Abstract
The human immunodeficiency virus type 1 (HIV-1) regulatory protein, Rev, mediates the nuclear export of unspliced and singly spliced viral mRNAs by bridging viral RNA and export receptor human CRM1 (hCRM1). Ribonucleoprotein complex formation, including the oligomerization of Rev proteins on viral RNA, must occur to allow export. We show here that Rev-Rev interactions, which are a basis of complex formation, can be initiated without cellular factors and are subsequently enhanced by hCRM1-Ran-GTP. Furthermore, we reveal functions for the Rev carboxy-terminal (C-terminal) region, which is well conserved among many HIV-1 strains, and for which no function has been reported. This region is required for the efficient binding of Rev to hCRM1 and consequently for nuclear export, Rev-Rev dimerization, and full Rev transactivator activity. Consistent with these results, a HIV-1 proviral plasmid that expresses a C-terminally truncated Rev mutant protein produces smaller amounts of the p24 antigen than does a plasmid that possesses an intact rev gene. These results indicate the functional importance of the C-terminal region for full Rev activity, which leads to efficient HIV-1 replication.
Human immunodeficiency virus type 1 (HIV-1) encodes the Rev protein, which localizes predominantly to the nucleus and/or nucleolus (4, 5, 10, 27) and actively shuttles between the nucleus and cytoplasm (35, 45, 50). Rev exports unspliced or incompletely spliced viral mRNAs to the cytoplasm, thereby supporting the efficient expression of the late viral enzymatic and structural proteins encoded by these mRNAs (9, 11, 14, 15, 16, 25, 52). To date, three functional domains indispensable for Rev function have been identified.
One is an arginine-rich basic domain located in the region of amino acids (aa) 35 to 50 of Rev, which has been shown to mediate the direct nuclear binding of Rev to the highly structured cis-acting Rev-responsive element in unspliced or incompletely spliced viral mRNAs (5, 12, 26, 28, 41, 60). This region also serves as a nuclear and/or nucleolar localization signal (5, 32, 42).
A second important domain was identified as a leucine-rich activation domain at aa 73 to 84 (30, 40, 44) which functions as a nuclear export signal (NES) (17, 59). This domain is recognized by a cellular transport receptor, human CRM1 (hCRM1; also known as exportin 1), which belongs to the importin β family, members of which act as carriers in the transport of cellular proteins and ribonucleoprotein complexes between the nucleus and cytoplasm (19, 21, 46, 48, 53). hCRM1 has been reported to bind with the aid of the small GTPase protein, Ran-GTP, to many cellular proteins possessing the leucine-rich NES motif and to convey these proteins into the cytoplasm. Since hCRM1 functions as an export receptor for proteins that are components of ribonucleoprotein complexes, consequently it also exports RNAs such as HIV-1 RNAs, human T-cell leukemia virus type 1 (HTLV-1) RNAs, and snRNAs.
Mutational analyses combined with functional assays of the Rev protein have revealed a third domain that mediates multimerization of this transactivator protein (2, 39, 58). It is generally agreed that the multimeric assembly of Rev on target mRNAs is critical for its function (6, 8, 13, 33, 38, 43). Two regions of Rev (aa 18 to 26 and 54 to 56) that flank the arginine-rich basic motif were originally defined as a multimerization domain (42, 39). Recent studies indicate that the more N-terminal of these two regions and specifically the residues tyrosine 23, serine 25, and asparagine 26 play the most essential role in multimerization (3).
Contradictory results concerning the involvement of hCRM1 in Rev-Rev interaction, which forms a basis for the multimerization of Rev on RNA, have been described in several reports. Two-hybrid analyses in mammalian cells indicate that a Rev activation domain mutant, which has little affinity for hCRM1, has a reduced ability to oligomerize (2, 38), suggesting the involvement of hCRM1 in Rev oligomerization. On the other hand, there are observations that wild-type Rev mislocalizes when coexpressed with the activation domain mutant, suggesting that the hetero-oligomer can form in the absence of hCRM1 (31, 54, 55). In vitro studies have firmly established that the purified recombinant Rev protein has an intrinsic tendency to multimerize and that it can form oligomers on the Rev-responsive element in the absence of any cellular factors and that these properties depend on the presence of an intact multimerization domain (34, 39). Furthermore, the Rex protein of HTLV-1, a functional homolog of Rev, has been indicated in vivo and in vitro to require hCRM1 for Rex multimerization (23, 24). Thus, it is of interest to examine whether hCRM1 takes part in the oligomerization of Rev.
In addition to these three important domains of Rev, a comparison of the sequences of a number of HIV-1 strains registered in the database has revealed that the amino acid sequence of the C-terminal region of Rev, spanning aa 87 to 116, is conserved, as well as the essential central region spanning aa 57 to 86. However, previous reports had shown that the C-terminal region is dispensable (13, 42).
In this study we investigate the well-conserved C-terminal region in more detail and describe important roles of this region. Our results show that the C terminus of Rev assures its efficient binding to hCRM1 and thereby affects the dynamic trafficking of Rev between the nucleus and the cytoplasm. Furthermore, we show that hCRM1 enhances Rev-Rev interactions, leading to promotion of Rev-Rev interaction by C-terminal region. Finally, we demonstrate that these properties of the C-terminal region are tightly linked to Rev activity.
MATERIALS AND METHODS
Plasmid construction.
To construct pSRαRev86, which expresses a mutant Rev protein lacking the region spanning aa 87 to 116, the corresponding region was amplified by PCR using pSRαRev as a template and the primer pair 5′-GAC TGC AGC CAC CAT GGC AGG AAG AAG CGG AG-3′ (primer 1) and 5′-GAG GTA CCT AGT TAC AAT CAA GAG TAA GTC T-3′ (primer 2). This PCR product was treated with Pfu DNA polymerase to create blunt ends, digested with PstI, and then cloned into pSRα296 (57), which had been treated with KpnI, Pfu polymerase, and PstI. The construction of pSRαRev100, which encodes a mutant Rev protein lacking aa 101 to 116, was performed by the same method used for pSRαRev86, except that primer 1 and 5′-GAG GTA CCT AAG GGC TTC CCA CCC CCT GCG T-3′ (primer 3) were used.
To construct pSRαRev-HA and pSRαRevM10-HA, the rev coding region from pSRαRev or pM10 (42) was amplified by PCR using the primer pair primer 1 and 5′-GGA ATT CTA AGC GTA GTC TGG GAC GTC GTA TGG GTA TTC TTT AGC TCC TGA CTC CA-3′, which encodes a hemagglutinin (HA) tag. The PCR products were digested with PstI and EcoRI and cloned into PstI- and EcoRI-digested pSRα296.
To construct pSRαRevM4-HA, PCR-based mutagenesis was carried out using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) in accordance with the manufacturer's instructions. For mutagenesis, pSRαRev-HA as a template and the primer pair 5′-CTC ATC AAG TTT CTC GAT CAA GAC CTC CCA CCT CCC AAT CCC G-3′ and its complement oligonucleotide were used.
To make pGAL-Rev86 or pGAL-Rev100, the corresponding rev sequences were amplified by PCR using pSRαRev as a template and the primer pair 5′-CTG CAA AGC TTA TGG CAG GAA GAA GCG GAG-3′ (primer 4) and primer 2 or primer 4 and primer 3, respectively. Each PCR product was cloned into pSGGALVP (20), which had been treated with BamHI and Pfu polymerase and was then digested with HindIII to remove the VP16 activation domain sequence.
To generate pGAL-RevM10, a corresponding region of the rev sequence was amplified by PCR using pSRαRevM10-HA as a template and the primer pair primer 4 and 5′-GAG GTA CCT ATT ATT TAG CTC CTG ACT C-3′ (primer 5). The PCR product was cloned into pSGGALVP as described above.
