Abstract
Glucagon-like peptide 2 (GLP-2) is a 33-aa proglucagon-derived peptide produced by intestinal enteroendocrine cells. GLP-2 stimulates intestinal growth and up-regulates villus height in the small intestine, concomitant with increased crypt cell proliferation and decreased enterocyte apoptosis. Moreover, GLP-2 prevents intestinal hypoplasia resulting from total parenteral nutrition. However, the mechanism underlying these actions has remained unclear. Here we report the cloning and characterization of cDNAs encoding rat and human GLP-2 receptors (GLP-2R), a G protein-coupled receptor superfamily member expressed in the gut and closely related to the glucagon and GLP-1 receptors. The human GLP-2R gene maps to chromosome 17p13.3. Cells expressing the GLP-2R responded to GLP-2, but not GLP-1 or related peptides, with increased cAMP production (EC50 = 0.58 nM) and displayed saturable high-affinity radioligand binding (Kd = 0.57 nM), which could be displaced by synthetic rat GLP-2 (Ki = 0.06 nM). GLP-2 analogs that activated GLP-2R signal transduction in vitro displayed intestinotrophic activity in vivo. These results strongly suggest that GLP-2, like glucagon and GLP-1, exerts its actions through a distinct and specific novel receptor expressed in its principal target tissue, the gastrointestinal tract.
Glucagon-like peptides (GLPs) encoded by the proglucagon gene play key roles in glucose homeostasis, gastric emptying, insulin secretion, and appetite regulation (1). Glucagon and GLP-1 exert their effects through distinct G protein-coupled receptors (GPCRs). In contrast, unique receptors for GLP-2, glicentin, and oxyntomodulin have not yet been identified, despite considerable attempts at receptor isolation via classical molecular biology approaches (2). Recent studies have shown that GLP-2 is a potent intestinal growth factor that stimulates crypt cell proliferation and inhibits epithelial apoptosis (3). GLP-2 promotes epithelial proliferation in both small and large intestine; however, the mechanisms utilized by GLP-2 for promotion of intestinal growth remain unclear.
To understand the mechanisms underlying GLP-2 action, we have carried out studies directed at the identification and cloning of the putative GLP-2 receptor. We now have isolated rat and human cDNAs encoding GLP-2-responsive GPCRs, which show highest similarity to receptors for glucagon and GLP-1. The GLP-2R is coupled to activation of adenylate cyclase, and the receptor is expressed selectively in rat hypothalamus and the gastrointestinal tract, known targets of GLP-2 action. These findings establish GLP-2 as a novel hormone that, like glucagon and GLP-1, exerts its actions through a distinct receptor expressed in a highly tissue-restricted manner. The GLP-2R should provide an important target for isolation of small molecules mimicking GLP-2 action and for future studies delineating specific mechanisms underlying GLP-2 action in the gut and central nervous system.
Methods and Materials
Primers, cDNA Libraries, and Cloning Strategy.
