Abstract
Estrogen receptor beta (ER-β) regulates diverse physiological functions in the human body. Current studies are confined to ER-β1, and the functional roles of isoforms 2, 4, and 5 remain unclear. Full-length ER-β4 and -β5 isoforms were obtained from a prostate cell line, and they exhibit differential expression in a wide variety of human tissues/cell lines. Through molecular modeling, we established that only ER-β1 has a full-length helix 11 and a helix 12 that assumes an agonist-directed position. In ER-β2, the shortened C terminus results in a disoriented helix 12 and marked shrinkage in the coactivator binding cleft. ER-β4 and -β5 completely lack helix 12. We further demonstrated that ER-β1 is the only fully functional isoform, whereas ER-β2, -β4, and -β5 do not form homodimers and have no innate activities of their own. However, the isoforms can heterodimerize with ER-β1 and enhance its transactivation in a ligand-dependent manner. ER-β1 tends to form heterodimers with other isoforms under the stimulation of estrogens but not phytoestrogens. Collectively, these data support the premise that (i) ER-β1 is the obligatory partner of an ER-β dimer, whereas the other isoforms function as variable dimer partners with enhancer activity, and (ii) a single functional helix 12 in a dimer is sufficient for gene transactivation. Thus, ER-β behaves like a noncanonical type-I receptor, and its action may depend on differential amounts of ER-β1 homo- and heterodimers formed upon stimulation by a specific ligand. Our findings have provided previously unrecognized directions for studying ER-β signaling and design of ER-β-based therapies.
Keywords: estrogen responsive element, heterodimerization, molecular modeling, steroid receptor coactivator, type I nuclear receptor
Since the discovery of estrogen receptor (ER)-β in 1996 (1), research efforts have been focused on defining its biological functions, which today remain poorly understood. ER-β is expressed in a variety of normal and malignant tissues, some of which express ER-α (2). The proposed functions of ER-β include antiproliferative action, regulation of apoptosis, control of antioxidant gene expression, and modulation of immune responses, anxiety-related behavior, and the risk of heart failure (2). In many respects, ER-β acts as an indispensable hormone receptor for maintenance of the proper functions of vital organs (2). Recent studies (3) have revealed the expression of this receptor to be under epigenetic regulation of a CpG island in exon 0N of its promoter region.
ER-β belongs to the nuclear receptor superfamily (4). It is classified as a type I nuclear receptor, because it resides in cytosol and undergoes nuclear translocation after ligand binding. It forms homodimers and binds to cognate-responsive elements consisting of a palindromic repeat. These characteristics are in contrast to type II nuclear receptors. For example, retinoic acid X receptor (RXR) only heterodimerizes with other nuclear receptors, resides in the nucleus in the absence of ligand, and binds to a cis-acting element composed of a direct repeat. Upon agonist binding, both types of nuclear receptors orientate their helix 12 to create a hydrophobic pocket for interaction with the nuclear receptor (NR) box (LXXLL motif) of coactivators such as the steroid receptor coactivator (SRC) protein family (5). Failure to form such a pocket results in recruitment of corepressors such as nuclear receptor corepressor (N-CoR) and silencing mediator for retinoic acid and thyroid hormone receptor (SMRT) (6). The bound coactivators facilitate acetylation of the receptor complex and its recruitment of additional essential comediators such as p300/cAMP response element binding protein binding protein (CBP) to form the transcriptional complex for initiation of transcription (7). Both ER-α and ER-β are categorized as type I receptors and are believed to possess properties common to all type I nuclear receptors. However, their known biological functions are different and often inverse. The underlying mechanisms of their functional differences remain largely unknown.
So far, published data on human ER-β function/signaling have been derived from studies on ER-β1, the originally cloned sequence (8). Sequencing data had suggested that multiple ER-β isoforms exist as a result of alternative splicing of the last coding exon (exon 8) (9). Limited studies, however, have revealed that ER-β2 (ER-βcx) functions as a dominant negative of ER-α (10), whereas ER-β3 expression appears to be restricted to the testis (9). Additionally, two truncated transcripts containing only part of the common exon 7 and different exon 8 sequences have been identified and named ER-β4 and ER-β5 (9). A recent report confirmed their existence as full-length transcripts (11). Presently the functional properties of the individual ER-β isoforms are still unclear, except for their differential expression in various tissues and cell lines (9). Without a comprehensive understanding of the functional uniqueness and similarity of these isoforms, the biological significance of ER-β signaling remains incomplete.
In this study, we successfully amplified the full-length transcripts of ER-β4 and ER-β5. The functional aspects of the newly cloned isoforms were examined, including size and ligand binding of the expressed proteins, dimerization, transactivation, and coactivator binding abilities, in parallel with in silico analyses of their molecular structural characteristics. We report here that ER-β1 is the only full-function isoform and that ER-β2, -β4, and -β5 do not have innate activities in their homodimeric forms but can heterodimerize with ER-β1 and enhance ER-β1-induced transactivation in a ligand-dependent manner. From this finding arises a concept in modeling the action of type I nuclear receptor that may be generally applicable to its members.
