Abstract
Iron-sulfur (Fe/S) cluster-containing proteins catalyze a number of electron transfer and metabolic reactions. The components and molecular mechanisms involved in the assembly of the Fe/S clusters have been identified only partially. In eukaryotes, mitochondria have been proposed to execute a crucial task in the generation of intramitochondrial and extramitochondrial Fe/S proteins. Herein, we identify the essential ferredoxin Yah1p of Saccharomyces cerevisiae mitochondria as a central component of the Fe/S protein biosynthesis machinery. Depletion of Yah1p by regulated gene expression resulted in a 30-fold accumulation of iron within mitochondria, similar to what has been reported for other components involved in Fe/S protein biogenesis. Yah1p was shown to be required for the assembly of Fe/S proteins both inside mitochondria and in the cytosol. Apparently, at least one of the steps of Fe/S cluster biogenesis within mitochondria requires reduction by ferredoxin. Our findings lend support to the idea of a primary function of mitochondria in the biosynthesis of Fe/S proteins outside the organelle. To our knowledge, Yah1p is the first member of the ferredoxin family for which a function in Fe/S cluster formation has been established. A similar role may be predicted for the bacterial homologs that are encoded within iron-sulfur cluster assembly (isc) operons of prokaryotes.
Keywords: ATP-binding cassette transporter, iron homeostasis
Iron-sulfur (Fe/S) proteins perform crucial roles in various electron transfer processes and in several enzymatic reactions (1–3). In eukaryotic cells, known Fe/S proteins are localized in mitochondria, the cytosol, and the nucleus (4). Only recently, the components and mechanisms of the biosynthesis of Fe/S proteins have been approached by employing genetic and biochemical methods. In a pioneering report, Culotta's group (5) demonstrated the involvement of the essential mitochondrial protein Nfs1p and the two mitochondrial heat shock proteins Jac1p and Ssq1p in the generation of Fe/S proteins of mitochondria of Saccharomyces cerevisiae. Nfs1p represents the functional ortholog of the bacterial cysteine desulfurase NifS/IscS, which initiates Fe/S cluster formation by producing elemental sulfur from cysteine (6, 7). Recently, we were able to show that mitochondrial Nfs1p, in addition to its role in generating mitochondrial Fe/S clusters, is necessary for the biosynthesis of extramitochondrial Fe/S proteins (7). Export of components required for assembly of cytosolic Fe/S proteins is mediated by the ATP-binding cassette transporter Atm1p of the mitochondrial inner membrane (7, 8). Different from the function of Nfs1p, this protein performs a specific role in the assembly of cytosolic Fe/S proteins, because no effects were observed on the generation of mitochondrial Fe/S proteins on depletion of Atm1p.
Mitochondria were proposed to contain a complex machinery catalyzing the formation of Fe/S clusters (7). Some of the components of this machinery have been inferred from bacterial homologs encoded by the nif (nitrogen fixation) and isc (iron-sulfur cluster assembly) operons, which contain genes required for assembly of the multinuclear Fe/S protein nitrogenase and for the biogenesis of “house-keeping” bacterial Fe/S proteins, respectively (9, 10). One of these proteins is a member of the [2Fe-2S] ferredoxin family. The yeast homolog termed Yah1p is an essential protein and has been shown to reside in mitochondria (11), but its function remained unclear.
Ferredoxins comprise a large family of low-molecular-mass proteins that are involved in various cellular redox processes (reviewed in ref. 12). The ferredoxin proteins contain an iron-sulfur cluster as their redox-active site. Regeneration of the reduced form of these proteins is accomplished by specific ferredoxin reductases, which gain the electrons by oxidation of NAD(P)H. Ferredoxins can be subdivided into several distinct classes according to sequence similarities and the presence of specific types of Fe/S clusters (12, 13). The [2Fe-2S] ferredoxins are characterized by a molecular mass of about 10–12 kDa, and they possess a high reduction potential of −400 mV. Multiple isoforms of [2Fe-2S] ferredoxins are present in cyanobacteria and chloroplasts. Other members of this family are involved in a number of biosynthetic reactions requiring redox steps such as the reduction of nitrite or sulfite, the synthesis of glutamate or unsaturated fatty acids, or the oxidoreduction of thioredoxin that is needed for formation and cleavage of disulfide bonds. Mitochondria of vertebrates contain a [2Fe-2S] ferredoxin homolog termed adrenodoxin (14). The protein transfers electrons to a mitochondrial cytochrome P450 that synthesizes pregnenolone, the common precursor of steroid hormones.