Initial attempts at low-stringency hybridization of intestine and brain cDNA libraries using GLP-1R/GlucagonR cDNA sequences were not successful. Two million cDNA clones from rat hypothalamus and rat duodenum/jejunum cDNA libraries subsequently were screened with degenerate oligonucleotides derived from conserved transmembrane II and VII GPCR coding sequences: C4–4 (5′-AACTACATCCACMKGM AYCTGTTYVYGTCBTTCATSCT-3′) (IUB nomenclature) and C9–2R (5′-TCYRNCTGSACCTCMYYRTTGASRAARCAGTA-3′) (for nomenclature, see ref. 4). First-round cDNA plugs (1,057) were isolated in this screen. In a complementary strategy, PCR was conducted on intestinal cDNA templates by using sets of degenerate PCR primers, based on conserved transmembrane amino acid motifs from family B GPCRs or from motifs conserved mainly within the glucagon/glucose-dependent insulinotropic polypeptide (GIP)/GLP-1 receptor subfamily. PCR products were Southern-blotted and probed with 32P-end-labeled C4–4 oligonucleotide. PCRs, amplified from rat neonatal intestine cDNA (Stratagene; catalog no. 936508) were chosen for cloning. These products had been amplified with the degenerate primers M2F (5′-TTTTTCTAGAASRTSATSTACACNGT SGGCTAC-3′) (based on conserved transmembrane domain I sequences) and M7R (5′-TTTTCTCGAGCCARCARCCASSWRTARTTGGC-3′) (based on conserved transmembrane III sequences). PCR products were cloned into pBluescript, screened by filter hybridization with the nested C4-4 oligonucleotide, and sequenced, leading to the identification of a sequence fragment from a novel GPCR family B member, designated WBR, that ultimately proved to be the GLP-2R. Two new GLP-2R-specific PCR primers, P23-F1 (5′-TCTGACAGATATGACATCCATCCAC-3′) and P23-R1 (5′-TCATCTCCCTCTTCTTGGCTCTTAC-3′), were used to screen the 1,057 cDNA plugs obtained by hybridization screening, leading to the identification of three independent clones, two from the duodenum/jejunum library and a third from hypothalamus, which, together, contained a 2,537-bp cDNA insert encoding full-length rat GLP-2R.
To clone the human GLP-2R, the coding region of the rat GLP-2R was used to screen a human hypothalamus cDNA library (CLONTECH; catalog no. 1172a), from which a single positive clone (HHT13) was isolated and sequenced. A full-length insert was ligated into pcDNA3 for expression studies.
Intestinotrophic Activity, cAMP Determination, and Radioligand-Binding Studies.
For cAMP assays, an episomal stable cell line was prepared by lipofection of 293-EBNA (Epstein–Barr nuclear antigen) cells (Invitrogen) with a pREP7-based (Invitrogen) construct containing the Met-42 → Ile-550 ORF of rGLP-2R. Parental 293-EBNA cells, as well as the stable cell line rG2R, expressed receptors for vasoactive intestinal polypeptide and pituitary adenylate cyclase-activating polypeptide. Therefore, the ligand specificity of GLP-2R was tested further in transiently transfected COS cells, which expressed no functional receptors for any of the ligands tested.
For cAMP assays, cells were treated at 80% confluency with GLP-2 peptide analogs at concentrations ranging from 10−12 to 10−5 M for 30 min in medium containing 3-isobutylmethylxanthine. The reaction was terminated with the addition of 95% ethanol and 5 mM EDTA. Aliquots of the ethanol extract were used to determine cAMP levels using an enzyme immunoassay kit (Amersham) as described by the manufacturer. Results were analyzed with graphpad prism software and expressed as pmol cAMP per well. For radioligand-binding assays, cells expressing GLP-2R were harvested and homogenized in 25 mM Hepes (pH 7.4) buffer containing 140 mM NaCl, 0.9 mM MgCl2, 5 mM KCl, 1.8 mM CaCl2, 17 mg/ml Diprotin A, and 100 μM phenanthroline. Homogenates were centrifuged for 10 min at 1,000 × g at 4°C to remove cellular debris. For saturation experiments, membranes containing 25 μg protein were incubated with increasing concentrations of 125I-[Tyr-34]GLP-2 (5–2,000 pM final concentration) in a volume of 0.5 ml for 2 hr at 4°C. Nonspecific binding was determined by the addition of 10 μM of native rat GLP-2 and subtracted from total binding to estimate specific binding to GLP-2R. Parallel experiments confirmed the lack of specific binding when the GLP-2R expression construct was not used. For competition-binding experiments, assays were initiated by the addition of 200 pM (final concentration) of 125I-[Tyr34]GLP-2 with increasing concentrations of competing peptide analogs (10−11 to 10−5 M) for 2 hr as described above. Reactions were terminated by centrifugation at 13,000 × g for 15 min at 4°C. The pellets were washed three times with cold 50 mM Tris buffer, and radioactivity was quantitated in a gamma counter. Results were analyzed by graphpad prism software.
