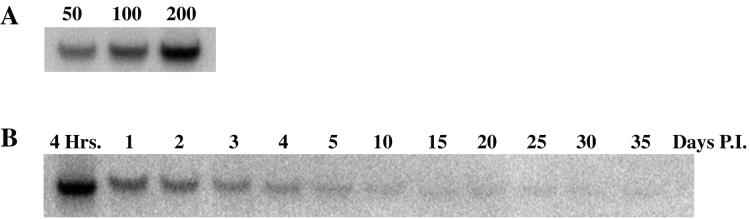
We next wanted to determine the level of baculovirus DNA present in HepG2 cells during the 35-day time course used to measure HBV replication. We selected a 332-bp fragment from pBlueBac4.5 to use as a probe for baculovirus DNA. To determine the validity of the assay, HepG2 cells were infected with HBV recombinant baculovirus at multiplicities of 50, 100, and 200 PFU/cell. At 4 h p.i. cells were harvested and analyzed for baculovirus DNA by Southern blot analysis (Fig. 3A). The results indicated that baculovirus DNA was successfully detected by the probe and that the intensity of the baculovirus DNA band increased with increasing multiplicity. It is important to note that this probe was designed to hybridize with baculovirus sequences and does not hybridize with HBV DNA sequences. This probe was then used to measure the levels of baculovirus DNA present in HepG2 cells with time after baculovirus infection (Fig. 3B). The level of baculovirus DNA in HepG2 cells infected at a multiplicity of 100 PFU/cell declined steadily after 4 h.
FIG. 3.
Effect of time on levels of baculovirus DNA in HBV baculovirus-infected HepG2 cells. HepG2 cells were seeded in 100-mm dishes at approximately 20 to 40% confluency, grown for 16 to 24 h, and then infected with 50, 100, or 200 PFU (panel A) or with 100 PFU (panel B) of HBV baculovirus/cell. At the indicated times p.i., cells were harvested and analyzed for baculovirus DNA by Southern blot analysis. The baculovirus DNA band was visualized with a PhosphorImager, and the digital images are shown. For panel A, cells were harvested at 4 h p.i. For panel B, cells were harvested at the indicated times p.i.
Analysis of antiviral agent-mediated loss of HBV production and subsequent rebound.
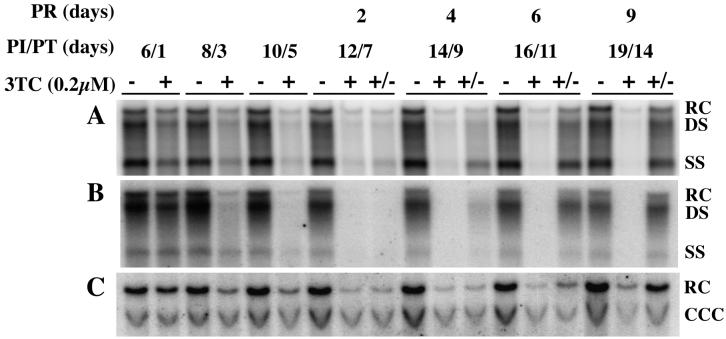
We next wanted to measure 3TC-mediated loss of HBV production and subsequent rebound of HBV replication after release from 3TC treatment. HepG2 cells were infected with HBV recombinant baculovirus at 100 PFU/cell. The protocol was designed so that one set of cultures remained as untreated controls throughout the course of the study, a second set of cultures were treated with 0.2 μM 3TC from days 5 to 19 p.i., and a third set of cultures were treated with 0.2 μM 3TC starting at day 5 and continuing until day 10 p.i. (5 days of treatment), at which time the cells were maintained in the absence of drug for an additional 9 days. Treatment with 3TC was initiated at 5 days p.i. because at that time point HBV replication had reached a level that remained reasonably constant through 35 days p.i. Conditioned media were collected, and cells were harvested at various time points p.i. and analyzed for RI, extracellular DNA, and non-protein-bound nuclear DNA (Fig. 4). A PhosphorImager system was used to detect HBV DNA.
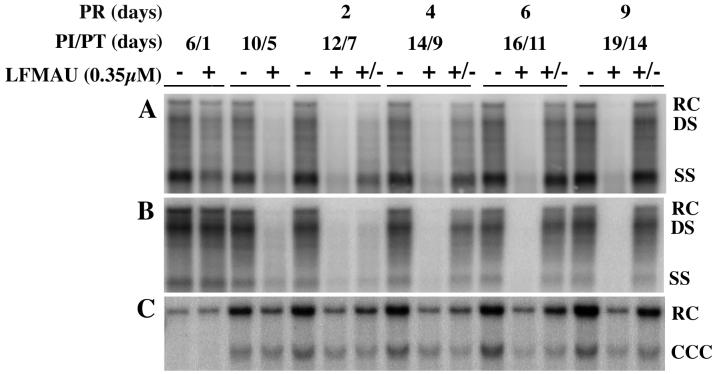
FIG. 4.
Disappearance of HBV DNA after 3TC treatment and reappearance of HBV DNA species after cessation of 3TC treatment. HepG2 cells were seeded in 60-mm dishes at approximately 20 to 40% confluency, grown for 16 to 24 h, and then infected with 100 PFU of HBV baculovirus/cell. Cells were either not treated with 3TC (−), treated with 3TC from days 5 through 19 p.i. (+), or treated with 3TC beginning on day 5 p.i. for 5 days, at which time they were fed medium without drug (+/−). Cultures were fed fresh 3TC-supplemented medium daily. Cells were harvested and analyzed by Southern blot analysis for (A) RI HBV DNA and (C) non-protein-bound HBV DNA. (B) Conditioned medium exposed to the infected cells for 24 h prior to harvest was collected and analyzed by Southern blot analysis for extracellular HBV DNA. The HBV DNA bands were visualized with a PhosphorImager, and the digital images are shown. PI, postinfection; PT, posttreatment; PR, postrelease.
