Abstract
Heterosis, or hybrid vigor, is a major genetic force that contributes to world food production. The genetic basis of heterosis is not clear, and the importance of loci with overdominant (ODO) effects is debated. One problem has been the use of whole-genome segregating populations, where interactions often mask the effects of individual loci. To assess the contribution of ODO to heterosis in the absence of epistasis, we carried out quantitative genetic and phenotypic analyses on a population of tomato (Solanum lycopersicum) introgression lines (ILs), which carry single marker-defined chromosome segments from the distantly related wild species Solanum pennellii. The ILs revealed 841 quantitative trait loci (QTL) for 35 diverse traits measured in the field on homozygous and heterozygous plants. ILs showing greater reproductive fitness were characterized by the prevalence of ODO QTL, which were virtually absent for the nonreproductive traits. ODO can result from true ODO due to allelic interactions of a single gene or from pseudoODO that involves linked loci with dominant alleles in repulsion. The fact that we detected dominant and recessive QTL for all phenotypic categories but ODO only for the reproductive traits indicates that pseudoODO due to random linkage is unlikely to explain heterosis in the ILs. Thus, we favor the true ODO model involving a single functional Mendelian locus. We propose that the alliance of ODO QTL with higher reproductive fitness was selected for in evolution and was domesticated by man to improve yields of crop plants.
Keywords: heterosis, hybrid vigor, domestication, reproductive barriers, breeding
Heterosis, or hybrid vigor, is defined as the ability of hybrids to outperform their best inbred parent with respect to growth, yield, and other quantitative traits (1). The agricultural benefits of cross-hybridization were realized by Shull (2) and East (3) who described the phenomenon of inbreeding depression and hybrid vigor in maize. These pioneers self-pollinated maize varieties for several generations and produced essentially homozygous lines, which showed considerable inbreeding depression and were then used to produce heterotic hybrids. These experiments contributed to the view that heterosis and inbreeding depression are two aspects of the same phenomenon, where the hybrid vigor lost during inbreeding is recovered by out-crossing (4). Indeed, the classic quantitative genetic explanation of heterosis revolved around a few main mechanisms (for a detailed review, see ref. 5), the first of which is the dominance hypothesis. The dominance hypothesis proposed that different sets of deleterious recessive alleles in each inbred parent are masked by dominant alleles in the F1 hybrid (6). More recent research on plant-genome evolution has had implications for the role of dominance in heterosis. The sequencing of the same genomic regions in different maize inbreds revealed that gene content differed as a consequence of transposon-induced gene shuffling (7). These findings highlight the dynamic structure of plant genomes where allelic variation between inbreds is often associated with deletions/additions of genes. From the standpoint of heterosis, these studies support the dominance theory, because inbred genomes containing different gene sets would contribute to a wider repertoire of ORFs upon hybridization. However, a review of the literature (5) suggests that there is more to heterosis than complementation of mutated alleles. For example, since the 1930s, there has been a continuous improvement in yield of maize hybrids mainly through intense breeding efforts to increase performance of parental inbreds by eliminating deleterious recessive alleles (8). However, the magnitude of heterosis has remained more or less unchanged. If heterosis was caused by complementation, then heterotic effects should have declined as the inbreds improved.
The second explanation of heterosis is the true overdominance (ODO) model, wherein a heterozygous combination of alleles at a single locus is superior to either homozygous line because of combined allelic expression (9, 10). ODO can also be explained by pseudoODO, where a chance linkage of loci with alleles having dominant advantageous effects are in repulsion–linkage phase (11, 12). Epistasis due to gene-by-gene interactions is a third genetic model to explain heterosis (13). For example, the architecture of a heterotic quantitative genetic locus (QTL) for growth in yeast identified three tightly linked epistatic genes that are neither necessary nor sufficient in isolation to confer the phenotype (14). Although it is highly likely that there are different genetic mechanisms that explain heterosis for specific traits in different organisms, it is noteworthy that, even for the same traits, in the same organism, different mechanisms were proposed. For instance, QTL studies in rice have mapped heterotic effects into specific genomic regions and characterized their modes of inheritance. Xiao et al. (15) found, based on marker-assisted QTL analysis, that dominance is the major genetic basis of heterosis for yield components; however, contradictory results in another study using a similar QTL approach in another rice population reported ODO and epistasis (16, 17) as the main constituents of heterosis. The above observations point to the complexity of the evidence regarding the genetic basis of heterosis.
