Abstract
The marine double-stranded DNA (dsDNA) bacteriophage PM2, studied since 1968, is the type organism of the family Corticoviridae, infecting two gram-negative Pseudoalteromonas species. The virion contains a membrane underneath an icosahedral protein capsid composed of two structural proteins. The purified major capsid protein, P2, appears as a trimer, and the receptor binding protein, P1, appears as a monomer. The C-terminal part of P1 is distal and is responsible for receptor binding activity. The rest of the structural proteins are associated with the internal phospholipid membrane enclosing the viral genome. This internal particle is designated the lipid core. The overall structural organization of phage PM2 resembles that of dsDNA bacteriophage PRD1, the type organism of the family Tectiviridae.
PM2, the only isolate in the family Corticoviridae, is the first bacteriophage for which the presence of lipids in the virion was firmly demonstrated (15, 24, 26). It is a lytic virus, infecting two Pseudoalteromonas species: marine gram-negative Pseudoalteromonas espejiana BAL-31 (originally Pseudomonas sp. strain BAL-31), isolated along with the virus (25, 32), and Pseudoalteromonas sp. strain ER72M2, obtained from the East River, New York, by Leonard Mindich (38). Originally, considerable interest was directed toward this virus system (about 200 references), but since 1983, only a few studies have been published. Recently, we decided to examine this virus system (38, 39), as our laboratory is focused on lipid-containing bacterial viruses.
The mass of the virion (∼4.5 × 107 Da) is distributed among nucleic acid (14%), lipid (14%), and protein (72%) constituents (15, 16, 17). The average diameter of the icosahedral particle is ∼60 nm (24, 34, 49), and the lipids are located internally (15, 34). The sedimentation coefficient (s20,w) of the particle is 293S, and buoyant densities in sucrose and cesium chloride are 1.26 and 1.28 g/cm3, respectively (16, 38). The stability of the virion is strongly dependent on sodium and especially on calcium ions, and the virion equilibrated in sucrose is inactive (38). Calcium ions are also essential in the final assembly process during PM2 infection (50).
The PM2 genome is a highly supercoiled circular double-stranded DNA (dsDNA) molecule (27) with the highest number of negative supercoils (−51) observed in a natural molecule (33). The organization of the genes and the transcriptional regulation of operons in the genome have been determined (39; R. H. Männistö, A. M. Grahn, D. H. Bamford, and J. K. H. Bamford, submitted for publication). The 10,079-bp genome replicates via a rolling-circle mechanism (18, 28, 39) while associated with the host cytoplasmic membrane (11).
The viral lipids are derived from the host cell membrane during the assembly process (26), and the composition is ∼64% phosphatidylglycerol, ∼27% phosphatidylethanolamine, ∼8% neutral lipids, and small amounts of asylphosphatidylglycerol (15, 53). The proportion of the major phospholipids in the virion is nearly the inverse of that found in the host, P. espejiana (8), and the phospholipid composition varies depending on the fatty acids used in the growth medium (51, 52). The distribution of the major phospholipids is asymmetric between the two membrane leaflets, phosphatidylglycerol occupying the outer lamella (45).
Previously, four structural proteins, I to IV (P1 to P4), were described (22), but observations of additional ones also have been reported (9). Recently, nine structural proteins, P1 to P9, were assigned to the PM2 virion (38; see also Results). P1, with a molecular mass of 37.5 kDa, has been identified as the spike protein located at the fivefold vertices, and P2, with a molecular mass of 30.2 kDa, is the major capsid protein. Proteins P3 and P4 are located internally (9, 10, 22, 36, 37, 38, 41, 45, 46), and P4 has been proposed to be an integral membrane protein interacting with the viral DNA (9, 40, 42).
Dissociation of the PM2 particle has been extensively studied. Treatment of the virion with different concentrations of urea enabled the separation of the previously described four structural proteins, lipid constituents, and the viral DNA, allowing the localization of the four originally described proteins in the virion (36, 45, 47). The previously described structural model presents PM2 as a particle with two protein shells with a membrane in between (45). According to Hinnen et al. (36) and Schäfer et al. (47), dissociation of the virion in the presence of 4.5 M urea results in particles (nucleocapsids) containing proteins P3 and P4 and the viral DNA, but no membrane. However, the presence of proteins in the membrane moiety also has been proposed. The electron density in the center of the bilayer (0.3 electron/Å3), obtained by low-angle X-ray diffraction, implies that about one-third of the PM2 membrane volume is occupied by protein (34). Proteins P3 and P4 and recently described proteins P5 to P8, with molecular masses of between 3.7 and 17.9 kDa, contain putative hydrophobic transmembrane domains (39). No such membrane signals can be predicted for proteins P1, P2, and P9.
Previous literature describes different subviral particles as well as infectious virions reconstituted from the separated viral components (36, 43, 44, 47, 54). It has also been proposed that no lipid-free nucleocapsids exist, as the dissociation products still contain the viral phospholipids (1, 41). Obviously, the virion structure is more complex than previously assumed, and the structure and assembly information is somewhat controversial. We have revisited the dissociation and association studies with PM2 and developed methods to produce material for further structural characterization. Our findings confirmed the previous observations of the outer capsid but provided new information about the composition and organization of the internal layers of the virion.
MATERIALS AND METHODS
Bacteria, bacteriophages, and purification of viruses.
Pseudoalteromonas sp. strain ER72M2 (38) was used for the propagation of PM2 (24). Bacteria and phages were cultured and purified as previously described (38). In brief, polyethylene glycol 6000-concentrated viral particles from the lysate were first purified in a linear rate zonal 5 to 17% (wt/vol) sucrose gradient in PM2 buffer (20 mM Tris-HCl [pH 7.2], 100 mM NaCl, 5 mM CaCl2) (Sorvall AH629 rotor, 24,000 rpm, 60 min, 15°C), yielding onefold-purified virus. Further purification to produce twofold-purified virus was done by isopycnic centrifugation either with a preformed 5 to 40% (wt/vol) iodixanol gradient (Opti-Prep; Nycomed Pharma) in PM2 buffer (Sorvall TH641 rotor, 35,000 rpm, 4 h, 10°C) or with a CsCl gradient in PM2 buffer with an average density of 1.3 g/cm3 (Sorvall AH629 rotor, 24,000 rpm, 20 h, 20°C [or, for large-scale purification, Sorvall T647.5 rotor, 26,000 rpm, 20 h, 20°C]) and with layering of the onefold-purified virus sucrose band over the CsCl. Before purification with iodixanol, the particles were concentrated by differential centrifugation (Sorvall T647.5 rotor, 32,000 rpm, 3 h 30 min, 15°C). The particles from the second purification gradients were collected by differential centrifugation.
The experiments described here were carried out with twofold-purified virus material (specific infectivity, 5 × 1012 to 1 × 1013 PFU/mg of protein), which was either used fresh or stored frozen at −80°C. The twofold-purified particles produced by the two different methods did not differ detectably, and both were used in all dissociation experiments. Reconstitution experiments were carried out only with virions purified with CsCl.
Protease treatments of phage particles.
Fresh viral particles (∼1 mg of protein/ml) were treated with trypsin (Boehringer Mannheim), pronase, proteinase K, or bromelain (ICN Biomedicals Inc.) in PM2 buffer at 30°C for 2 h. The specific infectivity was monitored before and after proteolysis, and the products were analyzed with a linear 5 to 20% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641, 24,000 rpm, 1 h 30 min, 15°C). The protein, DNA, and lipid (when appropriate) compositions of the gradient fractions were analyzed.
Dissociation of particles by low ionic strength.