Quantitative analyses of disappearance of HBV DNA species after 3TC treatment and reappearance of HBV DNA species after cessation of 3TC treatment.
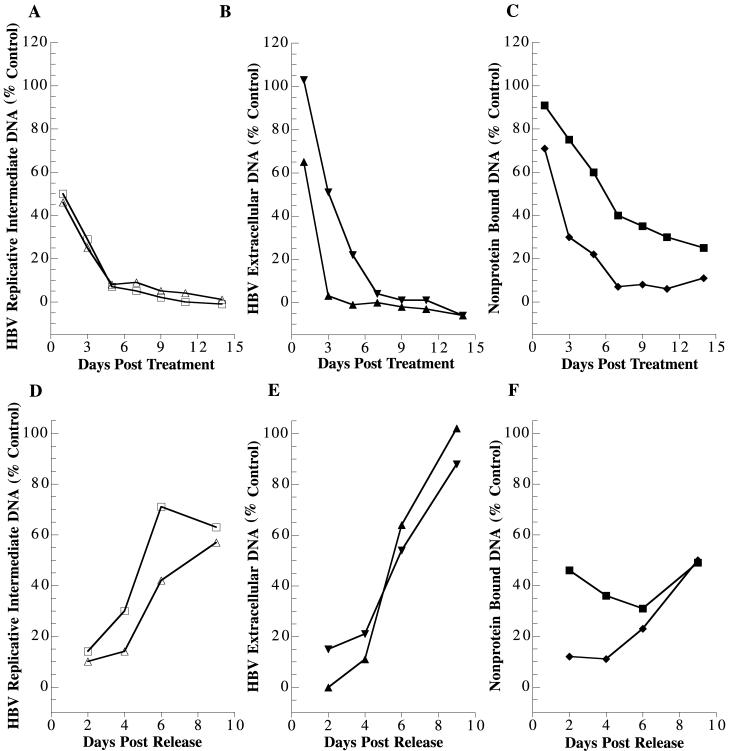
Two different approaches were taken to analyze 3TC-mediated reduction in HBV DNA species and rebound of these species after release from 3TC treatment. The first was to measure the percentage of each indicated HBV DNA species compared to that in the untreated control plotted against either days posttreatment (plus lane as a percentage of minus lane, Fig. 5) or days postrelease (plus/minus lane as a percentage of minus lane, Fig. 5).
FIG. 5.
Quantitative analyses of disappearance of HBV DNA after 3TC treatment and reappearance of HBV DNA species after cessation of 3TC treatment, given as a percentage of the control value. The experiment was carried out as described for Fig. 4. The percentage of indicated HBV DNA species compared to the untreated control cells (lanes − in Fig. 4) was plotted against the days posttreatment (panels A, B, and C) and days postrelease (panels D, E, and F). Panels A and D, replicative intermediates. RC/DS, Δ; SS, □. Panels B and E, extracellular HBV DNA. RC/DS, ▴; SS, ▾. Panels C and F, non-protein-bound DNA. RC, ♦; CCC, ▪.
This approach led to the following conclusions. The reduction in extracellular HBV DNA after 3TC treatment occurred rapidly, with levels reduced to 65% of that in the untreated control by 1 day, to 3% by 3 days, and to undetectable levels by 5 days of treatment (Fig. 5B). HBV RI levels declined more slowly than extracellular HBV DNA levels (Fig. 5A). The RC/DS band of HBV RI declined to 48% after 1 day of treatment and 27% by 3 days of treatment. The levels were reduced to 3% of the untreated control level by 9 days of treatment and became undetectable by 14 days of treatment. There was no difference in the time course or magnitude of reduction whether the analysis was carried out on the SS band or the RC/DS bands.
The effects of 3TC on non-protein-bound nuclear HBV DNA were considerably different than on HBV RI (Fig. 5C). In addition, the results were also markedly different when the effects on the RC form were compared to the effects on the CCC form of nuclear HBV DNA. The effects of 3TC treatment on the level of nuclear RC HBV DNA were slower by approximately 2 days than what was observed for HBV RI. Specifically, a reduction to 22% of the control level was observed after 5 days of treatment. In addition, a major difference in the magnitude of reduction was observed. The maximal level of reduction of non-protein-bound HBV RC levels was to 6 to 8% of the control level. This level of reduction was achieved by 7 days of treatment, and no further reduction was observed throughout the remaining 7 days of treatment. Resistance to 3TC treatment was even greater for the non-protein-bound CCC DNA species. Even after 14 days of treatment, the CCC DNA levels remained at 25% of control levels.
By 2 days after release from 3TC treatment, HBV RI were detectable and the magnitude of HBV RI DNA was greater than in 3TC-treated cells, indicating that the rebound in HBV DNA replication had been initiated (Fig. 5D). The time course for reappearance of the SS form of HBV RI preceded that for the RC/DS forms, as would be expected. By 2 days after release from 3TC treatment, the SS DNA levels were 14% of control levels. Extracellular RC/DS HBV DNA was not detectable by 2 days after the cells were fed fresh drug-free medium (Fig. 5E). By 4 days postrelease, extracellular HBV RC/DS DNA was detectable. The amount of extracellular HBV DNA at 4 days postrelease was 11% of the control HBV DNA level for the same time point. By 9 days postrelease, extracellular HBV DNA levels had returned to the levels of HBV produced by cells not subjected to antiviral treatment.
