Abstract
Insulin-like growth factor-binding proteins (IGFBPs) control bioavailability, activity, and distribution of insulin-like growth factor (IGF)1 and -2 through high-affinity IGFBP/IGF complexes. IGF-binding sites are found on N- and C-terminal fragments of IGFBPs, the two conserved domains of IGFBPs. The relative contributions of these domains to IGFBP/IGF complexation has been difficult to analyze, in part, because of the lack of appropriate three-dimensional structures. To analyze the effects of N- and C-terminal domain interactions, we determined several x-ray structures: first, of a ternary complex of N- and C-terminal domain fragments of IGFBP4 and IGF1 and second, of a “hybrid” ternary complex using the C-terminal domain fragment of IGFBP1 instead of IGFBP4. We also solved the binary complex of the N-terminal domains of IGFBP4 and IGF1, again to analyze C- and N-terminal domain interactions by comparison with the ternary complexes. The structures reveal the mechanisms of IGF signaling regulation via IGFBP binding. This finding supports research into the design of IGFBP variants as therapeutic IGF inhibitors for diseases of IGF disregulation. In IGFBP4, residues 1–38 form a rigid disulphide bond ladder-like structure, and the first five N-terminal residues bind to IGF and partially mask IGF residues responsible for the type 1 IGF receptor binding. A high-affinity IGF1-binding site is located in a globular structure between residues 39 and 82. Although the C-terminal domains do not form stable binary complexes with either IGF1 or the N-terminal domain of IGFBP4, in the ternary complex, the C-terminal domain contacts both and contributes to blocking of the IGF1 receptor-binding region of IGF1.
Keywords: structure, cell growth
The insulin-like growth factor-binding protein (IGFBP) family comprises six soluble proteins (IGFBP1–6) of ≈250 residues that bind to IGFs with nanomolar affinities (1–4). Because of their sequence homology, IGFBPs are assumed to share a common overall fold and are expected to have closely related IGF-binding determinants. Each IGFBP can be divided into three distinct domains of approximately equal lengths: highly conserved cysteine-rich N and C domains and a central linker domain unique to each IGFBP species. Both the N and C domains participate in the binding to IGFs, although the specific roles of each of these domains in IGF binding have not been decisively determined (1–13). The C-terminal domain may be responsible for preferences of IGFBPs for one species of IGF over the other (2, 3–7, 9–13); the C-terminal domain is also involved in regulation of the IGF-binding affinity through interaction with extracellular matrix components (1, 2, 14) and is most probably engaged in mediating IGF1-independent actions (1, 4, 14). The central linker domain is the least conserved region and has never been cited as part of the IGF-binding site for any IGFBP. This domain is the site of posttranslational modifications, specific proteolysis (4), and the acid-labile subunit (1) and extracellular matrix associations (1, 2, 14) known for IGFBPs. Proteolytic cleavage in this domain is believed to produce lower-affinity N- and C-terminal fragments that cannot compete with IGF receptors for IGFs, and, thus, the proteolysis is assumed to be the predominant mechanism for IGF release from IGFBPs (4, 9). However, recent studies indicate that the resulting N- and C-terminal fragments still can inhibit IGF activity and have functional properties that differ from those of the intact proteins (1, 3, 5, 9).
The structure of the N-terminal domain of IGFBP-5, free (15) and complexed to IGF1 (16), was solved some time ago. More recently, low-resolution structures of the C-terminal domain of IGFBP6 (12) and its binding surface on IGF2 (3, 12) have been determined with NMR spectroscopy. There is, however, no x-ray structure of a ternary complex of the C-terminal domain of any IGFBPs bound to both the N-terminal domain and IGF, although the C-terminal fragment of IGFBP4 was crystallized recently (9), and also the x-ray structure of the isolated C-terminal fragment of IGFBP1 has been solved (17). We recently reported the x-ray structure of the ternary complex of the N- and C-terminal domains of IGFBP4 bound to IGF1 (10) and described ordered structures for the N-terminal domain of IGFBP4 and IGF1. The C domain was represented by disconnected patches of electron density and could not be interpreted. We describe here the long-sought, high-resolution x-ray structure of a complex of the N- and C-terminal domains of IGFBP4 bound to IGF1. We also present the structure of the C-terminal domain of IGFBP1 bound to the N-terminal domain of IGFBP4 and IGF1 and the structure of the binary complex of the N-terminal domain of IGFBP4 (residues 1–92) and IGF1. These structures, together with isothermal titration calorimetry measurements of the IGFBPs binding to IGF1, explain how these proteins bind to IGFs and regulate their receptor binding. Inhibition of IGF/IGF-receptor binding interferes with cell growth and represents a strategy for the development of IGFBPs and variants as natural IGF antagonists in many common diseases that arise from disregulation of the IGF system, including diabetes, atherosclerosis, and cancer (1, 8, 18).
Results
Fold of the C-Terminal Domains of IGFBP4 and IGFBP1.