Results and Discussion
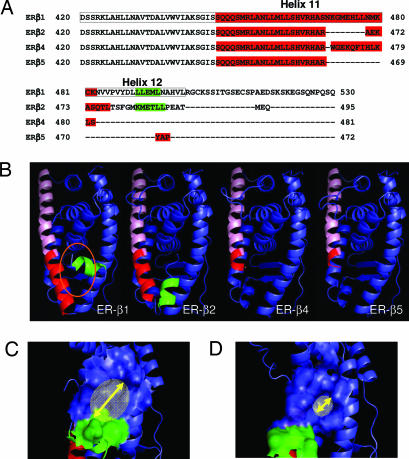
As the first step toward characterization of the ER-β isoforms, we cloned the full-length transcript of ER-β isoform 4 and 5 [National Center for Biotechnology Information (NCBI) accession nos. DQ838582 and DQ838583, respectively] from human prostate cancer cells (PC3). Acquisition of nucleotide sequences of these transcripts allowed us to precisely determine the amino acid composition of ER-β4 and -β5. Alignment of the amino acid sequences of the isoforms confirmed the previous prediction (9) that ER-β isoforms differ only in the last exon, which encodes an isoform-specific C terminus tail of reduced length (Fig. 1A). On the basis of these analyses, ER-β2, -β4 and, -β5 should have an AF-2 domain different from that of ER-β1 (9) and may not have a complete helix 12 (Fig. 1A).
Fig. 1.
Protein sequence analysis and molecular modeling of ER-β isoforms. (A) Protein sequence alignment of the C-terminal regions of ER-β1, -β2, -β4, and -β5 by using the Clustalw alignment program. The ligand binding domain of ER-β1 is boxed. The protein sequence forming helix 11 in each isoform is shown in red, whereas the protein sequence participating in helix 12 is in green. (B) Molecular models of ER-β isoforms. The common helix 11 region of each isoform is labeled in pink, whereas the isoform-specific region of helix 11 is highlighted in dark red. The orientation of helix 12 (green) in ER-β2 is different from that of ER-β1, which has “tight” configuration in the ER-β1 binding pocket (orange oval). (C and D) Molecular models of ER-β1 (C) and ER-β2 (D) show the coactivator binding pocket created by electrostatic potential of the amino acid residues in helices 3–5 and 12. The size of the coactivator binding pocket in ER-β2, which is indicated by a yellow arrow, was determined, by using PyMol software, to be smaller than that of ER-β1.
In silico modeling of ER-β isoform monomers with reference to the published ligand-induced ER-β1 ligand-binding domain (LBD) crystal [Protein Data Bank (PDB) ID codes 1x76 and 1x78] revealed that all isoforms share identical helices 3–10 (Fig. 1B). In comparison with ER-β1, each of the other isoforms has a shortened helix 11 with variable lengths (Fig. 1B). Helix 12 of ER-β1 was shown to pack against helices 3, 5/6, and 11, forming the reported agonist-directed position (12, 13), with the ligand-binding cavity, buried below helices 5 and 6. Only the C terminus of ER-β2 formed an α-helix, generating a helix 12 (Fig. 1B) with similar hydrophobicity (LLEML vs. KMETLL, Fig. 1A) as ER-β1. However, helix 12 of ER-β2 assumed a totally different orientation, likely due to the restriction imposed by a shorter loop between helices 11 and 12. The unusual orientation of helix 12 in the ER-β2 may hinder ligand access to the binding pocket. Furthermore, a comparison of the reconstructed molecular surface of the coregulator binding pocket (Fig. 1 C and D), which is formed by residues from helices 3, 4, 5, and 12 (14) between ER-β1 and -β2, showed a significant reduction in the size of the ER-β2 pocket. The molecular model of ER-β4 and -β5 are markedly different from that of ER-β1 and -β2. Use of an early stop codon in exon 8 of ER-β4 and -β5 transcripts resulted in a significantly truncated helix 11 and the complete absence of helix 12. These distinctive structural features of the ER-β isoforms portend variations in their functional properties.
To illuminate the functional properties of the individual ER-β isoforms, we overexpressed full-length human ER-β1, -β2, -β4, and -β5 transcripts in a human embryonic kidney cell line (HEK293) and Saccharomyces cerevisiae; both lack endogenous ER-β protein expression as determined by Western blot analysis and luciferase reporter assays (Figs. 2A and 3). All recombinant proteins matched the sizes expected from their primary sequences (Fig. 2A). The molecular weight of ER-β1, -β2, -β4, and -β5 was determined as 59, 56, 54 and 53 kDa, respectively (Fig. 2A). An in vitro estrogen receptor-binding assay (Fig. 2B) was used to assess the binding affinities of the yeast recombinant proteins. ER-β1 bound to estradiol (E2) with high affinities (Kd = 0.48 nM) comparable with values reported in refs. 15 and 16. ER-β2 exhibited no binding, as reported in ref. 16, whereas ER-β4 and -β5 bound to E2 with moderate affinities (9.87 and 23.45 nM, respectively) (Fig. 2B). These findings are in agreement with our molecular modeling data, which demonstrated a relatively open configuration of the ligand-binding pocket in ER-β4 and -β5 and an apparent restriction of ligand access to the pocket of ER-β2 because of its helix 12 positioning. Luciferase reporter assays indicated that, unlike ER-β1, ER-β isoforms (2, 4, and 5) could not transactivate a vitellogenin estrogen responsive element (ERE)-driven promoter in the presence of E2 or the steroid receptor coactivator-1 (SRC-1), nor in a combination of both (Fig. 2C). Yeast two-hybrid (Y2H) experiments further demonstrated that, except for ER-β1 (Fig. 2D), none of the isoforms (β2, β4, and β5) formed homodimers (data not shown) in the presence and absence of E2. Collectively, our data indicated that the isoforms 2, 4, and 5 have no intrinsic transactivation activity for two plausible reasons: (i) they cannot form homodimers because of weak/no ligand binding, and (ii) their inability to recruit coregulators as a result of either the lack of helix 12 in ER-β4 and -β5 or the shrinkage of the coregulator binding cleft in ER-β2. Yet another important conclusion drawn from these data is that ER-β1 is the only full-function receptor in the ER-β family currently identified, because it has displayed high E2 binding affinity, homodimer formation, SRC-1 interaction, and ligand-dependent transactivation by way of vitellogenin ERE. This discovery invalidates the general assumption that all ER-β isoforms are independently functional (17) and raises questions regarding the biological roles of ER-β isoforms (β2, β4, and β5).