Quantitative analysis clearly indicates that the RC form of non-protein-bound nuclear HBV DNA increased, although slowly, after release from 3TC treatment (Fig. 5F). At both 2 and 4 days postrelease, the levels of RC DNA were 12 and 11% of the control, respectively. By 6 days postrelease, the RC levels rose to 23% of control levels, and by 9 days they rose to 50% of control levels. These findings were highly significant because they indicate that new HBV RC was being delivered to the nucleus.
The effects of release from treatment with 3TC on the HBV CCC DNA were quite different from those observed for nuclear HBV RC DNA. The levels of HBV CCC DNA varied from 31 to 49% (average, 41%) over the 9-day period after release from 3TC treatment but did not follow any pattern that indicated rebound. The data indicate that the HBV CCC DNA levels remained reasonably stable when cells were released from 3TC treatment.
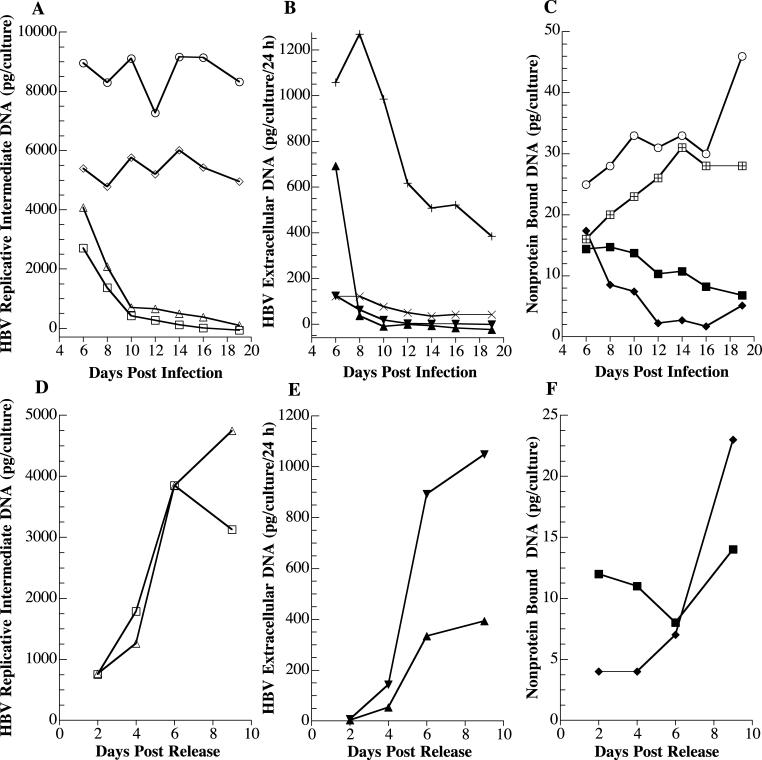
The second approach was to plot the actual amounts of each HBV DNA species for untreated control cultures, treated cultures, and cultures initially treated with 3TC and then released from 3TC treatment (Fig. 6). As would be expected, the overall conclusions regarding time course and magnitude of effect did not differ from what was observed when data were calculated as a percentage of the control. However, providing the data in terms of absolute amounts of HBV DNA further validates several critical points. (i) Treatment with 3TC causes a rapid reduction in extracellular HBV DNA from approximately 1,000 pg/culture/24 h prior to drug treatment to 36 pg/culture/24 h by 3 days of treatment with 3TC, which represents an approximately 30-fold decrease in HBV DNA. (ii) The amount of HBV CCC DNA per culture was reduced from 16 pg prior to treatment to 7 pg after 14 days of treatment, which represents only a little over a twofold reduction. (iii) The extracellular RC/DS HBV DNA species returned to 393 pg/culture/24 h by 6 days after release from 3TC treatment (14 days p.i.). This value was similar to the amount of RC/DS HBV DNA, 384 pg/culture/24 h, produced by non-3TC-treated HepG2 cells at 14 days p.i. with HBV recombinant baculovirus.
FIG. 6.
Quantitative analyses of disappearance of HBV DNA after 3TC treatment and reappearance of HBV DNA species after cessation of 3TC treatment. The experiment was carried out as described for Fig. 4. The amount of HBV DNA species for untreated cultures as well as treated cultures plotted against the days postinfection is shown in panels A, B, and C. The amounts of HBV DNA species after release from 3TC treatment are shown in panels D, E, and F. (A) Replicative intermediates in untreated cultures: RC/DS, ○; SS, ⋄; replicative intermediates in 3TC-treated cultures: RC/DS, ▵; SS, □. (B) Extracellular HBV DNA in untreated cultures; RC/DS, +; SS, ×; extracellular HBV DNA in 3TC-treated cultures: RC/DS, ▴; SS, ▾. (C) Non-protein-bound DNA in untreated cultures: RC, ⊙; CCC, ⊞; non-protein-bound DNA in 3TC-treated cultures: RC, ♦; CCC, ▪. (D) Replicative intermediates: RC/DS, ▵; SS, □. (E) Extracellular HBV DNA: RC/DS, ▴; SS, ▾. (F) Non-protein-bound DNA: RC, ♦; CCC, ▪.
Reduction in HBV replication following l-FMAU treatment.