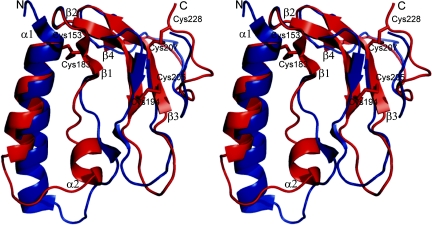
The C-terminal domain of IGFBP4 is a flat molecule without a pronounced hydrophobic core (Fig. 1). The secondary structure elements comprise two antiparallel helices α1 [Cys-153(C)-Ala-165(C)] and α2 [His-172(C)-Ile-177(C)], followed by four β strands: β1 [Asn-182(C)-Asp-184(C)], β2 [Asn-188(C)-His-195(C)], β3 [Lys-204(C)-Arg-210(C)], β4 [Val-214(C)-Leu-216(C)] that form a single, twisted β-sheet (Fig. 1). Strands β1 and β2, as well as β3 and β4, are connected by type I turns, whereas a hairpin-like elongated loop between Cys-194(C)-Gly-203(C) links β2 and β3. The loop has a conformation close to β, with a regular type I turn at the top of the loop but lacks a proper H-bonding pattern. The C terminus of the molecule, starting from Pro-217(C), does not have any regular secondary structure but is, instead, stabilized by the disulfide bridges Cys-207(C)-Cys-228(C), Cys-153(C)-Cys-183(C), and Cys-194(C)-Cys-205(C) and two H-bonds between main chains of Cys-194(C) and Glu-221(C).
Fig. 1.
Comparison of structures of the C-terminal domains of IGFBP1 (blue) (PDB ID code 1ZT5) (17) and IGFBP4 (red). Secondary structure elements discussed in Results are labeled. Disulfide bonds are shown as sticks.
Our structure of the C-terminal domain of IGFBP1 (CBP1) (residues 141–234), in complex with the N-terminal domain of IGFBP 4 (NBP4) (residues 1–92) and IGF1, is less well ordered than that of an isolated CBP1 (residues 147–221) recently published by Sala et al. (17). In addition, regions Gln-166(C)-Ile-173(C) and Asp-197(C)-Gly-198(C) in our CBP1 are fully disordered (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). We will therefore use the structure of Sala et al. (17) in comparing CBP1 with CBP4. The defined parts of our CBP1 are nearly identical to those of free CBP1, with the backbone rmsd of 0.96 Å.
The structures of the C-terminal domains of isolated IGFBP1 (PDB code 1ZT5) (17) and complexed IGFBP4 have a very similar fold (Fig. 1), as expected from their sequence identities (Fig. 5, which is published as supporting information on the PNAS web site). The rmsd for backbone atoms is 1.3 Å. A major difference is the region Ala-166(C)-Pro-179(C) of CBP4 that includes the end of helix α1 and the whole α2 helix. In contrast to CBP4, corresponding residues in CBP1 [Lys-164(C)-Tyr-177(C)] are not helical but form a short β-strand. Also in CBP4, the α1 helix is shorter by one turn, and the loop following it is bent to the opposite direction. The presence of a short β-strand after the α1 helix relocates the β3 strand of CBP1, which is longer by three residues and by ≈2.8 Å away from the binding surface of IGF compared with CBP4. The CBP4 region Pro-222(C)-Glu-225(C) has a 310 turn absent in the CBP1 structure. This element is, however, involved in crystal contacts, and its conformation might be artificial.
The C-Terminal Domain of IGFBP4 in the Ternary Complex NBP4(3–82)/IGF1/CBP4 (Residues 151–232).
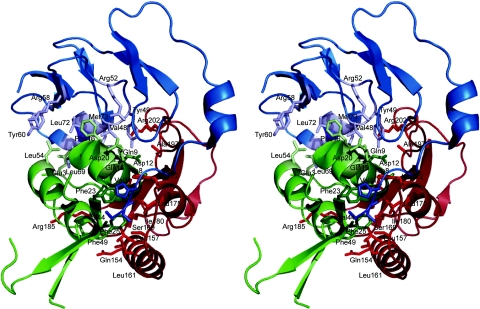
CBP4 interacts with both IGF1 and NBP4 in the complex (Fig. 2). The interaction surface encompasses a side of the CBP4 molecule that is built up by helices α1, α2, segment Ile-178(C)-Arg-185(C), and a hairpin-like loop between residues Cys-194(C)-Gly-203(C). We divide this CBP4 interface into three parts based on its interacting partners: the part that makes contacts with IGF1 only, the segment that interacts with both NBP4 and IGF1, and the stretch of residues Glu-168(C)-Glu-173(C) of CBP4 which interacts with the so-called “thumb” region of NBP4. The thumb region consists of a short stretch of the very first N-terminal residues of IGFBPs that precede the first N-terminal cysteine (amino acids 1–5 in IGFBP4) (10). It is worth noting that both CBP4 and NBP4 occupy one side, a dome part, of a pear-like IGF1 structure (Figs. 2 and 3).
Fig. 2.
Overall structure of the NBP4(3–82)/IGF1/CBP4 (residues 151–232) ternary complex. The NBP4 domain is blue, IGF1 is green, and CBP4 is red. The residues shown are important for binding among protein components (see Results).
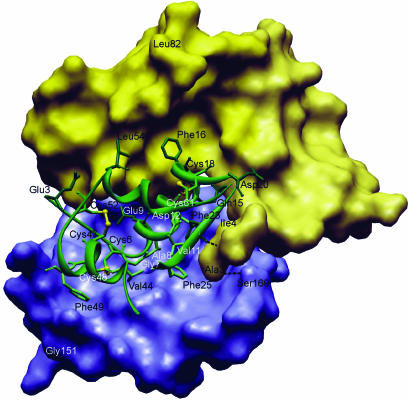
Fig. 3.