Here, we demonstrate that Solanum lycopersicum (tomato) is an ideal system to explore the genetic and molecular bases of heterosis, because inbreeding depression is low, and highly polymorphic crosses can be generated that facilitate analysis of best-parent heterosis (18). All of the species in the tomato clade are diploid (2n = 24) hermaphrodites and share a syntenic genome (http://sgn.cornell.edu). Hybrids between different cultivated inbreds of S. lycopersicum show heterosis for yield-related traits and dominate the fresh and processed tomato market. Tomato interspecific hybrids show strong heterotic vigor for vegetative growth, but seed set in early generations is generally low because of sterility, which is partially controlled by deleterious recessive alleles (19). In this study, we explored the genetic basis of heterotic traits using 76 introgression lines (ILs), each carrying a single chromosome segment derived from the wild species Solanum pennellii replacing the homologous segment of the cultivated tomato (S. lycopersicum) variety M82 (12). The ILs are nearly isogenic and thereby devoid of whole-genome epistatic interactions, which makes them uniquely suited to identify QTL with ODO effects (18). Furthermore, because the ILs are interspecific, both genetic and phenotypic diversity are maximized, thereby enabling comprehensive measurements on a wide range of diverse traits throughout plant development. Along these same lines, the fact that the ILs carry segments of wild-species DNA and were not selected for by breeders allowed us to explore whether heterotic phenotypes extend beyond agriculture and have a role in the evolution of plant fitness. Here, we demonstrate that specific genomic regions with ODO effects are overwhelmingly associated with yield-related phenotypes that define traits for increased reproductive fitness. We argue that this association reflects the ancestral origin for the increased yields in agricultural hybrids.
Results
Traits Showing Heterosis in the Cultivated Tomato.
To establish a reference framework for the role of agricultural heterosis in the cultivated tomato (S. lycopersicum) we crossed six tomato inbreds to our core inbred M82. The parents and hybrids were measured for a range of morphological and yield-associated traits. To assess the mean level of genome-wide heterosis, we pooled the six inbred varieties, the six hybrids, and M82 and compared these three groups across all traits. Best-parent heterosis was defined in cases where the hybrid outperformed the pool of inbred varieties and M82. As is shown in the ANOVA (Table 2, which is published as supporting information on the PNAS web site), four traits exhibited heterosis: seed number per plant, fruit number, total yield, and biomass, all yield-related traits. Other phenotypes, such as fruit weight, plant weight, brix, seed morphology, and other traits, showed no heterotic effects. These data show that heterosis is prevalent in the cultivated tomato and is associated with yield-related phenotypes.
Genome-Wide Heterosis in the Interspecific ILs.
Many studies investigated the relation between DNA-marker variation of parents and heterosis as expressed by their hybrid. For example, in maize, genetic distance between parents was strongly correlated to heterosis of their hybrid (20). It should be noted, however, that other studies reported low or no correlation at all between marker distance and yield, and it is well accepted that this association depends, at least in part, on the type of crop and the germ plasm being used. To maximize the genetic and phenotypic diversity, we focused our measurements on an interspecific IL population where the entire S. pennellii genome is represented in a cultivated background. The ILs and hybrids between IL and M82 (ILHs) and the common control (M82) were grown in the field in a completely randomized experiment (1 m2 per plant), and the mean phenotypic values for 35 traits representing the many attributes of plant development were compared (Fig. 3, which is published as supporting information on the PNAS web site). This phenomics approach allowed us to ask whether heterosis is generally prevalent, or is it specific to certain yield-related phenotypes. Among the cultivated hybrids, seed number per plant and fruit number exhibited the greatest heterotic effects. Because, in nature, seed production is a component of reproductive fitness, we defined the number of seeds produced per plant (the product of the number of seeds per fruit and the number of fruits per plant) as the phenotype that represents fitness.
To determine which traits were associated with reproduction, we performed a correlation analysis between seed number per plant and all other phenotypes (Tables 3 and 4, which are published as supporting information on the PNAS web site). Ten traits correlated strongly with seed number per plant (r > 0.55) and were thus considered reproductive. Five traits were considered as intermediates (0.31 > r > 0.26). The other 19 traits were independent of seed number per plant (r < 0.21) and classified as nonreproductive. In the cultivated tomato hybrid experiment (Table 2), we observed a bias of heterotic effects for reproductive yield-related phenotypes. To see whether this same trend occurred in the ILs, we pooled the data sets for homozygous lines, heterozygous lines, and M82 and compared among them (Table 3). The mean seed number produced per plant over all ILHs was significantly higher than the mean of all ILs and higher (although not significantly) than M82. For four additional reproductive traits, seed weight per plant, seed number per fruit, seed weight per fruit, and seed number per fruit unit, the ILH means were significantly higher than those of the ILs and M82. This genome-wide heterotic trend was not observed for any of the nonreproductive traits except fruit color, where the ILHs were, on average, less red than their parents. This whole-genome analysis on the IL population shows a strong bias for genome-wide heterosis, primarily in fitness-related phenotypes.