Fig. 2.
Characterization of ER-β isoforms. (A) Western blot analysis of ER-β isoforms overexpressed in HEK293 cells and yeast. N-terminal-specific polyclonal ER-β antibody (H150 from Santa Cruz Biotechnology), which would recognize all ER-β isoforms, was used in this study. An equal amount (50 μg) of protein was loaded to each lane. Mock-transfected cells or an untransformed yeast strain were set up as a control experiment (CTL). Samples expressing ER-β1, -β2, -β4, and -β5 were labeled as 1, 2, 4, and 5, respectively. The size of the ER-β isoforms was consistent with the predicted molecular size, ranging from 53 to 59 kDa. Coexpression of ER-β isoforms with ER-β1 was also performed in both cell line and yeast. Lanes 1 and 2, 1 and 4, and 1 and 5 represent the samples overexpressing ER-β1 and -β2, ER-β1 and -β4, and ER-β1 and -β5, respectively. (B) Tabulated results of in vitro estrogen receptor binding assay. Four hundred micrograms of total yeast lysate expressing ER-β isoforms was applied to each binding reaction as described in Materials and Methods. Binding data were calculated and analyzed with GraphPad Prism 4.0 software to determine the Bmax and Kd of each isoform. (C) Effects of SRC-1 on the transactivation activities of ER-β isoforms. SRC-1 expression vector was transfected into HEK293 cells carrying different ER-β isoform expression vectors with the reporter plasmid. Transactivation assays were performed as described in Materials and Methods in the presence or absence of 1 nM E2. Three independent experiments were performed and averaged. The standard deviation was calculated. (D) Dimerization of ER-β isoforms by Y2H experiment. E2 at two different concentrations (1 nM to 1 μM) was incubated overnight with different yeast strains. A Beta-Glo assay was performed to quantify the reporter (β-gal) activity. The higher the reporter activity, the stronger the interaction between two of the same ER-β (homodimer) or different (heterodimer) isoforms. Four types of homodimers (β1 + β1, β2 + β2, β4 + β4, and β5 + β5) and three kinds of heterodimers (β1 + β2, β1 + β4, and β1 + β5) were subjected to Y2H analyses. The background value was subtracted during data analyses. Experiments were performed in triplicate, and the standard deviation was calculated. All results were summarized in this figure, except for β2, β4, and β5 homodimers, in which activities were undetectable.
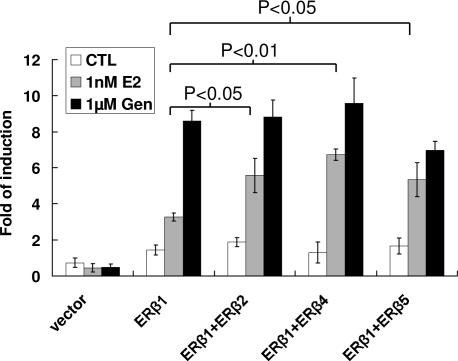
Fig. 3.
Transactivation activities of ER-β homo- and heterodimer in HEK293 cells. Single and double transfection of ER-β isoform expression vectors were performed to study the effects of ER-β homo- and heterodimers, respectively, on the transactivation of ERE reporter. E2, BPA, EE2, DES, genistein, and apigenin at different concentrations (100 pM to 1 μM) were incubated at 24 h after transfection. A control experiment was set up with vehicle. After 24 h of incubation, a Bright-Glo assay was used to measure the luciferase activity. Three independent experiments were performed and averaged. The standard deviation was calculated. A Student t test was applied to determine the significance between ER-β isoform coexpression with ER-β1 and ER-β1 alone with the same treatment. ∗∗, P < 0.01.