We next carried out the same kind of experiment on a second nucleoside analog, l-FMAU, that has been shown to have antiviral activity against HBV (10), WHV (37), and duck HBV (1). Both lamivudine and clevudine are pyrimidine analogs that belong to a new nucleoside family with a nonnatural levorotatory configuration (l-nucleoside analogs). The l-nucleoside analogs exhibit very potent activity against HBV without significant toxicity compared to their dextrorotatory counterpart (32). In general, the anti-HBV nucleoside analogs mainly interfere with the enzymatic function(s) of the HBV DNA polymerase. Their antiviral activity requires intracellular phosphorylation to the corresponding deoxynucleoside triphosphates (dNTP) equivalent. If the dNTP analog is accepted as a substrate by the HBV polymerase, it may be polymerized onto the 3′-hydroxy (OH) terminus of the nascent DNA chain. If the incorporated dNTP analog lacks the 3′-OH group, such as 3TC, the growing chain will terminate immediately. On the other hand, if the incorporated dNTP possesses a functional 3′-OH, such as l-FMAU, this may lead to aberrant, dysfunctional DNA and/or delayed chain termination (45).
Both the PhosphorImager detection of the Southern blot data (Fig. 7) and computer analysis of the imaged data (Fig. 8) revealed the following. (i) HBV extracellular DNA was rapidly reduced to below detectable levels after l-FMAU treatment. (ii) HBV RI levels were also markedly affected by l-FMAU treatment, although the time course was delayed relative to the effects on extracellular HBV DNA. It is interesting that the SS form of the HBV RI initially decreased more rapidly than the DS form as a result of l-FMAU treatment, but with time, the effects of l-FMAU treatment were greater with regard to the DS form than the SS form. (iii) Non-protein-bound nuclear RC and CCC DNA levels were reduced by l-FMAU treatment, but to a much lesser extent than RI or extracellular DNA. The CCC form was more resistant to l-FMAU treatment than the RC form.
FIG. 7.
Disappearance of HBV DNA after l-FMAU treatment and reappearance of HBV DNA species after cessation of l-FMAU treatment. HepG2 cells were seeded in 60-mm dishes at approximately 20 to 40% confluency, grown for 16 to 24 h, and then infected with 100 PFU of HBV baculovirus/cell. Cells were either not treated with l-FMAU (−), treated with l-FMAU from days 5 through 19 p.i. (+), or treated with l-FMAU beginning on day 5 p.i. for 5 days, at which time they were fed with medium without drug (+/−). Cultures were fed fresh l-FMAU-supplemented medium daily. Cells were harvested and analyzed by Southern blot analysis for (A) RI HBV DNA and (C) non-protein-bound HBV DNA. (B) Conditioned medium exposed to the infected cells for 24 h prior to harvest was collected and analyzed by Southern blot analysis for extracellular HBV DNA. The HBV DNA bands were visualized with a PhosphorImager, and the digital images are shown. PI, postinfection; PT, posttreatment; PR, postrelease.
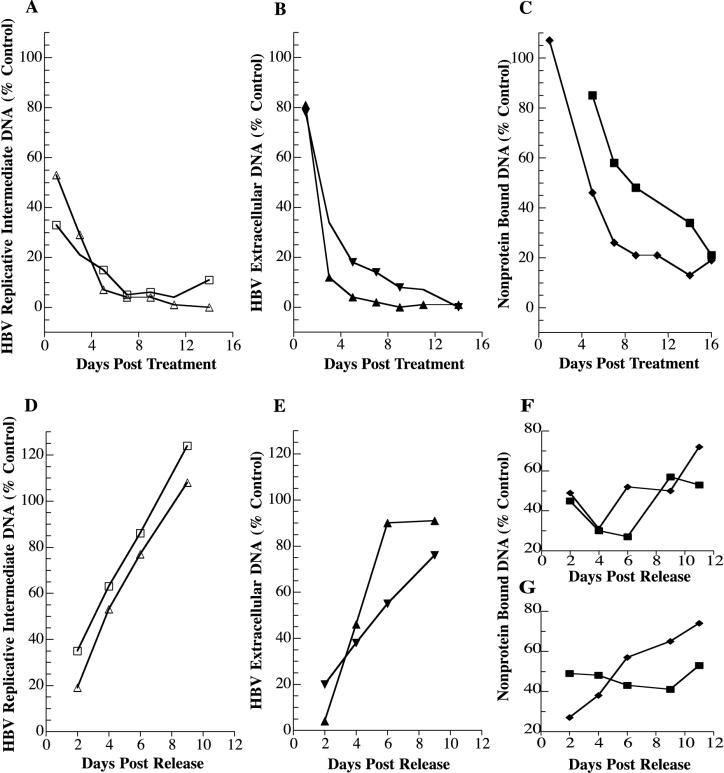
FIG. 8.
Quantitative analyses of l-FMAU-mediated reduction in HBV DNA and rebound of HBV DNA after release from l-FMAU treatment. Data in panels A through F were calculated from the experiment shown in Fig. 7. Data in panel G were calculated from an independent experiment carried out following the same protocol used for the experiment shown in Fig. 7. For quantitative analysis of l-FMAU-mediated reduction in HBV DNA, the percentage of indicated HBV DNA species compared to the untreated cells (lanes − in Fig. 7) was plotted against the days posttreatment (A, B, and C). For quantitative analysis of rebound of HBV DNA after release from l-FMAU treatment, the percentage of indicated HBV DNAs compared to the untreated cells (lanes − in Fig. 7) was plotted against the days postrelease (D, E, F, and G). Panels A and D, replicative intermediates: RC/DS, Δ; SS, □. Panels B and E, extracellular HBV DNA: RC/DS, ▴; SS, ▾. Panels C, F, and G, non-protein-bound DNA: RC, ♦; CCC ▪.