QTL Mapping.
To test whether heterotic phenotypes for reproductive fitness are associated with a particular mode-of-inheritance mechanisms at specific loci, the phenotypic database was subjected to a QTL analysis in which each IL and ILH was compared with the common M82 control. If one of the lines had a significant effect on a trait, the introgression was considered as harboring a QTL. Using a permissive significance level for the statistical analysis (see Materials and Methods), we resolved 841 QTL (Tables 5 and 6, which are published as supporting information on the PNAS web site, sorted by ILs and by traits, respectively), where the average number of QTL was significantly higher for the reproductive traits than for the nonreproductive. Of 841 QTL, 382 were in the reproductive group (≈35 QTL per trait), 118 for the five intermediate traits (≈24 QTL per trait), and 341 for the nonreproductive group (≈18 QTL per trait). This difference is likely because of the wider range of phenotypic extremes that are possible for reproductive traits, and, therefore, a larger number of QTL that can influence them. In agreement with this assumption, the genotypic coefficient of variation, which represents the influence of the genotype on the phenotype, is significantly greater for the reproductive traits compared with the nonreproductive (Table 3).
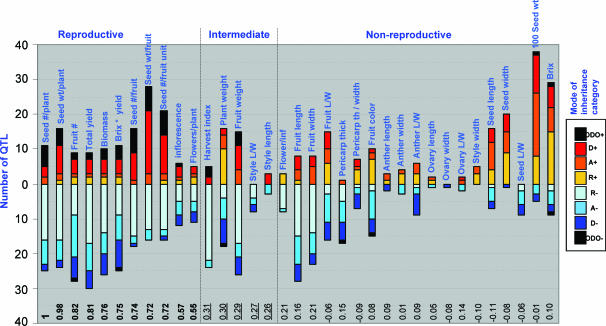
The inclusion of the ILHs enabled us to classify each wild-species QTL into the following mode-of-inheritance categories (Fig. 4 A and B, which is published as supporting information on the PNAS web site): recessive, additive, dominant, or ODO. This classification reflects a mode of inheritance in which the S. pennellii allele is compared with the M82 allele. For example, a QTL classified as dominant means that both the IL (homozygous for the S. pennellii allele) and the ILH (heterozygous) were very similar to each other and significantly different from M82. A recessive QTL means that only the IL is significantly different from M82, whereas the ILH is similar to M82. Additivity reflects a situation in which the ILH is in between its parents, which are significantly different from each other, and ODO is inferred where the ILH is significantly higher or lower than both its parents. The main question we addressed is whether there are differences in QTL distribution among the three groups of traits: reproductive, intermediate, and nonreproductive. Remarkably, the distribution of QTL numbers in each mode-of-inheritance category shows that the group of reproductive traits had many more increasing ODO QTL accompanied by more decreasing recessive QTL than in the nonreproductive group (Fig. 1); this trend was also evident in the analysis under more strict conditions, where a total of 507 QTL were identified (Table 7, which is published as supporting information on the PNAS web site). Similar results were obtained in a previous experiment of the same IL population, although fewer phenotypes were measured (Fig. 5, which is published as supporting information on the PNAS web site). In this analysis and the ones that follow, the intermediate group of traits was midway between the reproductive and the nonreproductive groups in the distribution trend of the mode-of-inheritance patterns of the QTL (Table 7). To focus on the differences between the reproductive and nonreproductive groups, we excluded the intermediate traits from subsequent analyses.
Fig. 1.
Distribution of QTL mode of inheritance for tomato traits. Each vertical bar represents the number of QTL for a specific trait, colored according to mode-of-inheritance categories; R, recessive; A, additive; D, dominant; and ODO. The bars above the 0 line represent the number of increasing QTL, whereas the negative bars represent the number of decreasing QTL relative to M82. The correlation value for each trait to seed no. per plant (Seed #/plant) is indicated at the bottom. Those that are >0.5 are in boldface type, and those between 0.25 and 0.5 are underlined. The reproductive and intermediate bars are organized in decreasing order according to their correlation coefficient to seed no. per plant. The remaining nonreproductive traits are ordered according to their type.
Pleiotropism-Corrected QTL.