The interaction of IGF1 (green) with NBP4 (residues 3–82) (the yellow surface) and CBP4 (residues 151–232) (the blue surface). Residues discussed in Results and Discussion are labeled and shown in stick representation; two hydrogen bonds from the thumb to IGF1and CBP4 are also shown.
Gln-154(C), Leu-157(C), and Leu-161(C) of one face of helix α1, most of the residues in α2, the Ile-178(C)-Arg-185(C) fragment, and Cys-194(C), Pro-196(C), Ala-197(C), Arg-202(C), and Gly-203(C) of the Cys-194(C)-Gly-203(C) segment of CBP4 make direct contacts with IGF1. Several residues of segment Cys-194(C)-Gly-203(C) also make contacts to NBP4 (see below). Most of these amino acids are hydrophobic, and the interaction of CBP4 and IGF1 is based principally on the hydrophobic contact. None of the CBP4 residues inserts deeply into IGF1, except Ile-180(C), whose side chain is in a hydrophobic cleft made up by the aromatic ring of Phe-25(I), the backbone of Cys-6(I) and Gly-7(I) on the sides, and, at the bottom, by Leu-10(I). Phe-25(I) is believed to be involved in binding to the IGF1 receptor (1–3, 7). This hydrophobic interaction is enhanced by Leu-161(C), close to Ile-180(C), which makes contacts to Phe-25(I), and it is further extended by side-chain interactions of Leu-157(C)/Val-44(I), His-172(C)/Val-11(I)/Glu-15(I), Leu-175(C)/Phe-25(I)/Ala-8(I), and Pro-196(C)/Cys-45(I). Residues His-172(C), Leu-175(C), Ile-180(C), Pro-181(C), Asn-182(C), Pro-196(C), and Ala-197(C) constitute the rim of the hole present in the CBP4 structure (Fig. 6, which is published as supporting information on the PNAS web site). This hole is filled by the N terminus of the IGF α-helix between Cys-6(I) and Gly-19(I) (i.e., Cys-6(I), Gly-7(I), Ala-8(I), Glu-9(I), and Leu-10(I)), with primary “inserting” residues being Gly-7(I) and Ala-8(I) (Fig. 3). The other main interactions on the IGF side are made by residues of a short helix Val-44(I) to Arg-50(I) of IGF1. Together with Phe-25(I), these two helical segments constitute the major binding sites of CBP4 on IGF1 (Figs. 2 and 3).
Asp-199(C), Gly-200(C), Gln-201(C), Arg-202(C), which are part of a hairpin-like loop between Cys-194(C)-Gly-203(C), constitute direct contacts of CBP4 to NBP4. On the NBP4 side, the N-terminal portion of NBP4, from Ala-3(N) up to Tyr-49(N), makes contacts with CBP4. Notable is the stacking of the side chain of Arg-202(C) with the aromatic ring of Tyr-49(N) (Figs. 2 and 3).
A CBP4 segment between residues 168 and 173 [Glu-168(C), Ser-169(C), backbone Arg 170(C), Thr-171(C), His-172(C), Glue173(C)] adopts mostly an extended conformation and interacts with the thumb region of the NBP4 (residues 1–5 of NBP4) (Figs. 2 and 3). The two fragments form a short parallel-like β-sheet (hydrogen bonds are formed between His-172(C)-N and His-5(N)-O, and between Ser-169(C)-OG and Ala-3(N)-N)). The core side-chain interactions are purely hydrophobic even for charged residues; for example, Pro-7(N) makes contact to the β-atoms of Glu-173(C).
The NBP4(1–92)/IGF1 Complex and the Ternary Complex of CBP1 (Residues 141–234) with NBP4(1–92)/IGF1.
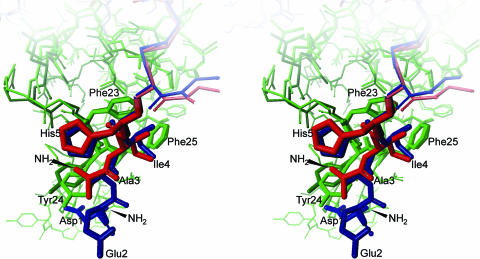
We also report here the structure of the binary complex of the “long form” of NBP4 (residues 1–92) and IGF1 solved to a 2.5-Å resolution (Fig. 4; and see Fig. 7, which is published as supporting information on the PNAS web site). NBP4 (residues 1–92) includes now the two first N-terminal thumb residues not present in our previously used NBP4 (residues 3–82) construct (10). Compared with the structure of NBP4(3–82)/IGF1 (10), to the NBP4(3–82)/IGF1 part of the NBP4(3–82)/IGF1/CBP4 (residues 151–232) and the NBP4(1–82)/IGF1 of the NBP4(1–92)/IGF1/CBP1 (residues 141–234), the four complexes reveal virtually no changes in positions of the backbone and most of the side-chain atoms of both NBP4 and IGF; heavy atom rmsds are 0.75, 0.71, and 0.96 Å respectively. The first two thumb residues are, however, defined only in the binary NBP4(1–92)/IGF1 complex (Fig. 4). Fig. 4 shows that these two residues cover Tyr-24(I), which is involved in binding to IGF1 receptor (IGF1R) (10, 15, 16).