We next examined whether the ER-β isoforms would heterodimerize with ER-β1 and modulate its function. Although ER-β2, -β4, and -β5 do not form homodimers in Y2H, they readily heterodimerize with ER-β1 in the presence of physiological concentrations of E2 in a dose-dependent manner (Fig. 2D). The propensity to dimerize follows the descending order of β1–β4 ≥ β1–β5 > β1–β1 > β1–β2 (Fig. 2D). The predilection of ER-β1–β2 formation was lower than that of ER-β1 homodimerization in the Y2H experiment. In contrast, heterodimerization between β1 and β4 or β5 was ≈50 times stronger than that of β1–β1 homodimerization, suggesting that β4 and β5 are the preferred dimerizing partners for β1 under physiological E2 concentrations. It should be noted, however, that although E2 did not trigger homodimerization of ER-β isoforms (β2, β4, and β5; data not shown), it strongly induced heterodimer formation between ER-β isoforms and ER-β1. Because ER-β2, -β4, and -β5 do not have a functional helix 12 or high-affinity ligand binding, these data imply that E2 binding to ER-β1, which induces an agonistic positioning of its helix 12, is sufficient to trigger heterodimer formation. A reporter assay in HEK293 cells showed that coexpression of ER-β2, -β4, or -β5 significantly enhanced transactivation activities induced by ER-β1 at physiological concentrations of E2 (Fig. 3), indicating that ER-β isoforms (ER-β2, -β4, and -β5) exert dominant positive effects on estrogen-induced ER-β1 transactivation. A comparison between Y2H and mammalian cell data suggests that ER-β2 is less efficient in forming heterodimers with ER-β1 in the Y2H system but exhibits equal potency as ER-β4/5 in enhancing the overall ER-β1-mediated transactivation. This discrepancy implies that additional mammalian coregulatory proteins, absent in yeast, may be involved in facilitating dimerization between ER-β1 and some of its isoforms.
We chose, in addition to natural estrogen, two synthetic estrogens, ethynylestradiol (EE2) and diethylstilbestrol (DES); an environmental estrogen, bisphenol A (BPA); and two phytoestrogens, genistein and apigenin, which humans encounter on a daily basis, to evaluate their abilities in promoting ER-β dimer formation in HEK293 reporter assays. In general, the transactivating activities of E2 either with ER-β1 alone or with heterodimer pairs is an order of magnitude higher than that observed for EE2, BPA, and DES (Fig. 4). All estrogens tested were shown to activate ER-β1 homodimers in a dose-dependent manner, except for genistein, which maximally transactivated the vitellogenin ERE-reporter even at the lowest concentration (100 nM) (Fig. 4). Consistent with Y2H results, none of the ligands examined induced homodimer formation for ER-β2, -β4, and -β5 (data not shown). Like the natural estrogen (E2), xenoestrogens (EE2, DES, and BPA) were more potent (>2-fold) in activating reporter transcription in cells coexpressing ER-β1 and an ER-β isoform than those expressing only ER-β1. However, the phytoestrogens (genistein and apigenin) did not exhibit this property. These results indicated that, whereas E2 and xenoestrogens preferentially promote ER-β heterodimerization, phytoestrogens do not. Estrogenicity of a ligand is originally defined on the basis of its stimulation of immature uterine weight gain in rodents (18). In this regard, EE2 used in contraceptives and DES used as a reproductive therapeutic are viewed as potent estrogens, whereas BPA, a contaminant leaching from polycarbonate plastic, is considered a weak estrogen. However, they all behave like E2, exhibiting a high potency in promoting heterodimer formation. In contrast, the phytoestrogens (genistein and apigenin) appear to favor only ER-β1 homodimerization. Real-time PCR assessment of pS2 gene transcription was used to further demonstrate the difference between estrogens and phytoestrogens in activating ER-β isoform interaction (Fig. 4). Consistent with transactivation studies, E2, although not genistein, induced higher levels of pS2 expression in the cells coexpressing ER-β1 and an isoform compared with its effects on cells expressing only ER-β1. Collectively, these data provide a dimension for assessing estrogenicity of various ligands, such as xenoestrogens, environmental estrogens, and phytoestrogens, based on their relative abilities to promote ER-β homo- and heterodimer formation.
Fig. 4.
Real-time PCR analysis of pS2 gene expression. HEK293 cells were transfected with ER-β1, a combination of ER-β1 with ER-β2/4/5, or an empty vector as a control. After 24 h of transfection, the cells were treated with 1 nM E2 or 1 μM genistein for another 24 h. The change in pS2 expression level was monitored by real-time PCR analysis. Three independent experiments were performed and averaged. The standard deviation was calculated. A Student t test was applied to determine the significance between ER-β isoforms coexpression with ER-β1 and ER-β1 alone with the same treatment.
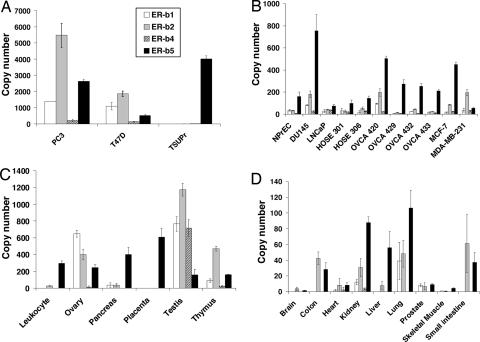
Expression of ER-β isoforms in human cell lines and tissue was evaluated through real-time PCR analyses (Fig. 5). The expression profiles between various tissues and cell lines were different. In established cell lines, levels of ER-β1 expression were low when compared with those of other ER-β isoforms (Fig. 5 A and B). In specific cell lines, such as PC3 and T47D, all ER-β isoforms are present at high levels, particularly ER-β2 (Fig. 5A). The bladder cancer cell line TSUPr expresses only ER-β5. ER-β5 seems to be the predominant isoform expressed in most tissues, followed by ER-β2 (Fig. 5 C and D). ER-β4 expression was notably expressed at a high level only in the testis. Compared with other isoforms, ER-β1 was expressed at relatively low levels in most tissues examined. Interestingly, ER-β5 was the only isoform that could be detected in placenta. Our data are in agreement with a recent finding that ER-β with a molecular weight (52 kDa) comparable with ER-β5 was dominantly expressed in the placenta (19). A wide distribution of ER-β5 in various tissues and cell lines raises an intriguing question of whether the current understanding of ER-β’s action based largely on ER-β1 is complete, because heteodimerization of ER-β5 with ER-β1 significantly enhances the overall activity in a ligand-dependent manner. Current in vivo data on the expression of ER-β isoforms are limited to a recent report suggesting that aberrant expression of ER-β isoforms may be involved in the development of breast cancer (17).