Intestinotrophic activities of various peptide analogs were determined by assessment of small bowel weight as described (5), after 14-day treatments with 2.5 μg of test peptide or PBS (vehicle-treated control) administered twice daily. Activity was defined as follows: active, small bowel wet weight 40–70% greater than in vehicle-treated control animals; partially active, 20–40% greater than controls; inactive, less than 20% greater than controls.
RNAse Protection Assay.
A fragment of GLP-2R cDNA was subcloned into pBluescript (Stratagene) for in vitro transcription with T3 or T7 RNA polymerases. The probe, called F1, spanned nucleotides encoding amino acids Met-1 → Arg-210. RNase protection assay was carried out essentially as described (6), using 50 μg of total RNA from adult rat tissues or 50 μg of yeast tRNA (negative control) or tRNA spiked with a known copy number of sense-strand cRNA (for standard curve construction). Each sample was hybridized with 100,000 cpm of [32P]CTP-labeled antisense cRNA and then digested with RNases T1 (140 units/ml) and A (8 μg/ml) at 30 C for 1 hr. The deproteinized, ethanol-precipitated probe was run on a 5% sequencing gel and analyzed after PhosphorImaging (Molecular Dynamics) with imagequant software. RNA copy number was calculated by interpolation relative to the standard curve after taking the lengths and specific activities of undigested and digested probes into account. A second RNase probe from a different region of the GLP-2R cDNA was used to confirm the quantitative results (data not shown).
RESULTS AND DISCUSSION
GLP-2 and the peptide hormones GLP-1, glucagon, and GIP have closely related amino acid sequences (7). Similarly, the sequences of cloned receptors for the latter three peptides form a cluster within the parathyroid hormone receptor-like GPCRs family B (8–11), suggesting that the GLP-2 receptor might also be found within this subfamily. Initial attempts at GLP-2 receptor cloning by conventional screening of cDNA libraries at low stringency with a combination of GLP-1 and glucagon receptor cDNA probes were not successful. Accordingly, we next used a combined approach of reverse transcription–PCR and hybridization screening followed by expression analysis of candidate cDNAs. As described in Methods and Materials, this strategy resulted in the isolation of a 2,537-bp rat GLP-2R cDNA insert encoding a 550-aa putative family B GPCR (Fig. 1). Hydropathy analysis of the GLP-2R amino acid sequence revealed a typical 7-transmembrane topology plus a hydrophobic amino-terminal signal peptide (data not shown). The GLP-2R gene product belongs to the GLP-1/glucagon/GIP receptor gene subfamily. Conserved features include a possible signal-peptide cleavage site between Val-64 and Thr-65, potential N-glycosylation sites within the amino-terminal putative extracellular domain, and six cysteine residues conserved in the mature GLP-1, GIP, and glucagon receptor amino-terminal domains (Fig. 1). Two putative alternative translation initiation codons, Met-1 and Met-42, were found amino-terminal to the first transmembrane domain in the rat GLP-2R. Functional analysis of Met-1 and Met-42 site-directed mutants showed they were functionally identical (unpublished results), consistent with signal-peptide removal predicted to yield an identical 486-aa mature polypeptide. Overlapping regions of the hypothalamus and duodenum/jejunum cDNA clones encoded homologous polypeptide fragments. Moreover, no evidence was obtained for differential splicing of intestinal GLP-2R RNA from an RNase protection assay employing two nonoverlapping probes derived from GLP-2R cDNA, which together spanned 387 of the 550 GLP-2R codons (data not shown). Additionally, sequencing of the full-length human GLP-2R cDNAs confirmed the identity of hypothalamic and gastrointestinal GLP-2Rs (Fig. 1).
Figure 1.