To make pRev86-VP or pRev100-VP, a corresponding region of the rev sequence was amplified by PCR using pSRαRev as a template and the primer pair 5′-GCC ATG GCA GGA AGA AGC GGA G-3′ (primer 6) and 5′-GGC CCA AGC TTG TTA CAA TCA AGA GTA AGT CT-3′ or primer 6 and 5′-GGC CCA AGC TTA GGG CTT CCC ACC CCC TGC GT-3′, respectively. Each PCR product was cloned into pSGGALVP, which had been treated sequentially with BamHI, Pfu polymerase, and HindIII to remove the GAL4 DNA binding domain sequence.
To generate pRevM10-VP, a corresponding region of the rev sequence was amplified by PCR using pSRαRevM10-HA as a template and the primer pair 5′-CCG AAT TCA CCA TGG CAG GAA GAA GCG GAG A-3′ and 5′-TTC CCA AGC TTT TCT TTA GCT CCT GAC TCC A-3′. The PCR product was cloned into pSGGALVP as described above.
The plasmids pSRαhCRM1, pGAL-hCRM1, pGAL4, pG5BCAT, pG5BLuc (23, 24), pSRαRev, pGAL-Rev, pRev-VP, pGAL-Rex, pRex-VP, pCDMβ-galactosidase (pCDMβ-Gal) (36), pGAL-RXRα, pRARα-VP (18), and pDM128 (32) have been previously described.
The recombinant RanQ69L expression plasmid pET3dRanQ69L and recombinant hCRM1 and the zz-Rev protein expression plasmids pET3αhCRM1 and p6z60Rev were kind gifts from Y. Yoneda (pET3dRanQ69L) and D. Görlich (hCRM1 and remaining plasmids) (29, 49).
pNL4-3.Luc.E−R− (7), which is derived from the HIV-1 proviral clone pNL4-3, is referred to as pNL4-3LucE−R−(Rev) in this study. To construct pNL4-3LucE−R−(Rev86), which expresses Rev86, PCR-based site-directed mutagenesis using pNL4-3LucE−R−(Rev) as a template and the primer pair 5′-CTC TTG ATT GTA ACG AGG ATT GTG GAA CTT CTG GGA CG-3′ and its complementary oligonucleotide was done. The insertion of a termination codon to delete the C-terminal region (aa 87 to 116) of Rev does not affect any other viral protein functions since pNL4-3LucE−R−(Rev) does not express the Env protein due to a frameshift near the 5′ end of the env gene.
The integrities of the newly constructed plasmids were ascertained by sequencing according to the manufacturer's instruction with an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
Cell culture and transfection techniques.
HeLa cells were cultured in Dulbecco modified Eagle medium, which was supplemented with 10% fetal bovine serum in a 5% CO2 atmosphere at 37°C. Transfection experiments were carried out using DOTAP (Boehringer Mannheim) or Lipofectamine Plus Reagent (GIBCO BRL Life Technologies) according to the manufacturer's recommendations. The cells were harvested 24 or 48 h posttransfection. In order to normalize for variations in transfection efficiency and nonspecific effects of various treatments, 0.1 μg of pCDMβ-Gal was included in transfection experiments and the total amount of transfected DNA was kept constant by adding various amounts of pSRα296.
In vivo assay of protein-protein interaction.
A two-hybrid system was used to analyze protein-protein interactions in mammalian cells as previously described (24). HeLa cells (105/well in six-well plates) were cotransfected with 0.2 μg of plasmids expressing the GAL4- and VP16-fused proteins, 0.1 μg of pCDMβ-Gal to monitor the transfection efficiency, and 0.6 μg of pG5BCAT or pG5BLuc as reporter plasmids that produce chloramphenicol acetyltransferase (CAT) or luciferase (Luc), respectively, when GAL4 fusion proteins interact with VP16 fusion proteins. In some experiments various amounts of pSRαhCRM1 were also cotransfected. Eight hours after transfection, the medium was replaced with fresh growth medium. At that time, leptomycin B (LMB) was added at the appropriate concentration in some experiments. At 24 h posttransfection cells were harvested and CAT, Luc, and β-Gal activities were quantitated with the CAT enzyme-linked immunosorbent assay (ELISA) Kit (the CAT sensitivity of this kit is >50 pg/ml according to the manufacturer's instruction; Roche), with the Steady-Glo Luciferase Assay System (Promega) followed by the Wallac 1420 ARVOsx system (WALLAC), and by standard colorimetric methods, respectively. The CAT/β-Gal or Luc/β-Gal ratios were calculated for all samples.
Measurement of HIV-1 Rev activity.
HeLa cells (105/well in six-well plates) were transfected with pRev-VP or pRev86-VP along with 0.1 μg of pCDMβ-Gal and 0.5 μg of pDM128 as a reporter. The cells were harvested 24 h after transfection, and the amount of CAT and the activity of β-Gal were quantitated and the CAT/β-Gal ratio was calculated for all samples.
When the HIV-1 proviral plasmids were being used, HeLa cells (106/10-cm-diameter dish) were transfected with 1.0 μg of pNL4-3LucE−R−(Rev) and pNL4-3LucE−R−(Rev86). At 48 h posttransfection, the culture supernatant was transferred to a new microcentrifuge tube and centrifuged to remove the cell debris, and then the level of the p24 antigen in the supernatant was evaluated by ELISA, as reported previously (22). The cells remaining at the bottom of the dish were collected, lysed, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis to estimate the amount of p24 antigen in the cells. As these proviral plasmids have the luc gene at the position of the nef gene, which is expressed in a Rev-independent manner, the efficiency of transfection was adjusted by normalizing to Luc activity.
Western blot analysis.
Cell lysates were resolved by SDS-PAGE, and the proteins were then transferred to nitrocellulose filters. Chicken anti-hCRM1 antibody (1:1,000 diluted), mouse anti-GAL4 monoclonal antibody (1:100 diluted; Santa Cruz Biotechnology), rabbit anti-VP antibody (1:200 diluted; Clontech;), rat anti-HA monoclonal antibody (1:1,000 diluted; Roche), mouse anti-Rev monoclonal antibody (8E7, 1:20 diluted [35]), or mouse anti-p24 monoclonal antibody (1:1,000 diluted) was used as primary antibody. Horseradish peroxidase- or alkaline phosphatase-conjugated anti-immunoglobulin G (IgG) antibodies (Promega) were used as secondary antibodies. Immunoreactive bands were visualized using ECL+ (Amersham Pharmacia Biotech) followed by the LAS-1000 plus system (Fujifilm) or 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium solution. For quantitation, the signal intensity on the Western blot membrane was evaluated by Image Gauge Version 3.4 software (Fuji Film) using the LAS-1000 plus system.
Indirect immunofluorescence analysis.
HeLa cells (105/well in six-well plates) were transfected with 0.1 μg of pSRαRev, pSRαRev100, pSRαRev86, or pSRαRevM10-HA with or without pSRαhCRM1. At 24 h posttransfection the cells were fixed with 2% paraformaldehyde solution. In some experiments, 10 μg of actinomycin D per ml and 50 μg of cycloheximide per ml were added to the culture at 20 h posttransfection. After 4 h of incubation, the cells were fixed. After perforation with 0.1% NP-40, the cells were incubated with rabbit anti-Rev antibody for 1 h followed by incubation with Cy3-conjugated goat anti-rabbit IgG antibody for 1 h (Jackson ImmunoResearch or Biosource). The stained cells were observed with an Axiovert 135 system (Zeiss) and classified into three categories based on the patterns of distribution of wild-type or mutant Rev proteins.
Expression and purification of recombinant proteins.
Recombinant RanQ69L and hCRM1 proteins were expressed and purified as described previously (23). After pilification, RanQ69L protein was charged by GTP.
