Abstract
In the cytochrome bc1 complex, the swivel motion of the iron–sulfur protein (ISP) between two redox sites constitutes a key component of the mechanism that achieves the separation of the two electrons in a substrate molecule at the quinol oxidation (Qo) site. The question remaining is how the motion of ISP is controlled so that only one electron enters the thermodynamically favorable chain via ISP. An analysis of eight structures of mitochondrial bc1 with bound Qo site inhibitors revealed that the presence of inhibitors causes a bidirectional repositioning of the cd1 helix in the cytochrome b subunit. As the cd1 helix forms a major part of the ISP binding crater, any positional shift of this helix modulates the ability of cytochrome b to bind ISP. The analysis also suggests a mechanism for reversal of the ISP fixation when the shape complementarity is significantly reduced after a positional reorientation of the reaction product quinone. The importance of shape complementarity in this mechanism was confirmed by functional studies of bc1 mutants and by a structure determination of the bacterial form of bc1. A mechanism for the high fidelity of the bifurcated electron transfer is proposed.
Keywords: crystal structures, electron transfer, inhibitor binding, mechanism
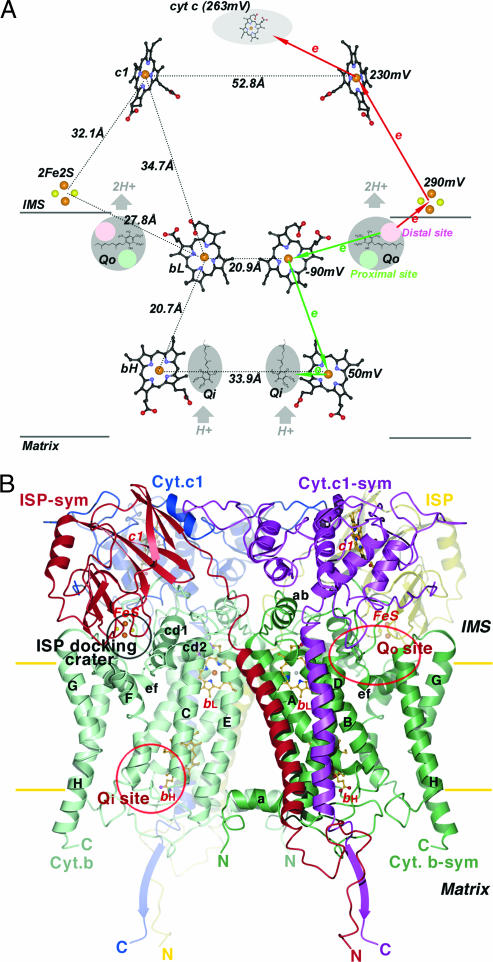
Within cellular energy-conserving membranes (the mitochondrial inner membrane in eukaryotes and the plasma membrane in prokaryotes), the only mobile carriers of redox equivalents are ubiquinone (Q), ubiquinol (QH2), and their derivatives. In the cytochrome (cyt) bc1 complex (cyt bc1 or bc1) of the respiratory chain, QH2 is oxidized to Q, and concomitantly, protons are pumped against the gradient across the membrane [from the matrix to the intermembrane space (IMS) in mitochondria and from the cytoplasm to the periplasm in prokaryotes], thus contributing to the electrochemical potential that drives ATP synthesis. The proposed mechanism by which bc1 performs this dual task, the Q-cycle hypothesis (1), has been widely accepted; it suggests that for the oxidation of two molecules of QH2, one molecule of Q is reduced at separate catalytic sites (Fig. 1A). According to this mechanism, the Q reduction site is located near the negative side of the membrane (Qi/Qn, the mitochondrial matrix or cytoplasmic side) and is electronically linked via two prosthetic groups [high-potential b heme (bH) and low-potential b heme (bL)] to the quinol oxidation site located close to the positive side of the membrane (Qo/Qp, the mitochondrial IMS side or periplasmic side in prokaryotes). At the Qo site, one electron from a substrate enters the energetically favorable path that leads via the iron–sulfur protein (ISP) and cyt c1 subunit (cyt c1) to the substrate cyt c, whereas the other electron proceeds via the bL and bH heme groups to Q (or semiquinone) bound at the Qi site. The half-reduction of a Q per oxidized quinol is achieved by the bifurcation of the electron pathway at the Qo site. In a complete catalytic cycle, two protons are taken up from the matrix, while the sequential oxidation of two QH2 molecules releases four protons into the IMS.
Fig. 1.