To capture as many phenotypic attributes of the plant life as possible, we measured a wide diversity of traits, some of which were highly correlated (Table 4). Consequently, we observed that certain QTL affected a number of phenotypes. For example, the phenotype of seed weight per plant is highly related to seed number per plant: There were 38 QTL for seed number per plant and 42 for seed weight per plant, and 34 of these QTL comapped to the same ILs, indicating pleiotropism for both traits. These traits were also highly correlated in the entire IL population (Table 4). Therefore, a global summation of QTL mode of inheritance over all traits generated a biased view of QTL distribution, because the same QTL would be counted multiple times for different highly correlated traits. To correct for pleiotropism and more realistically compare the distributions of inheritance modes between the reproductive and nonreproductive traits, we counted the number of QTL that affected highly correlated traits (where r > 0.5 or r < −0.5) with the same mode of inheritance as a single IL QTL (see Materials and Methods). This pleiotropism-corrected approach is more appropriate for comparing between the groups of traits as opposed to when we assumed no pleiotropism, because it eliminates redundant QTL (see Table 8, which is published as supporting information on the PNAS web site, for list of all pleiotropism-corrected QTL). In this way, we generated a set of nonredundant QTL by assuming that a single IL affecting several correlated traits with the same mode of inheritance was due to a single pleiotropic locus. Indeed, for the reproductive traits, there were a total of 382 QTL with only 136 nonredundant ones (35%), whereas, for the nonreproductive traits, the nonredundancy was higher (85%). This finding is not surprising, because the nonreproductive traits reflected a more diverse view of the phenotypic repertoire of the plant that included fruit, flower, and seed morphology characteristics. On the other hand, the reproductive traits are primarily yield related and, therefore, more integrated with each other.
Even with this pleiotropism-corrected approach, the mode-of-inheritance mechanisms among the nonredundant QTL showed some significant differences between the groups (Table 1). Similar to what we first observed when assuming no pleiotropism, the reproductive traits showed an excess of increasing ODO QTL. For instance, the reproductive group had 20 increasing ODO QTL of 136 (14.7%), whereas the nonreproductive had only 2 of 290 (0.7%). Moreover, whereas the nonreproductive traits showed a similar number of increasing and decreasing QTL in each mode-of-inheritance category, there was a bias toward recessive decreasing and ODO/dominant increasing QTL in the reproductive group. These significant differences were consistent with an analysis that was conducted under complete pleiotropism (Table 7). In this approach, if a QTL affected several traits from the same group with the same mode of inheritance, it was considered as a single QTL irrespective of the correlations among the traits. Even under these conservative assumptions and by using a more stringent statistical analysis, ODO of the reproductive traits was still 10 times higher than the nonreproductive traits (Table 7).
Table 1.
Qualitative mode-of-inheritance distribution
| Mode of inheritance | Reproductive, n (%) | Nonreproductive, n (%) | P χ2 |
|---|---|---|---|
| +ODO | 20 (14.7) | 2 (0.7) | 1.1E-09 |
| −ODO | 1 (0.7) | 3 (1) | 0.8 |
| +Dominant | 27 (19.9) | 39 (13.4) | 0.09 |
| −Dominant | 12 (8.8) | 33 (11.4) | 0.4 |
| +Additive | 10 (7.4) | 58 (20) | 0.0009 |
| −Additive | 24 (17.6) | 59 (20.3) | 0.5 |
| +Recessive | 6 (4.4) | 56 (19.3) | 4.80E-05 |
| −Recessive | 36 (26.5) | 40 (13.8) | 0.0014 |
| Total | 136 (100) | 290 (100) |
Qualitative distribution of mode of inheritance showing the numbers of QTL that were classified in each category in the reproductive and nonreproductive groups (the number in parentheses represents the percent of this category of all QTL in that group). The signs that precede the mode of inheritance indicate whether it is an increasing (+) or decreasing (−) QTL relative to M82. A statistical comparison between the reproductive and nonreproductive groups was conducted in each mode of inheritance by using a χ2 test (with one degree of freedom; by classifying the QTL into those that belong to this mode of inheritance and those that do not).
Mode of Inheritance on a Quantitative Scale.
