Abstract
Some of the mitochondrial tRNAs of higher plants are nuclearly encoded and imported into mitochondria. The import of tRNAs encoded in the nucleus has been shown to be essential for proper protein translation within mitochondria of a variety of organisms. Here, we report the development of an in vitro assay for import of nuclearly encoded tRNAs into plant mitochondria. This in vitro system utilizes isolated mitochondria from Solanum tuberosum and synthetic tRNAs transcribed from cloned nuclear tRNA genes. Although incubation of radioactively labeled in vitro-transcribed tRNAAla, tRNAPhe, and tRNAMet-e with isolated potato mitochondria resulted in importation, as measured by nuclease protection, the amount of tRNA transcripts protected at saturation was at least five times higher for tRNAAla than for the two other tRNAs. This difference in in vitro saturation levels of import is consistent with the in vivo localization of these tRNAs, since cytosolic tRNAAla is naturally imported into potato mitochondria whereas tRNAPhe and tRNAMet-e are not. Characterization of in vitro tRNA import requirements indicates that mitochondrial tRNA import proceeds in the absence of any added cytosolic protein fraction, involves at least one protein component on the surface of mitochondria, and requires ATP-dependent step(s) and a membrane potential.
It is currently well established that mitochondrial tRNA import is a process occurring in a number of evolutionary distinct organisms, such as the yeast Saccharomyces cerevisiae, protozoans, mammals, and plants (for reviews, see references 17 and 43). There are obvious differences between these organisms in the number and the identity of the imported tRNAs. For example, yeast mitochondria import only a single tRNA (35), as do marsupials (12), whereas kinetoplastid protozoan mitochondria import all of their tRNAs (20, 44). Other organisms fall somewhere between these two extremes. Mitochondria of the primitive bryophyte Marchantia polymorpha import only three tRNAs (3), there are at least nine nuclearly encoded mitochondrial tRNAs in Acanthamoeba castellanii (8), and higher plant mitochondria import one-third to one-half of their tRNAs (19, 26). These differences in the specificity and the number of imported tRNAs might reflect fundamental differences in the mechanisms underlying tRNA import in the various organisms.
While mitochondrial protein import has been studied in great detail (5, 23, 24), little is known about the pathway(s) of tRNA import into mitochondria. So far, most of the work defining mitochondrial tRNA import mechanisms has been performed either with S. cerevisiae or with protozoans. Using a combination of in vitro and in vivo studies, different models have been proposed to explain how such large, hydrophilic polyanions can cross the hydrophobic environment of the two mitochondrial membranes towards a negative compartment.
For S. cerevisiae, import of a single tRNALys (CUU) occurs via the formation of a complex with the precursor form of the mitochondrial lysyl-tRNA synthetase and requires an intact preprotein import machinery. As a prerequisite to import, the tRNALys (CUU) has to be aminoacylated by the cytosolic lysyl-tRNA synthetase. This cotranslocation of tRNALys (CUU) with a mitochondrial preprotein is ATP dependent and requires a transmembrane electrochemical potential (17).
For trypanosomatids, Trypanosoma brucei and different Leishmania species, tRNA import is an ATP-dependent process and involves outer membrane-bound and protease-sensitive receptors. In contrast to the situation for S. cerevisiae, none of the in vitro assays with isolated T. brucei and Leishmania mitochondria requires the addition of cytosolic factors. For Leishmania, it has been shown that tRNAs are imported as mature, fully processed molecules. The D domain appears to be recognized by RNA-specific receptor(s), as shown in an in vitro import study with Leishmania tropica tRNATyr (30). For Leishmania tarentolae, an exchange of the D arms between tRNAIle (naturally imported) and tRNAGln (mainly cytosol localized) also demonstrated that this part of the tRNA contains an import signal (29, 41). By contrast, Kapushoc et al. showed that in the case of tRNATrp, the anticodon contains import determinants (25). The situation for T. brucei remains contradictory. On one hand, it has been reported that in vivo import of heterologous tRNAs occurs independently of their genomic context, thus suggesting that mature tRNAs are the regular import substrates (21, 46). On the other hand, based on the detection of such molecular forms in T. brucei mitochondria and on in vitro import assays, it has been proposed that precursor tRNAs having a 5′ extension or dicistronic tRNA transcripts are the preferred import substrates (28, 52). However, Aphasizhev et al. did not detect 5′-extended tRNA precursors in mitochondrial RNA fractions (4). These analyses turned out to be complicated due to the presence of circularized mature tRNA molecules resulting from mitochondrial RNA ligase activity (4, 28).
Little is known about the mechanism of tRNA translocation across plant mitochondrial membranes. Up to now, in vivo import of mutated tRNAs into the mitochondria of transgenic plants has been the main approach used to study the import pathway. Using this approach, the implication of the aminoacyl-tRNA synthetases (aaRSs) in the tRNA import process has been suggested in the case of Arabidopsis thaliana tRNAAla, because a mutated tRNA that is not aminoacylatable by the alanyl-tRNA synthetase (AlaRS) is not imported into plant mitochondria (11). However, for Solanum tuberosum, there is a selective import of cytosolic tRNAGly(UCC) and tRNAGly (CCC), whereas cytosolic tRNAGly(GCC) is not imported (7). This indicates that if recognition of the different tRNAGlys by the same glycyl-tRNA synthetase is necessary, it is not sufficient to promote import. Although no common “import signature” has been identified so far in the sequences of imported tRNAs (40), these data suggest the existence of at least one additional import factor and/or of structural features shared by imported tRNA molecules.
In this paper, we describe a selective in vitro assay for import of tRNAAla transcript into isolated mitochondria of S. tuberosum. We show that tRNAAla ATP-dependent internalization does not require any added cytosolic fraction and includes at least one protein component on the surface of mitochondria.
MATERIALS AND METHODS
Plant materials.
Mitochondria were isolated from potato tubers (var. Bintje) by differential centrifugations and purification on Percoll gradients as described previously (18). By testing of the ability to reduce exogenously added cytochrome c (14), the integrity of the outer membrane of these mitochondria was estimated to be between 95 to 98%.