We noted a close sequence homology of the yeast mitochondrial ferredoxin Yah1p with bacterial ferredoxins that are encoded by the isc operons. This observation prompted us to investigate the potential involvement of Yah1p in the biosynthesis of cellular Fe/S proteins. By employing a direct method to follow the incorporation of an Fe/S cluster into both mitochondrial and cytosolic apoproteins, we were able to demonstrate an essential function of Yah1p in the generation of these Fe/S proteins. Depletion of the concentration of ferredoxin led to a drastic accumulation of iron within mitochondria similar to that reported for other proteins involved in cellular Fe/S protein synthesis. Together, our data identify the ferredoxin Yah1p as a central component of the mitochondrial Fe/S protein biosynthesis apparatus.
Materials and Methods
Yeast Strains and Cell Growth.
The following strains of S. cerevisiae were used: W303 (MATα, ura3-1, ade2-1, trp1-1, his3-11,15, leu2-3,112), which served as wild-type, and strains carrying mutations in the genes COR1 (15) and COX6 (16). Exchange of the promoter of the YAH1 gene for a galactose-inducible promoter (strain Gal-YAH1) was performed as described (7, 17). PCR fragments corresponding to the coding region (nucleotides −27 to 649) and the 5′ upstream region (nucleotides −412 to −8) of YAH1 were cloned into the BamHI/HindIII and HindIII/HpaI restriction sites, respectively, of the Yep51 vector carrying the GAL10 promoter. Deletion of the LEU1 gene in W303 and Gal-YAH1 cells was performed by a PCR-based method with the kanamycin resistance marker (18). The LEU1-deficient cells were transformed with the yeast expression vector pRS426-GPD (19) containing the coding sequence of a fusion protein comprised of the presequence of Neurospora crassa subunit 9 of Fo-ATPase and the entire yeast Leu1p. The resulting cells (termed ΔLWT-ML and ΔLGY-ML, respectively) expressed Leu1p that was exclusively localized to mitochondria. Cells were grown as detailed by using rich (yeast extract/peptone) or minimal medium (20–22) and lactate medium (23) containing the required carbon sources.
Miscellaneous Methods.
The following published methods were used: manipulation of DNA and PCR (24); transformation of yeast cells (25); isolation of plasmids from yeast (24); isolation of yeast mitochondria (23); import of radiolabeled precursor proteins into isolated mitochondria (26); preparation of whole-cell lysates by breaking cells with glass beads (27); and determining enzyme activities of citrate synthase, malate dehydrogenase, aconitase (22), succinate dehydrogenase (28), and isopropyl malate isomerase (29). The standard error of the determination of enzyme activities varied between 5% and 15%. The labeling of yeast cells with radioactive iron (55Fe) and the measurement of the incorporation of 55Fe into the cytosolic protein Leu1p by immunoprecipitation and liquid scintillation counting were described earlier (7). The amount of radioactive iron associated with the mitochondrial version of Leu1p was measured accordingly. The standard error for detection of 55Fe associated with cytosolic or mitochondrial Leu1p in a cell lysate was 15%; the cellular uptake of 55Fe varied by 30%.
Results
The S. cerevisiae gene YAH1 encodes a protein of 172 amino acid residues. The N-terminal 60 residues of Yah1p resemble a mitochondrial targeting signal (presequence). This part mediated the membrane potential-dependent import of the Yah1p precursor into the mitochondrial matrix and was cleaved off after import (not shown). The C-terminal 110 residues share considerable homology to adrenodoxin of mammalian mitochondria (11) and to the [2Fe-2S] cluster-containing class of bacterial ferredoxins (Fig. 1). The function of these bacterial proteins has not yet been elucidated. Some of these proteins are encoded in the isc and related operons in which genes with a (potential) function in Fe/S protein biosynthesis are clustered (10). This finding rendered it likely that the mitochondrial ferredoxin Yah1p performs a role in the generation of Fe/S proteins.
Figure 1.
The mitochondrial ferredoxin Yah1p of S. cerevisiae shares homology to bacterial [2Fe-2S] ferredoxins. The sequence alignment was prepared by using the multalin program (42). The four conserved cysteine residues (bold) are marked with arrows. Sc, S. cerevisiae; Hs, Homo sapiens; Rp, Rickettsia prowazekii; Rc, Rhodobacter capsulatus; Ec, Escherichia coli; Av, Azotobacter vinelandii; Hi, Haemophilus influenza; Adx, adrenodoxin; Fdx, ferredoxin.
Construction of a Yeast Strain Allowing Depletion of Yah1p by Regulated Gene Expression.