Fig. 4.
Interaction of the thumb region of NBP4 (residues 3–82) (dark red) and NBP4 (residues 1–92) (dark blue) complexed to IGF1; IGF1s are in green, with the residues important for the IGF–IR binding labeled.
The structure of the NBP4(1–92)/IGF1 complex served as a molecular replacement search model for the diffraction data of NBP4(1–92)/IGF1/CBP1 (residues 141–234). The refined model of NBP4/(1–92)/IGF1/CBP1, after removing part of NBP4 residues, was, in turn, used for molecular replacement with the experimental data of NBP4/(3–82)/IGF1/CBP4 (residues 151–232) (see Supporting Materials and Methods). The crystals of NBP4(1–92)/IGF1/CBP1 (residues 141–234) diffracted to 3.2 Å, and the model built is lacking several residues at the interface of CBP1 and NBP4 or IGF1. We therefore did not use this structure for a detailed analysis of the CBP complexes but describe NBP4(3–82)/IGF1/CBP4 (residues 151–232), which is identical with the rmsd of 0.96 Å for backbone atoms.
Fine Tuning of the N-Terminal Thumb Residues.
To assess the contribution of the thumb of the IGFBP to its IGF1-binding activity, a number of N-terminal mutants of NBP4 were produced, and their IGF1 binding activity was compared by using isothermal titration calorimetry (Table 1). Significantly, the removal of residues 1–4 and simultaneous substitution of His (5) by Gly [construct NBP4(G5–82)] reduces binding to IGF1 by 10-fold (Table 1). For BP1-NBP4, BP3-NBP4, and consBP3-NBP4, the differences in KD values, although not large, show extra contacts made by the longer thumb sequences.
Table 1.
N domain IGFBP constructs, their N-terminal sequences, and IGF1-binding constants
| Name | N-terminal sequences upstream from Cys6(N) of IGFBP4 | KD, nM |
|---|---|---|
| NBP4(1–82) | D E A I H C | 317 ± 19 |
| NBP4(3–82) | A I H C | 865 ± 78 |
| NBP4(G5–82) | G C | 3350 ± 590 |
| MiniNBP4(39–82) | – | 3020 ± 410 |
| BP1-NBP4* | A P W H C | 544 ± 45 |
| BP3-NBP4* | GA SSGGLGPVV HC | 268 ± 14 |
| ConsBP3-NBP4* | GA GAVGLGPVV RC | 316 ± 27 |
*Hybrid constructs of human IGFBPs that comprise the “core” of NBP4(6–82) and the N-terminally added sequences (in italics); ConsBP3 designates the consensus sequence found in NBP3s of different animal species (see Fig. 8, which is published as supporting information on the PNAS web site).
Discussion
The C-terminal domains of all IGFBPs show sequence homology with thyroglobulin type-1 domains (3, 12, 17, 19, 20) and share common elements of secondary structure: an α-helix and a 3- to 4-β-stranded β-sheet. The core of the molecule is connected by the consensus three disulfide pairings, has conserved Tyr/Phe amino acids and has the QC, CWCV motifs (20). These essential features are preserved in CBP1, CBP4, and CBP-6, the structures of C domains solved so far, although there are significant variations in detail. For example, CBP4 has helix α2, whereas the corresponding residues in CBP1 form a short β-strand seen in other structures of the thyroglobulin type-1 domain superfamily (17, 20). This particular region of CBPs has high sequence diversity and is involved in the IGF complex formation and thus may perform the role of an affinity regulator.
The ternary complexes of NBP4(3–82)/IGF1/CBP4 (residues 151–232) and NBP4(1–92)/IGF1/CBP1 (residues 141–234) provide an understanding of the roles of N- and C-terminal IGFBP domains in modulating IGF actions and show that the N- and C-terminal domains come into close proximity mutually in the complex to enhance or stabilize IGF binding. There has been a considerable body of work to delineate the determinants of IGFBPs binding to IGFs and vice versa (1–16, 21–29). The structural information presented in this work is broadly in agreement with these data, but disagrees with reports of a critical role of the completely conserved Gly-187(C) and Gln-193(C) (in the IGFBP4 sequence) for binding of C domains to IGFs (2, 6, 29, and references cited therein): These residues are not in contact with IGF1, although they are close to the IGF1/CBP4 interface surface. A second example is provided by the recent NMR mapping of the binding surfaces of IGFBP2 on IGF1 (7). These data, combined with previous mutational analyses (2), would suggest that the Gly-22(I)-Phe-25(I) region of IGF1 directly interacts with the C domain of IGFBP2. In our structure, this segment of IGF1 [with the exception of Phe-25(I)] clearly binds to the thumb region of NBP4. The thumb masks the IGF residues responsible for the IGF1R binding and, in turn, interacts with residues 168(C)–173(C) of the C domain. Thus, although our isolated CBP4 does not bind individually to either IGF1 or NBP4, in the ternary complex, CBP4 contacts both and contributes to blocking of the IGF1R-binding region of IGF1.