Prosthetic groups and subunit structures of the cyt bc1 complex. (A) Arrangement of prosthetic groups in the dimeric bc1 complex and illustration of the electron bifurcation at the Qo site. The bL, bH, and c1 heme groups are shown as ball-and-stick models, and the [2Fe2S] clusters are depicted as cpk models. Carbon atoms, black; nitrogen, blue; oxygen, red; sulfur, yellow; iron, brown. The Qo pockets near the IMS side of the membrane and the Qi pockets near the matrix side are labeled and shaded in gray. Cyt c is shown as a gray shaded oval. Distances between redox centers are given on the left half of the diagram, and the redox potential for each center is given on the right. The high- and low-potential ET paths are depicted with red and green arrows, respectively. Circles in pink and light green within the Qo pockets are hypothesized distal-QH2 and proximal-Q binding sites, respectively. (B) Ribbon diagram of the dimeric cyt b, cyt c1, and ISP subunit in the mitochondrial bc1 complex. Two symmetry-related cyt b subunits are shown (green and light green). The eight TM helices of cyt b are denoted with letters A–H. Helices A–E form one bundle in which the two b-type hemes (bL and bH in ball-and-stick models) reside; helices F–H form the other bundle. The ISP subunit (yellow and red for the symmetry pair) has an extrinsic soluble domain with a [2Fe2S] cluster at its tip, connecting to a TM segment by a flexible neck. The extrinsic domain of cyt c1 (blue and magenta for the symmetry pair) with its heme group is rigidly attached to its TM helix. The locations for the two active sites (Qo and Qi) per monomer in cyt b are labeled. The surface depression in cyt b at the IMS side of the membrane is labeled as the ISP-docking crater.
Knowledge of the reaction catalyzed by bc1 on a detailed molecular basis is a prerequisite to an understanding of mitochondrial myopathy and superoxide production (2, 3), and it forms the foundation on which it is possible to explain the modes of action of numerous antibiotics and fungicides as well as the phenotypes of mutations in cyt b that confer resistance (4, 5). However, the details of the mechanism that separates the electrons at the Qo site are a subject of much debate (see discussion in ref. 6 and references therein). Historically, the first x-ray structure of mitochondrial bc1 revealed an unusually long distance (31 Å) between the two-iron, two-sulfur cluster of ISP ([2Fe2S]) and the heme iron of cyt c1 (7); this gap is too large for a rapid electron transfer (ET) through space. However, the structure also suggests a solution to this problem: The extrinsic domain of ISP (ISP-ED) exhibited unexpectedly high flexibility, appeared at different locations, and was proposed to perform an oscillatory motion that would bridge the distance between the Qo site and cyt c1 (7, 8). Biochemical and genetic studies supported the importance of ISP mobility for the function of bc1 (9, 10).
However, the stoichiometric electron separation at the Qo site cannot be achieved with a randomly mobile ISP (11); the control of the ISP-ED conformational switch is critical. The fast rate exhibited by photoinduced ET between ISP and cyt c1 in the presence of myxothiazol precludes a control mechanism within the ISP (12), and therefore we believe such a mechanism must reside in the cyt b subunit (cyt b). Indeed, the ISP-ED was shown to undergo a binary switch: fixed or mobile conformation; specific yet different types of Qo site inhibitors can induce either state (13–15). Here, we present structural and biochemical evidence for an intrasubunit signaling pathway that establishes causality between the binding of an inhibitor and the capture or release of ISP-ED subunit and discuss its implication in the context of the bifurcated ET in a catalytic cycle of bc1.
Results and Discussion
Correlation Between Inhibitor-Induced Conformational Change in Cyt b and the Mobility Switch of ISP.
The bovine mitochondrial bc1 is a dimeric, integral membrane protein complex with 11 different subunits per monomer (7). Ubiquitously present in all bc1 complexes from various species are the three prosthetic group-containing subunits, cyt b, cyt c1, and ISP (Fig. 1B), which are crucial for the ET and proton translocation function. We cocrystallized bovine bc1 in the presence of eight Qo site inhibitors; all inhibitor complex crystals have the same symmetry as the native (apo) crystal, and their structures were refined independently starting with the native bc1 model (Protein Data Bank ID code 1NTZ; see Table 3, which is published as supporting information on the PNAS web site). For each data set, a corresponding anomalous difference Fourier map revealed the positions of the iron atoms in the prosthetic groups. This technique is particularly useful in tracking the position and occupancy of the highly mobile ISP that carries a [2Fe2S] group at its tip. This analysis (Table 1) demonstrated that there are two distinct types of Qo site inhibitors: One type (Pf) fixes the ISP head group, and the other (Pm) mobilizes it, each binding to a different site within the Qo pocket of cyt b (13, 15).
Table 1.
Conformational changes in cyt b and ISP subunits of the bc1 complex upon inhibitor binding
| Inhibitor bound structure | Inhibitor class | [2Fe2S]* anomalous peak | rmsd for Cα,†Å |
Displacement relative to the native,‡ Å |
Changes in binding of ISP to cyt b |
||||
|---|---|---|---|---|---|---|---|---|---|
| Cyt b (3–379) | Qo site§ 32 residues | bI146Cα | bP270Cα | bS151Cα | CA,¶ (Å2) | SC‖ | |||
| Native** | 0.52 | — | — | 0.0 | 0.0 | 0.0 | — | 0.55 | |
| Azoxystrobin | Pm | 0.36 | 0.26 | 0.72 | −0.76 | 1.81 | −0.71 | −86 | 0.39 |
| Myxothiazol | Pm | 0.46 | 0.26 | 0.81 | −0.57 | 1.66 | −0.71 | −69 | 0.38 |
| MOAS | Pm | 0.23 | 0.23 | 0.59 | −0.35 | 1.62 | −0.42 | −78 | 0.39 |
| Famoxadone | Pf | 1.02 | 0.33 | 0.91 | +0.87 | 1.96 | +0.58 | 65 | 0.60 |
| UHDBT | Pf | 0.96 | 0.40 | 1.11 | +1.73 | 0.72 | +0.60 | 129 | 0.60 |
| Stigmatellin | Pf | 1.20 | 0.33 | 1.32 | +1.61 | 1.58 | +0.96 | 92 | 0.60 |
| JG144†† | Pf | 1.17 | 0.28 | 0.81 | +1.06 | 1.86 | +0.52 | 79 | 0.59 |
| NQNO | PN | 0.83 | 0.33 | 0.59 | — | — | — | 56 | 0.61 |
| Stig-yeast‡‡ | Pf | — | — | — | — | — | — | — | — |
| Antimycin A§§ | N | 0.51 | 0.31 | 0.59 | +0.34 | 0.24 | −0.15 | — | — |
*Normalized anomalous difference Fourier peaks for the Fe atoms in the ISP subunit (15).