The expression of recombinant zz-Rev protein was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside for 15 h at 20°C in the Escherichia coli strain BL21-Gold(DE3)pLysS (Stratagene). E. coli cells were lysed in buffer A (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM MgCl2, 2 mM dithiothreitol, and 1 μg each of aprotinin, leupeptin, and pepstatin per ml) with a French press after three freeze-thaw cycles and clarified by centrifugation (100,000 × g, 1 h). The supernatant was applied to a Hi-Trap Q column (Amersham Pharmacia Biotech) that was equilibrated with buffer A and separated by a linear gradient of buffer A containing 50 to 500 mM NaCl. The fractions containing recombinant zz-Rev protein were applied to a Superdex 200 column (Amersham Pharmacia Biotech) equilibrated with the transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, 2 mM dithiothreitol, and 1 μg each of aprotinin, leupeptin, and pepstatin per ml) and then concentrated by ultrafiltration using Centricon YM10 (Amicon).
In vitro binding assay.
GAL-Rev, GAL-Rev86, and GAL-RevM10 proteins were synthesized by an in vitro transcription/translation reaction (TNT T7 Quick for PCR DNA; Promega). PCR using the plasmid pGAL-Rev, pGAL-Rev86, or pGAL-RevM10 as a template and the primer pair 5′-CAG ATT TAA TAC GAC TCA CTA TAG GGA AAA ACC ACC ATG AAG CTA CTG TCT TCT ATC GAA CAA GC-3′ (primer 7) and primer 5, primer 7 and primer 2, or primer 7 and primer 5, respectively, was done. Primer 7 contains the T7 promoter, the Kozak sequence, and a gal4 gene specific element. The amplified products were purified with a PCR purification kit (Qiagen) and used as a template for in vitro transcription/translation reactions performed according to the manufacturer's recommendations. The translated proteins were then incubated at 4°C for 2 h with 1 μg of mouse anti-GAL4 monoclonal antibody, which had been immobilized on 20 μl of protein G-Sepharose. The beads were washed several times with buffer B (50 mM HEPES, pH 7.9, 200 mM KCl, 2 mM β-mercaptoethanol, 0.4% Tween 20, and 0.4% skim milk). HeLa cells (8.0 × 106) were scraped off, washed twice with 10 ml of ice-cold PBS, and lysed in 200 μl of buffer B by sonication. After centrifugation at 20,000 × g for 10 min, the supernatants were added to the above washed beads. At that time, recombinant RanQ69L protein, GTP, and MgCl2 were also added at final concentrations of 2 μM, 2 mM, and 5 mM, respectively, to the beads, and they were then incubated at 4°C for 2 h and recovered by low-speed centrifugation. The supernatant was transferred to a new microcentrifuge tube and 4× sample buffer was added (unbound fraction). The recovered beads were washed three times with 1 ml of buffer C (50 mM HEPES, pH 7.9, 200 mM KCl, 2 mM β-mercaptoethanol, 0.4% Tween 20, and 5 mM MgCl2), and then sample buffer was added to the beads (bound fraction). hCRM1 proteins in unbound and bound fractions were analyzed by SDS-PAGE followed by Western blot analysis.
Pull-down assay.
HeLa or REF52 cells (n = 106) cultured on 10-cm-diameter dishes were transfected with 1.0 μg of pSRαRev-HA, pSRαRevM4-HA, or pSRαRevM10-HA. At 48 h posttransfection, the cells were washed with phosphate-buffered saline and lysed in 200 μl of buffer D (20 mM HEPES, pH 7.3, 150 mM NaCl, 3 mM MgCl2, and 0.1% NP-40) by sonication. After centrifugation at 20,000 × g for 10 min, 100 μl of supernatant was added to 4× sample buffer and stored as the input fraction. The remaining 100 μl of supernatant was transferred to a new microcentrifuge tube; recombinant RanQ69L, recombinant hCRM1, GTP, and LMB were added at final concentrations of 2 μM, 50 nM, 2 mM, and 10 to 100 nM, respectively; and the samples were then incubated at 4°C for 2 h with 1.5 μg of zz-Rev, which was previously immobilized on 20 μl of IgG-Sepharose. The beads were recovered by low-speed centrifugation and washed three times with 1 ml of buffer D, and sample buffer was then added to the beads (bound fraction). The HA-fused proteins in the input and bound fractions were analyzed by SDS-PAGE followed by Western blot analysis.
RESULTS
Deletion of the C-terminal region leads to weaker affinity of Rev for hCRM1.
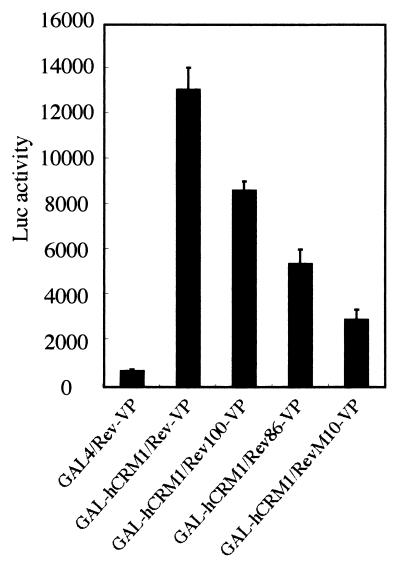
We first examined whether the well-conserved C-terminal region of the Rev protein affects its affinity for hCRM1 because this region is located just after the leucine-rich activation domain (NES), which is directly bound by hCRM1. Since Rev binds hCRM1 only in the nucleus, we utilized a mammalian two-hybrid assay in which hCRM1 was expressed as a fusion protein with the GAL4 DNA binding domain and in which wild-type or truncated Rev proteins were expressed as VP16 fusion proteins. The Rev100-VP and Rev86-VP proteins are VP16 fusion mutants that have a deletion of aa 101 to 116 and 87 to 116 of the Rev protein, respectively. The degree of affinity between pairs of proteins was evaluated by the extent of Luc activity expressed from a reporter plasmid, pG5BLuc (23). As shown in Fig. 1, very high Luc activity (more than 13,000) was detected for the wild-type Rev-VP-GAL-hCRM1 interaction while reduced Luc activity was detected for the interactions between the VP16-fused deletion mutants and GAL-hCRM1. Moreover, these decreases correlate with the extent of deletion. A negative control, the GAL4 expression plasmid, showed the lowest Luc activity (less than 1,000). Similar amounts of VP16-fused Rev proteins were produced in all the samples, as confirmed by Western blot analysis using anti-VP16 antibody (data not shown). These results indicate that the whole C-terminal region (aa 87 to 100 and 101 to 116) of Rev plays a role in enhancing the interaction of Rev with hCRM1. Only a residual affinity of the activation domain mutant RevM10 for hCRM1 was observed.
FIG. 1.
Affinity of Rev and C-terminally truncated mutants for hCRM1 in the nucleus. GAL4 represents the nonfused GAL4 DNA binding domain. The values for Luc activity after normalization represent the mean of three independent experiments, and the bars indicate the standard deviation.
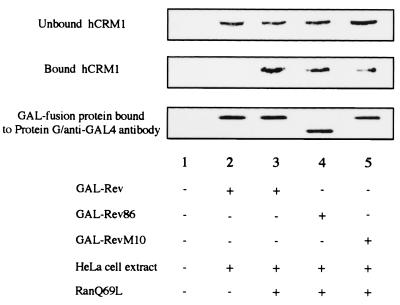
To further characterize the extent of binding of wild-type and mutant Rev proteins to hCRM1 as indicated by the two-hybrid method, an in vitro binding assay was done, in which GAL4 fusion Rev proteins synthesized by an in vitro transcription/translation system were complexed with hCRM1 present in HeLa cell lysates, in the presence or absence of recombinant RanQ69L protein, a GTPase-deficient Ran mutant. As shown in Fig. 2, the binding of Rev to hCRM1 requires the RanQ69L protein (lanes 2 and 3), which guarantees the specificity of this binding and confirms the previously well-documented formation of the trimeric complex of hCRM1, the leucine-rich NES motif, and Ran-GTP (19, 48, 53). RevM10 binds only a small amount of hCRM1, consistent with the two-hybrid data (Fig. 1), also suggesting that the binding of hCRM1 to Rev in this assay is specifically mediated by the Rev NES motif. The quantitation results indicated that the amount of hCRM1 bound to Rev86 or RevM10 is approximately 40 or 20% of that bound to Rev, respectively (data not shown). These results coincide with the data from the two-hybrid assay (Fig. 1). Therefore, we confirm a positive role for the Rev C-terminal region in the binding of Rev to hCRM1.