Immediately after purification, the virus pellets were carefully washed and resuspended overnight on ice either in 20 mM Tris-HCl (pH 7.2) or in sterilized water (∼1 mg of protein/ml). The infectivities of the dissociation products were determined before loading onto a linear 10 to 40% (wt/vol) sucrose gradient with appropriate conditions (Sorvall TH641 rotor, 32,000 rpm, 5 h, 15°C).
Dissociation of particles by freezing and thawing.
Viral particles in PM2 buffer (1 to 3 mg of protein/ml) were disrupted by one cycle of freezing (−80°C) and thawing. The outcome was analyzed with a linear 5 to 20% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C).
Chelation of calcium ions.
Fresh phages in PM2 buffer were diluted 1:5 in 20 mM Tris-HCl (pH 7.2)-100 mM NaCl, and EGTA was added to a final concentration of 20 mM (∼0.4 mg of protein/ml). After incubation at 4°C overnight, the infectivity was determined prior to analysis of the dissociation products with a linear 5 to 20% (wt/vol) sucrose gradient in 20 mM Tris-HCl (pH 7.2)-100 mM NaCl-10 mM EGTA (Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C).
Multimericity of proteins.
The sizes of PM2 proteins P1 and P2 obtained by freezing and thawing were determined (i) by gel filtration (Superdex 200 16/60 [Pharmacia], equilibrated with 20 mM Tris-HCl [pH 7.2]-150 mM NaCl) with blue dextran (2,000 kDa), catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (68 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa) as molecular mass markers; (ii) by sedimentation with a linear 10 to 40% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641 rotor, 35,000 rpm, 42 h, 15°C) and with phage PRD1 protein P3 (120 kDa) and P5 (85 kDa) trimers, bovine serum albumin (68 kDa), and lysozyme (14 kDa) as molecular mass markers; and (iii) by cross-linking proteins with glutaraldehyde (0.001 to 0.5% [vol/vol]) for 90 min at 22°C (250 μg of protein/ml in 20 mM potassium phosphate [pH 7.2]) and analyzing the samples by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (35) with phage φ6 structural proteins (10 to 85 kDa) and the phage PRD1 protein P3 trimer (120 kDa) as standards. Glutaraldehyde-cross-linked protein multimers were also identified by Western blotting with either polyclonal anti-P1 serum (1:20,000 dilution) or polyclonal anti-P2 serum (1:100,000 dilution) and anti-rabbit immunoglobulin G antibodies (Dako) as secondary antibodies detected by enhanced chemiluminescence reagents (Biological Industries). The multimericity of protein P2 was also determined by light scattering combined with size exclusion chromatography (Pharmacia analytical column equilibrated with 20 mM Tris-HCl [pH 7.2]-150 mM NaCl). The instrumentation, computer software, and ɛ280 value of 0.99 cm2/mg for P2 used for mass determinations are described elsewhere (14).
Production of polyclonal antisera against PM2 proteins P1 and P2.
For antiserum production, a mixture of P1 and P2 obtained from disrupted particles after freezing and thawing was purified by centrifugation and gel filtration (see above). After these steps, P1 was cut from the preparative SDS-polyacrylamide gel for immunization. P2 was used directly after gel filtration. Polyclonal antisera against P1 and P2 were raised by immunizing rabbits three times subcutaneously at 21-day intervals. Before immunizations, preimmune serum was obtained. Quantities of 100 μg of purified protein P2 and approximately 100 μg of protein P1, emulsified with complete Freund's adjuvant, were used for the first immunization. For subsequent immunizations, proteins were emulsified with incomplete Freund's adjuvant. Serum was collected 2 weeks after the last booster. The neutralization of PM2 virions with anti-P1 and anti-P2 sera was tested by treating approximately 150 PFU of twofold-purified virus in PM2 buffer with different dilutions of antisera (1:2 to 1:10,000) for 2 h at 23°C and then plating on strain ER72M2. Viruses treated with the corresponding preimmune serum (1:2 to 1:100) were used as controls. Immunoprecipitation of PM2 particles with anti-P2 serum was monitored by treating 50 μg of twofold-purified virus with different dilutions of anti-P2 serum (1:2 to 1:10,000) for 2 h at 23°C. Analysis was carried out by rate zonal centrifugation with a linear 5 to 20% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641 rotor, 24,000 rpm, 1 h 15 min, 15°C) followed by determination of virus material distribution in the gradient fractions and the pellet.
Dissociation of particles with urea.
Freshly made viral particles were treated with up to 6 M urea in PM2 buffer as described by Hinnen et al. (36) and Schäfer et al. (47). After the addition of urea, the samples (∼1 mg of protein/ml) were incubated at 25°C for 30 min prior to analysis with a linear 10 to 40% (wt/vol) sucrose gradient containing the corresponding concentration of urea (PM2 buffer; Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C). A urea concentration of 4.5 M was used to isolate soluble phage lipid cores (Sorvall TH641 rotor, 35,000 rpm, 4 h, 15°C). Proteolysis from the 4.5 M urea treatment was inhibited with 2 mM phenylmethylsulfonyl fluoride and a proteinase inhibitor cocktail (Complete Mini EDTA free; Boehringer Mannheim). The densities of phage lipid cores were determined with a linear 20 to 70% (wt/vol) sucrose gradient containing 2 M urea (PM2 buffer; Sorvall TH641 rotor, 32,000 rpm, 17 h, 15°C).
SDS treatment of lipid cores.
SDS treatment was carried out by disruption of fresh phage particles in 2 M urea (PM2 buffer; 25°C, 30 min) and purification with a linear 10 to 40% (wt/vol) sucrose gradient containing 2 M urea (PM2 buffer; Sorvall TH641 rotor, 35,000 rpm, 3 h, 15°C). The light-scattering lipid core zones were collected, dialyzed against PM2 buffer containing 2 M urea, and concentrated by using a Sartorius membrane concentrator (cutoff, 12 kDa). Concentrated lipid cores were treated with 0.1% SDS for 20 min at 25°C and centrifuged in a linear 10 to 40% (wt/vol) sucrose gradient containing 2 M urea, 0.1% SDS, and PM2 buffer (Sorvall TH660 rotor, 35,000 rpm, 16 h, 25°C).
Protease treatment of lipid cores.
For proteolysis of dissociated phage particles, fresh viruses were disrupted with 4.5 M urea in PM2 buffer (25°C, 30 min), and the urea was diluted to a final concentration of 2 M. The material was treated with trypsin (50 μg/ml) at 30°C, and the reactions were stopped with a 10-fold excess of soybean trypsin inhibitor (Boehringer Mannheim; 30°C, 30 min). The proteolysis products were analyzed with a linear 10 to 40% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641, 32,000 rpm, 1 h 30 min, 15°C).
Reconstitution.
Reconstitution of viral particles was carried out in principle as described by Schäfer and Franklin (43, 44). Viral particles in PM2 buffer with 20 mM 2-mercaptoethanol were disrupted with 4.5 M urea (0.7 mg of protein/ml). The urea concentration was reduced by dialysis (cutoff, 6 to 8 kDa) for 16 h at 4°C against PM2 buffer, 20 mM 2-mercaptoethanol, and different urea concentrations (0, 0.5, 1, or 2 M). The original dissociation mixture with 4.5 M urea was used as a control. In addition, purified separated components obtained from the virions after dissociation with 4.5 M urea (proteins P1 and P2 and lipid cores) were treated in a manner similar to that used for the reconstitution mixtures. The infectivity was determined at each step, and the reaction products were analyzed by rate zonal centrifugation with a linear 5 to 20% (wt/vol) sucrose gradient (dialysis buffer conditions; Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C). Gradients containing 4.5 M urea were centrifuged in similar conditions except at 35,000 rpm for 3 h.