Cytosolic protein extract was prepared from etiolated bean (Phaseolus vulgaris) hypocotyls according to reference 34.
In vitro transcription and labeling of synthetic RNAs.
Plasmids containing tRNA gene sequences directly fused to a T7 RNA polymerase promoter at the 5′ terminus and including a BstNI site at the 3′ terminus were used as substrates to synthesize in vitro transcripts with T7 RNA polymerase under conditions described previously (39, 45). To synthesize uniformly labeled in vitro transcripts, the conditions were identical except for the presence of [α-32P]UTP.
The corresponding constructs containing A. thaliana cytosolic tRNAAla or tRNAPhe gene sequences were obtained previously (9). The construct encoding mature A. thaliana tRNAMet-e was amplified by PCR with the relevant primers so that the intronless tRNA gene sequence was directly fused to the T7 RNA polymerase promoter at the 5′ end and to a BstNI site at the 3′ end (39). Since plant nuclear tRNAMet-e genes are interrupted by introns (2), we used a synthetic oligonucleotide corresponding to the complete intronless A. thaliana tRNAMet-e gene sequence as a substrate for PCR amplification with the two following primers: 5′-ATGAATTCGAATTGTAATACGACTCACTATAGGGGTGGTGGCGCAGTT-3′ (T7 promoter sequence underlined, EcoRI site in italics) and 5′-CAGGATCCCTGGTGGGGTGAGAGAGGCTC-3′ (BstNI site underlined and BamHI site in italics). PCR amplification, cloning, and sequencing were performed according to standard procedures.
Standard assay of in vitro tRNA import into isolated mitochondria.
The in vitro RNA import assays were performed in a 100-μl reaction volume containing mitochondria corresponding to 400 μg of proteins, uniformly labeled in vitro-transcribed tRNA (105 cpm/assay except when specified), 250 mM mannitol, 40 mM KCl, 1 mM dithiothreitol, 10 mM HEPES-KOH [pH 7.2], 1 mM K2HPO4, 2.5 mM MgCl2, 1 mM malate, 1 mM NADH, 2 μM ADP, and 5 mM ATP (except when specified). The tRNA import assay mixtures were incubated at 25°C for 20 min unless otherwise stated. Where appropriate, proteinase K was then added to a final concentration of 30 μg/ml, and digestion was carried out for 10 min at room temperature. This step, by removing proteins on the mitochondrial surface, eliminates tRNAs that may be nonspecifically associated with the mitochondrial outer membranes. To terminate the digestion, 100 μl of buffer A (250 mM mannitol, 10 mM KPO4 [pH 7.5], 0.1% [wt/vol] bovine serum albumin [BSA], 5 mM glycine) containing 2 mM phenylmethylsulfonyl fluoride was added, and the mixture was centrifuged at 15,000 × g for 10 min. To digest tRNAs that were not imported into the organelles, mitochondria were resuspended in 100 μl of buffer A containing RNase A and RNase T1 to final concentrations of 100 μg/ml and 750 U/ml (unless otherwise stated), respectively. This RNase treatment was carried out at room temperature for 10 min. One milliliter of stop buffer (250 mM mannitol, 10 mM KPO4 [pH 7.5], 5 mM EDTA, 5 mM EGTA, 0.1% [wt/vol] BSA, 5 mM glycine) was added, and the mixture was centrifuged at 15,000 × g for 10 min. This washing step was repeated twice. To isolate protected RNAs, the mitochondrial pellets were resuspended in a solution containing 100 μl of 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 1% (wt/vol) SDS and extracted with 100 μl of water-saturated phenol. The aqueous phase was submitted to ethanol precipitation.
The radioactively labeled tRNA samples were separated by electrophoresis through 7 M urea-15% polyacrylamide gels. After electrophoresis, the gels were dried and either submitted to autoradiography or exposed to a Phosphorimager plate, allowing quantification with a BAS 100 phosphorimager (Fujix) using the MacBAS v2.2 software (Fuji Photo Film).
Mitoplast preparation.
Mitoplast preparations were carried out according to a modified version of the efficient protocol described by Heins et al. (22). Briefly, the mitochondria were resuspended in a swelling buffer (5 mM KPO4 [pH 7.2]) (100 μl for 1 mg of proteins) and kept on ice for 10 min. The same volume of swelling buffer was added, and after 5 min mitochondria were ruptured by Dounce homogenization. Mitoplasts were separated from outer membranes and unbroken mitochondria by centrifugation through a sucrose step gradient (47/32/15% [wt/vol] sucrose in 2 mM EDTA, 10 mM KPO4 [pH 7.2], 0.2% [wt/vol] BSA) at 4°C for 18 min at 175,000 × g in a Beckman TLA-100 rotor. Outer membranes were present at the 15/32% interphase, whereas intact mitochondria were pelleted. Intact mitoplasts, collected from the 32/47% interphase, were diluted 10 times with washing buffer (300 mM mannitol, 10 mM KPO4 [pH 7.2], 1 mM EDTA, 0.1% [wt/vol] BSA) and pelleted by centrifugation at 135,000 × g for 15 min.
Protease pretreatment of mitochondria.
To remove proteins exposed on the mitochondrial outer membrane, purified mitochondria (1 mg of proteins/assay) resuspended in 100 μl of washing buffer (see above) were treated with different amounts of trypsin (Promega) (4, 8 and 16 μg) for 10 min at 25°C. After incubation, the action of the protease was stopped by the addition of 10 mg of trypsin inhibitor/ml. Mitochondria were then washed twice with washing buffer in the presence of the specific protease inhibitor.
Western blot analysis.