FIG. 2.
The binding of wild-type and mutant Rev proteins to hCRM1 in vitro. In vitro translated wild-type and mutant Rev proteins were immobilized on a resin and incubated with HeLa cell extracts in the absence (lane 2) or presence (lanes 3 to 5) of GTP-charged RanQ69L protein. The sample in lane 1 was incubated without cell extract and RanQ69L. After incubation, SDS-PAGE sample buffer was added to the supernatant (unbound fraction) and to the precipitated resin (bound fraction) for Western blot analysis. Chicken anti-hCRM1 antibody or mouse anti-GAL4 monoclonal antibody was utilized to detect hCRM1 or GAL4-fused proteins, respectively.
Reduced cytoplasmic accumulation of C-terminally truncated mutants.
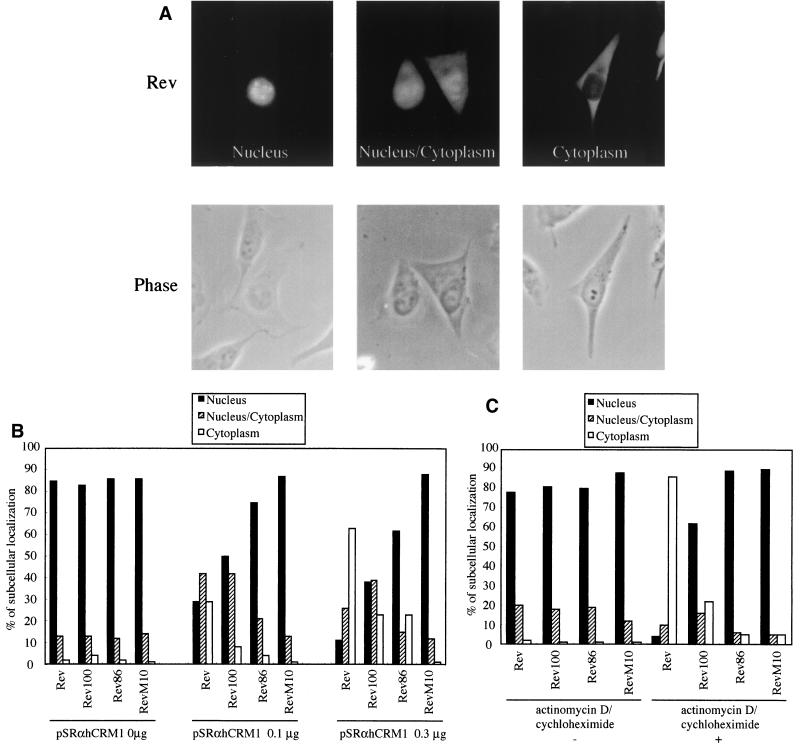
We investigated whether the reduced affinity of the C-terminal region-truncated mutants for hCRM1 directly affects the export process. Wild-type Rev is mainly localized in the nucleus at steady-state levels (Fig. 3A, left panel). However, the overexpression of hCRM1 by transfection changes the distribution pattern of Rev by enhancing the export rate; Rev is observed throughout the nucleus and the cytoplasm (Fig. 3A, center panel) or predominantly in the cytoplasm (Fig. 3A, right panel). The lower affinity of the deletion mutants for hCRM1 may affect the export rate, and consequently the tendency of Rev deletion mutants to accumulate in the cytoplasm may be reduced in comparison with that of wild-type Rev under conditions where hCRM1 is overexpressed. Therefore, we transfected wild-type Rev or deletion mutant expression plasmids in conjunction with the hCRM1 expression plasmid and evaluated the distribution pattern of wild-type or mutant Rev proteins by counting the number of cells, which are classified into the three categories nucleus, nucleus/cytoplasm, and cytoplasm as depicted in Fig. 3A. RevM10 was used as a negative control because this mutant has little affinity for hCRM1 (Fig. 1) and therefore cannot be transported to the cytoplasm. In the absence of the hCRM1 expression plasmid, the majority of wild-type and mutant Rev proteins are similarly localized in the nucleus (Fig. 3B). However, when hCRM1 is overexpressed, the deletion mutants exhibit a reduced tendency to accumulate in the cytoplasm, relative to the wild-type Rev. Furthermore, this lower rate of cytoplasmic accumulation is proportional to the degree of deletion of the C-terminal sequence, thereby correlating with the strength of the affinity of each protein for hCRM1. Under all conditions, most of the RevM10 proteins was seen in the nucleus.
FIG. 3.
The lower export rate of C-terminally truncated mutant proteins. (A) Representative staining pattern of cells transfected with Rev expression plasmids. The cells were classified into three categories based on the localization patterns of Rev: nucleus, nucleus/cytoplasm, and cytoplasm. Phase-contrast images are shown below the immunofluorescence images. (B) Quantitative analysis of the subcellular localization of wild-type and mutant Rev proteins under conditions of overexpression of hCRM1. HeLa cells were transfected with wild-type or mutant Rev expression plasmids along with various amounts of pSRαhCRM1. At 24 h posttransfection indirect immunofluorescence analysis was performed. The cells were classified into categories as described above, and more than 200 stained cells were counted, and percentages were calculated. The data presented are representative of two independent sets of experiments. (C) Subcellular localization of wild-type and mutant Rev proteins in the presence of actinomycin D and cycloheximide. HeLa cells were transfected with wild-type or mutant Rev expression plasmids. At 20 h posttransfection cells were treated with a solvent control (actinomycin D/cycloheximide −) or 10 μg of actinomycin D per ml and 50 μg of cycloheximide (actinomycin D/cycloheximide +) per ml for 4 h, and immunofluorescence staining was performed. These data are representative of two independent sets of experiments.
We next compared the cytoplasmic accumulation of the deletion proteins with that of wild-type Rev in the presence of actinomycin D, an inhibitor of RNA polymerases I and II. Actinomycin D treatment of Rev-expressing cells has been reported to favor the cytoplasmic accumulation of Rev by blocking its nuclear import. Thus, we can evaluate the rate of cytoplasmic accumulation of wild-type Rev and of the deletion mutants in the presence of endogenous levels of hCRM1 by using actinomycin D with the simultaneous addition of cycloheximide to inhibit the de novo synthesis of Rev proteins. As shown in Fig. 3C, the addition of actinomycin D and cycloheximide to the wild-type-Rev-expressing cells induces an efficient cytoplasmic accumulation of Rev, consistent with data from previous reports (35, 50). In contrast, the cytoplasmic accumulation of Rev100 is significantly reduced and Rev86 does not appear in the cytoplasm under these conditions. These results suggest that the C-terminal region of Rev clearly affects the export rate in a manner related to the degree of affinity of each protein for hCRM1.
The role of hCRM1 and the Rev C-terminal region in Rev-Rev interaction.