Reconstitution experiments were also carried out by using a combination of purified proteins P1 and P2 and purified lipid cores. The proteins were obtained from dissociated viral particles (see above). The top fraction of the gradient containing proteins P1 and P2 was collected, concentrated (Centrex UF-2, 10 kDa; Schleicher & Schuell), and filtered (Spin-X UF, 100 kDa; Costar) to remove the background of infectious particles. For lipid core isolation, fresh particles were dissociated with 2 M urea (PM2 buffer) prior to purification with a linear 10 to 40% (wt/vol) sucrose gradient in the same buffer conditions (Sorvall TH641 rotor, 35,000 rpm, 3 h, 15°C). The bands containing the lipid cores were collected, and the infectivity was assayed. Proteins P1 and P2 and the lipid cores were dialyzed separately against 20 mM Tris-HCl (pH 7.2), 1 M urea, 0.5 M NaCl, and 10 mM CaCl2. The lipid cores were concentrated by using a Sartorius membrane concentrator (cutoff, 12 kDa). The reconstitution reactions were initiated by combining the proteins and the lipid cores to obtain a final protein concentration of ∼0.5 mg/ml; this step was followed by incubation at room temperature for 20 min. Proteins P1 and P2 and the lipid core fractions were used separately as controls. The incubation was continued by dialysis (3 h, room temperature) against the same buffer containing either 0.75 M or no urea. A reconstitution reaction mixture dialyzed against the original buffer containing 1 M urea was used as a control. Analyses of the reconstitution mixtures and the controls were carried out by sedimentation with a linear 5 to 20% (wt/vol) sucrose gradient (dialysis buffer conditions; Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C). At each step, the infectivities were assayed.
Lipid analysis.
Phospholipids were extracted from gradient fractions of untreated virus material obtained from a linear 5 to 20% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C) and viruses treated with 4.5 M urea as described above and were separated with a linear 10 to 40% (wt/vol) sucrose gradient in 4.5 M urea-PM2 buffer (Sorvall TH641 rotor, 35,000 rpm, 4 h, 15°C). The extraction was carried out by the method of Bligh and Dyer (6) as modified by Davis et al. (23). In brief, after chloroform-methanol extraction (1:2, vol/vol) for 2 h at 28°C, the samples were centrifuged (Sorvall SS34 rotor, 3,000 rpm, 10 min), and water and chloroform were added to the supernatant to obtain two phases. The lower, solvent phase was washed once with 2 M KCl and once with water. The lipid fraction was dried under a stream of nitrogen, resuspended in chloroform-methanol (1:2, vol/vol), and stored at −20°C. Phospholipids were analyzed by thin-layer chromatography (HPTLC-aluminum silica gel 60 F254 precoated; Merck); development was done with chloroform-methanol-water (65:25:4). Phospholipids were visualized with iodine vapor. l-α-Phosphatidylethanolamine (PE; Sigma) and l-α-phosphatidyl-dl-glycerol (PG; Sigma) were used as standards.
Electron microscopy methods.
For negative-stain electron microscopy, freshly made virions resuspended in PM2 buffer were diluted to obtain a final buffer concentration of 20 mM Tris-HCl (pH 7.2)-20 mM NaCl-2 mM CaCl2. Protein P1-deficient particles (see above) were dialyzed against the same buffer. The effect of ion depletion was analyzed by rinsing fresh PM2 virions adsorbed on the grid with three consecutive drops of distilled water. Phage lipid cores isolated with 4.5 M urea were adsorbed to the grid and rinsed as described above. Trypsin-treated isolated lipid cores (see above) were dialyzed against 20 mM Tris-HCl (pH 7.2). The samples were negatively stained with 1% potassium phosphotungstic acid (pH 6.5). Micrographs were obtained with a JEOL 1200 EX electron microscope (60 kV). For thin-section electron microscopy, freshly made viruses were pelleted by using an Airfuge (Beckman A95 rotor, 200 kPa, 7 min, 18°C), and lipid cores were produced by dissociating viruses (1 mg/ml) with 4.5 M urea-PM2 buffer as described above and pelleted by using an Airfuge (Beckman A95 rotor, 207 kPa, 50 min, 18°C). Thin-section electron microscopy was carried out as previously described (4), except that the pellets were fixed with 3% (vol/vol) glutaraldehyde-potassium phosphate (pH 6.5)-100 mM NaCl (40 min, 22°C). Micrographs were obtained as described above.
Analytical methods.
Protein concentrations were measured by the Coomassie blue method (7) with bovine serum albumin as a standard. Proteins were separated by Tricine-SDS-PAGE (48) as described by Kivelä et al. (38). Proteins were stained with Coomassie brilliant blue or transferred to a polyvinylidene difluoride membrane (Millipore) and detected with antibodies described in this study (see above). Phage DNA in the upper gel was visualized by ethidium bromide staining. N-terminal amino acid sequences were determined from the protein bands separated by Tricine-SDS-PAGE, transferred to a polyvinylidene difluoride membrane, stained with Coomassie brilliant blue, and subjected to Edman degradation by using a Procise 494A protein sequencer (Perkin-Elmer/Applied Biosystems, Foster City, Calif.). The sedimentation behavior of viral particles that were untreated, protease treated, or dissociated by freezing and thawing was analyzed by centrifugation with a linear 5 to 20% (wt/vol) sucrose gradient in PM2 buffer (Sorvall TH641 rotor, 24,000 rpm, 1 h 30 min, 15°C).
RESULTS
PM2 virion.
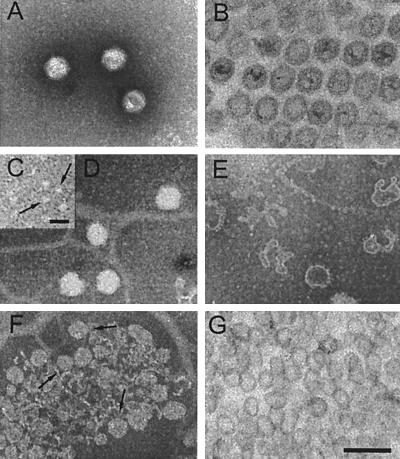
Negative-stain and thin-section electron microscopy of PM2 virions (Fig. 1A and B) revealed icosahedral particles with a diameter of ∼55 nm between the opposite facets and ∼60 nm between the vertices, consistent with previous structural studies (24, 34, 49). Viral DNA stained weakly in thin sections of viral pellets. This phenomenon has also been observed for intracellular particles (20, 21, 38). Particles with an open vertex or empty capsids with a collapsed membrane were occasionally observed in untreated negatively stained virus material. However, no seemingly intact unpackaged particles were observed. The internal diameter of the proteinaceous capsid was ∼45 nm, and the membrane appeared ∼4 nm thick. A spherical approximation of the PM2 membrane, which may follow the icosahedral shape of the capsid, as in the case of bacteriophage PRD1 (13), indicated an internal diameter of ∼35 nm and an enclosed volume of ∼2.3 × 104 nm3. The packing density for the 10,079-bp negatively supercoiled circular dsDNA genome was calculated to be ∼0.9 nt/nm3 (∼45 bp/100,000 Å3). A similar packaging density has also been observed for crystalline DNA and for other dsDNA bacteriophages, such as λ (19).
FIG. 1.
Electron micrographs of PM2 virions and subviral particles. (A and B) Negative-stain electron microscopy (A) and thin-section electron microscopy (B) of untreated PM2 virions. (C to E) Fresh PM2 virions disrupted in water prior to negative staining. (C) Soluble fraction containing capsid proteins P1 and P2. Released structures were identified as multimers of protein P2 (arrows) by negative staining of purified protein P2 (see the text). (D) Spherical lipid cores. (E) Pleomorphic PM2 membrane vesicles. (F and G) PM2 lipid cores obtained after dissociation of the virion with 4.5 M urea. (F) Negatively stained lipid cores (arrows) purified by rate zonal centrifugation (see Fig. 5B, lane 7). (G) Thin section of lipid cores. Bars, 100 nm in A, B, and D to G and 20 nm in C.