Fig. 5.
Tissue distribution of ER-β isoforms in various human cell lines (A and B) and human tissues (C and D) by real-time PCR analyses. Transcript levels of each isoform were determined by real-time PCR and expressed as “number of copy” (see Materials and Methods). The expression level of ER-β isoforms was normalized by human GAPDH gene level. Three independent experiments were performed and averaged. The standard deviations were calculated and shown as error bars.
In summary, ER-β1 is the only full-function ER-β, and it prefers to heterodimerize with ER-β isoforms, particularly ER-β4 and -β5, under the stimulation of estrogens, excluding phytoestrogens. All heterodimers have higher transactivation activities than the ER-β1 homodimer. Our data introduce a previously unrecognized concept for type I nuclear receptor signaling: ER-β1 serves as the “obligatory partner” of a functional dimeric complex, whereas ER-β2, -β4, or -β5 act as the “variable dimer partners” and serve as enhancers. This model differs from the original paradigm in which the two partners in a nuclear receptor dimer play identical roles in ligand binding and coactivator recruitment by way of helix 12. Hence, our data suggest that the ER-β heterodimer may recruit only one coactivator during transcriptional activation. In this regard, the model is somewhat analogous to the type II nuclear receptor mode of action, in which a heterodimer complex, such as retinoic acid receptor/retinoic acid X receptor (RAR/RXR) recruits only a single coactivator (20, 21). In this case, coactivators with more than one LXXLL motif, such as SRC, have been shown to interact simultaneously with the AF-2 domain in each helix 12 of the individual partner in the heterodimer (20, 21).
Because of the presence of only one AF-2 domain, the ER-β heterodimer can rely only on the functional helix 12 of ER-β1 for coactivator recruitment (Fig. 6). This model argues that the heterodimer could recruit a coactivator with one or more nuclear receptor (NR) boxes. In addition, we anticipate that the ER-β heterodimer, with a single coactivator, would have a set of coregulators binding asymmetrically around the ER-β heterodimer to a symmetrical palindromic ERE (Fig. 6). This model offers a plausible explanation of the greater efficiency of an ER-β heterodimer than an ER-β homodimer in transcriptional activation: They may recruit different sets of proteins to the complexes. Another reasonable speculation is that the ER-β heterodimer may experience less “allosteric hindrance” than the homodimer during coactivator recruitment. This notion is supported by a study on retinoic acid receptor/retinoic acid X receptor (RAR/RXR) dimers showing that the deletion of one AF-2 domain in one partner significantly enhanced coactivator recruitment to the complex (21). Also, the N-terminal half of the AF-2-disabled ER-β isoforms, lacking the ability to recruit a bulky coactivator, may have more accessibility to other regulatory factors, thus enhancing its transcriptional activity.
Fig. 6.
Model for ER-β isoforms interaction during transcription. (Upper) Putative protein recruitment by an ER-β1 homodimer and heterodimer. Comediators/cointegrators (M1–5) or coactivators (CoAct) with various numbers of nuclear receptor boxes (NR) recruited by ER-β homo- and heterodimers may be different. (Lower) The putative ultimate transcriptional machinery complex recruited by an ER-β heterodimer in a hypothetical promoter. A set of coregulators (M1–5), which maintains an active form of nucleosomes through acetylation (“Ac”-labeled purple spheres) and functions to form an active transcriptional complex with RNA polymerase II (RNA pol), is congregated in an asymmetrical manner around the ER-β1-βn heterodimer, which binds to a symmetrical palindromic ERE.
It is known that estrogen action by way of ER-α and -β mediates various aspects of human physiology and disease (22–24). Phytoestrogens are often reported to exert actions that are different from those of estrogens. The most common explanation is related to differential binding affinities of phytoestrogens and estrogens for ER-α and -β (25). A recent study also showed differential recruitment of coregulators induced by estrogens/phytoestrogens to ER-α and -β (26, 27). Our data have shed light on the differential actions of estrogens/phytoestrogens, indicating that they may be related to their respective abilities to induce ER-β heterodimer formation, with estrogens favoring heterodimer formation. Because the ER-β heterodimer exhibits higher transcriptional activity than its homodimer counterpart, we propose that this is an important consideration for the variations in their biological actions. We and others (9) have observed that each tissue/cell line has a unique pattern and/or ratio of expression between ER-β1 and its isoforms. This finding suggests that different estrogens induce the formation of different sets of heterodimers in a specific tissue/cell type, leading to widely varied biological responses. Last, our data are of relevance to the future design and selection of receptor subtype-specific therapeutics and nutraceutics, with undoubtedly higher specificities in their actions and/or in their targeted tissues.