Multiple alignment of the human (GL2R_HUMAN) and rat (GL2R_RAT) GLP-2R amino acid sequences with human GLP-1, GIP, and glucagon receptor sequences. Known receptor sequences are designated by Swiss-Prot identifiers. Alignment was performed with clustal w 1.60 and rendered with genedoc 2.2. Identities of rat and human GLP-2R sequences is shown in gray, and identities across all five receptor members are indicated by black shading. A predicted signal-peptide cleavage site in human and rat GLP-2R is indicated by an inverted triangle. Six conserved cysteine residues are indicated by arrows. Seven predicted transmembrane domains are shown as solid, black boxes labeled with Roman numerals, and asterisks are shown for spacing every 20 aa. The GenBank accession numbers for the human and rat GLP-2R sequences are AF105367 and AF105368, respectively.
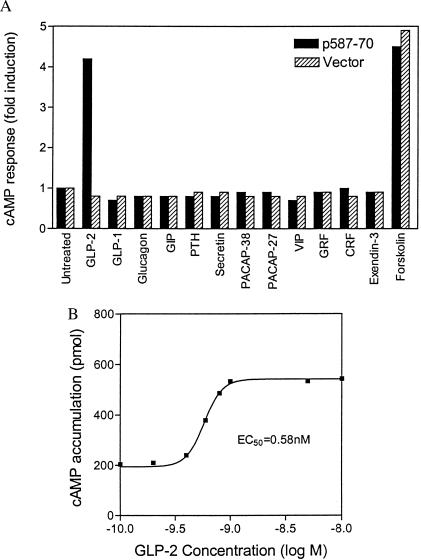
To assess whether the predicted GLP-2R sequence encodes a functional GLP-2 receptor, a GLP-2R expression construct, p587-70, was transiently transfected into COS cells. Because related family B GPCRs show functional coupling to cAMP production mediated by the heterotrimeric G protein Gs, cAMP accumulation was measured after incubation with GLP-2. Treatment of GLP-2R-transfected COS cells with 1 nM GLP-2 resulted in a 4-fold rise in cAMP levels relative to untreated cells, approximately equal to the response seen with 10 μM forskolin (Fig. 2A). Treatment with 10 nM GLP-1, glucagon, GIP, exendin-3, and seven other family B GPCR ligands did not induce cAMP production in p587-70-transfected cells. Control cells transfected with vector DNA alone failed to respond to any of the peptides tested, including GLP-2, though forskolin did induce cAMP production.
Figure 2.
Ligand-selective and concentration-dependent cAMP response to rat GLP-2 in transiently lipofected COS cells. (A) Ligand specificity of cAMP response to GLP-2. cAMP response to peptide analogs of family 2 GPCR ligands was determined in COS cells transiently lipofected with the GLP-2R expression vector p587-70 or the parental pcDNA3 expression vector. GLP-2 concentration was 1 nM; all other peptides were used at 10 nM. A similar profile of peptide specificity was observed with the human GLP-2R (data not shown). (B) Concentration–response curve for cAMP accumulation in response to synthetic rat GLP-2 in rG2R cells stably expressing GLP-2R.