The sedimentation behavior and the protein pattern of the untreated virion are shown in Fig. 2A and Fig. 2B, lane 1, respectively. A new PM2 structural protein, P9, was identified. This 24.7-kDa protein, not reported previously, had the N-terminal amino acid sequence MRTTTKKQIE and thus was encoded by gene IX. Gene IX (nucleotide coordinates 4615 to 5271) replaces the previously described open reading frames i and i∗ (for PM2 gene nomenclature, see reference 39). Accordingly, the virion is composed of nine structural proteins, P1 to P9.
FIG. 2.
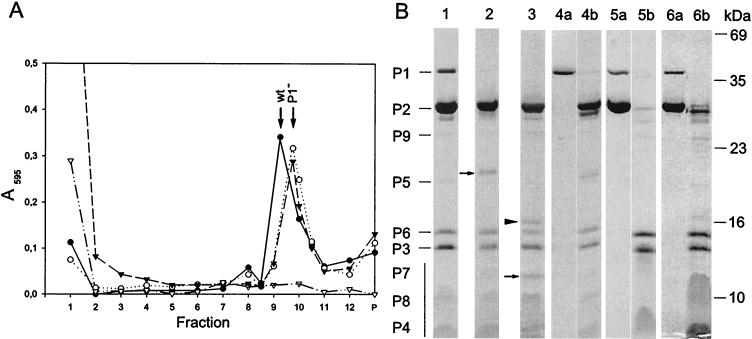
(A) Sedimentation analysis of fresh PM2 virions (filled circles), protein P1-deficient particles obtained by proteinase K treatment (50 μg/ml; open circles) or bromelain treatment (5 mg/ml; filled triangles), and particles after freezing and thawing (open triangles). Protein concentrations in the fractions and in the pellet (P) were assayed by the Coomassie blue method. Sedimentation positions of untreated virions (wt) and protease-treated particles (P1−) are indicated. (B) SDS-PAGE analysis of dissociated particles. Protein patterns of the peak fractions of PM2 virions (lane 1) and particles treated with either proteinase K (lane 2) or bromelain (lane 3) are shown. N-terminal amino acid sequences were determined for the new protein bands which appeared after protease treatments (arrows; see the text). The protein band marked by an arrowhead (lane 3) was derived from the bromelain enzyme. PM2 dissociation products either in 20 mM Tris-HCl (pH 7.2) or in sterilized water were separated in a sucrose gradient; the top fraction and the pellet are shown in lanes 4a and 4b, respectively. Calcium ions were removed from the virions with a 20 M excess of EGTA; the top fraction and the pellet are shown in lanes 5a and 5b, respectively. For particles after one cycle of freezing and thawing, the top fraction (lane 6a) and the pellet (lane 6b) are shown. The positions of the PM2 structural proteins are indicated on the left (the vertical bar denotes the small membrane proteins forming diffuse bands), and the molecular masses of the standard proteins are indicated on the right.
The C terminus of spike protein P1 is distally located at the virion vertices.
Freshly made virions were treated with trypsin, pronase, proteinase K, or bromelain. Intact virions were resistant to trypsin (50 μg/ml), but treatment with pronase (50 μg/ml), proteinase K (50 μg/ml), or bromelain (5 mg/ml) resulted in DNA-containing noninfectious (specific infectivity, ∼5 × 105 PFU/mg of protein) particles sedimenting faster than untreated virions (Fig. 2A). SDS-PAGE analysis of these particles revealed that protein P1 was absent but that proteins P2 to P9 were intact (pronase data not shown; proteinase K and bromelain treatments shown in Fig. 2B, lanes 2 and 3, respectively). Specific degradation products cosedimented with these purified protease-treated particles. Determination of the N-terminal amino acid sequences of these degradation products (resulting in the sequence MIVKKKLAAG) revealed that the ∼18-kDa protein fragment in proteinase K-treated particles and the ∼8-kDa fragment in bromelain-treated particles were N-terminal portions of spike protein P1 (Fig. 2B, arrows in lanes 2 and 3; pronase data not shown). These data indicate that the C-terminal fragment of the spike protein, removed by the protease treatments, contains the receptor binding activity.
The protein capsid of PM2 is quantitatively removed by freezing and thawing or by chelating calcium ions.
Virion integrity is strongly dependent on sodium and calcium ions, as previously reported (38, 50). Under low-ionic-strength conditions, soluble protein P1 was released quantitatively (Fig. 2B, lane 4a). Protein P2 was partly removed, and the rest of the virus material either sedimented as a diffuse zone or aggregated (Fig. 2B, lane 4b). Negative staining of released proteins P1 and P2 is shown in Fig. 1C. Aggregation of the internal lipid core particles (see below) occurred rapidly, but separate intact and pleomorphic lipid core particles (Fig. 1D and E) could occasionally be seen.
We observed that proteins P1 and P2 could be obtained in a soluble form either by removal of calcium ions from the particles with a 20 M excess of EGTA or by one cycle of freezing and thawing. In both situations, >99% of the particles were dissociated. Sedimentation analysis of particles after one cycle of freezing and thawing (Fig. 2A) and SDS-PAGE analyses of the dissociation products (Fig. 2B, lanes 5a to 6b) showed that the lipid cores were aggregative.
Isolation, immunogenicity, and multimericity of proteins P1 and P2.
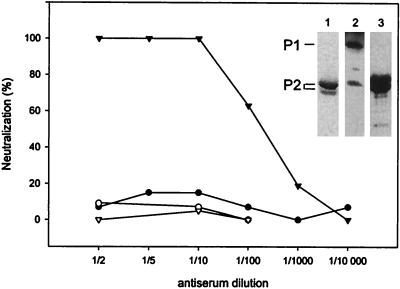
Soluble P1 and P2, removed from particles by one cycle of freezing and thawing (Fig. 2B, lane 6a), were purified by gel filtration. P2 after gel filtration was used for polyclonal anti-P2 serum production (Fig. 3, lane 1). P1 was further purified by preparative SDS-PAGE prior to antibody production. The specificities of the produced polyclonal anti-P1 and anti-P2 sera were assayed (Fig. 3, lanes 2 and 3). Anti-P2 serum was specific to P2 and induced virus neutralization (Fig. 3). However, anti-P1 serum also resulted in a low P2 signal. PM2 virions were also immunoprecipitated with anti-P2 serum, and sedimentation analysis showed that the virions were distributed in three positions: particles sedimenting as virions, material sedimenting as virus dimers, and aggregates (1:10 dilution of anti-P2 serum). However, even a 1:2 dilution of anti-P1 serum in the neutralization assay had no effect on the infectivity.
FIG. 3.
Neutralization of PM2 virions with anti-P1 serum (filled circles) and anti-P2 serum (filled triangles). P1 preimmune serum (open circles) and P2 preimmune serum (open triangles) were used as negative controls. Purified P2 protein used for the production of polyclonal anti-P2 serum is shown on a Coomassie brilliant blue-stained SDS-polyacrylamide gel (lane 1). The specificities of anti-P1 serum (1:20,000 dilution) and anti-P2 serum (1:100,000 dilution) are shown in lanes 2 and 3 (Western blots). The positions of proteins P1 and P2 determined from the corresponding Coomassie brilliant blue-stained gel are indicated on the left.