Proteins were separated by SDS-polyacrylamide gel electrophoresis, electrotransferred onto Immobilon-P membranes (Millipore, Bedford, Mass.), and submitted to immunological detection following classical protocols (42). Antisera were used at a 1/5,000 dilution. Antibodies against yeast mitochondrial cytochrome c1 (CYT C1) (provided by G. Schatz, Basel, Switzerland), subunit 9 of wheat NADH dehydrogenase (27) (a gift from J. M. Grienenberger, Strasbourg, France), and tobacco superoxide dismutase (6) (obtained from D. Inzé, Ghent, Belgium) were used as controls for mitochondrial inner membrane and matrix protein fractions, respectively. Antibodies raised against α-tubulin (Amersham Pharmacia Biotech) were used to evaluate the cytosolic contamination of our mitochondria preparations.
Northern blot analysis.
Transfer RNAs were extracted as described previously (45) from the mitochondrial pellet or the supernatant obtained after centrifugation at 9,000 × g for 10 min of a standard in vitro import assay. For Northern analysis, tRNAs were fractionated by electrophoresis on 15% polyacrylamide gels under denaturing conditions and transferred onto nylon membranes (Hybond-N; Amersham Pharmacia Biotech) by electroblotting (15 min at 150 mA and then 30 min at 500 mA) in 10 mM Tris-acetate (pH 8.0)-0.5 mM EDTA buffer. Hybridization was carried out overnight in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% (wt/vol) SDS at a temperature 5°C lower than the theoretical melting temperature of the duplex with the oligonucleotide used as a probe. The oligonucleotide (5′-CCAATGCCTTAAGCCACTCAG-3′), specific for plant mitochondrial tRNASer(GCU), was 32P labeled with T4 polynucleotide kinase according to classical procedures. After hybridization, the membranes were washed at the hybridization temperature successively in 2× SSC (twice for 10 min each wash) and in 2× SSC-0.1% (wt/vol) SDS (for 30 min). Radioactive hybridization was submitted to autoradiography or phosphorimager exposure.
Additional procedures.
Concentrated valinomycin, oligomycin, carbonylcyanide m-chlorophenylhydrazone (CCCP), and carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) stock solutions were prepared in 100% ethanol. KCN was dissolved in distilled water. Concentrated apyrase stock solution was prepared in 10 mM Tris-HCl (pH 7.5). Concentrated stock solutions were such that 1 μl of these inhibitors was added, when specified, to 100 μl of import mixture. These reagents were incubated with the mitochondria at 4°C for 10 min prior to addition of labeled tRNA transcripts in the relevant import assays. Their final concentration was 50 μM for valinomycin, oligomycin, CCCP, and FCCP, 0.5 mM for KCN, and 1 U/ml for apyrase. All these reagents were from Sigma.
RESULTS
ATP-dependent import of tRNAAla into isolated mitochondria from S. tuberosum occurs without any added cytosolic protein fraction.
The mature tRNA substrate used for in vitro import assays consisted of the 76-nucleotide cytosolic A. thaliana tRNAAla(UGC) with a fully processed 5′ end and a CCA at the 3′ end (Fig. 1A). It was previously demonstrated that in vivo this nuclearly encoded tRNAAla partitions between the cytosol and the mitochondria in all higher plants investigated so far (19, 26, 33). Percoll-purified potato mitochondria were previously shown to be suitable for various experiments, such as run-on transcription assays, in vitro tRNA processing assays, or in vitro protein import experiments (15, 18, 32).
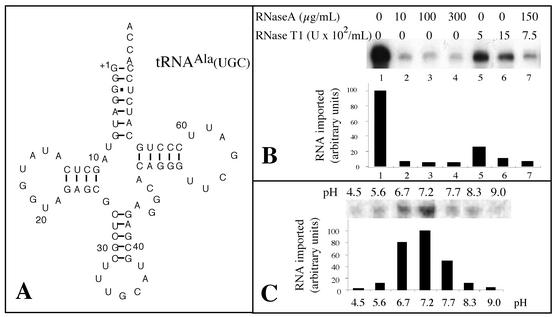
FIG. 1.
The mature tRNAAla is imported in vitro into potato mitochondria. (A) Cloverleaf structure of the mature A. thaliana cytosolic tRNAAla used as a substrate for standard in vitro import assays. (B) Protection of labeled tRNAAla transcripts from RNase degradation. Standard in vitro import assays of 100-μl volumes containing isolated mitochondria corresponding to 400 μg of proteins and 105 cpm of labeled tRNAAla substrate were performed. Reactions were carried out in the presence of 5 mM ATP. After incubation, RNase A and RNase T1 were added to various final concentrations ranging from 0 to 300 μg/ml and from 0 to 1,500 U/ml, respectively. (C) tRNA import is pH dependent. Standard in vitro import assays of 100 μl containing isolated mitochondria corresponding to 400 μg of proteins and 105 cpm of labeled tRNAAla substrate. Reactions were performed in the presence of 5 mM ATP but at different pHs obtained by adjusting the pH of the standard import mixture either with HCl or with NaOH. After import, the pH of all samples was adjusted to 7.2 with HCl or NaOH before addition of RNase A and RNase T1 to final concentrations of 300 μg/ml and 750 U/ml, respectively. Quantification of the amount of protected RNAs was done using a phosphorimager.
Incubation of labeled in vitro-transcribed tRNAAla with highly purified potato mitochondria resulted in protection from RNase A and RNase T1 digestion. RNase treatment (Fig. 1B) and pH conditions (Fig. 1C) were optimized. As shown on Fig. 1B, addition of RNase A to a final concentration of 10 μg/ml was sufficient to digest all the unprotected tRNAAla transcript. However, to be sure that nonimported tRNAs were always efficiently removed, mitochondria were usually digested with a mixture of RNase A at a final concentration of 150 μg/ml and RNase T1 at a concentration of 750 U/ml.
Efficient import of tRNAAla into isolated mitochondria requires the presence of ATP, and the existence of a correlation between the concentration of ATP and the amount of protected tRNAAla transcript is shown in Fig. 2A. It should be noted that the concentration of externally added ATP (5 to 10 mM) for which efficient tRNA import occurs is higher than the ATP concentration (about 1 mM) usually used for efficient in vitro protein import. Similar data were obtained in the case of tRNA import into isolated mitochondria from L. tarentolae (41), where the amount of protected tRNA was far higher in the presence of 5 to 10 mM ATP than in the presence of 1 mM ATP.