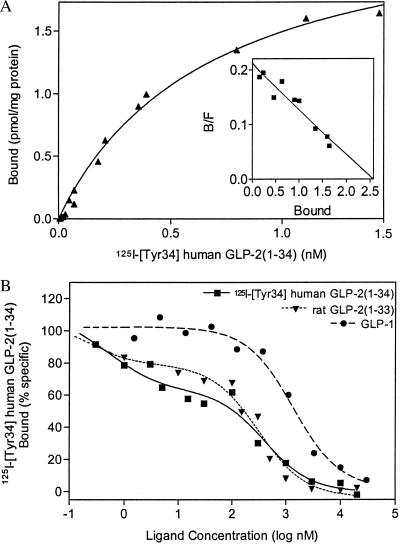
With a stable GLP-2R episomal expression cell line, rG2R, greater than 20-fold cAMP induction routinely was achieved with 1 or 10 nM GLP-2 (data not shown), confirming the potent induction of cAMP accumulation by GLP-2. In contrast, the nontransfected 293-EBNA cells showed no response to GLP-2. The EC50 of the cAMP response to GLP-2 in rG2R cells was 0.58 nM (Fig. 2B). The presence of saturable, specific, ligand-selective GLP-2-binding sites on these cells was shown by using a radioiodinated, C-terminally extended GLP-2 analog, 125I-[Tyr-34]GLP-2 (Fig. 3A). From Scatchard analysis of the saturation isotherms, a Bmax value of 1,839 fmol/mg of protein and a Kd of 0.57 nM were obtained for the radioligand. Mock-transfected cells showed no specific binding to the radioligand (data not shown). Competition-binding studies with rat GLP-2 revealed a high-affinity site (Ki = 0.06 nM) and a low-affinity site (Ki = 259 nM) (Fig. 3B). In contrast, no high-affinity GLP-1 sites were observed in the transfected GLP-2R/rG2R clone. Ki values determined for GLP-1, glucagon, and GIP peptides were 928, 500, and 765 nM, respectively. Although the binding and Scatchard data may reflect, in part, a degree of receptor overexpression, the functional studies and binding data provide firm evidence for a cDNA that encodes a functional high-affinity, ligand-selective GLP-2 receptor.
Figure 3.
Binding of 125I -[Tyr-34]GLP-2 to cell membranes prepared from rG2R cells stably transfected with GLP-2R cDNA. (A) Saturation isotherms of the specific binding of 125I -[Tyr-34]GLP-2 to membranes. Results shown are representative of six independent experiments, each conducted in triplicate. From Scatchard analysis (Inset), maximal-binding Bmax was estimated at 1,839 fmol/mg protein, and a Kd value of 0.57 nM was obtained. (B) Competition binding of 125I-[Tyr-34]GLP-2 binding to cell membranes in the presence of unlabeled peptides. Data are shown for the concentration-dependent inhibition of 125I-[Tyr-34]GLP-2 binding (200 pmol) to GLP-2R by various peptide analogs from at least two independent experiments conducted in triplicate. Inhibitory constants (Ki) were estimated by using graphpad prism.
The human GLP-2R polypeptide showed 81.6% similarity to rat GLP-2R (Fig. 1). Functional expression of the cloned human GLP-2R conferred a functional response to GLP-2 and the appearance of high-affinity ligand-binding sites on 293-EBNA cells, similar to data obtained with the rat GLP-2R (data not shown). Furthermore, the cloned human GLP-2 receptor exhibited the same profile of peptide-binding specificity (Fig. 2A and unpublished data) as the rat receptor. The gene encoding human GLP-2R was identified by screening an arrayed BAC library of human genomic DNA (Genome Systems, St. Louis), confirmed by sequencing, and mapped to chromosome 17p13.3 by fluorescence in situ hybridization analysis (data not shown).
A quantitative ribonuclease protection assay method was used to determine the tissue distribution of rat GLP-2R RNA because no signals were detected on multitissue Northern blots. GLP-2R RNA levels were highest in jejunum, followed by duodenum, ileum, colon, and stomach, whereas expression was undetectable in seven other tissues (Table 1). This expression pattern is clearly concordant with previously reported functional responses to GLP-2 in duodenum (12, 13), jejunum, ileum (5, 12, 14, 15), and colon (12, 16, 17); in contrast, no proliferative or histological changes were seen after GLP-2 treatment in spleen, heart, kidney, lung, or brain (18). Thus, GLP-2R expression is detected in known GLP-2 target tissues. This observation, together with the functional data from experiments with cloned GLP-2R cDNA, suggests that this receptor mediates the intestinotrophic actions of GLP-2.
Table 1.