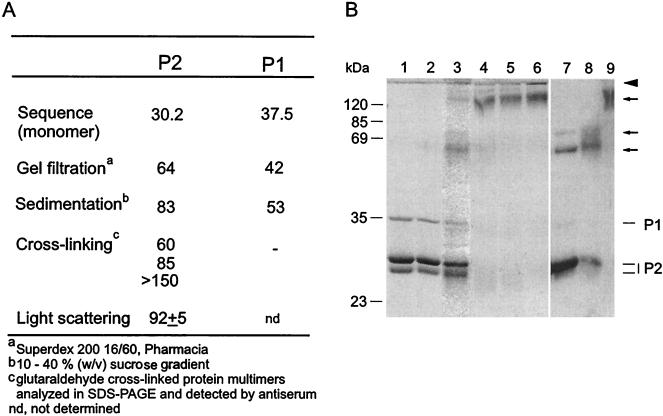
The multimericity of PM2 proteins P1 and P2 was analyzed by using a variety of methods (Fig. 4A). Protein P1 was released from the virion in a monomeric form, according to sedimentation and gel filtration assays (a molecular mass close to the mass predicted from the sequence [37.5 kDa]). In addition, a cross-linking experiment showed that no cross-linked products were recognized by the polyclonal anti-P1 serum in Western blots. Protein P2 behaved as approximately a dimer in the gel filtration assay but sedimented close to a trimer-size complex. Glutaraldehyde-cross-linked multimers of P2 (Fig. 4B, lane 3) were recognized by the polyclonal anti-P2 serum and showed sizes corresponding to dimers, trimers, and considerably larger multimers (arrows in Fig. 4B). Monomeric P2 (30.2 kDa) formed major and minor monomeric bands on SDS-PAGE analysis (Fig. 4B, lane 1). To confirm the oligomeric state of P2, the multimericity of the protein was also analyzed on an analytical size exclusion column coupled to detectors. The result of this light-scattering experiment indicated that the apparent molecular mass of eluted P2 was 92 ± 5 kDa, confirming that P2 is a trimer.
FIG. 4.
(A) Sizes of PM2 capsid proteins P1 and P2 (kilodaltons) obtained by different methods. (B) Cross-linking of proteins with glutaraldehyde. Lanes 1 to 6 show a Coomassie brilliant blue-stained polyacrylamide gel, and lanes 7 to 9 show a Western blot with anti-P2 serum. Non-cross-linked proteins P1 and P2 are shown in lane 1, and their positions are indicated on the right. Mixtures of proteins P1 and P2 (250 μg of protein/ml) were cross-linked with increasing glutaraldehyde concentrations (0.001% [vol/vol] [lane 2], 0.01% [lane 3], 0.05% [lane 4], 0.1% [lane 5], and 0.5% [lane 6]) as described in Materials and Methods. Multimeric forms detected by anti-P2 serum in samples cross-linked with 0.01% (lane 7), 0.05% (lane 8), and 0.1% (lane 9) glutaraldehyde are indicated by arrows. The boundary between the upper gel and the lower gel is indicated by an arrowhead. Numbers on the left indicate the molecular masses of the standard proteins.
Internal lipid core particles isolated with urea contain viral phospholipids, proteins P3 to P9, and the dsDNA genome.
A stepwise dissociation of bacteriophage PM2 with increasing concentrations of urea was previously reported (36, 47). To define the dissociation products, urea concentrations of between 0.5 and 6 M were used, and the products were analyzed by rate zonal centrifugation. Figure 5A shows untreated particles analyzed in a sucrose gradient immediately after virus purification. The peak fraction contained the major phospholipids, PE and PG, viral DNA, and structural proteins P1 to P9 (fraction 9 in Fig. 5A). Approximately 20% of the untreated particles were found already dissociated.
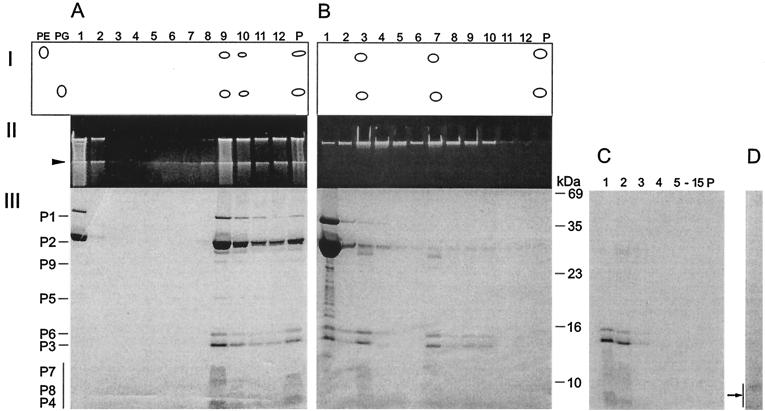
FIG. 5.
Dissociation of PM2 particles with urea and separation of the products by rate zonal centrifugation (see Materials and Methods). (A) Untreated particles. (B) Particles dissociated with 4.5 M urea. (Panel I) Major lipids separated by thin-layer chromatography with PE and PG as standards. (Panel II) Viral DNA in upper SDS-polyacylamide gel stained with ethidium bromide. (Panel III) Structural proteins in Coomassie brilliant blue-stained SDS-polyacrylamide gel. P, pellet. The boundary between the upper gel and the lower gel is indicated by an arrowhead. The positions of the PM2 structural proteins are indicated on the left (the vertical bar denotes the small membrane proteins forming diffuse bands). The numbers to the right of panel B indicate the molecular masses of the standard proteins. (C) Isolated phage lipid cores in 4.5 M urea were solubilized with 0.1% SDS and analyzed with a rate zonal sucrose gradient containing 2 M urea, 0.1% SDS, and PM2 buffer. Fifteen fractions and the pellet were collected and analyzed by SDS-PAGE. The area indicated by an arrow was subjected to N-terminal amino acid sequencing. (D) Isolated phage lipid cores in 4.5 M urea were treated with trypsin (50 μg/ml) and soybean trypsin inhibitor (500 μg/ml) and analyzed with a rate zonal sucrose gradient (PM2 buffer). One visible light-scattering zone was detected and analyzed by SDS-PAGE. No other PM2 proteins were detected in the gradient fractions.
The virus peak fraction disappeared when particles were treated with >1.5 M urea, while two new light-scattering zones were detected. With >2 M urea, virions were completely dissociated and the infectivity dropped irreversibly by 9 orders of magnitude. Released proteins P1 and P2 became more soluble; with 4.5 M urea, these proteins and the other phage components were completely solubilized (Fig. 5B). Proteins, DNA, and lipids in the gradient fractions were analyzed and compared with those in untreated particles. PE and PG cosedimented with the two light-scattering zones with viral proteins P3 to P9 (fractions 3 and 7 in Fig. 5B). In addition, viral DNA was associated with these fractions. These isolated subviral particles were designated lipid cores. The buoyant densities of the two distinct lipid core particles in sucrose were approximately 1.15 and 1.19 g/cm3, respectively. The ratio of these particles was about 1:3 (lighter/denser). The only difference detected between them was the smaller amount of protein P5 in the lighter fraction.
The protein compositions were verified by determining the N-terminal amino acid sequences of the proteins located in the diffuse area below protein band P3 in the SDS-PAGE analysis (38), resulting in the sequences MINKTTI (protein P7), MLGALMG (protein P8), and MQKPSGK (protein P4). The mobility of viral DNA in the upper gel of the SDS-PAGE analysis varied depending on the urea concentrations in the samples. With 4.5 M urea, viral DNA did not enter the gel. As shown in Fig. 5B, the gradient top fraction contained proteolytic fragments not observed in untreated particles. We noticed that proteolysis was reduced but not completely inhibited by use of either ultrapure or autoclaved sucrose in combination with protease inhibitors.