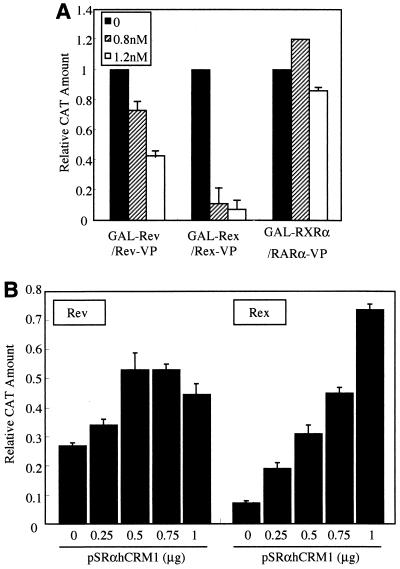
It was previously reported that the HTLV-1 Rex protein, a functional homolog of HIV-1 Rev, uses hCRM1 not only for nuclear export but also for Rex-Rex dimerization (23, 24). If hCRM1 is required for Rev-Rev dimerization, the extent of homodimer formation by Rev100 and Rev86 is predicted to be reduced since their respective affinities for hCRM1 are decreased, as described above. To examine this possibility, at first we investigated the effect of LMB, a specific inhibitor of hCRM1 (47), on Rev-Rev interactions. As Rev multimerization, which is necessary for the export of viral mRNAs, likely occurs in the nucleus, we assessed the involvement of hCRM1 in this process by mammalian two-hybrid assay. At the same time, Rex-Rex interaction was also monitored as a positive control, and the association between the retinoic acid receptors RXRα and RARα, which is not mediated by hCRM1, was also measured as a negative control. LMB inhibits both Rex-Rex and Rev-Rev interactions in a concentration-dependent manner, but its inhibition of Rev is less than that of Rex (Fig. 4A). On the other hand, the interaction between the retinoic acid receptors was almost unaffected by any concentration of LMB tested. These results suggest that hCRM1 may be involved in Rev-Rev interactions as well as in Rex-Rex interactions.
FIG. 4.
Involvement of hCRM1 in Rev-Rev interaction. (A) Decrease in Rev-Rev interaction in the presence of LMB. HeLa cells were transfected with pGAL-Rev and pRev-VP, pGAL-Rex and pRex-VP as a positive control, or pGAL-RXRα and pRARα-VP as a negative control. At 8 h posttransfection the medium was replaced with fresh growth medium containing LMB at final concentrations of 0, 0.8, and 1.2 nM. The cells were harvested, the amount of CAT and β-Gal activity was measured, and the CAT amount was normalized with respect to β-Gal activity (CAT/β-Gal). The CAT/β-Gal value of the sample without LMB in each experimental condition was arbitrarily set at 1. The actual amount of CAT produced by these basal transfection samples was more than 300 pg/ml. The results presented are representative of three independent experiments. (B) The overexpression of hCRM1 rescues the inhibition of Rev-Rev interaction by LMB. Transfection procedures were the same as for panel A except for the addition of various amounts of pSRαhCRM1. LMB was used at a concentration of 1.6 nM. The CAT/β-Gal value of the sample without either LMB or the hCRM1 expression plasmid was arbitrarily set at 1. The actual amount of CAT produced by these basal transfection samples was more than 300 pg/ml. The results are shown as the means and standard deviations of more than two independent experiments.
We next performed complementation experiments by overexpressing hCRM1 by cotransfection in the presence of LMB in the two-hybrid assay system. Overexpression of hCRM1 can relieve the inhibition of Rev-Rev dimerization by LMB, similar to its effect on Rex-Rex interactions (Fig. 4B). However, Rev-Rev and Rex-Rex interactions are only partially restored, in part because the overexpression of hCRM1 reduces the nuclear levels of Rev and Rex by enhancing their export (Fig. 3B).
In the place of LMB, the inhibitory effect of a plasmid expressing TAgRexM64, a dominant negative Rex mutant that sequesters CRM1 (23, 24, 36, 37, 51), was restored by hCRM1 overexpression, a result that also supports the involvement of hCRM1 in this interaction (data not shown).
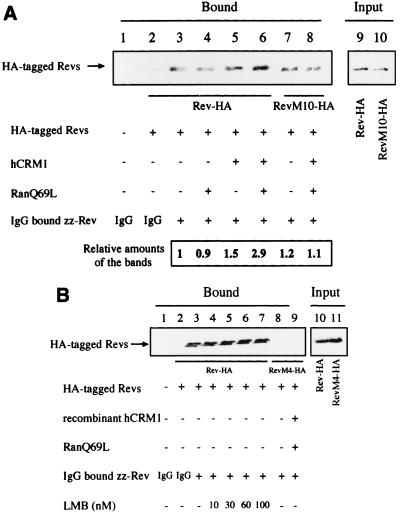
To confirm the above in vivo results, we carried out in vitro pull-down assays. HeLa cells were transfected with HA-tagged Rev and RevM10 expression plasmids, and extracts were incubated with recombinant zz-Rev immobilized on IgG-Sepharose in the presence or absence of RanQ69L and hCRM1. As shown in Fig. 5A, lane 3, Rev-HA binds zz-Rev in the absence of exogenous hCRM1 and RanQ69L. This binding is specific because RevM4-HA, a multimerization-deficient mutant, cannot interact with zz-Rev (Fig. 5B, lane 8). The addition of recombinant hCRM1 and RanQ69L proteins significantly enhances the interaction of Rev-HA with zz-Rev (Fig. 5A, lanes 3 and 6). Recombinant hCRM1 or RanQ69L protein alone was almost unable to enhance Rev-Rev interaction. Although RevM10-HA can be detected in the bound fraction in the absence of recombinant proteins at a level similar to that of Rev-HA (Fig. 5A, lanes 3 and 7), the association of RevM10-HA with zz-Rev is not enhanced by the addition of recombinant hCRM1 and RanQ69L (Fig. 5A, lanes 7 and 8). These results suggest that Rev-HA with a fully intact NES, hCRM1, and RanQ69L are all required for the enhanced interaction of Rev-HA with zz-Rev.
FIG. 5.
In vitro Rev-Rev interactions. (A) HeLa cell extracts were transfected with HA-tagged protein expression plasmids and incubated with recombinant zz-Rev protein, which had been immobilized on IgG-Sepharose, in the absence or presence of GTP-charged recombinant RanQ69L and/or hCRM1. After incubation, the precipitated proteins (Bound) were dissolved in sample buffer and subjected to Western blot analysis using rat anti-HA monoclonal antibody. The volume of the cell extract subjected to pull-down assay was 10 times that of the input fraction. In the quantitation, the ratios of the amounts of HA-tagged Rev proteins to the amount of Rev-HA detected in lane 3, which was arbitrarily set at 1, were calculated. (B) In vitro Rev-Rev interactions in the presence of LMB. RevM4-HA was used as a negative control (lanes 8 and 9). The volume of the cell extract subjected to pull-down assay was 20 times that of the input fraction.
With regard to the interaction of Rev-HA with zz-Rev in the absence of recombinant proteins (Fig. 5A, lane 3), two possibilities are conceivable. One is that this interaction is due to the presence of endogenous hCRM1 and Ran-GTP in cell extracts, and another is that self-association between Rev-HA and zz-Rev occurs independently of these cellular factors. To distinguish between these possibilities, we tested whether the Rev-Rev interaction is inhibited by LMB treatment of the cell extract (Fig. 5B). LMB at various concentrations has no inhibitory effect on this interaction (Fig. 5B, lanes 3 to 7), indicating that hCRM1 is not required for this core Rev-Rev interaction. These in vitro results suggest that there may be two mechanisms at work in establishing Rev-Rev interactions: one is independent of and the other is dependent on hCRM1-Ran-GTP. Moreover, these results suggest that efficient Rev-Rev interaction requires Ran-GTP in addition to hCRM1.
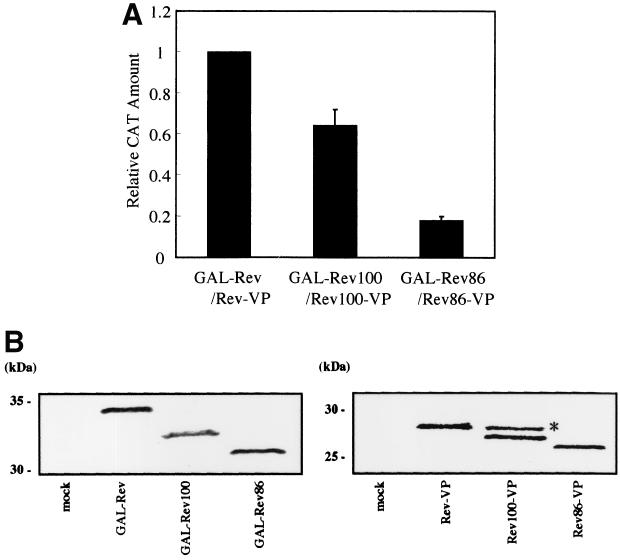
We next investigated the ability of the C-terminally truncated mutants of Rev to form homodimers in the nucleus. As shown in Fig. 6A, both Rev100-Rev100 and Rev86-Rev86 interactions are weaker than Rev-Rev interaction; specifically, the extent of Rev86-Rev86 interaction is less than 20% of the extent of Rev-Rev interaction. The inefficient dimerization of the deletion mutants correlates well with the degree of their affinity for hCRM1. The expression of these proteins in transfected cells was ascertained by Western blot analysis (Fig. 6B). These results additionally suggest that hCRM1 participates in the in vivo Rev-Rev interaction and also indicate that the C-terminal region of Rev is essential for efficient Rev-Rev interaction.