This study presents evidence that ER-β1 is obligatory to ER-β signaling that involves an ERE, whereas the other ER-β isoforms have no innate activities but play an enhancement role when dimerized with ER-β1. Our data indicate that ER-β1 prefers to form heterodimers with its isoforms and that the process is likely dependent on the type of ligand, with estrogens but not phytoestrogens acting as modulators. Our study further substantiates the ideas from Katzenellenbogen, Greene, and coworkers (28) that dimer formation can be triggered by a “phantom ligand” effect, which could involve a ligand-bound ER-β1 activating a nonliganded ER-β isoform to form a heterodimer with full functional activity. Our finding strongly suggests that an ER-β heterodimer with a single functional helix 12 is adequate for coactivator recruitment and transactivation. We conclude that ER-β functions as a noncanonical type I receptor and that its actions are likely mediated by multiple functional heterodimers, formed between ER-β1 and one of its isoforms in a ligand-dependent manner. Because the observed variation in tissue distribution of ER-β isoforms was pronounced, we speculate that the isoforms determine the tissue responsiveness. Our results have generated a previously unrecognized ER-β model to explain the diverse actions of estrogen, especially those involving the classical ERE, and open another dimension in the design of ER-β-based therapies.
Materials and Methods
Cell Culture.
DU145, LNCaP, PC3, MCF7, HEK293, and T47D cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured in standard conditions. The origin and culture conditions of HOSE and OVCA cell lines were published in ref. 29.
Real-Time PCR Analysis of ER-β Isoforms.
Multitissue human cDNA panel I and II were purchased from Clontech (Palo Alto, CA). Submicroliter (0.5 μl) of cDNA, ER-β isoform-specific primers (ER-β1 forward, 5′-GTC AGG CAT GCG AGT AAC AA-3′; ER-β1 reverse, 5′-GGG AGC CCT CTT TGC TTT TA-3′; ER-β2 forward, 5′-TCT CCT CCC AGC AGC AAT CC-3′; ER-β2 reverse, 5′-GGT CAC TGC TCC ATC GTT GC-3′; ER-β4 forward, 5′-GTG ACC GAT GCT TTG GTT TG-3′; ER-β4 reverse, 5′-ATC TTT CAT TGC CCA CAT GC-3′; ER-β5 forward, 5′-GAT GCT TTG GTT TGG GTG AT-3′; ER-β5 reverse, 5′-CCT CCG TGG AGC ACA TAA TC-3′; GAPDH-F: 5′-TCC CTG AGC TGA ACG GGA AG-3′; GAPDH reverse, 5′-GGA GGA GTG GGT GTC GCT GT-3′) and 1× iQ SYBR green Supermix (BioRad, Hercules, CA) were used for each reaction. A standard curve for each gene was constructed by serial dilutions of the PCR product. The copy number was determined according to the published formula in the instruction manual (Applied Biosystems, Foster City, CA). Loading control was normalized by using human GAPDH (hGAPDH) level.
Ectopic Expression of ER-β Isoforms in Yeast and Mammalian Cells.
Total RNA was isolated from PC3 cells and reverse transcribed as described in ref. 29. PCRs were performed using a Platinum Taq High Fidelity DNA polymerase system (Invitrogen, Carlsbad, CA) in the presence of 0.6× PCRx enhancer. A single forward primer (5′-TGG CCCCTT GAG TTA CTG AG-3′) was used. Reverse primers for ER-β4 and -β5 were 5′-CAA ATC TTT CAT TGC CCA CA-3′ and 5′-TGC AGA CAC TTT TCC CAA A-3′, respectively. Touchdown PCR was performed at 72°C to 60°C in the first 12 cycles. An additional 35 cycles were performed at 94°C for 3 min, 60°C for 30 sec, and 72°C for 2.5 min. PCR products were gel-purified and TA-cloned into a pCR2.1 vector (Invitrogen). DNA sequencing was performed by Macrogen (Seoul, Korea). Full-length sequences of ER-β4 and -β5 were subcloned into pcDNA4/HisMax (Invitrogen) and modified YEpc (30) vectors for mammalian and yeast expressions, respectively. Mammalian expression vectors of ER-β1 and -β2 isoforms were gifts from L. C. Murphy at the University of Manitoba (Winnipeg, Canada) (31). Plasmids expressing ER-β isoforms were transiently transfected into HEK293 cells by using Lipofectamine Plus reagent (Invitrogen). The cells were then lysed by using gel-loading buffer after 24 h of transient transfection. Yeast expression vectors were transformed into BJ2168 (ATCC). Transfected HEK293 cells were harvested, and yeast extracts were prepared according to Mak and coworkers (30). Proteins (≅100 μg) were resolved in SDS/7.5% PAGE and blotted as described in ref. 29. A human ER-β antibody (1:500, H150, Santa Cruz Biotechnology, Santa Cruz, CA) and IRDye800-labeled goat anti-rabbit antibody (1:5000; Rockland, Gilbertsville, PA) were respectively used as primary and secondary antibodies.
Estrogen Binding Assay.