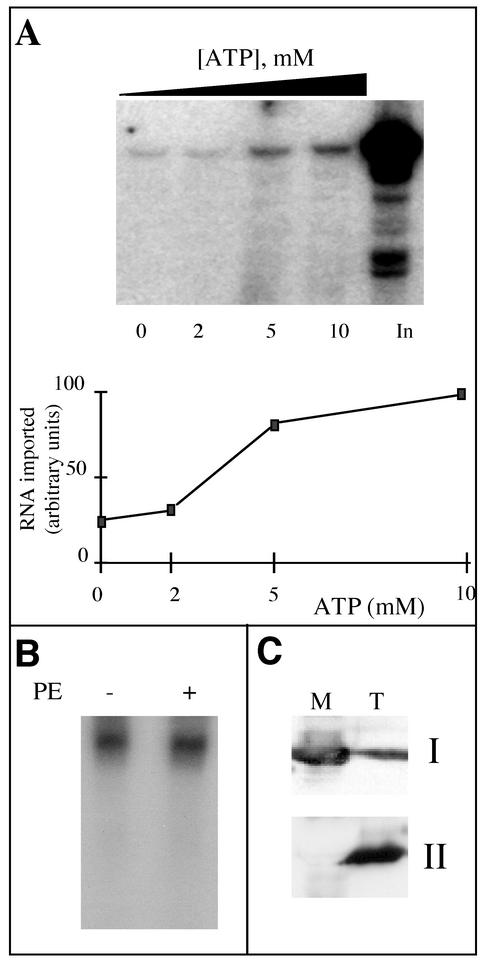
FIG. 2.
Mitochondrial incorporation of labeled tRNAAla transcripts is ATP dependent and does not require a cytosolic protein fraction. (A) Labeled in vitro-transcribed tRNAAla (105 cpm) was incubated with isolated mitochondria in the presence of increasing concentrations of ATP (0, 2, 5 and 10 mM). In, input RNA (7,500 cpm), no incubation with mitochondria. The import level of the labeled tRNAAla transcripts was quantified using a phosphorimager. (B) Standard in vitro import assays were performed in the presence of 5 mM ATP and in the absence (−) or presence (+) of a bean hypocotyl cytosolic protein extract (PE). (C) Immunodetection of mitochondrial superoxide dismutase (I) and α-tubulin (II) performed with total (T) and mitochondrial (M) extracts from potato tubers.
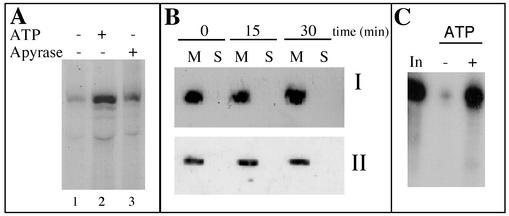
Even in the absence of externally added ATP, a residual amount of protected transcript was observed corresponding to 10 to 15% of the amount of protected tRNAAla in the presence of ATP. Removal of transcript protection in the absence of ATP could not be achieved by pretreating mitochondria with apyrase (1 U/ml) prior to import (Fig. 3A). Since apyrase hydrolyses endogenously produced ATP, it is unlikely that the low level of protected transcript still detected in the absence of exogenous ATP would be due to ATP leaking out of mitochondria. Similarly, pretreatment of mitochondria with atractyloside (400 μM), which blocks the adenine nucleotide translocator and thus inhibits the export of ATP, had no effect on transcript protection in the absence of added ATP (data not shown). This apparent ATP-independent tRNA incorporation might be due to a loss of integrity of the mitochondrial membranes during the import assay. However, the integrity of the outer membrane of the mitochondria, estimated to be between 95 to 98% at the end of the purification step, does not drop significantly during the import assays (data not shown). Furthermore, as shown on Fig. 3B, neither the mitochondrially encoded tRNASer(GCU) nor the soluble matrix superoxide dismutase protein could be detected on Northern and Western blots, respectively, of the supernatant obtained after centrifugation of a standard in vitro import assay (after 30 min of incubation). By contrast, they were both detected in the corresponding mitochondrial pellets analyzed before or after incubation. Thus, it seems that protection found in the absence of ATP is not due to engulfment of transcripts by leaky mitochondria during the import assay. Finally, 60 to 80% of this residual amount of protected transcript was eliminated by a mild posttreatment of mitochondria with proteinase K prior to RNase digestion, so that the background detected in the absence of ATP became very low (Fig. 3C). These data strongly suggest that most of the observed background might correspond to nonspecific association of labeled tRNA transcripts with the outer membrane rather than to import into the mitochondrial matrix.
FIG. 3.
The apparent incorporation of tRNAAla in the absence of added ATP is not due to ATP leaking out of mitochondria or to a partial loss of integrity of the mitochondrial membranes but rather to nonspecific protection at the mitochondrial surface. (A) Standard in vitro import assays were performed in the presence of 5 mM ATP (lane 2) and in the presence of apyrase (1 U/ml) (lane 3). After incubation, mild proteinase K digestion and RNase posttreatment were performed as for panel C. (B) Hybridization of a mitochondrial tRNASer-specific oligonucleotide probe to a Northern blot of tRNAs (I) or immunodetection of mitochondrial superoxide dismutase on a Western blot (II). tRNAs and proteins were extracted from the mitochondrial pellet (M) or from the supernatant (S) obtained after centrifugation at 9,000 × g for 10 min of a standard in vitro import assay after 0, 15, and 30 min of incubation. (C) Standard in vitro import assays were performed in the absence (−) or in the presence (+) of 5 mM ATP. After incubation, proteinase K was added to a final concentration of 30 μg/ml, and digestion was carried out for 10 min at room temperature. One hundred microliters of buffer (250 mM mannitol, 10 mM KPO4 [pH 7.5], 0.1% [wt/vol] BSA, 5 mM glycine) containing 2 mM phenylmethylsulfonyl fluoride was added, and the mixture was centrifuged at 15,000 × g for 10 min. RNase posttreatment was then performed as described in Materials and Methods. In, input RNA (1,000 cpm), no incubation with mitochondria.