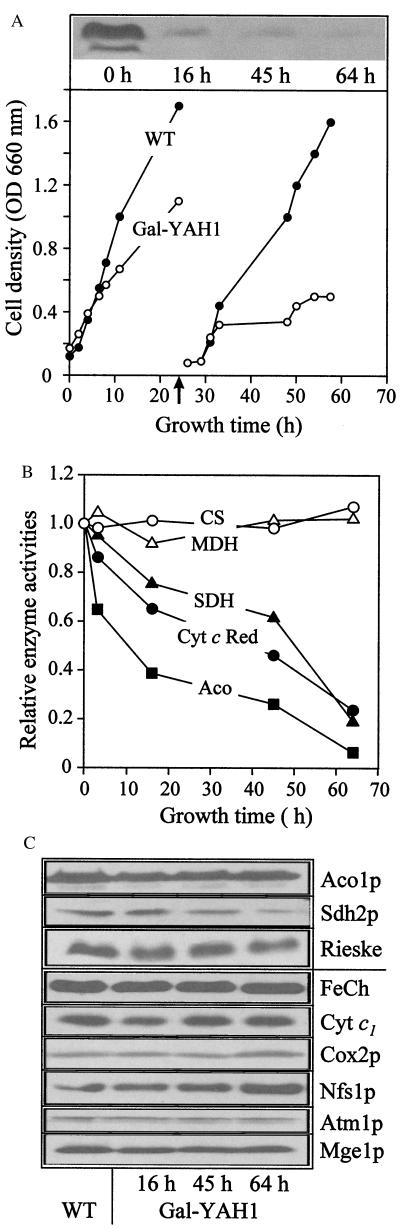
To facilitate the investigation of the function of Yah1p, a mutant yeast strain was constructed in which the expression of the essential YAH1 gene was under the control of a galactose-inducible promoter. The resulting strain, termed Gal-YAH1, grew at wild-type rates in the presence of galactose (Table 1). Without addition of this sugar, cells did not grow on medium containing the nonfermentable carbon-source glycerol. On fermentable carbon sources (glucose), cells were able to maintain viability. On cultivation on rich medium, they gave rise to small colonies comparable to what is seen for yeast pet mutant strains (30). On minimal medium, growth was retarded severely (Table 1 and Fig. 2A). The residual growth of the Gal-YAH1 cells in the presence of glucose is explained by the leaky character of the GAL10 promoter (see, e.g., ref. 7). Nevertheless, the more than 10-fold depletion of the mitochondrial concentration of Yah1p achieved in the Gal-YAH1 strain (Fig. 2A) allowed the investigation of the function of this protein.
Table 1.
Growth phenotype of Gal-YAH1 cells on various media
| Growth medium | Gal-YAH1 |
|---|---|
| MMG | − |
| MMG + 0.1% Gal | +++ |
| YPG | − |
| YPG + 0.1% Gal | +++ |
| MMD | + |
| YPD | ++ |
The growth of Gal-YAH1 cells was estimated on various media relative to the growth of wild-type cells. +++, same growth as wild-type cells; ++, retarded growth; +, strongly reduced growth; −, cells stop growing after 3 days. MMG, minimal medium containing 2% (vol/vol) glycerol; YPG, rich medium containing 2% (vol/vol) glycerol; MMD, minimal medium containing 2% (wt/vol) dextrose; YPG, rich medium containing 2% (wt/vol) dextrose; Gal, galactose.
Figure 2.
The ferredoxin Yah1p is required for normal activity of mitochondrial Fe/S proteins. (A) Depletion of Yah1p. Gal-YAH1 cells precultivated in galactose-containing medium or wild-type cells (WT) were grown for the indicated time periods in lactate medium containing 0.1% glucose at 30°C. After 24 h (arrow), cells were diluted 20-fold into fresh medium. Cell density was estimated from the OD at 600 nm. Mitochondria were isolated from Gal-YAH1 cells at the times indicated, and depletion of Yah1p was visualized by immunostaining (Upper). (B) The mitochondria described in A were used to measure the enzyme activities of several Fe/S cluster-containing proteins and of malate dehydrogenase (MDH) and citrate synthase (CS). The results are given relative to the activities measured for the mitochondrial enzymes of either wild-type or Gal-YAH1 cells grown in the presence of galactose. SDH, succinate dehydrogenase; Cyt c Red, cytochrome c reductase; Aco, aconitase. (C) Immunostaining of the indicated proteins with the mitochondria described in A. Aco1p, aconitase; Sdh2p, subunit 2 of complex II; Rieske, Rieske Fe/S protein; FeCh, ferrochelatase; cyt c1, cytochrome c1; Cox2p, subunit 2 of complex IV.
Yah1p Is Required for Fe/S Cluster Incorporation into Mitochondrial Apoproteins.