Recombinant ER-β1, -β2, -β4, and -β5 were extracted from yeast, and 400 μg of each were added to each binding reaction containing 1× TEDG buffer (10 mM Tris/1.5 mM EDTA/1.0 mM DTT/10% glycerol, pH 7.4), 0.1–6 nM [2,4,6,7,16,17-3H(N)]-E2 (Perkin–Elmer, Wellesley, MA), and with or without 10–600 nM cold E2 (Sigma, St. Louis, MO). After overnight incubation at 4°C, unbounded hot E2 was removed by charcoal-dextran (0.7%:0.07%). Bounded hot E2 was measured by scintillation counting (Beckman, Fullerton, CA). The Bmax and Kd of each isoform were calculated and analyzed by using GraphPad Prism 4.0 (San Diego, CA).
Transactivation Activities of ER-β Isoforms in the Mammalian System.
Luciferase reporter plasmid (pt109-ERE3-Luc) carrying 3× vitellogenin ERE was provided by Craig Jordan (Fox Chase Cancer Center, Philadelphia, PA) (32). HEK293 cells were seeded at 1.5 × 104 per well in 24-well plates. Mammalian vectors (200 ng) expressing an ER-β isoform together with an ERE reporter (50 ng) and β-galactosidase (25 ng) were transiently transfected into the cells cultured in phenol red-free DMEM with 5% charcoal-stripped serum for 48 h. The effects of other ER-β isoforms on ER-β1 transactivation were studied by cotransfecting vectors carrying ER-β isoforms (100 ng) with ER-β1 plasmid (100 ng). After 24 h of transfection, different concentrations of E2, EE2, DES, apigenin, genistein, and BPA were applied to the cultures. Transactivation activities were measured by using the Bright-Glo luciferase kit (Promega, Madison, WI) after 24 h of hormone treatment. The activities of β-galactosidase were measured with the β-gal assay kit (Promega) to normalize transfection efficiency.
Effects of SRC-1 on the Transactivation Activities of ER-β Isoforms.
The expression plasmid for SRC-1, pCR3.1SRC-1, was provided by Nancy Weigel at Baylor College of Medicine (Houston, TX) (33). Transfection and transactivation assays in the presence of various ER-β isoforms were performed as described in Transactivation Activities of ER-β Isoforms in the Mammalian System.
Dimerization of ER-β Isoforms by Y2H System.
The Y2H 3 system from Clontech was used in this study. Full-length ER-β isoforms were subcloned into Y2H vectors (pGADT7 and pGBKT7) and transformed into AH109 yeast strain to generate different combinations of homo- and heterodimers of ER-β isoforms. The double yeast transformants were incubated with different concentrations (0–1 μM) of E2 overnight at 30°C. The degree of dimerization upon ligand stimulation was measured and quantified by the Beta-Glo assay (Promega).
Real-time PCR Analysis of pS2 Gene Expression.
Twenty-four hours after transfection of ER-β1 or ER-β1 combined with ER-β2/4/5 to HEK293 cells, the cells were treated with 1 nM E2 or 1 μM genistein. Levels of pS2 were measured as described in ref. 34. Human GADPH was used as a housekeeping control, because its levels remained unchanged under either E2 or genistein treatment. Real-time PCR was performed with the iCycler IQ real-time PCR detection system (Bio-Rad) as described in ref. 35.
Bioinformatics and Molecular Modeling.
Sequence analyses were achieved by using NCBI BLAST programs (http://130.14.29.110/BLAST) and EBI Clustalw (www.ebi.ac.uk/clustalw). The prediction of the ORF of each transcript was performed using NCBI ORF Finder (www.ncbi.nlm.nih.gov/gorf/gorf.html). The ProModII algorithm in SWISS-MODEL was used to generate 3D structures of ER-β isoforms, and energy minimization was applied to each model by Gromos96 (swissmodel.expasy.org). Crystal structures of ER-β complexed with Way-697 (PDB ID code 1x76) and Way-244 (PDB ID code 1x78) were applied to generate molecular models for ER-β2, -β4, and -β5 isoforms. Each molecular model was evaluated using DeepView 3.7 software (GlaxoSmithKline, New York, NY) and the Swiss Institute of Bioinformatics. Evaluation of coregulator binding pocket was performed with PyMOL version 0.98 (DeLano Scientific, South San Francisco, CA).
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK61084, CA112532, CA15776, and ES013071 and Department of Defense Grant DAMD-W81XWH-04-1-0165.
Abbreviations
- BPA
bisphenol A
- DES
diethylstilbestrol
- E2
estradiol
- EE2
ethynylestradiol
- ER
estrogen receptor
- ERE
estrogen responsive element
- SRC
steroid receptor coactivator
- Y2H
yeast two-hybrid.