Both N- and C-terminal domains of IGFBPs were reported to bind to IGFs (1–16, 21–23, 29). Isolated N-terminal fragments of IGFBPs bind to IGF with 10- to 1,000-fold lower affinities than full-length IGFBPs. There is also no doubt that the C-terminal domains of IGFBPs increase the affinity of IGFBPs for IGFs (Table 1; and refs. 1–16). However, there are inconsistent reports in the literature regarding the strength of direct binding between isolated C-terminal domains and IGFs (1, 3, 6, 10, 11, 21, 22, 29). The C domain of bovine IGFBP2 (residues 136–279) was reported to bind IGF1 with a KD of 23.8 nM, whereas affinity of the N domain (residues 1–185) was 78.1 nM (11); e.g., the C domain had higher affinity than the N domain for IGF1. A similar trend was observed for human IGFBP3 in one report (21), but Vorwerk et al. (22) found that their C domain had a 3-fold weaker binding to IGF1 than the N domain, which had a KD of 163 nM. The C domains of IGFBP4 (9, 10), IGFBP5 (15), and IGFBP6 (13) showed lower binding affinities than their respective N counterparts. Features of the IGF-binding regions of CBP1 and CBP4 seen in current structures would support the latter trend. Although contact areas of NBP4 and CBP4 to IGF are approximately equal (758 Å2 and 670 Å2, respectively), a direct access of C domains to IGF is obscured in two sites by N-domain residues (in Fig. 3, for example, the C domain interacts through the N-terminal thumb residues with IGF). In addition, the hydrophobic interaction between IGF1 and the miniNBP subdomain of NBP4, consisting of interlaced protruding side chains of IGF1 and solvent-exposed hydrophobic side chains of the NBP4, seems to be more extensive than that seen in the CBP4/IGF binding. It is expected that the IGF-binding structure of our complexes will be shared among six IGFBPs, but the binding affinities of isolated N and C domains may differ among IGFBPs and between IGF1 and IGF2 because of sequence differences.
The NMR spectrum of the full-length IGFBP4 indicated that the central variable domain of IGFBP4 (Fig. 5) is unstructured and flexible, both when free and when in complex with IGF1 (10). Positions of the last C-terminal residue of NBP4 [Leu-82(N)] and the first N-terminal residue of CBP4 [Gly-151(C)] shown in Fig. 3 suggest the location of the central L domain of IGFBP4 in the full-length IGFBP. The central domain might act as a “mechanical flap” that covers the IGF not yet wrapped by N- and C-terminal domains. Proteolysis of the IGF/IGFBP capsule would first remove the central domain by degradation. This partial removal of the capsule exposes IGF but still maintains IGF inhibition toward IGF1R as long as the N-terminal thumb and/or CBP fragments of IGFBPs are not removed.
The first 92 residues of IGFBP4 are 59% identical to the corresponding N-terminal residues of IGFBP5, and the remaining residues are mostly functionally conserved. For miniNBP5 (residues 40–92), the last 9 aa showed no electron density in IGF complex structure (16) and were unstructured as determined by NMR (15). Equivalent residues were therefore not expressed in the construct NBP4 (residues 3–82) to aid crystallization of the complex. However, residues Glu-90 and Ser-91 of IGFBP4 were reported to be important for high-affinity binding with IGFs (23); therefore, we decided to include these residues in our extended N-terminal construct NBP4 (residues 1–92). The first two N-terminal residues were also added because the previous IGFBP4(3–82)/IGF1 structure (10) indicated the importance of the N-terminal hydrophobic residues conserved among IGFBPs. A possibility that eliminating the first two negatively charged residues, Asp-1 and Glu-2, at the N terminus in the IGFBP4, could have changed the properties of this N-terminal part, also existed. These residues were therefore added to the refined N-terminal construct, generating NBP4 (residues 1–92). It is evident from the structure of the current NBP4(1–92)/IGF1 binary complex that the sequence Ala-83-Leu-92, of which the fragment Glu-84-Glu-90 forms a short helix, does not contact IGF directly. In the study of Qin et al. (23) deletion of Glu-90 and Ser-91 led to the reduced IGF1- and -2-binding activity, suggesting the functional importance of these residues. The molecular structure, however, shows no direct contribution of these two residues to the formation of the IGF-binding site. The presence of the 10-aa-long fragment may, however, have an indirect influence on IGF binding: side chains of Ile-85(N), Ile-88(N), and Gln-89(N) shield the Tyr-60(N) side chain from the solvent and constrain its conformation, which would otherwise point away from the IGF surface, as can be seen in the NBP4(3–82)/IGF1 complex structure. Tyr-60(N), along with Pro-61(N), forms a small hydrophobic cleft, in which Leu-54(I) of IGF1 is inserted, thus extending a hydrophobic contact area of the two proteins.
The N-terminal domain of IGFBPs can be viewed as consisting of a globular base, corresponding to the miniBP5 that contains an important IGF-binding site (10, 15, 16), and an extended “palm” followed by a short hydrophobic thumb (Ala-3(N), Ile-4(N)) (Figs. 2–4 and 7). The thumb interacts with IGF residues Phe-23(I), Tyr-24(I), and Phe-25(I) upon complex formation. The palm is rigid because of four disulfide bonds arranged in a ladder-like plane and several intersubdomain H-bonds. Rigidity of the N-terminal domain of IGFBPs may be of significance where the competition with IGF1R for IGF binding is concerned. Previous studies revealed that Phe-23, Tyr-24, and Phe-25 of IGF1, and corresponding Phe-26, Tyr-27, and Phe-28 of IGF2, are important for binding to insulin and IGF type I receptors (1–3; 24–28). To displace the hydrophobic thumb that covers the primary IGF1R-binding site of IGFs [IGF1, Phe-23(I)-Phe-25(I)], the receptor also has to remove the rest of the N-terminal domain, which is bound to the opposite side of the IGF1 molecule, and does not prevent receptor binding on its own (15). Given the conserved arrangement of the N-terminal cysteine residues and the consistent presence of two hydrophobic residues at positions 2 and 3 with respect to the first N-terminal Cys residue, we expect that this mechanism is shared by all IGFBPs.