Rebound of HBV replication following release from l-FMAU treatment.
As we observed above for release from 3TC treatment, release from l-FMAU treatment also resulted in a rapid rebound in HBV replication. Temporally, the process occurred as would be expected. The SS intracellular RI form returned first, followed by the RC/DS intracellular RI, with both rebounding by 2 days after release from drug treatment (Fig. 7 and 8). Extracellular HBV DNA did not rebound until 4 days postrelease from l-FMAU treatment.
The non-protein-bound CCC DNA levels fluctuated after release from l-FMAU treatment, but no steady increase or decrease in CCC DNA levels was observed. The increase in non-protein-bound HBV RC DNA after release from l-FMAU treatment was not as apparent as was observed after release from 3TC treatment. By 11 days after release, the non-protein-bound RC levels had returned to 72% of control levels. One possible reason for this difference was that the RC levels at the time of release (5 days treatment) were twice as high in cells treated with l-FMAU (46% of the control) as in those treated with 3TC (22% of the control). To further address the question of differential rebound of the RC and CCC forms of non-protein-bound nuclear HBV DNA, a second rebound experiment identical in protocol to that shown in Fig. 7 with l-FMAU was independently carried out and analyzed (Southern blot data not shown). Quantitative analysis of rebound of non-protein-bound nuclear HBV DNA revealed a pattern quite similar to what was observed for rebound after release from 3TC treatment (Fig. 8G). The CCC form fluctuated with time after release from treatment, but no steady trend upward was observed. In contrast, the RC form clearly demonstrated rebound.
DISCUSSION
One nucleoside analog, 3TC, is currently approved for chronic HBV therapy, and numerous others are in some stage of development. Although it is clear from patient studies as well as studies with woodchucks that virions return to pretreatment levels after nucleoside analog therapy is stopped, no in vitro cell culture system has been used to examine rebound of HBV replication in detail. We demonstrate in this study that it is possible to use HepG2 cells in which HBV replication has been initiated by gene delivery with an HBV recombinant baculovirus (13) to study the time course of antiviral agent-mediated loss of individual species of HBV DNA involved in HBV replication as well as the relative time course and magnitude of production of these HBV products after release from antiviral therapy.
We can draw several conclusions from this study. (i) HBV replication in the HBV recombinant baculovirus HepG2 system can proceed at high levels for at least 35 days p.i., and for a 30-day period (from days 5 to 35 p.i.), the magnitude of replication is essentially at a plateau level. (ii) Following treatment with antivirals, all HBV DNA species decrease in a time-dependent fashion, but the magnitude of decline differs for each HBV DNA species; the CCC form of nuclear non-protein-bound HBV DNA is the most resistant to drug therapy. (iii) When drug treatment ceases, HBV DNA species reappear with a pattern that recapitulates the initiation of HBV replication, but with a different time course. In addition, the magnitude of production of extracellular HBV DNA returns to the level found in untreated control cultures.
Data reported in this study confirm and extend our initial studies with the HBV recombinant baculovirus/HepG2 system to study HBV replication (13). First, we have shown that high plateau levels of HBV replication can be extended to at least 35 days. Second, we showed that the recombinant baculovirus DNA is only transiently present at high levels in the HepG2 cell cultures and the levels decline rapidly with time, as would be expected. Third, at a time when cultures demonstrate no production of HBV virions for several days as a result of antiviral treatment, reinitiation of HBV production to the levels seen prior to drug treatment can be achieved simply by feeding cells fresh drug-free medium. Fourth, the nuclear non-protein-bound CCC DNA form of HBV DNA is very stable even after extended treatment with either nucleoside analog. These findings, taken together, strongly suggest that the baculovirus is responsible for initiation of HBV replication in this system, but with time, renewal of the replication process proceeds from the HBV CCC DNA present in the nucleus.
To our knowledge, the HBV recombinant baculovirus/HepG2 system is the only in vitro cell culture model in which it is possible to quantify individual HBV DNA species, including the RC and CCC forms of nuclear HBV DNA. In our previous study, we reported that the magnitude of HBV replication is dependent on the multiplicity of infection and that, at a multiplicity of 100 PFU/cell, 90 to 100% of the cells express HBV core antigen and the levels of HBV replication are in 100-fold excess of what was observed with HepG2.2.15 cells (13). We also observed that the levels of replication were sufficiently high that both the RC and CCC forms of nuclear HBV DNA could be readily detected by Southern blot analysis coupled with autoradiographic detection of HBV DNA bands (13). More recently, we have shown that the technology can be extended to include PhosphorImager detection and computer analysis of digital images to quantify levels of individual HBV DNA species (unpublished data).