Whereas the direct or indirect implication of the aaRS in the in vivo mitochondrial tRNA import process has been suggested in the case of A. thaliana tRNAAla (11), the experiments presented in Fig. 1B and C and 2A were performed in the absence of any added cytosolic protein extract. Indeed, the addition of a bean cytosolic enzymatic extract to the import reaction mixture had no effect on the amount of protected tRNAAla transcript (Fig. 2B). Mitochondria purified from potato tubers on Percoll gradients are, by contrast to those from A. thaliana, usually virtually free of cytosolic, nuclear, or plastidic contamination (15, 38). As shown on Fig. 2C, cross-contaminations between cytosolic and mitochondrial fractions were checked by Western blot analyses: in the mitochondrial fraction, the mitochondrial superoxide dismutase was strongly detected, whereas no cytosolic α-tubulin was immunodetected. Furthermore, systematic N-terminal sequencing of the membrane proteins from isolated potato tuber mitochondria allowed us to identify about 25 mitochondrial proteins, none of them corresponding to a cytosolic contaminant (L. Delage and L. Maréchal-Drouard, unpublished data). Although we cannot completely exclude a low level of cytosolic contamination in our mitochondrial preparations, these results suggest that in vitro tRNA import does not require cytosolic factors, but they do not rule out the possibility that cytosolic factors play a role in vivo.
tRNAAla is imported into the matrix of mitochondria.
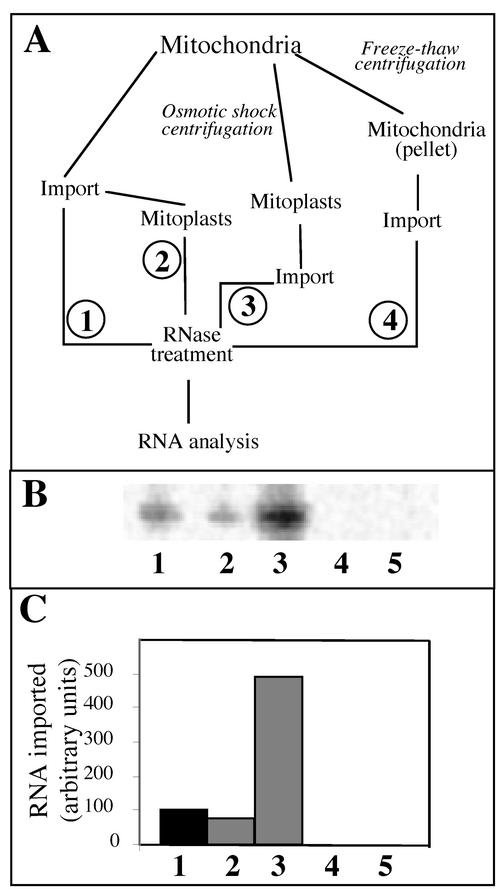
Since mitochondrial translation occurs in the matrix, the nuclearly encoded tRNAs imported into mitochondria have to reach this subcompartment. As shown on Fig. 4B and C (lane 2), about 75% of labeled tRNAAla transcript remains protected after disruption of the outer membrane and purification of mitoplasts on a sucrose gradient, compared to the amount of protected transcript in the absence of membrane disruption (Fig. 4B and C, lane 1). These results suggest that the externally added labeled tRNAAla transcripts were internalized by the mitochondria. Interestingly, when tRNAAla transcripts were incubated in the presence of mitoplasts, the amounts of protected labeled transcripts were estimated to be about five times higher than when the import assay was performed with mitochondria (Fig. 4B and C, lanes 3). It seems, therefore, that once the outer membrane has been eliminated, it is easier for a tRNA to penetrate into the matrix, or in other words, the outer membrane constitutes a stronger barrier than the inner membrane for the internalization of RNA molecules. When mitochondria were subjected to freeze-thaw cycles, broken organelles were obtained after centrifugation. Incubation of labeled tRNAAla transcript with this mitochondrial fraction under import conditions did not lead to protection of this RNA against RNase digestion (Fig. 4B and C, lanes 4), thus showing that detection of labeled tRNAAla transcripts after treatment with nucleases cannot be explained by a protection with mitochondrial membrane proteins but rather by its internalization into mitochondria. Furthermore, in the absence of any mitochondrial fraction, this labeled transcript was completely digested by RNases A and T1 (Fig. 4B and C, lanes 5). This indicates that protection from RNase digestion was not due to partial resistance of tRNAAla transcripts to these nucleases but resulted from import into isolated mitochondria.
FIG. 4.
Labeled tRNAAla transcript is imported into the matrix of isolated mitochondria. (A) Fractionation and import scheme. (B) Labeled in vitro-transcribed tRNAAla (2 × 105 cpm) was incubated under standard import conditions with intact mitochondria (1); with intact mitochondria, but mitoplasts generated before RNase treatment (2); with mitoplasts (3); with broken mitochondria (4); and without mitochondria (5). (C) The amounts of protected tRNAAla transcript were quantified using a phosphorimager.
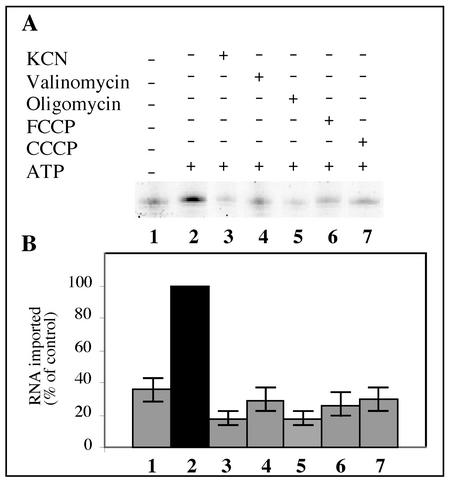
Import of tRNAAla requires a membrane potential and a functional respiratory chain.