FIG. 6.
Restricted Rev100-Rev100 and Rev86-Rev86 interactions in vivo. (A) HeLa cells were transfected with pGAL-Rev and pRev-VP, pGAL-Rev100 and pRev100-VP, or pGAL-Rev86 and pRev86-VP for two-hybrid assay. The cells were harvested, and the CAT/β-Gal values were calculated. The CAT/β-Gal value obtained by transfection of pGAL-Rev and pRev-VP was arbitrarily set at 1. The actual amount of CAT produced by this basal transfection sample was more than 300 pg/ml. The results represent the means and standard deviations of three independent experiments. (B) Western blot analysis to determine the expression level of GAL4- and VP16-fused proteins. The mock sample (lane “mock”) received pSRα296 alone. As the first antibodies, mouse anti-GAL4 monoclonal antibody and rabbit anti-VP antibody were used, respectively. The band (∗) that migrates more slowly than the authentic Rev100-VP band may represent modified Rev100-VP protein.
The C-terminally truncated Rev mutant is a weaker activator than the wild-type Rev protein.
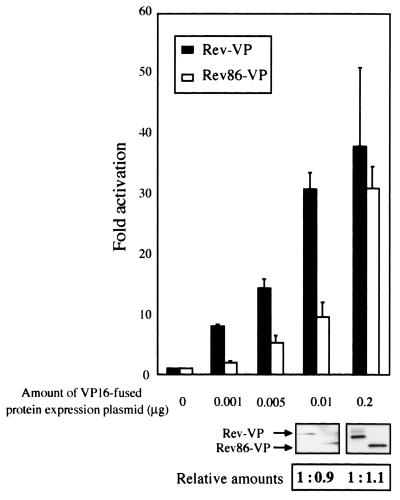
Previous studies reported that the C-terminally truncated Rev mutant is fully active compared to wild-type Rev (13, 42). However, we detected differences between wild-type and mutant Rev proteins in their affinities for hCRM1 and in their ability to engage in Rev-Rev interactions. Thus, we reexamined the activity of Rev86 in comparison with that of wild-type Rev. To evaluate wild-type Rev or deletion mutant activity, the plasmid pDM128, which expresses the CAT protein in a Rev-dependent manner (32), was employed as a reporter. We used VP16-fused Rev and Rev86 derivatives instead of nonfusion Rev and Rev86 proteins since the VP16-fused proteins were shown to have an affinity for hCRM1 (Fig. 1) and since a good antibody against the VP16 moiety was available to quantitate Rev derivatives. Various amounts of the Rev-VP or Rev86-VP expression plasmids were transfected along with pDM128, and the activities of Rev proteins were evaluated by measuring the level of CAT activity. At small amounts (less than 0.01 μg of the Rev expression plasmid), the activity of Rev86-VP is less than half that of Rev-VP (Fig. 7). On the other hand, when 0.2 μg of the expression plasmids is used, the activities of Rev-VP and Rev86-VP are not significantly different. We ascertained by Western blot analysis that similar amounts of Rev-VP and Rev86-VP proteins are synthesized in cells transfected with the same amount of each expression plasmid. Using the nonfusion Rev or Rev86 expression plasmid, we also obtained similar results (data not shown). These results indicate that Rev86 has lower activity than does wild-type Rev but that overexpression of the deletion mutant overcomes this deficiency, explaining the previous studies (13, 42). Our results suggest that, at low levels of expression of the Rev protein, such as in the early phase of HIV-1 infection, the C-terminal region may be required for full Rev activity.
FIG. 7.
Lower activity of a Rev deletion mutant. HeLa cells were transfected with pDM128 and pCDMβ-Gal together with various amounts of pRev-VP or pRev86-VP. CAT/β-Gal values were calculated. The results are presented as n-fold activation relative to the basal CAT/β-Gal value obtained by transfection of pDM128 and pCDMβ-Gal without any effector plasmids. The results represent the means and standard deviations of three independent experiments. Western blotting and the quantitation data under the graph indicate the levels of the VP16-fused protein expression in this functional assay in which 0.01 μg and 0.2 μg of VP16-fused Rev derivative expression plasmids were transfected. In the quantitation data, the levels of Rev86-VP expression relative to that of Rev-VP were calculated.
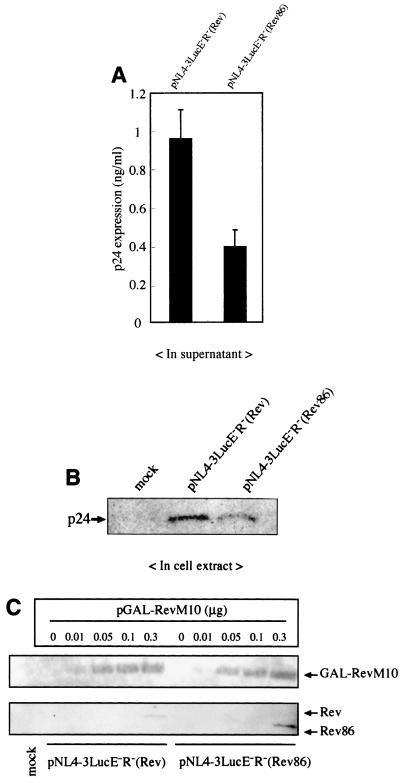
Next we compared the activity of wild-type Rev with that of the deletion mutant in the context of the HIV-1 viral genome. The HIV-1 proviral plasmids pNL4-3LucE−R−(Rev) and pNL4-3LucE−R−(Rev86), which express the intact Rev and the C-terminally truncated Rev86 protein, respectively, were transfected into HeLa cells, and the levels of p24 antigen in the supernatant and cellular fractions were determined by ELISA (Fig. 8A) and Western blot analysis (Fig. 8B) at 2 days posttransfection. Lesser amounts of p24 were detected in both the supernatant and cellular fraction of cells transfected with pNL4-3LucE−R−(Rev86) than in those transfected with pNL4-3LucE−R−(Rev). At 1 day posttransfection, similar results were obtained (data not shown). However, we could not detect the expression of wild-type Rev and Rev86 in lysates of the same cells, indicating that very little Rev or Rev86 is expressed under these conditions. To evaluate whether the lesser expression of p24 from pNL4-3LucE−R−(Rev86) is due to the level of Rev86 protein being lower than that of wild-type Rev, we cotransfected this plasmid and the plasmid pGAL-RevM10, which expresses a Rev protein that exerts a dominant negative effect. It has been reported that the coexpression of a trans-dominant Rev increases the amount of wild-type Rev protein because it inhibits the transition from the early phase to the late phase of the infection cycle (56). As shown in Fig. 8C, when 0.3 μg of pGAL-RevM10 was transfected, slightly higher levels of Rev86 than of wild-type Rev were detected, suggesting that pNL4-3LucE−R−(Rev86) can potentially produce at least as much Rev86 as pNL4-3LucE−R−(Rev) can produce of wild-type Rev. Therefore, the restricted expression of p24 from pNL4-3LucE−R−(Rev86) is not due to a difference in the levels of expression of the wild-type Rev and the Rev86 proteins but is rather due to the lower activity of the Rev86 protein. These results, in conjunction with those obtained from the reporter assay using pDM128 (Fig. 7), suggest that the C-terminal region is important for the full activity of Rev when it is present at low levels, such as during the early phase of HIV-1 infection.