Although the major focus of this study was to analyze rebound of HBV replication after cessation of treatment with antiviral agents, the inherent design of the rebound protocol also made it possible to measure the disappearance of each HBV DNA species after antiviral treatment. The antiviral treatment studies with 3TC or l-FMAU showed that the time course and magnitude of disappearance are quite different for the various HBV products generated during the replication process. The time course for antiviral agent-mediated reduction in extracellular HBV DNA is very rapid. The amount of extracellular HBV DNA being measured reflects the effects of antiviral treatment on the levels of virions secreted during the preceding 24-h period because the medium is changed daily. By 3 to 4 days of treatment with either 3TC or l-FMAU, the HBV DNA bands became undetectable. The rapidity and extent of the loss are particularly impressive taking into consideration the magnitude of the viral load in HepG2 cells infected with HBV recombinant baculovirus at 100 PFU/cell. Many antiviral studies are carried out with the HepG2.2.15 cell line, which produces considerably lower levels of virus. In fact, quantitation of extracellular HBV DNA with HepG2.2.15 cells is routinely done by dot blot analysis because the quantities of HBV DNA are not sufficient to carry out Southern blot analysis.
The loss of intracellular RI from HepG2 cells treated with either 3TC or l-FMAU was slower than was observed for extracellular HBV DNA. To achieve a greater than 95% reduction in total HBV RI levels required 9 days of treatment with 3TC. Reduction in total HBV RI to greater than 90% of control levels required 11 days of l-FMAU treatment. Because HBV RI accumulate within the cells, it is important to realize that the level of HBV RI being analyzed at any one time after treatment with either antiviral agent includes the effects of the drug on blocking newly synthesized forms of RI as well as the half-life of existing forms of RI over the treatment period. It is important to note that whether 3TC or l-FMAU was used for antiviral treatment, intracellular capsid DNA levels declined and maintenance of constant levels of immature capsid DNA was not observed. These findings suggest that DNA-containing capsids are reasonably unstable.
One of the unique aspects of the recombinant HBV baculovirus/HepG2 system is the ability to readily detect nuclear non-protein-bound HBV DNA, and therefore this system can be used to quantify the effects of antivirals on nuclear HBV DNA. Although it has been assumed that the nuclear forms of HBV DNA are highly stable and also resistant to nucleoside analog therapy, we were able to clearly demonstrate in this study not only that nuclear HBV is more resistant than intracellular RI to treatment with 3TC or l-FMAU, but also that the time course and magnitude of reduction differ for the two different forms of nuclear HBV DNA. We observed that the CCC form was considerably more resistant to treatment with either drug than the RC form.
Two independent events contribute to the magnitude of decline of the RC form of HBV DNA in the nucleus. During HBV replication, it is generally accepted that nuclear HBV RC DNA is converted to the CCC DNA form by the host machinery. Therefore, treatment with an antiviral such as 3TC or l-FMAU should have no effect on this conversion, and RC levels should decline as the RC form continues to be converted into the more stable CCC DNA form. In the absence of antiviral treatment, the nuclear RC is continually replenished. However, when RI synthesis is blocked by antiviral treatment, the number of nucleocapsids that recycle HBV DNA back to the nucleus decreases, and hence, the level of HBV RC DNA in the nucleus also declines.
The data in this report clearly show that the rebound phenomenon which has been observed in chronic HBV patients after cessation of nucleoside analog treatment can be reproduced in an in vitro cell culture. Examination of the pattern, time course, and magnitude of rebound of HBV replication compared to the initial replication process revealed some interesting similarities and differences. The stepwise pattern of replication seen at early times after gene transfer-mediated initiation of HBV replication was reproduced when treatment with 3TC or l-FMAU was stopped. Specifically, SS RI was seen first, followed by RC/DS RI, followed by extracellular HBV DNA. However, the time course for rebound of each of these HBV DNA species was slower than was seen during the initial steps of replication. Specifically, RI levels did not approach maximal levels until 9 days after release from treatment with 3TC, whereas they were maximal at 4 to 5 days after initiation of replication by HBV recombinant baculovirus.
Comparative analysis of the non-protein-bound nuclear HBV DNA forms after release from treatment with either 3TC or l-FMAU yielded particularly interesting but not unexpected results. After release from treatment with either drug, an increase in the levels of the RC form of nuclear non-protein-bound HBV DNA was seen. These findings indicate that not only were new virions being synthesized and secreted to the medium, but nucleocapsids were also recycling back to the nucleus to perpetuate the replication process. Although we found almost complete rebound of HBV after cessation of l-FMAU treatment in vitro, Peek et al. (37) found that viremia remained significantly suppressed in at least half of chronically infected woodchucks treated with a high dose of l-FMAU (10 mg/kg of body weight/day) for 10 to 12 weeks posttreatment. This difference between the in vivo and in vitro results could be due to differences in biophysiology of human HepG2 cells compared to woodchucks, differences in l-FMAU pharmacokinetics in human and woodchuck cells, or differences in HBV relative to WHV.
The data reported in this study with an in vitro cell culture system support the concept that the nucleoside analogs 3TC and l-FMAU behave more as virostatic agents than viricidal agents. Although HBV was highly efficiently removed from the medium after 3TC or l-FMAU treatment, the presence of a highly stable HBV CCC DNA form in the nucleus led to a rapid return of HBV replication and secretion of virions to high levels. It would be ideal to identify or develop an anti-HBV agent that removes CCC DNA from infected hepatocytes by degrading CCC DNA, blocking its synthesis, or altering its stability. In the absence of such technology, removal of HBV CCC DNA can only be accomplished by clearance of the infected hepatocyte. Nevertheless, nucleoside analogs still represent significant advances in HBV therapy because many of these compounds have very low levels of toxicity and rebound can be prevented by long-term continuous therapy.
Acknowledgments
We thank Chris Tseng (NIH, NIAID Antiviral Research and Antimicrobial Chemistry Program) and Robert Rando of BioChem Therapeutic Inc. for providing the 3TC used in these studies. We also thank John Bilello for helpful discussions during preparation of the manuscript.