†rmsds in Å between cyt b subunits of inhibitor-bound and native bc1 complex.
‡Distance between residues either in the same subunit or in different subunits. Superscript letter on the left denotes subunit; subscript on the right is for atom type.
§Residues included in the calculation are 141–160, 167–170, and 263–270 of the cyt b subunit.
¶CA between ISP and cyt b. The CA is calculated by using the ISP extrinsic domain only (71–196). The native (apo) protein has a CA of 356 Å2.
‖Surface complementarity (16).
**Native coordinates are from Protein Data Bank ID code 1NTM (17).
††The chemical structure of JG144 is S-3-anilino-5-methyl-5-(4,6-difluorophenyl)-1,3-oxazolidine-2,4-dione.
‡‡Obtained from ref. 18.
§§Obtained from ref. 17.
Among the cocrystallized Qo inhibitors, three belong to class Pm [azoxystrobin, methoxyacrylate–stilbene (MOAS), and myxothiazol], and four are class Pf inhibitors [stigmatellin, 5-undecyl-6-hydroxy-4,7-dioxobenzothiazo (UHDBT), JG144, and famoxadone]. Pairwise superposition of Cα atoms between cyt b subunits with various inhibitors and the native gave rise to small rms deviations (rmsds) in the range between 0.234 and 0.402 Å (Table 1), indicating relatively small overall changes of cyt b upon complex formation. However, further analyses of the residues constituting the immediate environment of the inhibitor-binding pocket showed significantly larger rmsds of Cα atoms in the range between 0.588 and 1.316 Å. We therefore conclude that this conformational switch seen in the ISP subunit is correlated with structural changes in the immediate environment of the inhibitor-binding pocket.
Interactions Between ISP-ED and Its Docking Site in Cyt b.
To understand the mechanism that controls the switch of ISP-ED conformation, one needs to analyze the forces that hold it in place at the binding site (the ISP docking crater). The ISP assembles into the bc1 dimer with its N-terminal transmembrane (TM) helix anchored in one monomer, while its C-terminal extrinsic domain interacts with the other (Fig. 1B). A linker region (residues 62–74), whose flexibility is essential for the electron-shuttling function of ISP (9, 10), provides the connection between the TM helix and the ISP-ED. The latter binds to the ISP docking crater on cyt b through the small tip area that surrounds the [2Fe2S] cluster and thus forms a part of the Qo site. The total contact area (CA) between the two subunits in the complex is ≈350 Å2 (Table 1). It is conceivable for energetic reasons that the formation of a large number of strong interactions between the docked ISP and residues in the binding site would be unfavorable for rapid switching of ISP conformational states. Indeed, in the structures where the ISP-ED is in the fixed conformation as seen in the complexes of famoxadone, stigmatellin, JG144, NQNO, or UHDBT, as few as seven hydrogen bonds (H-bonds; see Table 4, which is published as supporting information on the PNAS web site) are formed between ISP and cyt b. Although two of them are permanent and found in the neck region of ISP in all structures, the remaining five H-bonds form only when the ISP is docked with bonding distances ranging from 2.5 to 3.0 Å. Most notably, the ISP employs only backbone oxygen atoms for nonpermanent H-bonds, suggesting that the docking of ISP is guided to precisely position the head group with respect to the substrate bound in the Qo pocket. The tip of the ISP features a smooth, rigid, and largely hydrophobic surface suited to fit into a well defined docking site. As shown in Table 4, those nonpermanent H-bonds disappear when azoxystrobin, MOAS, or myxothiazol is bound. Besides the ISP-cyt b H-bonding interactions, inhibitors like stigmatellin and UHDBT form an additional H-bond with the protonated H161 of ISP (Table 4).
Most residues in cyt b that contribute to the formation of the ISP binding crater are hydrophobic in nature, and only 16 of them have side chains facing the ISP. Of these residues, all but one are located on the CD and EF loops and are highly conserved with a mean identity of >99% (Fig. 4, which is published as supporting information on the PNAS web site), a fact that has been known for a long time but had no clear explanation. Of particular interest are those residues on the cd1 helix, which interact with the ISP-ED.