The very first N-terminal residues of IGFBPs have been neglected to date in mutagenesis studies that aimed at delineation of these proteins' structure/function relationships (reviewed in ref. 2). Truncation of the thumb (residues 1–5) reduces the IGF1 binding of NBP4 to that of miniNBP4 (residues 39–82) (Table 1), suggesting that the palm (residues 6–39) does not contribute directly to IGF binding. The segment appears, therefore, to serve a solely mechanical purpose as a rigid linker between primary binding sites, the base and the thumb residues. The central element of the palm consists of a GCGCCXXC consensus motif, around which the polypeptide chain (residues Cys-6-Cys-23) is bent, forming a disulfide ladder and assuring a proper spatial relationship between the base and the thumb (Figs. 3, 4, and 7). Substitution of the IGFBP4 thumb with a corresponding region from two other IGFBPs does not influence the strength of IGF1 binding markedly (Table 1). Interestingly, available sequences of IGFBPs from different species show a remarkably high degree of interspecies conservation of the thumb residues within a single type of IGFBP (Fig. 8), implying that the sequence of the thumb may confer unique properties to each IGFBP.
The structural studies carried out here provide important information to aid in the design of IGFBP-based therapeutics (1, 8, 30). The involvement of the IGF system in tumor cell growth and survival makes it an excellent target for anticancer treatment; especially in view of the fact that IGF1R is not absolutely necessary for normal growth (1, 8, 18, 30). Indeed, recent data have shown that targeting the IGF system produces impressive antineoplastic activity in many in vitro and in vivo models of human cancers. The most advanced are the strategies based on the direct disruption of receptor function that are based on small-molecule inhibitors of the tyrosine kinase domain of IGF1R and on antibodies directed against IGF1R (1, 18, 30). The possibility that these therapies will lack sufficient specificity to avoid cotargeting the insulin receptor must, however, be considered, and, therefore, carefully designed clinical trails will be required to assess the effect on host glucose metabolism (18). Neutralization of IGF ligands through IGFBPs (which target only the IGFs) would avoid this problem, because insulin action would be mostly unaffected (1, 8, 18, 30). The design of the therapeutic IGFBPs or their fragments would have to take into account the building characteristics of the IGFBP/IGF structure presented here. For example, our structures revealed the importance of the N-terminal thumb hydrophobic residues for blocking the IGF/IGF1R interaction, and, thus, IGFBP constructs should include “long” N-terminal thumb segments.
Conclusion
The crystal structures described provide a molecular basis for the IGFBPs' regulation of IGF signaling. Key features of the IGFBP/IGF complexes include: (i) a disulfide bond ladder in the N-terminal domain and the first five N-terminal thumb residues, which bind to IGF and partially mask the IGF residues responsible for type 1 IGF receptor binding; (ii) a high-affinity IGF1 interaction site formed by residues 39–82 in a globular fold; (iii) although CBP4 does not bind individually to either IGF1 or NBP4, in the ternary complex, CBP4 contacts both, stabilizes the complex, and contributes to blocking of the IGF1R-binding region of IGF1; (iv) the central domain, which is unstructured and flexible, even when IGFBPs are bound to IGFs, covers the IGF not yet covered by the N- and C-terminal domains and additionally blocks the access of IGF1R to IGF through steric hindrance.
Materials and Methods
Protein Cloning and Purification.
DNA fragments corresponding to the NBP4 (residues 1–92 of IGFBP4), BP1-NBP4 (residues 1–3 of IGFBP1, followed by residues 5–82 of IGFBP4), BP3-NBP4 (residues 1–11 of IGFBP-3, followed by residues 5–82 of IGFBP4), BP3cons-NBP4 (consensus residues 1–12 of IGFBP-3, followed by residues 6–82 of IGFBP4), and NBP4(G5–82) were generated by PCR amplification using human IGFBP4 cDNA (vector BP4–2/pFDX500, Roche Diagnostics, Penzberg, Germany) as a template and the following primer sets: 1 and 9 [NBP4 (residues 1–92)]; 2 and 8 (BP1-NBP4); 3, 4, and 8 (BP3-NBP4); 5, 6, and 8 (BP3cons-NBP4); 7 and 8 [NBP4(G5–82)]. The sequences of these primers are shown in Table 2, which is published as supporting information on the PNAS web site. The resultant PCR products were subcloned into the BamHI and HindIII restriction sites of the pET 28a vector (Novagen, Mississauga, ON, Canada). MiniNBP5 (residues 40–92) and NBP4 (residues 3–82) were cloned as described (10, 15).