Different treatments were used to inhibit mitochondrial functions: (i) protonophores, CCCP, or FCCP, which permits proton diffusion across the mitochondrial inner membrane and thus dissipates the proton motive force (chemical gradient [ΔpH] and electrical gradient [Δψ]); (ii) a potassium ionophore, valinomycin, which dissipates the membrane potential (Δψ); (iii) respiratory chain uncouplers, KCN, which inhibits electron transport from complex III to complex IV, and oligomycin, which blocks the activity of the F1F0-ATPase. The action of these different inhibitors on the membrane potential or the respiratory functions is well established (e.g., see reference 13). All of these different treatments led to a significant loss of internalization of tRNAAla transcript into isolated mitochondria (Fig. 5A). Note that such a decrease in tRNA import was not due to the ethanol used to dissolve most of these inhibitors, since the efficiencies of tRNA internalization were comparable between import assays containing no ethanol and import assays containing 1% ethanol (data not shown). As an average for three independent experiments, we estimated an inhibition of about 70% (Fig. 5B). These results indicate that tRNAAla import requires an intact mitochondrial membrane electrochemical potential, as well as a functional respiratory chain. However, a residual amount of protected tRNA transcripts is still observed after pretreatment of mitochondria with any of these inhibitors (Fig. 5). This residual amount of protected tRNA transcript is in the same range as that observed in the absence of ATP when mitochondria were not posttreated with proteinase K (Fig. 2C).
FIG. 5.
Mitochondrial membrane potential and functional respiration are essential for tRNAAla import. Protonophores (FCCP, lane 6; CCCP, lane 7), an ionophore (valinomycin, lane 4), or specific inhibitors (KCN, lane 3; oligomycin, lane 5) were added to the standard import mixture and incubated for 10 min before adding the labeled tRNAAla transcripts (2 × 105 cpm). As controls, the labeled transcripts were incubated with mitochondria under standard import conditions without (lane 1) or with (lane 2) 5 mM ATP. (B) Histogram showing the import level of the labeled tRNAAla transcript quantified using a phosphorimager. The RNA import level in the presence of ATP and without any inhibitor was taken as 100%.
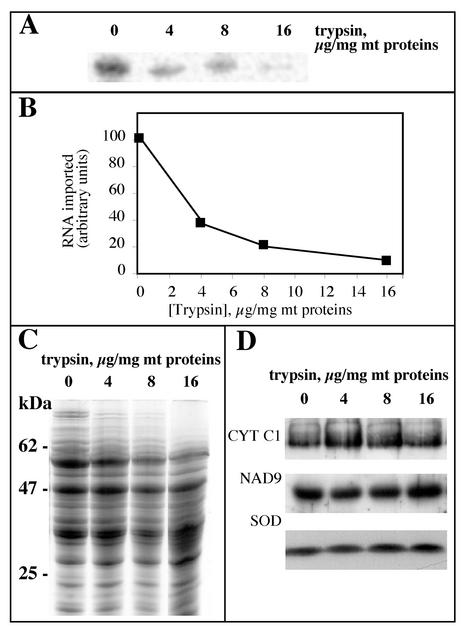
Dependence of the in vitro mitochondrial import of tRNAAla on outer membrane receptor protein(s).
In all systems studied so far, in vitro mitochondrial import of tRNAs depends on mitochondrial proteins associated with the outer membrane (1, 37, 47, 52). To determine whether a protein component on the surface of potato mitochondria is required for tRNA import, we treated isolated mitochondria with increasing amounts of trypsin and tested them for their ability to import labeled tRNAAla transcripts (Fig. 6A). Protease treatment of isolated mitochondria resulted in a dramatic decrease of their ability to import the RNA (Fig. 6B). To ensure that the pretreatment with trypsin did not affect the integrity of mitochondria but only degraded protease-sensitive outer membrane receptors, Western analyses were performed. Proteins of protease-treated mitochondria were probed with polyclonal antibodies directed against a matrix protein, the mitochondrial superoxide dismutase (SOD), an intrinsic protein of the inner membrane, the mitochondrial subunit 9 of NADH dehydrogenase (NAD9), and an extrinsic mitochondrial protein of the inner membrane largely exposed to the intermembrane space, cytochrome c1. Whereas a number of proteins appeared to be sensitive to trypsin digestion (this is particularly obvious for high-molecular-weight proteins [Fig. 6C]), Western profiles demonstrated that SOD, NAD9, and CYT C1 proteins were fully resistant, even for a trypsin concentration as high as 16 μg/mg of mitochondrial proteins (Fig. 6D), showing that the integrity of the mitochondria was not affected by trypsin treatment. Altogether, the results indicate that at least one protease-sensitive outer membrane protein receptor is required for mitochondrial tRNA import.
FIG. 6.
In vitro mitochondrial import of tRNAAla transcripts is dependent on surface-accessible proteins. (A) To remove proteins exposed on the outer membrane, isolated mitochondria were treated with increasing concentrations of trypsin (0, 4, 8, and 16 μg of mitochondrial proteins/mg). Labeled in vitro-transcribed tRNAAla (2 × 105 cpm) was then incubated with these different mitochondrial samples under standard import conditions, and protected RNAs were analyzed. (B) The extent of nuclease protection obtained in panel A was quantified using a phosphorimager. (C) Coomassie-blue-stained protein profiles of potato mitochondria treated with increasing concentrations of trypsin as shown in panel A. (D) Corresponding Western blots probed with antibodies directed against CYT C1, NAD9, and SOD.