It had been noted (15) that when Pm inhibitors bind, the ISP-ED remains mobile, as evident from the relatively weak anomalous signal of the [2Fe2S] cluster. In contrast, certain Pf inhibitors (stigmatellin and UHDBT) not only immobilized the ISP but also increased its midpoint potential (Em7) (19, 20). Both observations were explained by the formation of a direct H-bond between an oxygen atom of the inhibitor and the protonated H161: a ligand of the [2Fe2S] cluster (8). Structural studies of a number of bc1 complexes appear to support this notion (18). Surprisingly, the bc1–famoxadone complex showed the ISP-ED in the same fixed state as observed in the stigmatellin complex, yet there was no direct ISP–inhibitor interaction (14). That this observation is not a singularity was shown by the structure solution of bc1 with another oxazolidinedione-based inhibitor, JG144, which once again demonstrated the fixation of the ISP-ED without a direct H-bond between the inhibitor and the ISP (Table 1 and Fig. 5, which is published as supporting information on the PNAS web site). It is therefore clear that direct H-bonding between ISP and the bound inhibitor may contribute to, but not cause, the conformational fixation of the ISP. Instead, the conformational switch of the ISP is an intrinsic property of the bc1 complex.
Bidirectional Motion of the cd1 Helix in Response to Binding of Different Types of Inhibitors and Its Function as an ISP Motion Switch.
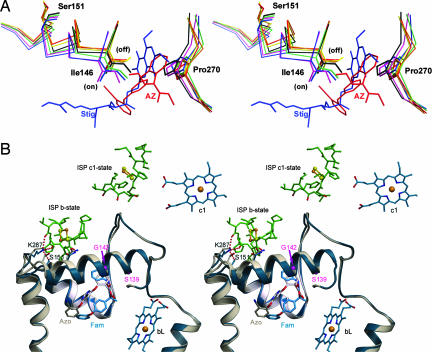
It was previously observed that the volume of the Qo pocket changes upon binding of inhibitors (14, 21). From our analysis, this change is due to a large movement of two structural motifs of cyt b, the PEWY motif and the cd1 helix. Inhibitor binding generally causes Pro-270 of the conserved PEWY motif to back away from the Qo pocket by >1.5 Å in a unidirectional fashion (Fig. 2A), whereas the direction in which the cd1 helix moves clearly depends on the type of the inhibitor. Table 1 lists the relative shifts in the Cα positions of Ile-146. This residue is of special importance because it is both on the cd1 helix and in contact with all bound inhibitors (15). Invariably, the inhibitors that are known to promote the mobile state of the ISP-ED cause a negative shift (up to −0.76 Å), whereas all inhibitors that cause the capture of the ISP at the Qo site lead to a positive displacement (up to +1.7 Å) of Ile-146; here a + or − sign denotes the direction of the shift relative to the native. More precisely, the positional shifts of the cd1 helix are roughly parallel to the membrane surface; + signifies a displacement of the cd1 helix outward away from the Qo pocket leading to an expanded Qo pocket, and − indicates a shift inwards. Although only a limited number of residues on the cd1 helix such as G142, A143, L145, and I146 are in contact with Qo pocket inhibitors (15), the entire helix undergoes an inhibitor-type dependent translation that can be as much as 1 Å (Cα of S151 in Table 1; see also Fig. 2B).
Fig. 2.
Motions observed in the cd1 helix and in the ef loop in the presence of two types of inhibitors. (A) Impact of inhibitors on the conformation of the cd1 switch at the Qo site are illustrated by this stereoscopic pair. Part of the cd1 helix and the P270 of the PEWY motif with and without bound inhibitors are shown by the stick models. Colors are as follows: native (black), stigmatellin (blue), famoxadone (green), UHDBT (magenta), azoxystrobin (red), myxothiazol (orange), and MOAS (yellow). Upon binding of different types of inhibitors, such as stigmatellin (Stig; blue molecule) or azoxystrobin (AZ; red molecule), I146 along with the entire cd1 helix moves either into the on or off position. (B) Stereo pair showing the structural features of the cd1 switch in ribbon form. The structure with famoxadone (Fam) is in blue, and that with azoxystrobin (Azo) is in gray. The inhibitors azoxystrobin and famoxadone are shown as stick models and as labeled. The side chains of K287 and S151 are shown in stick models with the dashed lines indicating H-bonds. H-bonds are also formed between K287 and residues from the ISP-ED when the cd1 switch is at the on position (blue model). In the native structure, or in inhibitor structures in which the ISP-ED is in a free or c1-state (8), no H-bond is formed between K287 and S151 (gray model).
Based on the direction of the positional shift of I146, we define an “on” position of the cd1 helix when class Pf inhibitors are bound and an “off” position when class Pm inhibitors are bound (Fig. 2A). The importance of the cd1 helix in bc1 function is also manifest in its high degree of sequence conservation that reaches 93.1% identity (averaged over 16 residues) as compared with the neighboring cd2 helix (68.2% for 10 residues) or with the whole subunit (74.6% for 379 residues) based on 5,355 aligned sequences (see Table 5, which is published as supporting information on the PNAS web site). Some residues on the cd1 helix are particularly well conserved (Fig. 4); I146, for example, is 98.7% conserved or 99.5% when similar residue types are included. Two residues on the cd1 helix were subject to extensive mutagenesis studies: S155 of R. sphaeroides (Rsbc1) (22) and G158 of R. capsulatus (23) (S139 and G142 in bovine cyt b, respectively; Fig. 2B). In both cases, the enzyme activities strongly depend on the size of the substituting amino acid, suggesting that, among other possible explanations, large amino acid substitutions would sterically hinder the free motion of the cd1 helix.
Origin of ISP Binding Affinity: H-Bonding vs. Shape Complementarity.