FIG. 8.
Lower expression of Gag from an HIV-1 proviral plasmid harboring the C-terminally truncated Rev mutant. HeLa cells were transfected with pNL4-3LucE−R−(Rev) or pNL4-3LucE−R−(Rev86). (A) At 48 h posttransfection the supernatants were subjected to p24 ELISA. At the same time the cells were lysed and Luc activity was measured to monitor the efficiency of transfection. p24 and Luc values were calculated. The results are shown as the means with standard deviations of several experiments. (B) The normalized volume of cell lysates was subjected to Western blot analysis to detect intracellular levels of p24. The mock sample received pSRα296 alone. (C) The expression of Rev by HIV-1 proviral plasmids in the presence of a dominant negative RevM10. HeLa cells were transfected with 1.0 μg of each HIV-1 proviral plasmid along with various amounts of pGAL-RevM10. At 48 h posttransfection, the cells were lysed and subjected to Western blot analysis. 8E7 anti-Rev monoclonal antibody was used as the primary antibody.
DISCUSSION
In the present study we addressed the functions of the well-conserved C-terminal region of Rev protein in relation to hCRM1. We demonstrate that this region is required for the efficient binding of Rev to hCRM1, for the precise nuclear export of Rev, and for the efficient formation of Rev-Rev dimers (which is a prerequisite for Rev oligomerization on RNA), and that it ultimately assures the full activity of Rev at low levels of expression, such as during the early phase of HIV-1 infection.
Askjaer et al. reported that the cysteine residue at position 89 (Cys89) of Rev, but not Cys85, can be cross-linked to hCRM1 in the presence of Ran-GTP, indicating that the distance between the Cys89 and hCRM1 is reduced in a Ran-GTP-dependent manner (1). Our results consistently indicate that the C-terminal region outside the NES motif contributes to a tight association of Rev with hCRM1. The observation that Rev100 has a weaker affinity for hCRM1 than for wild-type Rev suggests that a number of amino acids in the C-terminal region other than Cys89 also functionally associate with hCRM1. The binding of the Rev NES to hCRM1 in concert with Ran-GTP might induce a conformational change of hCRM1 and/or Rev, leading to a close association of the C-terminal region of Rev with hCRM1 and then to a stabilization of this interaction.
The residual ability of RevM10 to bind to hCRM1 in the presence of Ran-GTP was detected both in vivo and in vitro (Fig. 1 and 2). On the other hand, RevM10 cannot be exported to the cytoplasm even when hCRM1 is overexpressed or when import is inhibited with actinomycin D and cycloheximide (Fig. 3B and C). It is conceivable that, although a RevM10-hCRM1 interaction may exist, these proteins may form an aberrant ternary complex with respect to nuclear export. Alternatively, it is also possible that RevM10 cannot be exported because the RevM10-hCRM1 interaction is appropriate but too weak.
The involvement of hCRM1 as a cellular factor in the oligomerization of Rev has been under debate, as described in the introduction (2, 31, 38, 54, 55). Our in vivo and in vitro results support the involvement of hCRM1 in the oligomerization of Rev. First, LMB reduces in vivo Rev-Rev interactions in a concentration-dependent manner in the mammalian two-hybrid assay (Fig. 4A). Second, the overexpression of hCRM1 relieves this LMB inhibitory effect (Fig. 4B). Third, the restricted ability of C-terminally truncated proteins to form homodimers correlates well with their reduced affinity for hCRM1 (Fig. 6A). Fourth, the addition of recombinant hCRM1 augments the in vitro interaction of Rev-HA with zz-Rev, the specificity of which was ascertained by its requirement for Ran-GTP and by the inability of RevM4-HA to interact with zz-Rev. However, it is clear that Rev-Rev interactions can occur in vitro in the absence of hCRM1, because RevM10-HA can associate with zz-Rev as efficiently as can wild-type Rev-HA in the absence of exogenous hCRM1 and RanQ69L (Fig. 5A) and because LMB cannot inhibit this core Rev-Rev interaction at all (Fig. 5B). In light of a series of experiments about the involvement of hCRM1 in Rev-Rev interactions, we propose that the multimerization of Rev consists of at least two steps: an initial step involves the formation of a “core Rev-Rev interaction” via the multimerization domain, which is independent of hCRM1; in a second step, this interaction is enhanced or stabilized through association of a Rev-Rev multimer with hCRM1-Ran-GTP. This proposal unifies previous contradictory results. We do not know the mechanism by which enhancement or stabilization occurs, but it might be linked to structural changes in Rev induced by the association of hCRM1 and Ran-GTP. These protein-protein interactions, which can occur in the absence of RNA, may provide the basis for complex formation, including Rev oligomerization on viral RNA in vivo.
Scientists previously reported the involvement of hCRM1 in Rex multimerization in vivo and in vitro (23, 24). Rex apparently has a more stringent requirement for hCRM1 than does Rev, since RexM90, a Rex NES mutant, cannot form heterodimers with wild-type Rex in vitro (23); in contrast, RevM10 can form heterodimers with wild-type Rev (Fig. 5A). This difference is consistent with the observation that LMB suppresses Rex-Rex interactions more efficiently than Rev-Rev interactions, as measured in two-hybrid assays (Fig. 4A). Therefore, unlike Rev, Rex may require hCRM1 for the initial interaction between two Rex molecules to create a core Rex-Rex interaction. Alternatively, an initial Rex dimer, like an initial Rev dimer, could be formed in the absence of hCRM1, but it may be too unstable to be detected.
Rev86 induces expression of a reporter gene less efficiently than does wild-type Rev when it is present at low levels, but it has an activity similar to that of wild-type Rev when either is expressed at high levels from transfected plasmids (Fig. 7 and 8). This observation explains why previous studies did not reveal roles for the C-terminal region in Rev activity: large amounts of Rev expression plasmids were used in these assays (13, 42). Moreover, transfection with HIV-1 proviral plasmids (which produce very low levels of Rev), a situation that therefore mimics the early phase of HIV-1 replication, revealed that the C-terminal region of Rev has an obvious and important role in producing Gag protein (Fig. 8).
The C-terminal region of Rev overlaps a part of the Env reading frame. Thus, we cannot exclude the possibility that the good conservation of the C-terminal region of Rev may be due to the overlapping of the Env reading frame. However, our results showing that the C-terminal region of Rev has a significant role in full Rev activity should provide at least one explanation for its conservation among HIV-1 isolates.
Acknowledgments
We thank M. Yoshida for his gift of LMB, K. Umesono for providing pGAL-RXRα and pRARα-VP, N. R. Landau for providing pNL4-3.Luc.E−R−, A. M. Szilvay and K. H. Kalland for providing the anti-Rev monoclonal antibody (8E7), S. Ueda for providing the anti-p24 monoclonal antibody, T. Kimura for his helpful information on the manuscript, F. Kokusen and T. Nakashima for technical assistance, and A. Okuhara and A. Kanayama for excellent secretarial assistance.
These investigations were supported by grants from the Ministry of Education, Science and Culture, Tokyo, Japan, and the Ministry of Health and Welfare, Tokyo, Japan. Y. Hakata is a JSPS Research Fellow.