Quantitative GLP-2R RNA distribution in various rat tissues determined by RNase protection
| Tissue | F1 quantitation, copies per μg total RNA | β-Actin quantitation, copies per μg total RNA | GLP-2R/-actin, ratio |
|---|---|---|---|
| Jejunum | 11,900 | 15,500,000 | 76.8 × 10−5 |
| Duodenum | 9,150 | 85,700,000 | 10.7 × 10−5 |
| Ileum | 7,490 | 51,400,000 | 14.6 × 10−5 |
| Colon | 4,150 | 19,800,000 | 21.0 × 10−5 |
| Stomach | 1,530 | 23,600,000 | 6.48 × 10−5 |
| Brain | <600 | 40,600,000 | <1.48 × 10−5 |
| Heart | <600 | 6,600,000 | <9.09 × 10−5 |
| Kidney | <600 | 14,900,000 | <4.03 × 10−5 |
| Liver | <600 | 16,700,000 | <3.59 × 10−5 |
| Lung | <600 | 38,500,000 | <1.56 × 10−5 |
| Muscle | <600 | 4,600,000 | <13.0 × 10−5 |
| Spleen | <600 | 44,800,000 | <1.34 × 10−5 |
Total RNA (50 μg) from rat tissues or sense-strand cRNA standards was hybridized to radiolabeled antisense cRNA probes prepared in vitro from GLP-2R cDNA (F1) or actin cDNA. After RNase digestion as described in Methods and Materials, protected probe was precipitated, electrophoresed, and quantitated by PhosphorImage analysis relative to the standard curve obtained from sense-strand cRNA. RNA quantitation is expressed as copy number per μg of total RNA. GLP-2R RNA copy number was detectable to a lower limit of 30,000 copies per 50 μg sample, setting the limit of detection shown above for nongastrointestinal tissues.
Pharmacological support for this hypothesis was obtained from parallel in vivo/in vitro studies of GLP-2 analogs containing simple changes in sequence and length (Table 2). Carboxyl-terminal extension analogs bound and activated GLP-2R and retained in vivo activity whereas those with amino-terminal extensions lost both activities. Analogs with blocked amino- or carboxyl-terminal residues displayed diminished in vivo activity and GLP-2R activation. Insertion of a Thr residue between GLP-2 residues 6 and 7 resulted in loss of activity in vivo and in vitro. Truncation of the carboxyl-terminus to a 29-residue peptide (analogous to glucagon) reduced but did not eliminate in vivo or in vitro activities. Interestingly, truncation of one or two amino-terminal residues abolished in vivo activity but did not completely eliminate binding or the GLP-2R cAMP response. Taken together, a clear correspondence was revealed between the structural requirements for GLP-2R binding and activation and the in vivo intestinotrophic activity of GLP-2, providing additional evidence that the GLP-2R isolated here and the intestinotrophic GLP-2 receptor mediating GLP-2 action in vivo are synonymous.
Table 2.
In vitro and in vivo activity profiles of selected peptide analogs of GLP-2
| Peptide |
Ki,
nMa*
|
EC50, nM† | Emac, %†‡ | In vivo activity§ | |
|---|---|---|---|---|---|
| High-affinity | Low-afinity | ||||
| rGLP-2(1-33)¶ | 0.06 ± 0.00 | 259 ± 46 | 1.00 ± 0.2 | 100 ± 0 | Active |
| N-Ac-rGLP-2(1-33) | — | 140 ± 2 | 20.8 ± 0.1 | 80.1 ± 7 | Partially active |
| [Arg-1]rGLP-2(−1-33) | NA | n* | 901 ± 41 | 69.0 ± 2 | Inactive |
| [Arg-34]rGLP-2(1-34) | ND | n§ | 3.1 ± 0.3 | 105 ± 10 | Active |
| [Tyr-34]hGLP-2 | 0.56 ± 0.3 | 255 ± 7 | 1.4 ± 0.1 | 113 ± 5 | Active |
| rGLP-2(2-33) | — | 876 ± 147 | 210 ± 22 | 109 ± 8 | Inactive |
| rGLP-2(3-33) | — | 251 ± 8 | 10.7 ± 0.8 | 115 ± 5 | Inactive |
| rGLP-2(1-29) | 0.30 ± 0.00 | 584 ± 19 | 3.60 ± 0.4 | 102 ± 8 | Partially active |
| [Thr-7 insertion] | — | 977 ± 470 | 1,100 ± 30 | 76 ± 4.0 | Inactive |
| [Gly-2]GLP-2(1-33) | — | 126 ± 5 | 2.0 ± 0.2 | 103 ± 6 | Active |
| hGLP-2(1-33)¶ | 1.7 ± 0.4 | 596 ± 9 | 1.3 ± 0.20 | 99 ± 10.6 | Active |
| Glucagon | — | 500 ± 332 | NA | NA | Inactive |
| GLP-1(7-36)amide | — | 928 ± 1 | NA | NA | Inactive |
| GIP | — | 765 (n = 1) | NA | NA | Inactive |
NA, not active—no detectable binding; ND, not determined.