To measure ISP binding to cyt b, unbiased omit maps were computed and systematically searched for the ISP-ED by using a high-resolution model (24). The position of ISP-ED that gave the highest linear correlation was used to calculate some of the parameters in Tables 1 and 4, which provide quantitative measures for binding of ISP and the motion of the cd1 helix. These calculations raise the question as to which mechanism induces this dramatic transition and whether it is possible to trace the signal from the source (the binding of inhibitor to the Qo site) to the target residue(s) that carry out the capture of the ISP-ED. We looked at two possible factors: (i) changes in H-bonding patterns, and (ii) adjustments in van der Waals interactions expressed in changes of surface complementarity (SC).
First, the ISP-ED makes five new H-bonds to four cyt b residues when bound to the Qo site. The residue K287 in cyt b, which is doubly H-bonded to ISP and singly H-bonded to S151 (see Fig. 6, which is published as supporting information on the PNAS web site, and Table 4), presents itself as the most likely participant in a mechanism that controls the motion of ISP for the following reasons. Both residues are highly conserved: K287 is nearly 100% conserved, and S151 has 92% identity and 97% similarity (Ser+Thr). Furthermore, S151 undergoes significant positional adjustments in response to the displacements experienced by the cd1 helix (Table 1). It is therefore conceivable that a proper positioning of S151 causes the Lys–Ser (KS) dyad to form. As it happens, K287 is in turn stabilized and able to form two H-bonds with the backbone oxygen atoms of the ISP. This possible mechanism is attractive because it offers a signal amplification effect: One H-bond forms in the KS dyad, and two H-bonds are formed in turn with the ISP-ED.
Assuming that H-bonding represents a dominant force in ISP fixation and that the role of S151 is to either form or break the interaction with K287, a disruption of the KS dyad would be predicted to have a reduced turnover rate because of imprecise positioning of the ISP-ED. Interestingly, in the WT sequence of Rsbc1, a Gly residue (G167) is found at this position (bovine S151; Fig. 4). Furthermore, a G167S mutant of Rsbc1 showed virtually the same activity as the WT enzyme (Table 2), a result that is against the expectation of an enhanced activity for the mutant.
Table 2.
Enzymatic activity of WT and mutants bc1
| bc1 mutants | Bovine equivalent | Photosynthetic growth | Specific activity* |
Km | Note | Source | |
|---|---|---|---|---|---|---|---|
| Purified Rsbc1 | Chromatophore | ||||||
| Bovine bc1 | — | 20.0 | — | N/A | Having a KS dyad in cyt b with two H-bonds to ISP | This work | |
| Wt Rsbc1 | +++++ | 2.5 | 2.2 | 1.56 | Having no KS dyad still with two H-bonds to ISP | This work | |
| KS dyad mutants | |||||||
| G167S | S151 | +++++ | 2.4 | 1.9 | 1.50 | Forming a KS dyad, presumably leading to two H-bonds | This work |
| S322A | ++++† | 0.76 | 0.67 | 2.00 | Single mutation in the ef1 insertion in rsbc1 | This work | |
| Δ(309–326) | ++++† | 0.48 | 0.46 | 1.45 | Deletion mutant of the ef1 insertion in rsbc1 | This work | |
| Δ(309–326)/ G167S | ++++† | 0.46 | 0.34 | 1.39 | Double mutations mimicking bovine bc1 with the KS dyad | This work | |
| SC mutant | |||||||
| W142R,K,T,S | W141 | — | — | — | — | Respiration incapable in yeast, c+c1 reduction kinetics severely affected | Ref. 25 |
| T160S,Y | T144 | — | — | — | — | 70–80% lower activity in rsbc1 | Ref. 26 |
| 292F | I268 | — | — | — | — | Incapable of photosynthetic growth in R.s., affecting QH2 oxidation | Ref. 27 |
| L282F | L281 | — | — | — | — | Respiration incapable in yeast | Ref. 28 |
| L305A,D | L281 | — | — | — | — | Slow QH2 oxidation in rsbc1 | Ref. 27 |
| K329A | K287 | ++++† | 0.9 | 0.8 | 1.40 | Mutant that has lowered QH2 oxidation activity in rsbc1 and changes the shape complementarity | This work and ref. 27 |
*Enzymatic activity is expressed as μmol of cyt c reduced per min/nmol cyt b at room temperature. The concentration of cyt c in assay mixture is 50 μM.
†Delayed by >12 h.