The Gal-YAH1 cells were employed to test the requirement of Yah1p for the activity of mitochondrial Fe/S proteins. Gal-YAH1 cells were cultivated in the absence of galactose to deplete the concentration of Yah1p selectively. After various times, cells were harvested; mitochondria were isolated; and the activities of several enzymes were measured. A strong decay in the activities of the soluble Fe/S protein aconitase and the Fe/S cluster-containing complexes II (succinate dehydrogenase) and III (cytochrome c reductase) was observed on depletion of Yah1p (Fig. 2B). After 2.5 days, the activities were reduced 5- to 10-fold compared with wild-type cells. No changes were detectable in the activities of control proteins that do not contain Fe/S clusters (malate dehydrogenase and citrate synthase). The loss of activity of the Fe/S cluster-containing proteins did not result from impaired synthesis or import of these proteins into mitochondria. Hardly any changes in the levels of aconitase and the Rieske Fe/S protein of complex III and only a 3-fold decrease in subunit 2 of complex II (Sdh2p) were detectable by immunostaining analysis (Fig. 2C). No alterations were seen for non-Fe/S proteins except for the amount of Nfs1p, which was increased 3-fold on depletion of Yah1p. Taken together, these data indicate a requirement of the ferredoxin Yah1p for the activity of mitochondrial Fe/S proteins and suggest a function in the generation of these proteins.
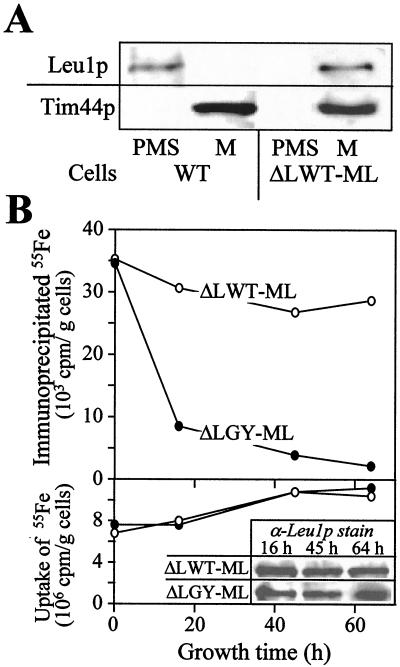
To measure the incorporation of an Fe/S cluster into a mitochondrial apoprotein directly, we attempted to follow the assembly of iron with the protein by immunoprecipitation (see ref. 7). Because our antibodies directed against aconitase did not immunoprecipitate this protein, the cytosolic Fe/S protein Leu1p was directed to mitochondria. A DNA segment encoding a mitochondrial presequence was introduced at the 5′ end of the coding sequence of LEU1. The DNA was inserted into a yeast expression vector and transformed into wild-type or Gal-YAH1 yeast cells. To ease subsequent analyses, the genomic copy of the LEU1 gene was deleted in these cells. The resulting strains (termed ΔLWT-ML and ΔLGY-ML, respectively) expressed Leu1p protein that was exclusively localized within mitochondria (Fig. 3A and data not shown).
Figure 3.
Depletion of Yah1p results in a defect in the assembly of an Fe/S cluster into a mitochondrial apoprotein. (A) Mitochondria (M) and postmitochondrial supernatants (PMS) of wild-type (WT) and ΔLWT-ML cells expressing a mitochondrial version of Leu1p were subjected to SDS/PAGE and immunostaining with antisera against Leu1p and matrix-exposed Tim44p. (B) ΔLWT-ML and ΔLGY-ML cells expressing Leu1p in mitochondria were grown in iron-free minimal medium containing glucose for the indicated times. Cells were radiolabeled with (55Fe) iron chloride in the presence of 1 mM ascorbate for 1 h; cell lysates were prepared; and the uptake of 55Fe was quantitated by liquid scintillation counting (Lower). Aliquots of the lysates were subjected to SDS/PAGE and immunostaining with antisera directed against Leu1p (α-Leu1p stain; Bottom Insert). Immunoprecipitation with anti-Leu1p antibodies was performed, and coprecipitated 55Fe was estimated by liquid scintillation counting (Upper; ref. 7). The background signal obtained for immunoprecipitation performed with preimmune serum (usually less than 2 × 103 cpm per g of cells) was subtracted.
The incorporation of an Fe/S cluster into mitochondrial Leu1p was followed in vivo. ΔLWT-ML and ΔLGY-ML cells were radiolabeled with (55Fe) iron chloride for 1 h, and total cell lysates were prepared. Mitochondrial Leu1p was immunoprecipitated under native conditions with specific antibodies, and the 55Fe radioactivity associated with Leu1p was estimated by liquid scintillation counting. The amount of 55Fe bound to Leu1p serves as a direct measure for the incorporation of the Fe/S cluster (cf. ref. 7). In cells containing Yah1p, significant amounts of 55Fe were immunoprecipitated with the Leu1p-specific antiserum (Fig. 3B Upper), whereas hardly any radioactive 55Fe was precipitated by employing antibodies derived from preimmune serum (not shown; ref. 7). In contrast, on depletion of Yah1p by growth of ΔLGY-ML cells in the absence of galactose, the amount of 55Fe bound to Leu1p became rapidly decreased by a factor of 5–10. The uptake of radioactive 55Fe iron into the cells was hardly affected during the experiment (Fig. 3B Lower). The slight increase in 55Fe content of the yeast cells is related to the depletion of residual levels of nonradioactive iron on cultivation in “iron-free” minimal medium (not shown). The amounts of mitochondrial Leu1p remained unchanged on depletion of Yah1p (Fig. 3B Insert), clearly showing that the decrease of Yah1p levels caused a defect in the incorporation of the Fe/S cluster into the Leu1p apoprotein. In conclusion, these data attribute a crucial function to the ferredoxin Yah1p in the biosynthesis of mitochondrial Fe/S proteins.