n = 2, except where indicated.
n = 3.
Relative to 100 nM rGLP-2(1-33), n = 3.
Relative to vehicle-treated control animals, n = 4 or greater. In vivo activity is based on changes in small bowel wet weight after 14-day treatment as described in Methods and Materials.
rGLP-2(1-33) is native rat GLP-2 peptide; hGLP-2(1-33) is native human GLP-2 peptide.
Enteroglucagon synthesis long has been associated with a humoral adaptive response to massive small bowel resection, in which hyperplasia and elongation of jejunal villi are seen (19–22). Proglucagon-derived GLP-2 is detectable in plasma from fasted rats and humans and rises 1.5- to 3.6-fold after feeding (23). Moreover, the intestinotrophic efficacy of GLP-2 has been shown after administration by i.p., i.m., or s.c. routes (14), as well as by coinfusion in parenterally fed rats (12). Thus, it is likely that circulating GLP-2 mediates adaptive changes in the villus-absorptive area in the small intestine. The cloning and characterization of a GLP-2 receptor expressed in the gastrointestinal tract qualifies GLP-2, like GLP-1, glucagon, and GIP, as a bona fide endocrine hormone and should facilitate the discovery of novel pharmacologic agents with similar functional activity. The expression of GLP-2R in hypothalamus also raises the possibility of as yet undescribed role(s) for this intestinotrophic hormone in the central nervous system.
Acknowledgments
We thank P. Khanna, Y.-D. Huang, R. Mathieson, Y.-P. Zhang, and E. Fan for technical assistance; and J. W. Dietrich, R. Zastawny, and D. Lee for advice and critical reading of this manuscript. D.J.D. is a consultant to Allelix Biopharmaceuticals Inc.
ABBREVIATIONS
- EBNA
Epstein–Barr nuclear antigen
- GPCR
G protein-coupled receptor
- GLP-1 and -2
glucagon-like peptide 1 and 2, respectively
- GLP-2R
GLP-2 receptor
- GIP
glucose-dependent insulinotropic polypeptide
Footnotes
References
- 1.Drucker D J. Diabetes. 1998;47:159–169. doi: 10.2337/diab.47.2.159. [DOI] [PubMed] [Google Scholar]
- 2.McGregor G P, Goke R, Goke B. Exp Clin Endocrinol Diabetes. 1998;106:25–28. doi: 10.1055/s-0029-1211945. [DOI] [PubMed] [Google Scholar]
- 3.Tsai C-H, Hill M, Asa S L, Brubaker P L, Drucker D J. Am J Physiol. 1997;273:E77–E84. doi: 10.1152/ajpendo.1997.273.1.E77. [DOI] [PubMed] [Google Scholar]
- 4.Nomenclature Committee of the International Union of Biochemistry (NC-IUB) J Biol Chem. 1986;261:13–17. [PubMed] [Google Scholar]
- 5.Drucker D J, Ehrlich P, Asa S L, Brubaker P L. Proc Natl Acad Sci USA. 1996;93:7911–7916. doi: 10.1073/pnas.93.15.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 7.Fehmann H-C, Goke R, Goke B. Endocrine Rev. 1995;16:390–410. doi: 10.1210/edrv-16-3-390. [DOI] [PubMed] [Google Scholar]