Although the phenotype of the G167S mutant marginalizes the contribution of the KS dyad to the ISP fixation in certain bacteria, we cannot eliminate certain roles this dyad may play in bc1 complexes of the vast majority of other species because these bacterial bc1 may possess some as of yet unknown compensatory structural features. To address this issue, we looked at the crystal structure of the Rsbc1 recently determined in our laboratory at 3.2-Å resolution. The statistics for the x-ray diffraction data and the model quality are given in Table 3; a full account of this work will be given elsewhere (L.E., C.-A.Y., and D.X., unpublished data). Like its mitochondrial counterpart, the Rsbc1 is a symmetric dimer but contains only four subunits per monomer: cyt b, cyt c1, ISP, and subunit IV. The overall structure of the three-subunit Rsbc1 is similar to the corresponding subunits of mitochondrial bc1 but differs in a number of insertions and deletions (see Fig. 7A, which is published as supporting information on the PNAS web site). The cyt b structures of mitochondria and R. sphaeroides are superimposable with an rmsd of 0.99 Å for 369 residues (Fig. 7B). Specific to this work, there is an insertion at the end of the EF loop in the cyt b of Rsbc1 consisting of a short helix (312–322) and connecting coils (Helix ef1 in Fig. 4), which provide a few hydrophobic and electrostatic interactions with the ISP-ED at the tip (Fig. 7C). It is possible that the ef1 helix in bacterial bc1 is a functional equivalent of the KS dyad of mitochondrial bc1 and may serve to control the motion of the ISP subunit. As expected, mutants [S322A and Δ(309–326) in Table 2] designed to remove the ef1 insertion but otherwise keep the enzyme structurally intact significantly reduced the activity of Rsbc1. However, the double-mutant enzyme with the ef1 deletion and the G167S point mutation [Δ(309–326)/G167S] meant to resemble the mitochondrial bc1 exhibited only a very low activity. This result suggests that the KS dyad is a consequence of the ISP fixation. Even more convincingly, in the absence of the KS dyad, the two H-bonds between K329 and carbonyl oxygen atoms of T130 and H131 of ISP are formed as seen in the Rsbc1 structure (see Fig. 8, which is published as supporting information on the PNAS web site).
Next, we looked at the van der Waals interactions in terms of changes in CA and SC of the surfaces between the cyt b and the ISP-ED in various complexes (Table 1). It becomes clear that the changes observed are correlated to the motion of the cd1 helix: As one goes from Pf- to Pm-type inhibitors, there is a significant reduction in the CA and most notably the near-40% loss in SC, ≈0.620 for Pf-type and ≈0.385 for Pm-type inhibitors, respectively. Because the ISP-ED has shown no sign of internal plasticity, and the cd1 helix is known to undergo a shear motion as inhibitors bind to the Qo site, the change in shape complementarity caused by the cd1 motion represents another possible mechanism that could account for the change in ISP dynamics.
Among the 16 residues in the binding crater that are in direct contact with the fixed ISP, a number of mutants were made, including K329A, and their phenotypes were described (Table 2). A common characteristic of these mutants is that although they did not interfere with the binding of substrate, the Qo activity was significantly reduced. In the cases of W141, the cyt c/c1 reduction kinetics was severely affected. These data support the importance of SC in controlling the motion of ISP.
Control Mechanism of the ISP Conformation Switch and Electron Bifurcation.
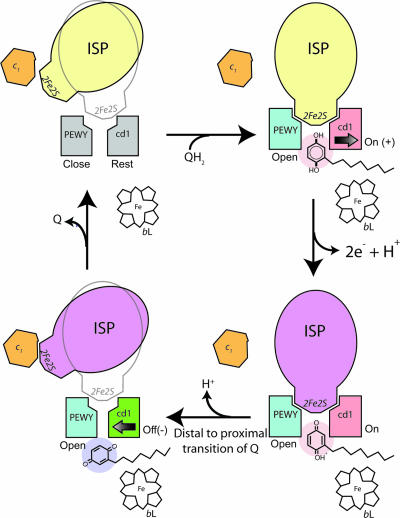
We showed experimentally that the inhibitor binding at the Qo site correlates to the direction of the cd1 helix motion, to the change in shape complementarity between ISP-ED and its binding site on cyt b, and to the eventual motion switch of ISP. The flexibility of the ISP-ED has been proven critical for the function of bc1, but a bc1 complex with a randomly mobile ISP-ED does not function either (11). The key to the bifurcated ET at the Qo site is the control of the ISP mobility. One possible mechanism was to suggest that the ISP-ED oscillates in an optimal frequency range; oscillation mutagenesis in the conserved neck region would change the oscillation frequency and lead to either short-circuit or slow turnover (10). Here, our results suggest that by modulating the shape of the binding surface, cyt b effectively controls its affinity for the ISP-ED, and thereby the motion of the ISP (Fig. 3).
Fig. 3.
Control of the ISP-ED motion switch and the proposed mechanism for electron bifurcation at the Qo pocket. The structural components necessary for the control of ISP conformational switch are illustrated in this cartoon rendition of the Qo pocket. The PEWY motif and cd1 helix in gray represent a native (Rest) configuration. The ISP in yellow and magenta are of oxidized and reduced form, respectively. The PEWY in blue stands for the open configuration with a bound Qo site inhibitor. The cd1 helix in red symbolizes the conformation (On/+) in the presence of a Pf inhibitor occupying the distal site (pink), and the cd1 helix in green shows the conformation (Off/−) when a Pm inhibitor is taking the proximal site (purple). Cyt c1 and heme bL are also shown.