This work was supported in part by research grants from the National Institutes of Health (CA73045 and CA23931 to H.C.I.).
REFERENCES
- 1.Aguesse-Germon, S., S. H. Liu, M. Chevallier, C. Pichoud, C. Jamard, C. Borel, C. K. Chu, C. Trepo, Y. C. Cheng, and F. Zoulim. 1998. Inhibitory effect of 2′-fluoro-5-methyl-β-l-arabinofuranosyl-uracil on duck hepatitis B virus replication. Antimicrob. Agents Chemother. 42:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, M. I., M. Deslauriers, C. W. Andrews, G. A. Tipples, K. A. Walters, D. L. Tyrrell, and N. C. L. D. Brown. 1998. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology 27:1670-1677. [DOI] [PubMed] [Google Scholar]
- 3.Bartholomew, M. M., R. W. Jansen, L. J. Jeffers, K. R. Reddy, L. C. Johnson, H. C. L. D. Bunzendahl, A. G. Tzakis, E. R. Schiff, and N. A. Brown. 1997. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet 349:20-22. [DOI] [PubMed] [Google Scholar]
- 4.Beasley, R. P. 1988. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61:1942-1956. [DOI] [PubMed] [Google Scholar]
- 5.Bilello, J. P., W. E. Delaney IV, F. M. Boyce, and H. C. Isom. 2001. Transient disruption of intracellular junctions enables baculovirus entry into nondividing hepatocytes. J. Virol. 75:9857-9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 93:2348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruss, V., and D. Ganem. 1991. The role of envelope proteins in hepatitis B virus assembly. Proc. Natl. Acad. Sci. USA 88:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruss, V., and R. Thomssen. 1994. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J. Virol. 68:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, C. K., F. D. Boudinot, S. F. Peek, J. H. Hong, Y. Choi, B. E. Korba, J. L. Gerin, P. J. Cote, B. C. Tennant, and Y. C. Cheng. 1998. Preclinical investigation of l-FMAU as an anti-hepatitis B virus agent. Antivir. Ther. 3:113-121. [PubMed] [Google Scholar]
- 10.Chu, C. K., T. Ma, K. Shanmuganathan, C. Wang, Y. Xiang, S. B. Pai, G. Q. Yao, J. P. Sommadossi, and Y. C. Cheng. 1995. Use of 2′-fluoro-5-methyl-β-l-arabinofuranosyluracil as a novel antiviral agent for hepatitis B virus and Epstein-Barr virus. Antimicrob. Agents Chemother. 39:979-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colacino, J. M., and K. A. Staschke. 1998. The identification and development of antiviral agents for the treatment of chronic hepatitis B virus infection. Prog. Drug Res. 50:259-322. [DOI] [PubMed] [Google Scholar]
- 12.Dane, D. S., C. H. Cameron, and M. Briggs. 1970. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet i:695-698. [DOI] [PubMed]
- 13.Delaney, W. E., IV., and H. C. Isom. 1998. Hepatitis B replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 28:1134-1146. [DOI] [PubMed] [Google Scholar]
- 14.Delaney, W. E., IV., T. G. Miller, and H. C. Isom. 1999. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (−)-β-2′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob. Agents Chemother. 43:2017-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dienstag, J. L., R. P. Perrillo, E. R. Schiff, M. Bartholomew, C. Vicary, and M. Rubin. 1995. A preliminary trial of lamivudine for chronic hepatitis B infection. N. Engl. J. Med. 333:1657-1661. [DOI] [PubMed] [Google Scholar]
- 16.Dienstag, J. L., E. R. Schiff, T. L. Wright, R. P. Perrillo, H. W. Hann, Z. Goodman, L. C. L. D. Crowther, M. Woessner, M. Rubin, and N. A. Brown. 1999. Lamivudine as initial treatment for chronic hepatitis B in the United States. N. Engl. J. Med. 341:1256-1263. [DOI] [PubMed] [Google Scholar]
- 17.Ganem, D., and H. E. Varmus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 18.Gerelsaikhan, T., J. E. Tavis, and V. Bruss. 1996. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 70:4269-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honkoop, P., H. G. Niesters, R. A. de Man, A. D. Osterhaus, and S. W. Schalm. 1997. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns. J. Hepatol. 26:1393-1395. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle, J. H., and A. M. di Bisceglie. 1997. The treatment of chronic viral hepatitis. N. Engl. J. Med. 336:347-356. [DOI] [PubMed] [Google Scholar]
- 21.Hurwitz, S. J., B. C. Tennant, B. E. Korba, J. L. Gerin, and R. F. Schinazi. 1998. Pharmacodynamics of (−)-β-2′,3′-dideoxy-3′-thiacytidine in chronically virus-infected woodchucks compared to its pharmacodynamics in humans. Antimicrob. Agents Chemother. 42:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis, B., and D. Faulds. 1999. Lamivudine: a review of its therapeutic potential in chronic hepatitis B. Drugs 58:101-141. [DOI] [PubMed] [Google Scholar]
- 23.Kamimura, T., A. Yoshikawa, F. Ichida, and H. Sasaki. 1981. Electron microscopic studies of Dane particles in hepatocytes with special reference to intracellular development of Dane particles and their relation with HBeAg in serum. Hepatology 1:392-397. [DOI] [PubMed] [Google Scholar]
- 24.Kane, M. 1995. Global programme for control of hepatitis B infection. Vaccine 13:S47-S49. [DOI] [PubMed]