The sets of primers shown in Table 2 were designed for cloning of CBP4 (residues 151–232) and CBP1 (residues 141–234) into the BamHI and HindIII and EcoRI and XhoI restrictions sites, respectively, of the pET 28a-vector in-frame to a His-T7-tag. Standard procedures were used for construction and verification of vectors pET28a (NBP) and pET28a (CBP), which were transformed finally into the Escherichia coli strain Bl21(DE3)pLysS (Novagen) for overexpression. The proteins were produced and purified as described (10, 15).
Isothermal Titration Calorimetry.
Binding of amino-terminal IGFBP constructs to IGF1 was carried out as described (10).
X-Ray Crystallography.
The ternary complexes of NBP4(1–92)/IGF1 (GroPep, Adelaide, Australia)/CBP1 (residues 141–234), NBP4/(3–82)/IGF1/CBP4 (residues 151–232), and the binary complex of NBP4 (residues 1–92)/IGF1 were prepared by mixing equimolar amounts of the components. The complexes were separated from any excess of free proteins by gel-filtration chromatography on the Superdex 75 16/60XR column (Amersham Pharmacia, Uppsala, Sweden). Crystallization of the complexes was carried out with the sitting-drop vapor-diffusion method by mixing equal volumes (2 μl) of protein (10 mg/ml) and reservoir solution. The best diffracting crystals of NBP4/(1–92)/IGF1/CBP1 (residues 141–234) were obtained from 20% PEG 3350, 0.2 M lithium acetate, pH 7.3, after 2 days at 20°C. The crystals had a space group P21, with unit cell parameters a = 71.3 Å, b = 43.7 Å, c = 81.1 Å, and β = 91.7° There are two complexes per asymmetric unit. Crystals of NBP4/(3–82)/IGF1/CBP4 (residues 151–232) (space group C2, a = 74.4 Å, b = 50.2 Å, c = 64.3 Å, and β = 115.3°) grew after 4 months at 4°C in 1 M lithium sulfate monohydrate and 2% PEG 8000. The crystals of the binary complex NBP4 (residues 1–92)/IGF1 were obtained from 23% PEG 1500 and 25 mM Tris, pH 7, after 3 weeks, in the form of plates measuring ≈0.5 × 0.3 × 0.1 mm. The crystals belong to the space group P21 (a = 32.3 Å, b = 39.0 Å, c = 61.3 Å, and β = 99.9°) and contained one complex per asymmetric unit. Before plunge-freezing, all crystals were soaked for ≈30 s in a drop of a reservoir solution containing 20% vol/vol glycerol or 20% ethylene glycol as a cryoprotectant. The structure of NBP4(1–92)/IGF1 was solved by molecular replacement by the Molrep program of the CCP4 suite. An initial model was rebuilt and completed manually. The resulting structure was then used as a molecular replacement probe for the NBP4/(1–92)/IGF1/CBP1 (residues 141–234) crystal data. The missing CBP1 part was built manually. This model served subsequently as a probe for molecular replacement for the NBP4/(3–82)/IGF1/CBP4 (residues 151–232) diffraction data. A molecular replacement solution was clear, and the model was improved by Arp/wArp and manual building. Detailed crystallographic procedures are described in Supporting Materials and Methods. Data collection and refinement statistics are summarized in Table 3, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Kinga Brongel (Max-Planck-Institut für Biochemie) for purified IGFBP1.
Abbreviations
- IGF
insulin-like growth factor
- IGFBP
IGF-binding protein
- NBP4(1–92)
the N-terminal domain of IGFBP4 (residues 1–92)
- CBP4
the C-terminal domain of IGFBP4
- IGFR
IGF receptor
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2DSP, 2DSQ, and 2DSR).
References
- 1.Firth S. M., Baxter R. C. Endocr. Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 2.Clemmons D. R. Endocr. Rev. 2001;22:800–817. doi: 10.1210/edrv.22.6.0449. [DOI] [PubMed] [Google Scholar]
- 3.Bach L. A., Headey S. J., Norton R. S. Trends Endocr. Metabol. 2005;16:228–234. doi: 10.1016/j.tem.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Bunn R. C., Fowlkes J. L. Trends Endocr. Metabol. 2003;14:176–181. doi: 10.1016/s1043-2760(03)00049-3. [DOI] [PubMed] [Google Scholar]
- 5.Payet L. D., Wang X. H., Baxter R. C., Firth M. Endocrinology. 2003;144:2797–2806. doi: 10.1210/en.2003-0102. [DOI] [PubMed] [Google Scholar]
- 6.Allan G. J., Tonner E., Szymanowska M., Shand J. H., Kelly S. M., Phillips K., Clegg R. A., Gow I. F., Beattie J., Flint D. J. Endocrinology. 2006;147:338–349. doi: 10.1210/en.2005-0582. [DOI] [PubMed] [Google Scholar]