To date, quinol (the substrate), semiquinol (the intermediate), or Q (the product) has not been identified experimentally in the Qo pocket of bc1. For structural analyses, bc1 inhibitors were used instead because many of them are structurally and chemically similar to Q, bind to the Qi site in a similar way as Q, and produce similar spectroscopic effects in EPR spectra of reduced ISP (7, 8, 13–15, 18, 29, 30). Furthermore, mutations in cyt b that render the complex inhibitor resistant frequently cause a penalty in catalytic activity of the enzyme, suggesting that substrate and inhibitors bind in the same pocket (31). The fact that QH2/Q has a low binding affinity at the Qo site prompts the speculation that the same mechanism for ISP motion switch observed during the inhibitor binding could be used by the natural substrate to control the motion of the ISP-ED by undergoing a translocation from the distal to the proximal site in the Qo pocket during catalysis (Figs. 1A and 3). In summary, the following events in the QH2 oxidation may take place. (i) In the absence of any substrate at the Qo site, the ISP-ED is capable of sampling the Qo site frequently. The binding surface on the side of cyt b is in a resting state in a sense that its shape matches that of the ISP-ED sufficiently well to keep it in proximity but not enough to hold it in a fixed conformation. This state is characterized by the ef helix in the “close” position and the cd1 helix in the “rest” position. (ii) When a substrate QH2 molecule enters the catalytic site, it proceeds to the distal part of the Qo pocket. The accommodation of the QH2 molecule widens the ISP-binding crater by moving the cd1 switch toward the “on” position and by pushing the ef helix to the “open” position. The reshaping of the ISP-binding site increases its affinity for the ISP-ED to the point of fixation. As a result, a transient cyt b-QH2-ISP complex forms, which features an H-bond from H161 of ISP to the substrate. (iii) As the first electron is being transferred to the ISP that remains fixed in place, the second electron in the Q radical has to enter the low-potential chain, a process that likely occurs simultaneously with the first ET and possibly via the residue Y131 that undergoes a large conformational change when bound with UHDBT (15). The notion of a concerted ET is supported by the inability to detect an ubisemiquinone radical on the EPR time scale and by the observation of simultaneous reduction of the ISP and bL heme in pre-steady-state kinetic analysis of the ET at the Qo site. We speculate that as the second electron moves toward the bH heme, the proton translocation is completed. (iv) The Q molecule exits via the proximal binding site in the Qo pocket; this event would reverse the pressure on the cd1 helix, retract it to the “off” position, and cause the release of the ISP-ED. The ef helix stays at the open position until the product exits the Qo pocket. This model of ensuring that the ISP-ED stays fixed at the Qo site until both ET and proton transfer are completed readily explains the high fidelity of the bifurcation of the electron pathway.
Materials and Methods
Crystallization of bc1 with Qo Site Inhibitors and Structure Determination.
The preparation of bovine bc1 complex and its crystallization follow the published procedures (7). Crystals were stable in synchrotron radiation when cryocooled to 100 K, allowing data collection for several hours. Crystallographic treatment including data processing, phasing, structure refinement, and Fourier map calculations are given in ref. 15. For details on crystallization, structure determination, and purification of Rsbc1, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Structure Alignment and SC Calculation.
The available native structure of bc1 exhibits a Qo site that is free of substrate QH2 or product Q and thus serves as the reference of the apo enzyme with regard to this catalytic site. Cα atoms of residues 3–379 of the respective cyt b subunits of native- and inhibitor-bound structures were superimposed by using the unit-weighted least-squares techniques (32). The search method to determine orientation and position of the ISP-ED was developed in-house (13). Calculations of CA and shape complementarity are as described by Lawrence and Coleman (16).
Supplementary Material
Acknowledgments
We thank staff members of the SERCAT beamline at Advanced Photon Source, Argonne National Laboratory for their assistance in data collection; Drs. M. M. Gottesman and X. Ji of the National Cancer Institute (NCI) and Dr. W. Yang of the National Institute of Diabetes and Digestive and Kidney Diseases for their valuable comments; and Dr. S. Pamber (Dupont, Wilmington, DE) for providing the JG144. This work was supported in part by the National Institutes of Health (NIH) Intramural Research Program, the NCI, the Center for Cancer Research, and NIH Grant GM 30721 (to C.-A.Y.).
Abbreviations
- ET
electron transfer
- IMS
intermembrane space
- ISP
iron–sulfur protein
- ISP-ED
extrinsic domain of ISP
- MOAS
methoxyacrylate–stilbene
- Q
ubiquinone
- QH2
ubiquinol
- Qo
quinol oxidation
- Qi
Q reduction
- [2Fe2S]
two-iron, two-sulfur cluster of ISP
- TM
transmembrane
- UHDBT
5-undecyl-6-hydroxy-4,7-dioxobenzothiazo
- cyt c
cytochrome c
- QH2
ubiquinol
- bc1
QH2 cyt c oxidoreductase
- Rsbc1
Rhodobacter sphaeroides bc1
- bH
high-potential b heme
- bL
low-potential b heme
- cyt b
cyt b subunit
- cyt c1
cyt c1 subunit
- CA
contact area
- SC
surface complementarity
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. W.A.C. is a guest editor invited by the Editorial Board.
Data deposition: Atomic coordinates for the bovine bc1 structures with bound inhibitors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 1SQB (azoxystrobin), 1SQP (myxothiazol), 1SQQ (MOAS), 1SQX (stigmatellin), 1SQV (UHDBT), 1NU1 (NQNO), 2FYU (JG144), and 2FYN (Rsbc1)].