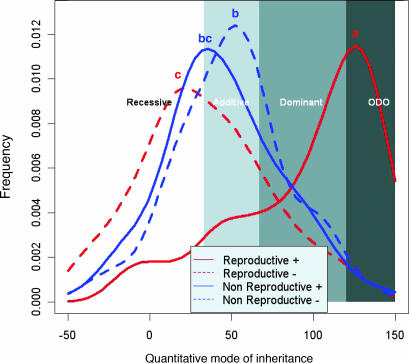
Thus far, the QTL mode-of-inheritance classification was analyzed on a qualitative scale and included the recessive, additive, dominant, and ODO categories. However, our large data set of quantitative measurements allowed us to attach a quantitative index to the mode of inheritance of each QTL. Toward this end, we repeated the analysis using a quantitative index, which more accurately represents each QTL’s mode of inheritance (see Materials and Methods). This index is equivalent to the more conventional measure of d/| a | (4), where a is half the distance between IL and M82, and d is the distance between ILH and the midparent value. Our index presents few modifications to the d/| a | measure, such that it is not dramatically affected by very small a values and that it does not differentiate between increasing and decreasing QTL (see Materials and Methods). Fig. 2 shows the frequency distribution of the mode-of-inheritance index for QTL in the reproductive and nonreproductive groups. The “reproductive+” curve (which corresponds to the distribution of quantitative mode of inheritance of QTL for increasing reproductive traits) has a peak in the ODO domain, indicating that many of the QTL fall within this mode of inheritance. In contrast, most of the QTL for the nonreproductive group and for the decreasing reproductive phenotypes resided in the recessive–additive domain. Thus, Fig. 2 presents the difference in mode of inheritance among the four distribution curves in a quantitative manner, where the overrepresentation of increasing reproductive QTL begins at the dominant part of the spectrum and increases substantially toward the ODO domain. This trend was also consistent when we repeated the analysis with stricter statistical thresholds and under three pleiotropism assumptions (Fig. 6, which is published as supporting information on the PNAS web site) and is also consistent with the qualitative analysis (Fig. 1 and Table 1). The overrepresentation of ODO QTL for reproductive traits in the ILs is in agreement with the genome-wide heterosis for yield-related traits observed in the cultivated tomato intraspecific inbred crosses (Table 2). According to our interpretation, heterosis is partitioned, in part, into small genomic regions that convey advantage in the heterozygous state (ODO QTL), and, together, they contribute to the genome-wide effect.
Fig. 2.
Distribution of quantitative mode-of-inheritance pleiotropism-corrected QTL for reproductive and nonreproductive groups. The results obtained here were based on the permissive QTL analysis and by using a correlation threshold of | r | > 0.5 (pleiotropism-corrected). The x axis represents the region (−50 to +150) of the quantitative index of a QTL, and the y axis is the frequency of these indices. Each of the four curves shows the distribution of either increasing or decreasing QTL for traits in the reproductive or nonreproductive groups. The range is divided into four partitions (recessive, additive, dominant, and ODO), representing the classification of the qualitative analysis. The qualitative mode of inheritance was compared among the four curves by using a multiple-comparison Tukey test (P < 0.01). Significant differences are indicated by the letters “a,” “b,” etc., above each curve; curves that do not share the same letter are significantly different.
Discussion
The complex nature of heterosis makes it difficult to partition it into individual components, particularly in F2, back-cross, and recombinant inbred populations because of the epistatic interactions among the many segregating loci throughout the genome (16, 17). These limitations have contributed to the difficulty in defining specific heterotic phenotypes and the individual genomic loci that control them. We addressed both these issues by taking a phenomic approach to heterosis by measuring a large number of traits on ILs carrying single chromosome segments from the wild species S. pennellii. Using this nearly isogenic population, we partitioned heterosis into defined genomic regions, eliminating a major part of the genome-wide epistasis. A total of 841 QTL for 35 fitness-related and -unrelated phenotypes were mapped, revealing a fundamental difference in mode of inheritance between increasing QTL for reproductive traits as opposed to reducing reproductive QTL and QTL for nonreproductive traits. The wealth of QTL data indicates that dominant complementation is, in part, involved in heterosis as is shown by the many increasing dominant and decreasing recessive QTL, the hallmarks of the complementation model (15). Because genome-wide epistasis is largely eliminated in our population, the other major component of heterosis according to this study is ODO as is indicated by our detection of many ODO QTL.
To explain the genetic basis of ODO, the one-gene hypothesis was proposed, in which two alleles in the heterozygous state are superior (true ODO). Alternatively, ODO phenotypes can result from the linkage of two or more loci where the dominant alleles are in repulsion (pseudoODO). Comparing between the reproductive and nonreproductive groups reveals that both groups contain many decreasing recessive and increasing dominant QTL (Table 1), which, if linked may result in pseudoODO. If pseudoODO was the major component of ODO QTL, and assuming that increasing dominant and decreasing recessive QTL are randomly distributed throughout the genome, then we would have expected to observe a similar expression of ODO QTL for both reproductive and nonreproductive groups. The fact that we detected only two ODO QTL for nonreproductive traits suggests that pseudoODO (resulting from randomly linked genes) is not a major player in the observed best-parent heterosis of the IL hybrids. Instead, single loci exhibiting ODO may be responsible, and, although further work is needed to resolve this issue, there are examples of single-gene heterosis (21).