Yah1p Is Essential for the Assembly of Cytosolic Fe/S Proteins.
Mitochondria have been shown to perform an essential function also in the biogenesis of extramitochondrial Fe/S proteins (7). We therefore examined the potential involvement of Yah1p in the biosynthesis of cytosolic Fe/S proteins. Gal-YAH1 cells were grown in medium lacking galactose; after various times, cells were lysed, and the enzyme activity of cytosolic Leu1p was measured. A strong decrease in the enzyme activity of Leu1p was observed on depletion of Yah1p in comparison to wild-type cells and to Gal-YAH1 cells grown in the presence of galactose (Fig. 4A). Other cytosolic enzymes were not significantly altered in their activities (not shown; see ref. 7). Apparently, Yah1p is required either for the synthesis of this cytosolic Fe/S protein or for the maintenance of its function. The fact that the amount of Leu1p in the cytosol remained unchanged on diminishing Yah1p (Fig. 4B) suggests an involvement of this ferredoxin in the incorporation of the Fe/S cluster into the Leu1p apoprotein.
Figure 4.
Mitochondrial ferredoxin Yah1p is required for the activity of the cytosolic Fe/S protein Leu1p. (A) Wild-type (WT) cells were grown in minimal medium in the presence of galactose (Gal) or glucose (Glu). For depletion of Yah1p, the Gal-YAH1 cells were cultivated in minimal medium in the presence of glucose for the indicated times. A cell lysate was prepared by breaking the cells with glass beads. The enzyme activities of isopropyl malate isomerase (Leu1p) were measured immediately (29). (B) Aliquots of the cell extracts obtained as described in A were analyzed for the amount of Leu1p by immunostaining.
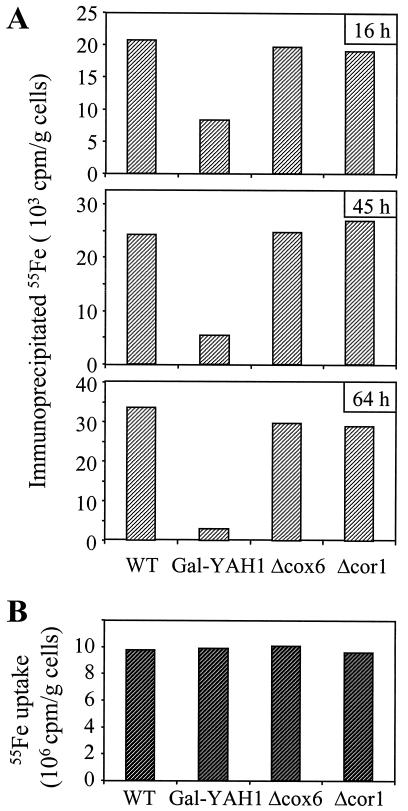
To investigate directly whether a defect in Yah1p affects the assembly of the Fe/S cluster in cytosolic Leu1p, Gal-YAH1 cells were depleted in Yah1p, labeled with radioactive (55Fe) iron chloride, and assayed for the specific association of 55Fe with Leu1p by immunoprecipitation (see above; ref. 7). The deficiency of Yah1p caused an up to 10-fold decrease in the amount of 55Fe associated with Leu1p (Fig. 5A). No such effect was noted for cells in which complexes III and IV were inactivated by deletion of the COR1 and COX6 genes, respectively. Iron uptake into the yeast cells was unaffected by depletion of Yah1p, excluding the possibility that a defective supply of iron caused the impaired iron incorporation into Leu1p (Fig. 5B). Together, these experiments illustrate a requirement of mitochondrial Yah1p for the incorporation of an Fe/S cluster into cytosolic Leu1p. In addition to mitochondrial Nfs1p and Atm1p, the ferredoxin Yah1p is the third component with a known function in cytosolic Fe/S cluster biogenesis.
Figure 5.