Footnotes
References
- 1.Kuiper G. G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J. A. Proc. Natl. Acad. Sci. USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koehler K. F., Helguero L. A., Haldosen L. A., Warner M., Gustafsson J. A. Endocr. Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X., Leav I., Leung Y. K., Wu M., Liu Q., Gao Y., McNeal J. E., Ho S. M. Am. J. Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans R. M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraichely D. M., Sun J., Katzenellenbogen J. A., Katzenellenbogen B. S. Endocrinology. 2000;141:3534–3545. doi: 10.1210/endo.141.10.7698. [DOI] [PubMed] [Google Scholar]
- 6.Fleming F. J., Hill A. D., McDermott E. W., O’Higgins N. J., Young L. S. J. Clin. Endocrinol. Metab. 2004;89:375–383. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- 7.McKenna N. J., Xu J., Nawaz Z., Tsai S. Y., Tsai M. J., O’Malley B. W. J. Steroid Biochem. Mol. Biol. 1999;69:3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 8.Mosselman S., Polman J., Dijkema R. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 9.Moore J. T., McKee D. D., Slentz-Kesler K., Moore L. B., Jones S. A., Horne E. L., Su J. L., Kliewer S. A., Lehmann J. M., Willson T. M. Biochem. Biophys. Res. Commun. 1998;247:75–78. doi: 10.1006/bbrc.1998.8738. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa S., Inoue S., Watanabe T., Orimo A., Hosoi T., Ouchi Y., Muramatsu M. Nucleic Acids Res. 1998;26:3505–3512. doi: 10.1093/nar/26.15.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poola I., Abraham J., Baldwin K., Saunders A., Bhatnagar R. Endocrine. 2005;27:227–238. doi: 10.1385/ENDO:27:3:227. [DOI] [PubMed] [Google Scholar]
- 12.Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engstrom O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 13.Pike A. C., Brzozowski A. M., Hubbard R. E. J. Steroid Biochem. Mol. Biol. 2000;74:261–268. doi: 10.1016/s0960-0760(00)00102-3. [DOI] [PubMed] [Google Scholar]
- 14.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 15.Jisa E., Dornstauder E., Ogawa S., Inoue S., Muramatsu M., Jungbauer A. Biochem. Pharmacol. 2001;62:953–961. doi: 10.1016/s0006-2952(01)00731-6. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa S., Inoue S., Watanabe T., Hiroi H., Orimo A., Hosoi T., Ouchi Y., Muramatsu M. Biochem. Biophys. Res. Commun. 1998;243:122–126. doi: 10.1006/bbrc.1997.7893. [DOI] [PubMed] [Google Scholar]
- 17.Chi A., Chen X., Chirala M., Younes M. Anticancer Res. 2003;23:211–216. [PubMed] [Google Scholar]
- 18.Rooks W. H., Kugler S. L., Dorfman R. I. Fertil. Steril. 1968;19:419–423. doi: 10.1016/s0015-0282(16)36671-7. [DOI] [PubMed] [Google Scholar]
- 19.Bukovsky A., Caudle M. R., Cekanova M., Fernando R. I., Wimalasena J., Foster J. S., Henley D. C., Elder R. F. Reprod. Biol. Endocrinol. 2003;1:36–55. doi: 10.1186/1477-7827-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 21.Westin S., Kurokawa R., Nolte R. T., Wisely G. B., McInerney E. M., Rose D. W., Milburn M. V., Rosenfeld M. G., Glass C. K. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 22.Lindsay R., Cosman F. Osteoporos. Int. 1997;7(Suppl. 1):S40–S42. doi: 10.1007/BF01674812. [DOI] [PubMed] [Google Scholar]
- 23.McEwen B. S. J. Appl. Physiol. 2001;91:2785–2801. doi: 10.1152/jappl.2001.91.6.2785. [DOI] [PubMed] [Google Scholar]
- 24.Rissman E. F., Wersinger S. R., Fugger H. N., Foster T. C. Brain Res. 1999;835:80–90. doi: 10.1016/s0006-8993(99)01452-3. [DOI] [PubMed] [Google Scholar]
- 25.Kuiper G. G., Lemmen J. G., Carlsson B., Corton J. C., Safe S. H., van der Saag P. T., van der B. B., Gustafsson J. A. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- 26.Hong T., Nakagawa T., Pan W., Kim M. Y., Kraus W. L., Ikehara T., Yasui K., Aihara H., Takebe M., Muramatsu M., et al. Biochem. Biophys. Res. Commun. 2004;317:259–264. doi: 10.1016/j.bbrc.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 27.Routledge E. J., White R., Parker M. G., Sumpter J. P. J. Biol. Chem. 2000;275:35986–35993. doi: 10.1074/jbc.M006777200. [DOI] [PubMed] [Google Scholar]
- 28.Nettles K. W., Sun J., Radek J. T., Sheng S., Rodriguez A. L., Katzenellenbogen J. A., Katzenellenbogen B. S., Greene G. L. Mol. Cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- 29.Leung Y. K., Lau K. M., Mobley J., Jiang Z., Ho S. M. Cancer Res. 2005;65:3726–3734. doi: 10.1158/0008-5472.CAN-04-3771. [DOI] [PubMed] [Google Scholar]
- 30.Salerno A. J., He Z., Goos-Nilsson A., Ahola H., Mak P. Nucleic Acids Res. 1996;24:566–572. doi: 10.1093/nar/24.4.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng B., Lu B., Leygue E., Murphy L. C. J. Mol. Endocrinol. 2003;30:13–29. doi: 10.1677/jme.0.0300013. [DOI] [PubMed] [Google Scholar]
- 32.Catherino W. H., Jordan V. C. Cancer Lett. 1995;92:39–47. doi: 10.1016/0304-3835(95)03755-l. [DOI] [PubMed] [Google Scholar]
- 33.Agoulnik I. U., Vaid A., Bingman W. E., III, Erdeme H., Frolov A., Smith C. L., Ayala G., Ittmann M. M., Weigel N. L. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 34.Lau K. M., LaSpina M., Long J., Ho S. M. Cancer Res. 2000;60:3175–3182. [PubMed] [Google Scholar]
- 35.Tam N. N., Ghatak S., Ho S. M. Prostate. 2003;55:1–8. doi: 10.1002/pros.10169. [DOI] [PubMed] [Google Scholar]