Negative-stain and thin-section micrographs of soluble PM2 lipid cores isolated with 4.5 M urea and containing viral DNA (fractions 3 and 7 in Fig. 5) are shown in Fig. 1F and G. These spherical isolated lipid cores were soluble in >1 M urea. Thus, the solubility of the lipid cores with negative staining was decreased due to the removal of the urea prior to staining. Lipid cores isolated with 4.5 M urea could be further dissociated with the addition of 0.1% SDS (Fig. 5C). Analysis after extended rate zonal centrifugation revealed soluble PM2 proteins at the top of the gradient. Free viral DNA sedimented in fractions 11 to 15 and was also found in the pellet (data not shown).
We were not able to produce other previously described substructures by published methods with 1 M urea (36). With this urea concentration, the infectivity was reduced only ∼30% and released proteins P1 and P2 were mostly aggregative, while the rest of the phage components were found in two light-scattering zones and in the pellet (data not shown).
Protease-treated and isolated lipid cores contain intact protein P4.
The susceptibility of membrane-associated proteins P3 to P9 to proteases was tested by treating lipid cores (obtained with 4.5 M urea) with trypsin (50 μg/ml) for 5, 10, 20, or 60 min and analyzing the resultant material by rate zonal centrifugation under conditions where the lipid cores were aggregative (no urea). After 60 min of trypsin treatment, a visible light-scattering zone was detected. The only observed nondegraded protein had the N-terminal amino acid sequence MQKPSGKGLK, identifying it as protein P4 (Fig. 5D). With negative staining, trypsin-treated membranes containing P4 appeared as pleomorphic structures with low contrast and were devoid of DNA (data not shown).
Reconstitution of bacteriophage PM2.
Dissociation of the PM2 virion to separate subunits was previously reported (36, 47). The subviral particles were used in reconstitution experiments as described by Schäfer and Franklin (43, 44). We repeated these experiments with the PM2 subunits defined in this investigation.
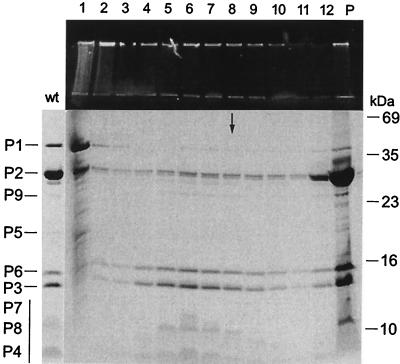
Soluble PM2 proteins P1 and P2 and viral lipid cores containing DNA and proteins P3 to P9 could be obtained by dissociation of virions with 4.5 M urea (Fig. 5B). Dissociation and the following reconstitution were carried out in the presence of 2-mercaptoethanol as described by Schäfer and Franklin (43, 44). The addition of 2-mercaptoethanol to a final concentration of 20 mM, either to virions in PM2 buffer or to 4.5 M urea-treated samples, did not have a measurable effect on the infectivity or the outcome of the disruption experiments. PM2 particles were reconstituted from the virus material dissociated with 4.5 M urea (0.7 mg of protein/ml; PM2 buffer and 20 mM 2-mercaptoethanol) followed by dialysis against a reduced urea concentration (PM2 buffer and 20 mM 2-mercaptoethanol). During dialysis, the reconstitution reactions turned turbid; the reaction undergoing dialysis against the buffer without urea was the most turbid. With 4.5 M urea, the control material stayed soluble. The dialyzed material was analyzed by sedimentation, and the protein and DNA contents of the collected fractions were assayed. The infectivities were determined after each step.
Figure 6 shows PM2 reconstitution with dialysis against 0.5 M urea. Three distinct light-scattering zones were detected and collected (fractions 4, 5, and 6 in Fig. 6). Below these fractions, there was a diffuse light-scattering zone (corresponding to fractions 7 to 10 in Fig. 6). Viral DNA cosedimented with the formed particles (fractions 4 to 10; see the upper gel in Fig. 6). However, reduced stoichiometry of proteins P1 and P2 in these noninfectious particles was observed compared to the results for intact virions. Approximately 90% of the input material showed assembly behavior. Separately treated P1 and P2 and the majority of the lipid cores remained soluble under the corresponding conditions. Increasing the urea concentration in the reconstitution reaction resulted in a lower yield of particles, and with a 2 M urea concentration, no particle assembly was detected. The outcome of the sedimentation analysis in the presence of 2 M urea resembled the results obtained with 4.5 M urea (Fig. 5B). After the complete removal of urea, the virus material was quantitatively found aggregated.
FIG. 6.
Reconstitution of bacteriophage PM2. PM2 virions were dissociated with 4.5 M urea-20 mM 2-mercaptoethanol in PM2 buffer. The separation of dissociated phage components by rate zonal centrifugation is shown in Fig. 5B. Reconstitution was carried out by dialyzing the dissociation mixture against a reduced urea concentration with 20 mM 2-mercaptoethanol in PM2 buffer overnight at 4°C and was followed by rate zonal centrifugation (see Materials and Methods). Viral DNA and protein compositions of collected gradient fractions and the pellet (P) were analyzed by SDS-PAGE with an ethidium bromide-stained upper gel and a Coomassie brilliant blue-stained separation gel. The figure shows the outcome of the experiment after dialysis against 0.5 M urea (for results obtained with other concentrations, see the text). The sedimentation position of PM2 virions is indicated by an arrow. The protein pattern of intact virions (wt) is indicated on the left (the vertical bar denotes the small membrane proteins forming diffuse bands), and the molecular masses of the standard proteins are indicated on the right.
The assembly behaviors of separated proteins P1 and P2 obtained by freezing and thawing (Fig. 2B, lane 6a) and lipid cores isolated with urea (Fig. 5B, lanes 3 and 7) were also tested. The reconstitution of particles was initiated by mixing the components, which had been dialyzed separately against 1 M urea (20 mM Tris-HCl [pH 7.2], 1 M urea, 0.5 M NaCl, 10 mM CaCl2) (43) at a final protein concentration of 0.5 mg/ml, with stoichiometric amounts of the proteins. A concentration higher than 1 M urea could not be used due to the precipitation of P1 and P2. Intact virions incubated under the corresponding conditions were not completely dissociated, and the infectivity was reduced only ∼30%. Sedimentation analysis of the reconstitution reactions revealed that already with the initial 1 M urea concentration, combinations of P1 and P2 with the lipid cores formed noninfectious aggregates. Reconstitution reactions with the same components were also tested in the presence of either bovine serum albumin (43) or 6% polyethylene glycol 4000, but no soluble particles were detected. Nor was there any increase in the infectivity in any of these experiments.
DISCUSSION
It was previously shown that the outer protein layer of the icosahedral PM2 virion is composed of major coat protein P2 and cell attachment protein P1 located at the fivefold vertices (9, 34, 36, 46). The protease treatment experiments indicated that the N-terminal portion of spike protein P1 is associated with the capsid vertex. Soluble protein P1 could be quantitatively removed from the virion by lowering the ionic strength. A monomeric status for isolated P1 was determined based on the sedimentation behavior, the gel filtration analysis, and the cross-linking experiments.