ODO in Sexually Reproducing Organisms.
The association of ODO with reproductive QTL in the tomato interspecific ILs is supported by a similar trend observed in intraspecific segregating populations of rice and maize, albeit very few traits were measured (16, 17). For example, Lu et al. (22) studied four traits in two back-cross populations of maize. Based on the absolute value of d/a, 24 of 28 QTL (86%) for grain yield showed ODO. For three other nonreproductive traits, the ODO level was much lower: 2 ODO QTL of 16 (12.5%) for grain moisture; 1 ODO QTL of 8 (12.5%) for stalk lodging, and 4 ODO QTL of 11 (36%) for plant height. Interestingly, the association of ODO with reproductive traits was also observed in animals. In a QTL-mapping study in an F2 cross between two mouse strains, 17 body composition and growth traits were measured and 139 QTL mapped (23, 24); 9% were ODO (d/a > 1 or d/a < −1), whereas for five reproductive traits (25), 47% of the 15 QTL were ODO. In a study of chromosomal substitution lines in Drosophila, 12 quantitative characters were investigated (26). All of the measures of reproductive-fitness components, such as egg hatchability, showed strong directional dominance and ODO, whereas other nonreproductive traits, such as body size and bristle number, showed little or no dominance.
The observation that diverse taxonomic groups associate reproductive fitness with ODO leads us to propose that this is a general characteristic of sexually reproducing organisms. Importantly, the role of heterosis in the elevation of agricultural yields (e.g., fruit number, seed number), which represents reproductive traits, is consistent with the ODO for reproductive fitness found in our study involving introgressions from a wild species. Another major difference between the reproductive and nonreproductive groups is that the reproductive traits have much higher phenotypic variation, which is reflected by the larger number of QTL per trait and higher average phenotypic effect of a single QTL (Table 3). It is not clear whether this is the reason for the differences in QTL mode of inheritance between the reproductive and nonreproductive groups, but the aforementioned studies in Drosophila and mice, in which reproductive traits have lower phenotypic variation than other traits, show essentially the same differences in mode of inheritance as we observed in our study. This finding indicates that there is a more fundamental reason for mode-of-inheritance difference between the groups of traits than variation alone.
ODO in Evolution.
The fact that ODO QTL were primarily associated with reproductive traits and not for nonreproductive indicates that this association may have a role in evolution. Reproductive traits represent major components of fitness and are under strong directional evolutionary selection for higher values (4). Nonreproductive traits represent phenotypes that experience stabilizing natural selection, where individuals with extreme phenotypic values are less fit than those at or near the mean. It is possible that there was selection for ODO QTL for reproductive fitness during evolution irrespective of whether such QTL comprise single genes or multiple linked genes residing as a complex Mendelian locus. The classic example for such complex loci is the self-incompatibility (SI) system in plants, which comprises tightly linked “male”- and “female”-specific genes that are inherited as a single locus because of lack of recombination in their intervening sequences. The SI system increases diversity by promoting gene migration between more distantly related genotypes (27). It is important to note that similar SI multigene-locus structures evolved independently in a number of plant lineages that recruited different linked male/female-specific genes. The evolutionary role of the SI system is similar to that of the ODO QTL, because both promote heterozygosity in populations; hence, it is temping to speculate that the genetic organization of some of the ODO QTL is similar to that of the SI system.
Natural populations, particularly small and isolated ones, often suffer from the loss of genetic variation in a manner that reduces their persistence (inbreeding depression) (28). Reproductive isolation limits gene flow between species, races, and even varieties and reduces the potential benefit of the infusion of novel genetic variation into recipient gene pools. Theoretical studies supported by experimental observations indicate that the trajectory of population fitness can be significantly improved as a result of the migration of new alleles from divergent genotypes; this infusion of exotic alleles was recently termed “genetic rescue” (29). A limiting factor for such gene flow is that the more divergent the migrating genotype, the higher the probability is that its descendents will be sterile or partially sterile as a result of karyotype differences (30) and the breakdown of coadapted gene complexes (i.e., sets of independent genes that were selected in concert to have favorable epistatic interactions that improve fitness). Nevertheless, despite the barriers to gene flow, a recent survey has indicated that at least 25% of plant and 10% of animal species in nature are involved in hybridization and potential introgressions with other species (31).