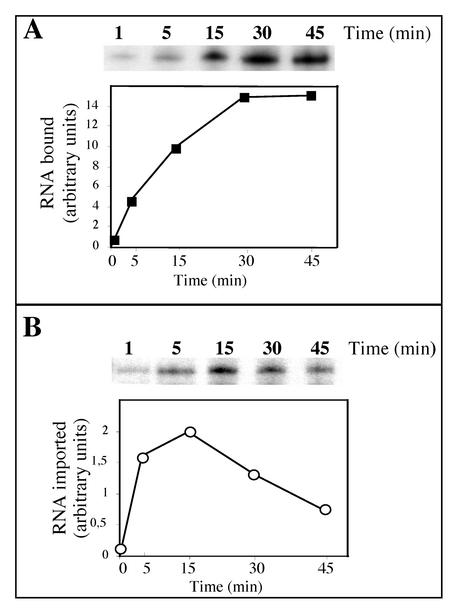
Time course of tRNAAla binding and import.
Isolated potato mitochondria were also tested for their ability to bind tRNAAla transcripts. The binding test was performed under the same conditions as for tRNA import except that the RNase digestion step was omitted. The time course of binding of tRNAAla transcripts to mitochondria and the time course of import were analyzed in parallel. Under standard assay conditions, binding of labeled tRNAAla transcripts to the outer membrane of mitochondria increases constantly to reach a plateau at 30 min (Fig. 7A). The estimated amount of complex formed after 30 min is about 15% of the input RNA present in the assay. The rate of RNA import is rather high, since protected RNA can already be detected after a few min of incubation and RNA import reaches a maximum after 15 min (Fig. 7B). Since ATP was not depleted in the binding experiment, the internalization of tRNA transcripts was possible. As a consequence, for incubation times shorter than 5 min, the amount of detected transcripts reflects not only the binding step but also the internalization. For longer incubations, the amount of transcripts recovered mainly reflects the binding step. This fact is also illustrated in Fig. 1B, where after a 20-min incubation of a standard import assay, the amount of imported tRNAs corresponds to about 10% of the amount of tRNA transcripts detected when the RNase treatment was omitted. In the experiment presented in Fig. 7B, the maximum amount of tRNA imported after 15 min was estimated to be about 20% of the bound tRNA, but in most assays the ratio of imported tRNA/bound tRNA was estimated to be between 0.1 and 0.15. Incubation longer than 15 to 20 min resulted in a decrease in the level of internalized RNA. A similar time frame was already reported for tRNA import into isolated mitochondria of T. brucei (52).
FIG. 7.
Time course of binding or import of tRNAAla transcripts. (A) Kinetics of binding of labeled tRNAAla (105 cpm) onto isolated mitochondria and graphical representation of the results. To test for binding, tRNAAla transcripts were incubated with isolated mitochondria under the same conditions as for tRNA import except that the RNase digestion step was omitted. (B) Kinetics of import of labeled tRNAAla (105 cpm) into isolated mitochondria and graphical representation of the results. Note that between panels A and B, exposure times of the autoradiography were very different (about 10 times longer exposure for import than for binding). For graphical representation of the results, the amounts of bound (squares) or protected (circles) RNAs were quantified using a phosphorimager.
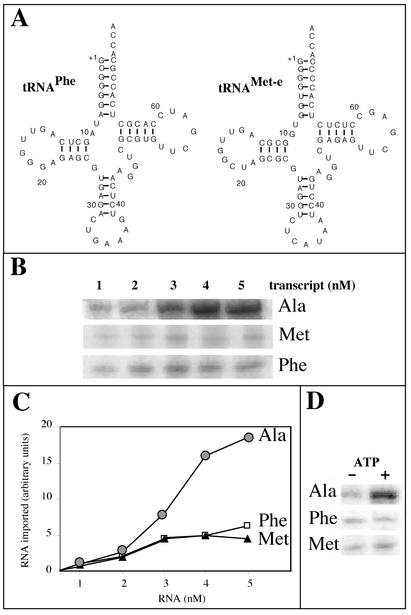
Specificity of tRNA import into isolated mitochondria.
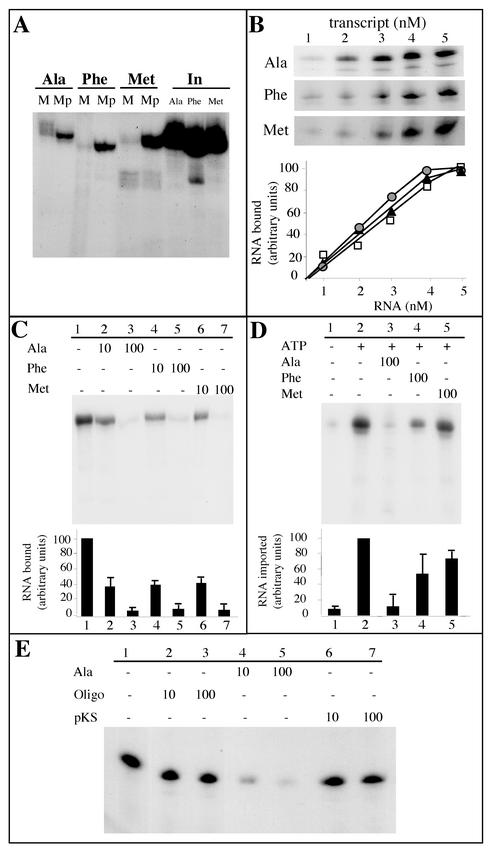
Two other tRNA substrates were used in our in vitro import assays: mature cytosolic tRNAPhe and tRNAMet-e (Fig. 8A). Previous in vivo analyses have shown that these two nuclearly encoded tRNAs remain in the cytosol and are not imported into potato mitochondria (26). As in the case of tRNAAla, radioactively labeled tRNAPhe and tRNAMet-e were prepared by in vitro transcription using T7 RNA polymerase. When these three labeled in vitro transcripts were incubated with isolated potato mitochondria under standard import conditions, RNase-protected transcripts were observed in the three cases (Fig. 8B). However, whereas the amounts of tRNAAla transcript protected from RNases increased continuously for increasing RNA input concentrations, the amounts of tRNAPhe and tRNAMet-e transcripts protected from RNases remained almost constant, even for high RNA input concentrations (Fig. 8C). We estimated that for an RNA input of 5 nM, about 2 to 3% of the input tRNAAla and about 0.5% of the input tRNAPhe or tRNAMet-e were protected from RNases after incubation with isolated mitochondria under these conditions. This difference is consistent with the in vivo localization of these tRNAs, where tRNAAla partitions between the cytosol and mitochondria, whereas tRNAPhe and tRNAMet-e are cytosolic in most species. Moreover, the ATP dependence of this process is observed only in the case of tRNAAla (Fig. 8D), that is, for the in vivo imported tRNA. By contrast, the efficient uptake of transcript into mitoplasts, compared to the import level into intact mitochondria, obtained for tRNAAla (Fig. 4B) was also observed for both tRNAPhe and tRNAMet-e (Fig. 9A). For these last two transcripts, the level of internalization into mitoplasts was in the same range as for tRNAAla, thus suggesting that in vitro, the passage through the inner membrane does not constitute a strong and selective barrier.