Depletion of mitochondrial Yah1p causes a defect in the incorporation of an Fe/S cluster into the cytosolic Fe/S protein Leu1p. (A) Wild-type (WT), Gal-YAH1, Δcox6, and Δcor1 cells were grown in iron-free minimal medium for the indicated times. Radiolabeling, cell lysis, and analysis of 55Fe incorporation into cytosolic Leu1p by immunoprecipitation was performed as described for Fig. 3B. (B) The uptake of radioactive iron into the various yeast cells grown for 64 h in minimal medium was quantitated by liquid scintillation counting from the amount of 55Fe in the extracts.
Depletion of Yah1p Results in Iron Accumulation Within Mitochondria.
Mutants in components of Fe/S cluster biosynthesis (e.g., of ATM1, SSQ1, and NFS1) were shown to accumulate high levels of iron within mitochondria (7, 22, 31). A similar increase of the mitochondrial iron concentration was noted on depletion of Yah1p (Fig. 6). A significant rise of the mitochondrial iron concentration was seen as early as 3 h after turning off the expression of the YAH1 gene. After 2.5 days of cultivation of the Gal-YAH1 cells in medium lacking galactose, up to 30-fold higher levels of iron compared with the levels in mitochondria of wild-type cells were found. Thus, the accumulation of mitochondrial iron in Yah1p-depleted cells seems to be a distinct phenotypic feature that is common for components involved in Fe/S cluster biosynthesis.
Figure 6.
Mitochondria depleted of Yah1p accumulate high levels of iron. Wild-type (WT) and Gal-YAH1 cells were grown as described for Fig. 2A. At the indicated times, mitochondria were isolated. The amount of nonheme non-Fe/S (free) iron associated with the mitochondria was measured by the bathophenantroline method (43).
Discussion
In our study, we have identified a function of a member of the ferredoxin protein family. The mitochondrial ferredoxin Yah1p is required for the generation of Fe/S proteins inside the organelle and plays a crucial role in the formation of Fe/S proteins in the cytosol. The assembly of the Fe/S clusters into mitochondrial and cytosolic apoproteins was followed by a recently developed immunoprecipitation procedure (7). The amount of iron associated with Leu1p as a model Fe/S protein served as a direct measure of the conversion of this protein to its holoform. This experimental approach excludes that the observed effects of the Yah1p depletion on the function of Fe/S proteins may result from a decreased stability of the Fe/S clusters or from their oxidative damage. Rather, these data suggest a predominant function of Yah1p in the de novo assembly of Fe/S proteins. Nevertheless, the findings do not rule out an auxiliary function of ferredoxin in the repair of damaged Fe/S proteins.
At least one step in the biosynthesis of Fe/S clusters within mitochondria requires the input of reducing equivalents by Yah1p. Several possibilities for an involvement of Yah1p can be envisioned. First, the generation of sulfide by the cysteine desulfurase Nfs1p may necessitate reduction. In the course of the in vitro analysis of the function of bacterial NifS, reduction during the generation of sulfide was facilitated by either DTT or cysteine (32). Second, iron may require continuous reduction after its import into the mitochondrial matrix to avoid precipitation of insoluble ferric ions. Third, formation of intermediates of the Fe/S cluster assembly may depend on the supply of electrons. Excellent candidates for such a function are the bacterial NifU/IscU proteins and their eukaryotic homologs Isu1p/Isu2p and Nfu1p (33, 34). For NifU, a function in iron binding has been demonstrated recently (44). The enzymatic action of NifS leads to the production of an NifU-bound “intermediate” [2Fe-2S] cluster that can be released in vitro on reduction by dithionite. In vivo, such a reducing function may be contributed by ferredoxin. Clearly, a more detailed picture of the molecular events leading to Fe/S cluster assembly is needed to clarify the reaction(s) requiring electron supply by ferredoxin.
Yah1p has considerable homology to adrenodoxin of mammalian mitochondria (11) and to the bacterial [2Fe-2S] ferredoxins (see Fig. 1). It seems likely from our data that some of the bacterial homologs of mitochondrial ferredoxin also perform a function in Fe/S cluster biosynthesis, explaining why their genes are encoded within the isc operon (10). The well known function of adrenodoxin in the biosynthesis of steroid hormones raises the interesting question whether in mammalian mitochondria this protein is also involved in the generation of cellular Fe/S proteins or whether a second (still unknown) mitochondrial ferredoxin may fulfill this crucial biochemical task. Human adrenodoxin did not functionally complement the deletion of YAH1 in yeast (11). However, this observation does not readily rule out a function in Fe/S cluster biogenesis. The activity of human adrenodoxin may depend on the specific interaction with the human NADPH-dependent adrenodoxin reductase that regenerates the reduced form of adrenodoxin. The human protein may not cooperate efficiently with the yeast ferredoxin reductase Arh1p (35, 36), which is the putative reducing partner of Yah1p. An involvement of Arh1p in Fe/S cluster biosynthesis has not yet been reported.