References
- 1.Trumpower B. L. J. Biol. Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 2.DiMauro S., Schon E. A. N. Engl. J. Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 3.Staniek K., Gille L., Kozlov A. V., Nohl H. Free Radical Res. 2002;36:381–387. doi: 10.1080/10715760290021225. [DOI] [PubMed] [Google Scholar]
- 4.Brasseur G., Saribas A. S., Daldal F. Biochim. Biophys. Acta. 1996;1275:61–69. doi: 10.1016/0005-2728(96)00051-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett D. W., Clough J. M., Godwin J. R., Hall A. A., Hamer M., Parr-Dobrzanski B. Pest Manag. Sci. 2002;58:649–662. doi: 10.1002/ps.520. [DOI] [PubMed] [Google Scholar]
- 6.Crofts A. R., Barquera B., Gennis R. B., Kuras R., Guergova-Kuras M., Berry E. A. Biochemistry. 1999;38:15807–15826. doi: 10.1021/bi990962m. [DOI] [PubMed] [Google Scholar]
- 7.Xia D., Yu C. A., Kim H., Xia J. Z., Kachurin A. M., Zhang L., Yu L., Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Huang L., Shulmeister V. M., Chi Y. I., Kim K. K., Hung L. W., Crofts A. R., Berry E. A., Kim S. H. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 9.Tian H., Yu L., Mather M. W., Yu C. A. J. Biol. Chem. 1998;273:27953–27959. doi: 10.1074/jbc.273.43.27953. [DOI] [PubMed] [Google Scholar]
- 10.Darrouzet E., Valkova-Valchanova M., Moser C. C., Dutton P. L., Daldal F. Proc. Natl. Acad. Sci. USA. 2000;97:4567–4572. doi: 10.1073/pnas.97.9.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurung B., Yu L., Xia D., Yu C. A. J. Biol. Chem. 2005;280:24895–24902. doi: 10.1074/jbc.M503319200. [DOI] [PubMed] [Google Scholar]
- 12.Sadoski R. C., Engstrom G., Tian H., Zhang L., Yu C. A., Yu L., Durham B., Millett F. Biochemistry. 2000;39:4231–4236. doi: 10.1021/bi000003o. [DOI] [PubMed] [Google Scholar]
- 13.Kim H., Xia D., Yu C. A., Xia J. Z., Kachurin A. M., Zhang L., Yu L., Deisenhofer J. Proc. Natl. Acad. Sci. USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao X., Wen X., Yu C., Esser L., Tsao S., Quinn B., Zhang L., Yu L., Xia D. Biochemistry. 2002;41:11692–11702. doi: 10.1021/bi026252p. [DOI] [PubMed] [Google Scholar]
- 15.Esser L., Quinn B., Li Y., Zhang M., Elberry M., Yu L., Yu C. A., Xia D. J. Mol. Biol. 2004;341:281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence M. C., Coleman P. M. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 17.Gao X., Wen X., Esser L., Yu L., Yu C. A., Xia D. Biochemistry. 2003;42:9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 18.Hunte C., Koepke J., Lange C., Rossmanith T., Michel H. Structure (London) 2000;15:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 19.Bowyer J. R., Dutton P. L., Prince R. C., Crofts A. R. Biochim. Biophys. Acta. 1980;592:445–460. doi: 10.1016/0005-2728(80)90091-2. [DOI] [PubMed] [Google Scholar]
- 20.von Jagow G., Ohnishi T. FEBS Lett. 1985;185:311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]
- 21.Crofts A. R., Guergova-Kuras M., Huang L., Kuras R., Zhang Z., Berry E. A. Biochemistry. 1999;38:15791–15806. doi: 10.1021/bi990961u. [DOI] [PubMed] [Google Scholar]
- 22.Tian H., Yu L., Mather M. W., Yu C. A. J. Biol. Chem. 1997;272:23722–23728. doi: 10.1074/jbc.272.38.23722. [DOI] [PubMed] [Google Scholar]
- 23.Atta-Asafo-Adjei E., Daldal F. Proc. Natl. Acad. Sci. USA. 1991;88:492–496. doi: 10.1073/pnas.88.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata S., Saynovits M., Link T. A., Michel H. Structure (London) 1996;4:567–579. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 25.Bruel C., di Rago J., Slonimski P. P., Lemesle-Meunier D. J. Biol. Chem. 1995;270:22321–22328. doi: 10.1074/jbc.270.38.22321. [DOI] [PubMed] [Google Scholar]
- 26.Mather M. W., Yu L., Yu C. A. J. Biol. Chem. 1995;270:28668–28675. doi: 10.1074/jbc.270.48.28668. [DOI] [PubMed] [Google Scholar]
- 27.Crofts A. R., Barquera B., Bechmann G., Guergova M., Salcedo-Hernandez R., Hacker B., Hong S., Gennis R. B. Photosynthesis: From Light to Biosphere. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 493–500. [Google Scholar]
- 28.Lemesle-Meunier D., Brivet-Chevillotte P., di Rago J. P., Slonimski P. P., Bruel C., Tron T., Forget N. J. Biol. Chem. 1993;268:15626–15632. [PubMed] [Google Scholar]
- 29.Palsdottir H., Lojero C. G., Trumpower B. L., Hunte C. J. Biol. Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 30.Samoilova R. I., Kolling D., Uzawa T., Iwasaki T., Crofts A. R., Dikanov S. A. J. Biol. Chem. 2002;277:4605–4608. doi: 10.1074/jbc.C100664200. [DOI] [PubMed] [Google Scholar]
- 31.Robertson D. E., Daldal F., Dutton P. L. Biochemistry. 1990;29:11249–11260. doi: 10.1021/bi00503a014. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. Acta Crystallogr. A. 1978;34:827–828. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.