REFERENCES
- 1.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273:33414-33422. [DOI] [PubMed] [Google Scholar]
- 2.Bogerd, H., and W. C. Greene. 1993. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J. Virol. 67:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brice, P. C., A. C. Kelley, and P. J. G. Butler. 1999. Sensitive in vitro analysis of HIV-1 Rev multimerization. Nucleic Acids Res. 27:2080-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cochrane, A., R. Kramer, S. Ruben, J. Levine, and C. A. Rosen. 1989. The human immunodeficiency virus rev protein is a nuclear phosphoprotein. Virology 171:264-266. [DOI] [PubMed] [Google Scholar]
- 5.Cochrane, A. W., A. Perkins, and C. A. Rosen. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 64:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. L., J. D. Gehman, J. A. Shafer, and L. C. Kue. 1993. Solution oligomerization of the Rev protein of HIV-1: implications for function. Biochemistry 32:11769-11775. [DOI] [PubMed] [Google Scholar]
- 7.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 8.Cook, K. S., G. J. Fisk, J. Hauber, N. Usman, T. J. Daly, and J. R. Rusche. 1991. Characterization of HIV-1 Rev protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 19:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 1991. Regulation of human immunodeficiency virus replication. Annu. Rev. Microbiol. 45:219-250. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, B. R., J. Hauber, K. Campbell, J. G. Sodroski, W. A. Haseltine, and C. A. Rosen. 1988. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J. Virol. 62:2498-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, B. R., and M. H. Malim. 1991. The HIV-1 Rev protein: prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem. Sci. 16:346-350. [DOI] [PubMed] [Google Scholar]
- 12.Daly, T. J., K. S. Cook, G. S. Gray, T. E. Maione, and J. R. Rusche. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342:816-819. [DOI] [PubMed] [Google Scholar]
- 13.Daly, T. J., R. C. Doten, P. Rennert, M. Auer, H. Jaksche, A. Donner, G. Fisk, and J. R. Rusche. 1993. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev responsive element. Biochemistry 32:10497-10505. [DOI] [PubMed] [Google Scholar]
- 14.Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155-1165. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg, M. B., R. F. Jarrett, A. Aldovini, R. C. Gallo, and F. Wong-Staal. 1986. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 46:807-817. [DOI] [PubMed] [Google Scholar]
- 16.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 18.Forman, B. M., K. Umesono, J. Chen, and R. M. Evans. 1995. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541-550. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 19:1051-1060. [DOI] [PubMed] [Google Scholar]
- 20.Fujii, M., H. Tsuchiya, T. Chuhjo, T. Akizawa, and M. Seiki. 1992. Interaction of HTLV-1 Tax1 with p67SRF causes the aberrant induction of cellular immediate early genes through CArG boxes. Genes Dev. 6:2066-2076. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 20:308-311. [DOI] [PubMed] [Google Scholar]
- 22.Giavedoni, L. D., M. C. Velasquillo, L. M. Parodi, G. B. Hubbard, and V. L. Hodara. 2000. Cytokine expression, natural killer cell activation, and phenotypic changes in lymphoid cells from rhesus macaques during acute infection with pathogenic simian immunodeficiency virus. J. Virol. 74:1648-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakata, Y., M. Yamada, and H. Shida. 2001. Rat CRM1 is responsible for the poor activity of human T-cell leukemia virus type 1 Rex protein in rat cells. J. Virol. 75:11515-11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakata, Y., T. Umemoto, S. Matsushita, and H. Shida. 1998. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 72:6602-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammarskjold, M. L., J. Heimer, B. Hammarskjold, I. Sangwan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63:1959-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammerschmid, M., D. Palmeri, M. Ruhl, H. Jaksche, I. Weichselbraun, E. Bohnlein, M. H. Malim, and J. Hauber. 1994. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans activator. J. Virol. 68:7329-7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauber, J., M. Bouvier, M. H. Malim, and B. R. Cullen. 1988. Phosphorylation of the rev gene product of human immunodeficiency virus type 1. J. Virol. 62:4801-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heaphy, S., C. Dingwall, I. Ernberg, M. J. Gait, S. M. Green, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 60:685-693. [DOI] [PubMed] [Google Scholar]
- 29.Hieda, M., T. Tachibana, F. Yokoya, S. Kose, N. Imamoto, and Y. Yoneda. 1999. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol. 144:645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hope, T. J., N. P. Klei, M. E. Elder, and T. G. Parslow. 1992. trans-dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J. Virol. 66:1849-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hope, T. J., X. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev trans activator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwai, S., C. Pritchard, D. A. Mann, J. Karn, and M. J. Gait. 1992. Recognition of the high affinity binding site in rev-response element RNA by the human immunodeficiency virus type-1 rev protein. Nucleic Acids Res. 20:6465-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain, C., and J. G. Belasco. 2001. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell 7:603-614. [DOI] [PubMed] [Google Scholar]
- 35.Kalland, K. H., A. M. Szilvay, K. A. Brokstad, W. Saetrevik, and G. Haukenes. 1994. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell. Biol. 14:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katahira, J., T. Ishizaki, H. Sakai, A. Adachi, K. Yamamoto, and H. Shida. 1995. Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J. Virol. 69:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyokawa, T., T. Umemoto, Y. Watanabe, S. Matsushita, and H. Shida. 1997. Two distinct pathways for intronless mRNA expression: one related, the other unrelated to human immunodeficiency virus Rev and human T cell leukemia virus type I Rex functions. Biol. Signals 6:134-142. [DOI] [PubMed] [Google Scholar]
- 38.Madore, S. J., L. S. Tiley, M. H. Malim, and B. R. Cullen. 1994. Sequence requirements for Rev multimerization in vivo. Virology 202:186-194. [DOI] [PubMed] [Google Scholar]
- 39.Malim, M. H., and B. R. Cullen. 1991. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241-248. [DOI] [PubMed] [Google Scholar]
- 40.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 65:4248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 42.Malim, M. H., S. Böhnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 43.Mann, D. A., I. Mikaelian, R. W. Zemmel, S. M. Green, A. D. Lowe, T. Kimura, M. Singh, P. J. G. Butler, M. J. Gait, and J. Karn. 1994. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 241:193-207. [DOI] [PubMed] [Google Scholar]
- 44.Mermer, B., B. K. Felber, M. Campbell, and G. N. Pavlakis. 1990. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 18:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 46.Neville, M., F. Stutz, L. Lee, L. Davis, and I. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 47.Nishi, K., M. Yoshida, D. Fujiwara, M. Nishikawa, S. Horinouchi, and T. Beppu. 1994. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269:6320-6324. [PubMed] [Google Scholar]
- 48.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 49.Paraskeva, E., E. Izaurralde, F. R. Bischoff, J. Huber, U. Kutay, E. Hartmann, R. Lührmann, and D. Görlich. 1999. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richard, N., S. Incampo, and A. Cochrane. 1994. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology 204:123-131. [DOI] [PubMed] [Google Scholar]
- 51.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sodroski, J., W. C. Goh, C. Rosen, A. Dayton, E. Terwilliger, and W. Haseltine. 1986. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature (London) 321:412-417. [DOI] [PubMed] [Google Scholar]
- 53.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 54.Stauber, R., G. A. Gaitanaris, and G. N. Pavlakis. 1995. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology 10:439-449. [DOI] [PubMed] [Google Scholar]
- 55.Szilvay, A. M., K. A. Brokstad, S. O. Boe, G. Haukenes, and K. H. Kalland. 1997. Oligomerization of HIV-1 Rev mutants in the cytoplasm and during nuclear import. Virology 235:73-81. [DOI] [PubMed] [Google Scholar]
- 56.Szilvay, A. M., S. O. Boe, and K. H. Kalland. 1999. Co-expression of a trans-dominant negative mutant of the human immunodeficiency virus type 1 (HIV-1) Rev protein affects the Rev-dependent splicing pattern and expression of HIV-1 RNAs. J. Gen. Virol. 80:1965-1974. [DOI] [PubMed] [Google Scholar]
- 57.Takebe, Y., M. Seiki, J.-I. Fujisawa, P. Hoy, K. Yokota, K.-I. Arai, M. Yoshida, and N. Arai. 1988. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas, L. S., M. Oft, H. Jaksche, G. Casari, P. Herger, M. Dobrovnik, D. Bevec, and J. Hauber. 1998. Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J. Virol. 72:2935-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 60.Zapp, M. L., and M. R. Green. 1989. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature (London) 342:714-716. [DOI] [PubMed] [Google Scholar]