A conspicuous phenotypic consequence of defects in the biosynthesis of Fe/S proteins is the drastic accumulation of iron within mitochondria. To date, increases in mitochondrial iron concentration have been reported for Atm1p, Ssq1p, Nfs1p, Yah1p, and a combined inactivation of Isu1p and Nfu1p (refs. 7, 22, 31, and 34; this study). Further, elevated iron levels have been observed for mutants in frataxin, a mitochondrial matrix protein that is defective in patients with the neurodegenerative disease Friedreich's ataxia (37, 38). A regulatory role of frataxin in Fe/S cluster assembly has been suggested based on defects in the mitochondrial aconitase activity (39, 40). Clearly, in any of these cases, the molecular reason for the iron accumulation within mitochondria and the sequence of events leading to the unregulated iron uptake remain to be defined. Based on the specific function of Atm1p in the biosynthesis of extramitochondrial but not intramitochondrial Fe/S proteins (7), it is tempting to propose a decisive role of an extramitochondrial Fe/S protein in the regulation of the iron uptake by the organelles.
It has become clear during the past year that mitochondria contain a complex machinery for the biosynthesis of cellular Fe/S proteins. Our study adds the ferredoxin Yah1p to the list of components of this apparently complex apparatus. Yah1p seems to perform a central task in biogenesis, because the protein is essential for viability of yeast cells. An essential function has been shown for other components of Fe/S cluster biogenesis. These proteins include the cysteine desulfurase Nfs1p and the chaperone Jac1p (5, 7). Further, simultaneous deletion of the genes of the two homologs of the bacterial proteins NifU/IscU, termed Isu1p and Isu2p, is lethal (34). The essential character of these mitochondrial components underlines the importance of mitochondria in the biogenesis of Fe/S proteins. The present investigation lends further support to the suggestion of a function of mitochondria in the biosynthesis of Fe/S proteins outside the organelle. There is growing evidence for a general requirement of at least the central components of mitochondrial Fe/S protein biogenesis in the formation of extramitochondrial Fe/S proteins (ref. 7; this study; A. K. and G. Kispal, unpublished data). The elemental sulfur required for cytosolic Fe/S clusters is produced by Nfs1p inside mitochondria (7). Likely, iron also moves though mitochondria before incorporation into cytosolic Fe/S clusters. We therefore speculate that Fe/S clusters become assembled within mitochondria and are exported to the cytosol, presumably via the ATP-binding cassette transporter Atm1p. At present, it is unclear how the preassembled Fe/S clusters might be stabilized during transport.
At present, it is unknown why the generation of cellular Fe/S proteins is so crucial for viability and why its failure is more detrimental than mutations in components of oxidative phosphorylation, the well known primary function of mitochondria. The essential character of Fe/S protein biosynthesis could be explained by an indispensable task of at least one Fe/S protein in the yeast cell. Alternatively, the combined inactivation of the entire set of cellular Fe/S proteins might lead to the loss of viability. Ferredoxins are Fe/S cluster-containing proteins. The involvement of the ferredoxin Yah1p in Fe/S cluster assembly predicts the requirement of the functional ferredoxin holoprotein in its own biosynthesis. This situation resembles the essential role of the mitochondrial chaperonin Hsp60 in its own assembly to the functional oligomer (41). Both cases explain why mitochondria cannot be formed de novo but rather have to be segregated faithfully from the parent to the daughter cells.
The identification of the components of cellular Fe/S protein biosynthesis in eukaryotes (refs. 5, 7, and 34; this study) will open the avenue to detailed investigations of the molecular mechanism of this process. The close similarity of the mitochondrial machinery of Fe/S cluster formation to the apparatus identified in prokaryotes will boost the elucidation of this fundamental process in a way similar to that encountered for analysis of the bacterial and mitochondrial machineries of protein folding.