Thus, two opposing forces mold the genetic architecture and evolutionary fate of populations and genes. On one hand, reproductive-isolation mechanisms retard gene flow between divergent populations (30) and maintain coadapted gene complexes that improve fitness. On the other hand, metapopulation theory and practice, focusing on networks of small local populations connected by migration corridors, has established a central role for immigrant alleles in survival (32). An attractive hypothesis is that ODO QTL, which have been selected to improve fitness through heterozygous advantage, can bring balance between the opposing isolation and diversity needs of gene pools. These ancestral ODO QTL were domesticated by men to improve yield of agricultural organisms via heterosis.
Materials and Methods
Plant Material and Phenotyping.
The IL population, composed of 152 lines in the genetic background of M82 (http://sgn.cornell.edu), was planted in the field at the Akko Experimental Station (Akko, Israel) in the year 2004 in a completely randomized design. Twelve seedlings of each homozygous IL and heterozygous ILH (IL*M82) were transplanted as well as 70 seedlings of M82. Eight ILs were not included in the analysis because of poor germination (ILH2–4, IL3–1, ILH3–4, ILH6–2, ILH6–2-2, ILH6–4, ILH7–2, and ILH9–3-2). The experiment was planted at a density of 1 plant per m2 and harvested when 80–100% of the tomatoes were red (12). Five weeks before harvest date, three replicates for each genotype were destructively measured for the following traits: inflorescence number per plant, flowers per inflorescence, and flowers per plant. The rest of the traits were measured on nine replicates at harvest time as detailed in Fig. 3
Statistical Analyses and QTL Mapping.
Statistical analyses were performed by using the R statistical language (www.R-project.org). The broad sense heritability (h2), which is the σ2G/σ2G+E, was calculated for each trait where the genotype was defined as a factor with random effect, and the genetic variation was calculated as the percent from the total variation (genetic + environmental).
For the QTL mapping, each IL or ILH was compared (by t test) to M82 as well as to each other. If either of them was significantly different from the reference genotype M82, the introgression was considered as harboring a QTL. Because the number of replications for the ILs and ILHs was similar, but the number of replications of M82 was higher, two significance levels were used for the QTL identification. The first level was used to compare IL and ILH to M82 (a1) and the second to compare between IL and ILH (a2). The QTL analysis was performed by using two thresholds: (i) a1 = 1%, a2 = 5% (permissive analysis) and (ii) a more stringent threshold of a1 = 0.1%, a2 = 1%. We used the above permissive statistical stringency because our objective was to compare trends in the abundance of QTL characteristics between the reproductive and nonreproductive traits. Results of the stringent analysis are presented in Table 7 and Fig. 6.
Qualitative Mode-of-Inheritance Classification.
The phenotypic effect of a QTL was considered to be the effect of the significant line (IL or ILH) and was presented as percent of M82 (positive values for increasing QTL in which the introgression was higher than M82 and negative values for decreasing ones). If both the IL and the ILH had a significant effect in the same direction, the higher value was considered the QTL phenotypic effect. If both the IL and ILH were significant but in opposite directions relative to M82, the introgression was considered as harboring two QTL: One is increasing, and the other is decreasing. The mode of inheritance of a QTL was determined according to a decision tree (Fig. 4). In cases in which the IL was significantly different from M82 and the ILH phenotype was in between the IL and M82, there were three possibilities: (i) If the ILH was significantly different from the IL but not from M82, it was considered recessive; (ii) If the ILH differed from both parents or did not differ from either of them, it was considered additive; and (iii) If the ILH differed from M82 but not from the IL, the QTL was assigned as dominant. The last possibility is where the ILH was significantly higher or lower than both its parents, in which case it was considered ODO.
Quantitative Mode-of-Inheritance Classification.
In addition to the qualitative classification, each QTL was scored for a quantitative index of mode of inheritance. This scoring was done as follows: If the IL was significantly different from M82, then the phenotypic interval between the IL and M82 was regarded to be of 100 mode-of-inheritance units, where the M82 is located on the 0 coordinate, and the IL is located on the 100 coordinate. Then, the quantitative index of the QTL was determined according to the relative location of the ILH on this interval (Fig. 6).
Pleiotropism-Corrected QTL.
The study here is based on the comparison of QTL mode-of-inheritance between reproductive and nonreproductive traits. However, counting the number of QTL for each group of traits may be biased, because a single QTL can be counted twice for two highly correlated traits (pleiotropism). To correct for this possibility, a genomic region that affected several correlated traits (| r | > 0.5) with the same mode of inheritance was considered as a single QTL, as described in Fig. 6.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Israel Science Foundation. Z.L. was supported by a Long-Term Fellowship from the Human Frontier Science Program.
Abbreviations
- IL
introgression line
- ILH
hybrid between IL and M82
- ODO
overdominant or overdominance
- QTL
quantitative trait locus or loci.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 12957.