- 25.Knowles, B. B., C. C. Howe, and D. P. Aden. 1980. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 209:497-499. [DOI] [PubMed] [Google Scholar]
- 26.Kocic, I. 2000. Clevudine. University of Georgia/Abbott/Bukwang/Triangle/Yale University. Curr. Opin. Investig. Drugs 1:308-313. [PubMed] [Google Scholar]
- 27.Lai, C. L., C. K. Ching, A. K. Tung, E. Li, J. Young, A. Hill, B. C. Wong, J. Dent, and P. C. Wu. 1997. Lamivudine is effective in suppressing hepatitis B virus DNA in Chinese hepatitis B surface antigen carriers: a placebo-controlled trial. Hepatology 25:241-244. [DOI] [PubMed] [Google Scholar]
- 28.Lenhoff, R. J., and J. Summers. 1994. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J. Virol. 68:4565-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liaw, Y. F., R. N. Chien, C. T. Yeh, S. L. Tsai, and C. M. Chu. 1999. Acute exacerbation and hepatitis B virus clearance after emergence of YMMD motif mutation during lamivudine therapy. Hepatology 30:567-572. [DOI] [PubMed] [Google Scholar]
- 30.Marion, P. L., L. S. Oshiro, D. C. Regnery, G. H. Scullard, and W. S. Robinson. 1980. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA 77:2941-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason, W. S., G. Seal, and J. Summers. 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 36:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair, V., and T. Jahnke. 1995. Antiviral activities of isomeric dideoxynucleosides of d- and l-related stereochemistry. Antimicrob. Agents Chemother. 39:1017-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevens, F., J. Main, P. Honkoop, D. L. Tyrrell, J. Barber, M. T. Sullivan, J. Fevery, R. A. De Man, and H. C. Thomas. 1997. Lamivudine therapy for chronic hepatitis B: a six-month randomized dose-ranging study. Gastroenterology 113:1258-1263. [DOI] [PubMed] [Google Scholar]
- 34.Niederau, C., T. Heintges, S. Lange, G. Goldmann, C. M. Niederau, L. Mohr, and D. Haussinger. 1996. Long-term follow-up of HBeAg-positive patients treated with interferon alfa for chronic hepatitis B. N. Engl. J. Med. 334:1422-1427. [DOI] [PubMed] [Google Scholar]
- 35.Ou, J. H., H. Bao, C. Shih, and S. M. Tahara. 1990. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J. Virol. 64:4578-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkin, D. M., P. Pisani, and J. Ferlay. 1999. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 80:827-841. [DOI] [PubMed] [Google Scholar]
- 37.Peek, S. F., P. J. Cote, J. R. Jacob, I. A. Toshkov, W. E. Hornbuckle, B. H. Baldwin, F. V. Wells, C. K. Chu, J. L. Gerin, B. C. Tennant, and B. E. Korba. 2001. Antiviral activity of clevudine [l-FMAU, 1-(2-fluoro-5-methyl-β-l-arabinofuranosyl) uracil] against woodchuck hepatitis virus replication and gene expression in chronically infected woodchucks (Marmota monax). Hepatology 33:254-266. [DOI] [PubMed] [Google Scholar]
- 38.Peters, M. G., G. Singer, T. Howard, S. Jacobsmeyer, X. Xiong, C. S. Gibbs, P. Lamy, and A. Murray. 1999. Fulminant hepatic failure resulting from lamivudine-resistant hepatitis B virus in a renal transplant recipient. Transplantation 68:1912-1914. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, p. 9.1-9.62. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sandig, V., C. Hofmann, S. Steinert, G. Jennings, P. Schlag, and M. Strauss. 1996. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum. Gene Ther. 7:1937-1945. [DOI] [PubMed] [Google Scholar]
- 41.Santantonio, T., M. Mazzola, T. Iacovazzi, A. Miglietta, A. Guastadisegni, and G. Pastore. 2000. Long-term follow-up of patients with anti-HBe/HBV DNA-positive chronic hepatitis B treated for 12 months with lamivudine. J. Hepatol. 32:300-306. [DOI] [PubMed] [Google Scholar]
- 42.Summers, J., P. M. Smith, and A. L. Horwich. 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Summers, J., J. M. Smolec, and R. Snyder. 1978. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA 75:4533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tine, F., A. Liberati, A. Craxi, P. Almasio, and L. Pagliaro. 1993. Interferon treatment in patients with chronic hepatitis B: a meta-analysis of the published literature. J. Hepatol. 18:154-162. [DOI] [PubMed] [Google Scholar]
- 45.Torresi, J., and S. Locarnini. 2000. Antiviral chemotherapy for the treatment of hepatitis B virus infections. Gastroentrology 118:S83-S103. [DOI] [PubMed]
- 46.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 47.Wei, Y., J. E. Tavis, and D. Ganem. 1996. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 70:6455-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong, D. K., A. M. Cheung, K. O'Rourke, C. D. Naylor, A. S. Detsky, and J. Heathcote. 1993. Effect of alpha-interferon treatment in patients with hepatitis B e antigen-positive chronic hepatitis B. A meta-analysis. Ann. Intern. Med. 119:312-323. [DOI] [PubMed] [Google Scholar]
- 49.Yamada, G., Y. Sakamoto, M. Mizuno, T. Nishihara, T. Kobayashi, T. Takahashi, and H. Nagashima. 1982. Electron and immunoelectron microscopic study of Dane particle formation in chronic hepatitis B virus infection. Gastroenterology 83:348-356. [PubMed] [Google Scholar]