- 7.Carrick F. E., Hinds M. G., McNeil K. A., Wallace J. C., Forbes B. E., Norton R. S. J. Mol. Endocrinol. 2005;34:685–698. doi: 10.1677/jme.1.01756. [DOI] [PubMed] [Google Scholar]
- 8.Kibbey M. M., Jameson M. J., Eaton E. M., Rosenzweig S. A. Mol. Pharmacol. 2006;69:833–845. doi: 10.1124/mol.105.016998. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Tornero C., Lozano R. M., Rivas G., Jimenez M. A., Ständker L., Diaz-Gonzalez D., Forssmann W. G., Cuevas P., Romero A., Giménez-Gallego G. J. Biol. Chem. 2005;280:18899–18907. doi: 10.1074/jbc.M500587200. [DOI] [PubMed] [Google Scholar]
- 10.Siwanowicz I., Popowicz G. M., Wisniewska M., Huber R., Kuenkele K. P., Lang K., Engh R. A., Holak T. A. Structure (London) 2005;13:155–167. doi: 10.1016/j.str.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Carrick F. E., Forbes B. E., Wallace J. C. J. Biol. Chem. 2001;276:27120–27128. doi: 10.1074/jbc.M101317200. [DOI] [PubMed] [Google Scholar]
- 12.Headey S. J., Keizer D. W., Yao S., Brasier G., Kantharidid P., Bach L. A., Norton R. S. Mol. Endocrinol. 2004;18:2740–2750. doi: 10.1210/me.2004-0248. [DOI] [PubMed] [Google Scholar]
- 13.Headey S. J., Leeding K. S., Norton R. S., Bach L. A. J. Mol. Endocrinol. 2004;33:377–386. doi: 10.1677/jme.1.01547. [DOI] [PubMed] [Google Scholar]
- 14.Xu Q., Yan B., Li S., Duan C. J. Biol. Chem. 2004;279:4269–4277. doi: 10.1074/jbc.M311586200. [DOI] [PubMed] [Google Scholar]
- 15.Kalus W., Zweckstetter M., Renner C., Sanchez Y., Georgescu J., Grol M., Demuth D., Schumacher R., Dony C., Lang K., Holak T. A. EMBO J. 1998;17:6558–6572. doi: 10.1093/emboj/17.22.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeslawski W., Beisel H. G., Kamionka M., Kalus W., Engh R. A., Huber R., Lang K., Holak T. A. EMBO J. 2001;20:3638–3644. doi: 10.1093/emboj/20.14.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sala A., Capaldi S., Campagnoli M., Faggion B., Labo S., Perduca M., Romano A., Carrizo M. E., Valli M., Visai L., et al. J. Biol. Chem. 2005;280:29812–29819. doi: 10.1074/jbc.M504304200. [DOI] [PubMed] [Google Scholar]
- 18.Pollak M. N., Schernhammer E. S., Hankinson S. E. Nat. Rev. Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 19.Kiefer M. C., Masiarz F. R., Bauer D. M., Zapf J. J. Biol. Chem. 1991;266:9043–9049. [PubMed] [Google Scholar]
- 20.Novinec M., Kordis D., Turk V., Lenarcic B. Mol. Biol. Evol. 2006;23:744–755. doi: 10.1093/molbev/msj082. [DOI] [PubMed] [Google Scholar]
- 21.Galanis M., Firth S. M., Bond J., Nathanielsz A., Kortt A. A., Hudson P. J., Baxter R. C. J. Endocrinol. 2001;169:123–133. doi: 10.1677/joe.0.1690123. [DOI] [PubMed] [Google Scholar]
- 22.Vorwerk P., Hohmann B., Oh Y., Rosenfeld R. G., Shymko R. M. Endocrinology. 2002;143:1677–1685. doi: 10.1210/endo.143.5.8760. [DOI] [PubMed] [Google Scholar]
- 23.Qin X. Z., Strong D. D., Baylink D. J., Mohan S. J. Biol. Chem. 1998;273:23509–23516. doi: 10.1074/jbc.273.36.23509. [DOI] [PubMed] [Google Scholar]
- 24.Sakano K., Enjoh T., Numata F., Fujiwara H., Marumoto Y., Higashihashi N., Sato Y., Perdue J. F., Fujita-Yamaguchi Y. J. Biol. Chem. 1991;266:20626–20635. [PubMed] [Google Scholar]
- 25.Hodgson D. R., May F. E. B., Westley B. R. Regul. Pept. 1996;66:191–196. doi: 10.1016/S0167-0115(96)00102-4. [DOI] [PubMed] [Google Scholar]
- 26.Cascieri M. A., Chicchi G. C., Applebaum J., Hazes N. S., Green B. C., Bayne M. L. Biochemistry. 1988;27:3229–3233. doi: 10.1021/bi00409a016. [DOI] [PubMed] [Google Scholar]
- 27.Bayne M. L., Applebaum J., Chicchi G. G., Miller R. E., Cascieri M. A. J. Biol. Chem. 1990;265:15648–15652. [PubMed] [Google Scholar]
- 28.Perdue J. F., Bach L. A., Hashimoto R., Sakano K., Fujita-Yamaguchi Y., Fujiwara H., Rechler M. M. In: The Insulin-Like Growth Factors and Their Regulatory Proteins. Baxter R. C., Gluckman P. D., Rosenfeld R. G., editors. Amsterdam: Elsevier Science; 1994. pp. 67–76. [Google Scholar]
- 29.Shand J. H., Beattie J., Song H., Phillips K., Kelly S. M., Flint D. J., Allan G. J. J. Biol. Chem. 2003;278:17859–17866. doi: 10.1074/jbc.M300526200. [DOI] [PubMed] [Google Scholar]
- 30.Yee D. Br. J. Cancer. 2006;94:465–468. doi: 10.1038/sj.bjc.6602963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.