- 8.Thorens B. Proc Natl Acad Sci USA. 1992;89:8641–8645. doi: 10.1073/pnas.89.18.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacNeil D J, Occi J L, Hey P J, Strader C D, Graziano M P. Biochem Biophys Res Commun. 1994;198:328–334. doi: 10.1006/bbrc.1994.1046. [DOI] [PubMed] [Google Scholar]
- 10.Jelinek L J, Lok S, Rosenberg G B, Smith R A, Grant F J, Biggs S, Bensch P A, Kuijper J L, Sheppard P O, Sprecher C A, et al. Science. 1993;259:1614–1616. doi: 10.1126/science.8384375. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda K, Inagaki N, Yamada Y, Kubota A, Seino S, Seino Y. Biochem Biophys Res Commun. 1994;205:1556–1562. doi: 10.1006/bbrc.1994.2844. [DOI] [PubMed] [Google Scholar]
- 12.Chance W T, Foley-Nelson T, Thomas I, Balasubramaniam A. Am J Physiol. 1997;273:G559–G563. doi: 10.1152/ajpgi.1997.273.2.G559. [DOI] [PubMed] [Google Scholar]
- 13.Brubaker P L, Izzo A, Hill M, Drucker D J. Am J Physiol. 1997;272:E1050–E1058. doi: 10.1152/ajpendo.1997.272.6.E1050. [DOI] [PubMed] [Google Scholar]
- 14.Tsai C-H, Hill M, Drucker D J. Am J Physiol. 1997;272:G662–G668. doi: 10.1152/ajpgi.1997.272.3.G662. [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman C I. Am J Physiol. 1997;273:R1965–R1971. doi: 10.1152/ajpregu.1997.273.6.R1965. [DOI] [PubMed] [Google Scholar]
- 16.Drucker D J, Deforest L, Brubaker P L. Am J Physiol. 1997;273:G1252–G1262. doi: 10.1152/ajpgi.1997.273.6.G1252. [DOI] [PubMed] [Google Scholar]
- 17.Litvak D A, Hellmich M R, Evers B M, Banker N A, Townsend C M., Jr J Gastrointest Surg. 1998;2:146–150. doi: 10.1016/s1091-255x(98)80005-x. [DOI] [PubMed] [Google Scholar]
- 18.Tsai C-H, Hill M, Asa S L, Brubaker P L, Drucker D J. Am J Physiol. 1997;273:E77–E84. doi: 10.1152/ajpendo.1997.273.1.E77. [DOI] [PubMed] [Google Scholar]
- 19.Buchan A M J, Griffiths C J, Morris J F, Polak J M. Gastroenterology. 1985;88:8–12. doi: 10.1016/s0016-5085(85)80125-6. [DOI] [PubMed] [Google Scholar]
- 20.Rountree D B, Ulshen M H, Selub S, Fuller C R, Bloom S R, Ghatei M A, Lund P K. Gastroenterology. 1992;103:462–468. doi: 10.1016/0016-5085(92)90835-m. [DOI] [PubMed] [Google Scholar]
- 21.Gornacz G E, Ghatei M A, Al-Mukthar M Y T. Dig Dis Sci. 1984;29:1041–1049. doi: 10.1007/BF01311257. [DOI] [PubMed] [Google Scholar]
- 22.Taylor R G, Fuller P J. Baillieres Clin Endocrinol Metab. 1994;8:165–183. doi: 10.1016/s0950-351x(05)80230-7. [DOI] [PubMed] [Google Scholar]
- 23.Brubaker P L, Crivici A, Izzo A, Ehrlich P, Tsai C-H, Drucker D J. Endocrinology. 1997;138:4837–4843. doi: 10.1210/endo.138.11.5482. [DOI] [PubMed] [Google Scholar]