References
- 1.East E. M., Jones D. F. Inbreeding and Outbreeding. Philadelphia: Lippincott; 1919. [Google Scholar]
- 2.East E. M. Connecticut Agr. Exp. Sta. Rpt. 1908:419–428. [Google Scholar]
- 3.Shull G. H. Am. Breed. Assn. 1908;4:296–301. [Google Scholar]
- 4.Falconer D. S., Macay T. F. C. Quantitative Genetics. New York: Longman; 1996. [Google Scholar]
- 5.Birchler J. A., Auger D. L., Riddle N. C. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce A. B. Science. 1910;32:627–628. doi: 10.1126/science.32.827.627-a. [DOI] [PubMed] [Google Scholar]
- 7.Fu H., Dooner H. K. Proc. Natl. Acad. Sci. USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duvick D. N. Nat. Rev. Genet. 2001;2:69–74. doi: 10.1038/35047587. [DOI] [PubMed] [Google Scholar]
- 9.Crow J. F. Genetics. 1948;33:477–487. doi: 10.1093/genetics/33.5.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.East E. M. Genetics. 1936;21:375–397. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow J. F. Heterosis. Ames, IA: Iowa State College Press; 1952. pp. 282–297. [Google Scholar]
- 12.Eshed Y., Zamir D. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu S. B., Li J. X., Xu C. G., Tan Y. F., Gao Y. J., Li X. H., Zhang Q., Maroof M. A. Proc. Natl. Acad. Sci. USA. 1997;94:9226–9231. doi: 10.1073/pnas.94.17.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., McCusker J. H., Davis R. W. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 15.Xiao J., Li J., Yuan L., Tanksley S. D. Genetics. 1995;140:745–754. doi: 10.1093/genetics/140.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z. K., Luo L. J., Mei H. W., Wang D. L., Shu Q. Y., Tabien R., Zhong D. B., Ying C. S., Stansel J. W., Khush G. S., Paterson A. H. Genetics. 2001;158:1737–1753. doi: 10.1093/genetics/158.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo L. J., Li Z. K., Mei H. W., Shu Q. Y., Tabien R., Zhong D. B., Ying C. S., Stansel J. W., Khush G. S., Paterson A. H. Genetics. 2001;158:1755–1771. doi: 10.1093/genetics/158.4.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gur A., Zamir D. PLoS Biol. 2004;2:e245. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyle L. C., Graham E. B. Genetics. 2005;169:355–373. doi: 10.1534/genetics.104.029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi S. P., Bhave S. G., Chowdari K. V., Apte G. S., Dhonukshe B. L., Lalitha K., Ranjekar P. K., Gupta V. S. Biochem. Genet. 2001;39:179–200. doi: 10.1023/a:1010293325482. [DOI] [PubMed] [Google Scholar]
- 21.Redei G. P. Z. Verebungsl. 1962;93:164–170. [Google Scholar]
- 22.Lu H., Romero-Severson J., Bernardo R. Theor. Appl. Genet. 2003;107:494–502. doi: 10.1007/s00122-003-1271-7. [DOI] [PubMed] [Google Scholar]
- 23.Rocha J. L., Eisen E. J., Van Vleck L. D., Pomp D. Mamm. Genome. 2004;15:100–113. doi: 10.1007/s00335-003-2308-6. [DOI] [PubMed] [Google Scholar]
- 24.Rocha J. L., Eisen E. J., Van Vleck L. D., Pomp D. Mamm. Genome. 2004;15:83–99. doi: 10.1007/s00335-003-2312-x. [DOI] [PubMed] [Google Scholar]
- 25.Rocha J. L., Eisen E. J., Siewerdt F., Van Vleck L. D., Pomp D. Mamm. Genome. 2004;15:878–886. doi: 10.1007/s00335-004-2364-6. [DOI] [PubMed] [Google Scholar]
- 26.Kearsey M. J., Kojima K. I. Genetics. 1967;56:23–37. doi: 10.1093/genetics/56.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takayama S., Isogai A. Annu. Rev. Plant Biol. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 28.Dudash M. R., Carr D. E. Nature. 1998;393:682–684. [Google Scholar]
- 29.Tallmon D. A., Luikart G., Waples R. S. Trends Ecol. Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Burke J. M., Arnold M. L. Annu. Rev. Genet. 2001;35:31–52. doi: 10.1146/annurev.genet.35.102401.085719. [DOI] [PubMed] [Google Scholar]
- 31.Mallet J. Trends Ecol. Evol. 2004;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Molofsky J., Ferdy J. B. Proc. Natl. Acad. Sci. USA. 2005;102:3726–3731. doi: 10.1073/pnas.0404576102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.