Both freezing and thawing or removal of calcium ions quantitatively released both proteins P1 and P2 from the virion. Based on the sedimentation, cross-linking, and light-scattering analyses, we propose that protein P2 is a trimer, although it eluted from the size exclusion chromatography as a particle with a mass closer to that of a dimer. This conclusion was recently confirmed by cryoelectron microscopy-based three-dimensional image reconstruction of the virion (J. Huiskonen, H. M. Kivelä, D. H. Bamford, and S. Butcher, unpublished data). We assume that the gel filtration behavior is due to the compact shape of the protein complex. In negatively stained micrographs, P2 multimers appeared as donut-shaped structures approximately 7 nm in diameter. As deduced from the amino acid sequence, the molecular mass of the P2 trimer is 90.6 kDa, a value close to the mass obtained by light scattering (92 ± 5 kDa). It was previously proposed that protein P2 binds calcium (45). The dissociation experiments support the idea that the P2 lattice is stabilized by calcium ions most likely located at the trimer interfaces.
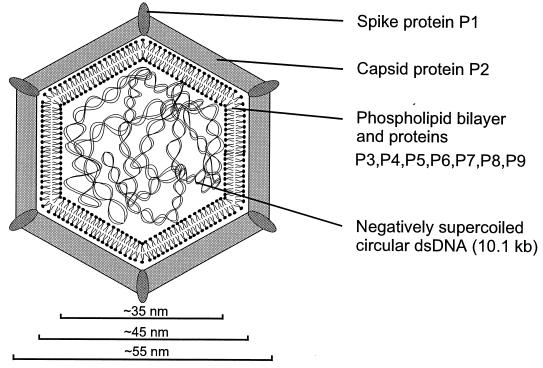
The previous structural model proposed that the lipid bilayer is sandwiched between the outer capsid and an inner protein shell composed of one protein species (29, 30, 31). However, the observation that the number of structural proteins is much higher than previously expected (38) and the results obtained here considerably change the idea of how the inner part of the virion is organized. It appears that the viral membrane vesicle, the lipid core, with associated proteins P3 to P9 (where proteins P3 to P8 contain a putative transmembrane region), encloses the genome. When the outer protein shell is removed, the remaining lipid core is very aggregative and soluble only in the presence of urea. Protease treatment of lipid cores digested all the membrane-associated proteins, except for P4, and solubilized the remaining vesicle. The digested proteins, however, were protected against proteolysis in the intact virion. Basic protein P4 may interact with viral DNA, being in close association with the membrane inside the vesicle (2, 12, 36, 40, 41). This view is supported by the in vitro interaction of protein P4 with DNA (42). DNA release leads to the formation of pleomorphic lipid vesicles, indicating that the presence of the genome is a prerequisite for the regular spherical shape of the lipid core (Fig. 1F and G). The PM2 lipid core is reminiscent of the internal membrane vesicle of another lipid-containing bacteriophage, PRD1, in which a number of small viral structural proteins are associated with the bilayer (3). In PRD1, the genome (a linear dsDNA molecule) is also enclosed in a membrane vesicle and is very aggregative when the outer protein shell is removed (5). Our results are in agreement with results obtained by Satake et al. (41) and Akutsu et al. (1), who proposed that the lipids are arranged in the form of a bilayer in the lipid core as well as in the virion. A schematic model of the structural organization of PM2 is depicted in Fig. 7.
FIG. 7.
Schematic representation of PM2 virion organization.
It was previously proposed that infectious viral particles can be reconstituted from viral subunits (43, 44, 54). We repeated systematically these experiments and extended the analysis by using more defined purified constituents. We were not able to measure any increase in infectivity in any of the reconstitution experiments. However, it was possible to reconstitute DNA-containing particles where substoichiometric amounts of spike protein P1 and major coat protein P2 were assembled on the lipid cores. The sedimentation of these particles was slower than that of intact virions.
Acknowledgments
We thank Roman Tuma for help in the light-scattering experiments. We thank Marja-Leena Perälä, Sari Korhonen, and Riitta Tarkiainen for skilled technical assistance. We thank Tuire Koro for thin sectioning.
This study was supported by Academy of Finland grants 168694, 172621, and 164298 (to D.H.B; Finnish Center of Excellence Program, 2000 to 2005). H.M.K. is a fellow of the Helsinki Graduate School of Biotechnology and Molecular Biology.
REFERENCES
- 1.Akutsu, H., H. Satake, and R. M. Franklin. 1980. Phosphorus nuclear magnetic resonance studies on the lipid-containing bacteriophage PM2. Biochemistry 19:5264-5270. [DOI] [PubMed] [Google Scholar]
- 2.Armour, G. A., and G. J. Brewer. 1990. Membrane morphogenesis from cloned fragments of bacteriophage PM2 DNA that contain the sp6.6 gene. FASEB J. 4:1488-1493. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., J. Caldentey, and J. K. H. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology 107:222-228. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. 27:911-917. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein, S. N., and R. M. Franklin. 1971. Structure and synthesis of a lipid-containing bacteriophage. V. Phospholipids of the host BAL-31 and of the bacteriophage PM2. Virology 43:685-695. [DOI] [PubMed] [Google Scholar]
- 9.Brewer, G. J., and S. J. Singer. 1974. On the disposition of the proteins of the membrane-containing bacteriophage PM2. Biochemistry 13:3580-3588. [DOI] [PubMed] [Google Scholar]
- 10.Brewer, G. J. 1976. Control of membrane morphogenesis in bacteriophage PM2. J. Supramol. Struct. 5:73-79. [DOI] [PubMed] [Google Scholar]
- 11.Brewer, G. J. 1978. Membrane-localized replication of bacteriophage PM2. Virology 84:242-245. [DOI] [PubMed] [Google Scholar]
- 12.Brewer, G. J. 1979. In vivo assembly of a biological membrane of defined size, shape, and lipid composition. J. Virol. 30:875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butcher, S. J., D. H. Bamford, and S. D. Fuller. 1995. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14:6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldentey, J., R. Tuma, and D. H. Bamford. 2000. Assembly of bacteriophage PRD1 spike complex: role of the multidomain protein P5. Biochemistry 39:10566-10573. [DOI] [PubMed] [Google Scholar]
- 15.Camerini-Otero, R. D., and R. M. Franklin. 1972. Structure and synthesis of a lipid-containing bacteriophage. XII. The fatty acids and lipid content of bacteriophage PM2. Virology 49:385-393. [DOI] [PubMed] [Google Scholar]
- 16.Camerini-Otero, R. D., and R. M. Franklin. 1975. Structure and synthesis of a lipid-containing bacteriophage. The molecular weight and other physical properties of bacteriophage PM2. Eur. J. Biochem. 53:343-348. [DOI] [PubMed] [Google Scholar]
- 17.Camerini-Otero, R. D., P. N. Pusey, D. E. Koppel, D. W. Schäfer, and R. M. Franklin. 1974. Intensity fluctuation spectroscopy of laser light scattered by solutions of spherical viruses: R17, Q beta, BSV, PM2, and T7. II. Diffusion coefficients, molecular weights, solvation, and particle dimensions. Biochemistry 13:960-970. [DOI] [PubMed] [Google Scholar]
- 18.Canelo, E., O. M. Phillips, and R. N. del Roure. 1985. Relating cistrons and functions in bacteriophage PM2. Virology 140:364-367. [DOI] [PubMed] [Google Scholar]