Acknowledgments
The expert technical assistance of M. Weidgans is gratefully acknowledged. We thank Dr. A. Tzagoloff for the Δcor1 and Δcox6 yeast strains and Dr. F. Nobrega for anti-Yah1p antibodies. Our work was supported by grants of the Sonderforschungsbereich 286 of the Deutsche Forschungsgemeinschaft, the Volkswagen Stiftung, the Fonds der Chemischen Industrie, the Hungarian Funds Országos Tudományos Kutatási Alap (Grants T6378, T020079, and T022581), and the Alexander von Humboldt Stiftung.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cammack R. In: Iron-Sulfur Proteins. Cammack R, editor. Vol. 38. San Diego: Academic; 1992. pp. 281–322. [Google Scholar]
- 2.Beinert H, Holm R H, Münck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 3.Beinert H, Kiley P J. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 4.Lill R, Diekert K, Kaut A, Lange H, Pelzer W, Prohl C, Kispal G. Biol Chem. 1999;380:1157–1166. doi: 10.1515/BC.1999.147. [DOI] [PubMed] [Google Scholar]
- 5.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 6.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kispal G, Csere P, Prohl C, Lill R. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leighton J, Schatz G. EMBO J. 1995;14:188–195. doi: 10.1002/j.1460-2075.1995.tb06989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters J W, Fisher K, Dean D R. Annu Rev Microbiol. 1995;49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 10.Zheng L, Cash V L, Flint D H, Dean D R. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 11.Barros M H, Nobrega F G. Gene. 1999;233:197–203. doi: 10.1016/s0378-1119(99)00137-7. [DOI] [PubMed] [Google Scholar]
- 12.Matsubara H, Saeki K. In: Iron-Sulfur Proteins. Cammack R, editor. Vol. 38. San Diego: Academic; 1992. pp. 223–280. [Google Scholar]
- 13.Sweeney W V, Rabinowitz J C. Annu Rev Biochem. 1980;49:139–161. doi: 10.1146/annurev.bi.49.070180.001035. [DOI] [PubMed] [Google Scholar]
- 14.Lambeth J D, Seybert D W, Lancaster J R J, Salerno J C, Kamin H. Mol Cell Biochem. 1985;45:13–31. doi: 10.1007/BF01283159. [DOI] [PubMed] [Google Scholar]
- 15.Crivellone M D, Wu M A, Tzagoloff A. J Biol Chem. 1988;263:14323–14333. [PubMed] [Google Scholar]
- 16.Koerner T J, Homison G, Tzagoloff A. J Biol Chem. 1985;260:5871–5874. [PubMed] [Google Scholar]
- 17.Sirrenberg C, Bauer M F, Guiard B, Neupert W, Brunner M. Nature (London) 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- 18.Wach A, Brachat A, Poehlmann R, Phillipsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 19.Mumberg D, Müller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 20.Sherman F. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 21.Kispal G, Steiner H, Court D A, Rolinski B, Lill R. J Biol Chem. 1996;271:24458–24464. doi: 10.1074/jbc.271.40.24458. [DOI] [PubMed] [Google Scholar]
- 22.Kispal G, Csere P, Guiard B, Lill R. FEBS Lett. 1997;418:346–350. doi: 10.1016/s0014-5793(97)01414-2. [DOI] [PubMed] [Google Scholar]
- 23.Daum G, Böhni P C, Schatz G. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steiner H, Zollner A, Haid A, Neupert W, Lill R. J Biol Chem. 1995;270:22842–22849. doi: 10.1074/jbc.270.39.22842. [DOI] [PubMed] [Google Scholar]
- 27.Woonter M, Jaehning J A. J Biol Chem. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 28.Robinson K M, Lemire B D. Methods Enyzmol. 1995;260:34–51. doi: 10.1016/0076-6879(95)60128-7. [DOI] [PubMed] [Google Scholar]
- 29.Kohlhaw G B. Methods Enzymol. 1988;166:423–429. doi: 10.1016/s0076-6879(88)66055-1. [DOI] [PubMed] [Google Scholar]
- 30.Tzagoloff A, Dieckmann C L. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight S A, Sepuri N B, Pain D, Dancis A. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 32.Zheng L, Dean D R. J Biol Chem. 1994;269:18723–18726. [PubMed] [Google Scholar]
- 33.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 34.Schilke B, Voisine C, Beinert H, Craig E. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacour T, Achstetter T, Dumas B. J Biol Chem. 1998;273:23984–23992. doi: 10.1074/jbc.273.37.23984. [DOI] [PubMed] [Google Scholar]
- 36.Manzella L, Barros M H, Nobrega F G. Yeast. 1998;14:839–846. doi: 10.1002/(SICI)1097-0061(19980630)14:9<839::AID-YEA283>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 37.Babcock M, De Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 38.Foury F, Cazzalini O. FEBS Lett. 1997;411:373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 39.Rötig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 40.Foury F. FEBS Lett. 1999;456:281–284. doi: 10.1016/s0014-5793(99)00961-8. [DOI] [PubMed] [Google Scholar]
- 41.Cheng M Y, Hartl F-U, Horwich A L. Nature (London) 1990;348:455–458. doi: 10.1038/348455a0. [DOI] [PubMed] [Google Scholar]
- 42.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tangeras A, Flatmark T, Bäckström D, Ehrenberg A. Biochim Biophys Acta. 1980;589:162–175. doi: 10.1016/0005-2728(80)90035-3. [DOI] [PubMed] [Google Scholar]
- 44.Yuvaniyama, P., Agar, J. N., Cash, V. L., Johnson, M. K. & Dean, D. R. (2000) Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]