- 19.Casjens, S. 1997. Principles of virion structure, function, and assembly, p. 3-37. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 20.Cota-Robles, E., R. T. Espejo, and P. W. Haywood. 1968. Ultrastructure of bacterial cells infected with bacteriophage PM2, a lipid-containing bacterial virus. J. Virol. 2:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahlberg, J. E., and R. M. Franklin. 1970. Structure and synthesis of a lipid-containing bacteriophage. IV. Electron microscopic studies of PM2-infected Pseudomonas BAL-31. Virology 42:1073-1086. [DOI] [PubMed] [Google Scholar]
- 22.Datta, A., R. D. Camerini-Otero, S. N. Braunstein, and R. M. Franklin. 1971. Structure and synthesis of a lipid-containing bacteriophage. VII. Structural proteins of bacteriophage PM2. Virology 45:232-239. [DOI] [PubMed] [Google Scholar]
- 23.Davis, T. N., E. D. Muller, and J. E. Cronan, Jr. 1982. The virion of the lipid-containing bacteriophage PR4. Virology 120:287-306. [DOI] [PubMed] [Google Scholar]
- 24.Espejo, R. T., and E. S. Canelo. 1968. Properties of bacteriophage PM2: a lipid-containing bacterial virus. Virology 34:738-747. [DOI] [PubMed] [Google Scholar]
- 25.Espejo, R. T., and E. S. Canelo. 1968. Properties and characterization of the host bacterium of bacteriophage PM2. J. Bacteriol. 95:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espejo, R. T., and E. S. Canelo. 1968. Origin of phospholipid in bacteriophage PM2. J. Virol. 2:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1969. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc. Natl. Acad. Sci. USA 63:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espejo, R. T., E. S. Canelo, and R. L. Sinsheimer. 1971. Replication of bacteriophage PM2 deoxyribonucleic acid: a closed circular double-stranded molecule. J. Mol. Biol. 56:597-621. [DOI] [PubMed] [Google Scholar]
- 29.Franklin, R. M. 1974. Structure and synthesis of bacteriophage PM2, with particular emphasis on the viral lipid bilayer. Curr. Top. Microbiol. Immunol. 1974:107-159. [DOI] [PubMed] [Google Scholar]
- 30.Franklin, R. M. 1977. In vitro and in vivo assembly of bacteriophage PM2: a model for protein-lipid interactions. Cell Surf. Rev. 1977:803-827. [Google Scholar]
- 31.Franklin, R. M., R. Hinnen, R. Schäfer, and N. Tsukagoshi. 1976. Structure and assembly of lipid-containing viruses, with special reference to bacteriophage PM2 as one type of model system. Philos. Trans. R. Soc. Lond. B 276:63-80. [DOI] [PubMed] [Google Scholar]
- 32.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 33.Gray, H. B. J., W. B. Upholt, and J. Vinograd. 1971. A buoyant method for the determination of superhelix density of closed circular DNA. J. Mol. Biol. 62:1-19. [DOI] [PubMed] [Google Scholar]
- 34.Harrison, S. C., D. L. Caspar, R. D. Camerini-Otero, and R. M. Franklin. 1971. Lipid and protein arrangement in bacteriophage PM2. Nat. New Biol. 229:197-201. [DOI] [PubMed] [Google Scholar]
- 35.Helin, J., J. Caldentey, N. Kalkkinen, and D. H. Bamford. 1999. Analysis of the multimeric state of proteins by matrix assisted laser desorption/ionization mass spectrometry after cross-linking with glutaraldehyde. Rapid Commun. Mass Spectrom. 13:185-190. [DOI] [PubMed] [Google Scholar]
- 36.Hinnen, R., R. Schäfer, and R. M. Franklin. 1974. Structure and synthesis of lipid-containing bacteriophage. Preparation of virus and localization of the structural proteins. Eur. J. Biochem. 50:1-14. [DOI] [PubMed] [Google Scholar]
- 37.Hinnen, R., R. Chassin, R. Schäfer, R. M. Franklin, H. Hitz, and D. Schäfer. 1976. Structure and synthesis of a lipid-containing bacteriophage. Purification, chemical composition, and partial sequences of the structural proteins. Eur. J. Biochem. 68:139-152. [DOI] [PubMed] [Google Scholar]
- 38.Kivelä, H. M., R. H. Männistö, N. Kalkkinen, and D. H. Bamford. 1999. Purification and protein composition of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:364-374. [DOI] [PubMed] [Google Scholar]
- 39.Männistö, R. H., H. M. Kivelä, L. Paulin, D. H. Bamford, and J. K. H. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 40.Marcoli, R., V. Pirrotta, and R. M. Franklin. 1979. Interaction between bacteriophage PM2 protein IV and DNA. J. Mol. Biol. 131:107-131. [DOI] [PubMed] [Google Scholar]
- 41.Satake, H., H. Akutsu, M. Kania, and R. M. Franklin. 1980. Structure and synthesis of a lipid-containing bacteriophage. Studies on the structure of the bacteriophage PM2 nucleocapsid. Eur. J. Biochem. 108:193-201. [DOI] [PubMed] [Google Scholar]
- 42.Satake, H., M. Kania, and R. M. Franklin. 1981. Structure and synthesis of a lipid-containing bacteriophage. Amphiphilic properties of protein IV of bacteriophage PM2. Eur. J. Biochem. 114:623-628. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer, R., and R. M. Franklin. 1975. Structure and synthesis of a lipid-containing bacteriophage. XIX. Reconstitution of bacteriophage PM2 in vitro. J. Mol. Biol. 97:21-34. [DOI] [PubMed] [Google Scholar]
- 44.Schäfer, R., and R. M. Franklin. 1978. Structure and synthesis of a lipid-containing bacteriophage. Total reconstitution of bacteriophage PM2 in vitro. Eur. J. Biochem. 92:589-596. [DOI] [PubMed] [Google Scholar]
- 45.Schäfer, R., R. Hinnen, and R. M. Franklin. 1974. Structure and synthesis of a lipid-containing bacteriophage. Properties of the structural proteins and distribution of the phospholipid. Eur. J. Biochem. 50:15-27. [DOI] [PubMed] [Google Scholar]
- 46.Schäfer, R., R. Hinnen, and R. M. Franklin. 1974. Further observations on the structure of the lipid-containing bacteriophage PM2. Nature 248:681-682. [DOI] [PubMed] [Google Scholar]
- 47.Schäfer, R., P. Kunzler, A. Lustig, and R. M. Franklin. 1978. Structure and synthesis of a lipid-containing bacteriophage. Dissociation of bacteriophage PM2 into its morphological subunits. Eur. J. Biochem. 92:579-588. [DOI] [PubMed] [Google Scholar]
- 48.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 49.Silbert, J. A., M. Salditt, and R. M. Franklin. 1969. Structure and synthesis of a lipid-containing bacteriophage. III. Purification of bacteriophage PM2 and some structural studies on the virion. Virology 39:666-681. [DOI] [PubMed] [Google Scholar]
- 50.Snipes, W., J. Cupp, J. A. Sands, A. Keith, and A. Davis. 1974. Calcium requirement for assembly of the lipid-containing bacteriophage PM2. Biochim. Biophys. Acta 339:311-322. [DOI] [PubMed] [Google Scholar]
- 51.Tsukagoshi, N., M. H. Petersen, and R. M. Franklin. 1975. Structure and synthesis of a lipid-containing bacteriophage. XVIII. Modification of the lipid composition in bacteriophage PM2. Virology 66:206-216. [DOI] [PubMed] [Google Scholar]
- 52.Tsukagoshi, N., M. H. Petersen, and R. M. Franklin. 1975. Effect of unsaturated fatty acids on the lipid composition of bacteriophage PM2. Nature 253:12512-12516. [DOI] [PubMed] [Google Scholar]
- 53.Tsukagoshi, N., M. N. Kania, and R. M. Franklin. 1976. Identification of acyl phosphatidylglycerol as a minor phospholipid of Pseudomonas BAL-31. Biochim. Biophys. Acta 450:131-136. [DOI] [PubMed] [Google Scholar]
- 54.Tsukagoshi, N., R. Schäfer, and R. M. Franklin. 1977. Structure and synthesis of a lipid-containing bacteriophage. Effects of lipids containing cis or trans fatty acids on the reconstitution of bacteriophage PM2. Eur. J. Biochem. 73:469-476. [DOI] [PubMed] [Google Scholar]