FIG. 8.
RNase protection of in vitro-transcribed tRNAAla, tRNAPhe, and tRNAMet incubated with isolated mitochondria. (A) Cloverleaf structures of the mature A. thaliana cytosolic tRNAPhe and tRNAMet-e used as substrates for standard in vitro import assays. (B) Increasing concentrations of labeled tRNAAla, tRNAPhe, and tRNAMet-e were incubated with isolated mitochondria in standard import assays. (C) The amounts of protected RNAs (circles, tRNAAla; squares, tRNAPhe; triangles, tRNAMet-e) were quantified using a phosphorimager, and the graph presented here is the result of three independent experiments. (D) Incubation of 3 nM concentrations of labeled in vitro-transcribed tRNAAla (Ala), tRNAPhe (Phe), and tRNAMet-e (Met) with isolated mitochondria in the absence (−) or in the presence (+) of 5 mM ATP under standard import conditions.
FIG. 9.
Analysis of tRNAAla, tRNAPhe, and tRNAMet-e binding to mitochondria or import into mitochondria. (A) Labeled in vitro-transcribed tRNAAla, tRNAPhe, and tRNAMet (2 × 105 cpm each) were incubated under standard import conditions with intact mitochondria (M) or with mitoplasts (Mp). In, input RNAs (5,000 cpm). (B) Bound tRNAAla, tRNAPhe, and tRNAMet-e as a function of tRNAAla, tRNAPhe, and tRNAMet-e concentration, respectively. Increasing concentrations of labeled tRNA transcripts (from 1 to 5 nM) were incubated with isolated mitochondria (corresponding to 100 μg of proteins). Incubation was performed for 10 min under the same conditions as for tRNA import, except that the RNase digestion step was omitted. (C) Competition experiments of tRNAAla binding onto isolated mitochondria using unlabeled tRNAAla, tRNAPhe, or tRNAMet-e as competitors. For these experiments, isolated mitochondria (corresponding to 100 μg of proteins) were preincubated, for 10 min on ice, in the absence of competitor (−) or in the presence of a 10-fold or a 100-fold excess of competitor. Labeled tRNAAla (105 cpm) was then added, and incubation was prolonged for 10 min under the same conditions as for tRNA import, except that the RNase digestion step was omitted. (D) Competition experiments of mitochondrial tRNAAla import into isolated mitochondria using unlabeled tRNAAla, tRNAPhe, or tRNAMet-e as competitor. For these experiments, isolated mitochondria (corresponding to 400 μg of proteins) were preincubated, for 10 min on ice, in the absence of competitor (−) or in the presence of a 100-fold excess of competitor. Then, 2 × 105 cpm of labeled tRNAAla was added, and incubation was prolonged for 15 min under the same conditions as for tRNA import. The experiments were performed in the absence (−) or in the presence (+) of 5 mM ATP. After incubation, a proteinase K treatment was performed as described in Materials and Methods prior to digestion with RNases. (E) Competition experiments of tRNAAla binding to isolated mitochondria using an unlabeled oligodeoxyribonucleotide (Oligo) corresponding to the complete sequence of the cytosol-specific tRNAMet-e (Fig. 8A) or AluI-digested pBluescript vector (pKS) as competitors. The competition experiments were conducted essentially as described for panel C. For graphical representation of the results in panels B, C, and D, the amounts of bound or protected RNAs were quantified using a phosphorimager. Bars represent standard deviations from two and three independent experiments for panels C and D, respectively, and mean values were used to plot the graph.
Quantitative analysis of tRNA binding.
Quantitative binding experiments performed at different tRNAAla transcript concentrations showed that binding of tRNAAla transcript was concentration dependent (Fig. 9B). Binding increased almost linearly and then reached a maximum for a tRNAAla transcript concentration of 4.5 to 5 nM that might correspond to a saturation of the receptor molecules on the surface of mitochondria. Similar binding experiments were performed with tRNAPhe and tRNAMet-e transcripts. As shown in Fig. 9B, the binding capacity of tRNAMet-e or tRNAPhe on the mitochondrial surface is essentially identical to that estimated for tRNAAla, thus suggesting that the specificity of import seen in vitro (Fig. 8) very likely is not due to a specific binding of the naturally imported tRNAAla versus the cytosol-localized tRNAPhe and tRNAMet-e on the mitochondrial outer membrane.
Unlabeled tRNAPhe and tRNAMet-e are efficient competitors for tRNAAla binding but weak competitors for tRNAAla import.
In view of the results presented above, competition experiments between the naturally imported tRNAAla and the nonimported tRNAPhe or tRNAMet-e were performed both at the level of binding and at the level of import. Binding of labeled tRNAAla transcript on the mitochondrial surface was almost completely competed out when a 100-fold molar excess of unlabeled tRNAAla transcript was added to the assay (Fig. 9C, line 3). Furthermore, the unlabeled tRNAPhe or tRNAMet-e transcripts were competing as efficiently as the tRNAAla transcript for binding of labeled tRNAAla (Fig. 9C, lines 4 to 7). These results are in good agreement with those of the binding experiment presented in Fig. 9B, where the behavior of the imported tRNAAla was roughly the same as the behavior of the two nonimported tRNAs. Altogether, these data suggest that the selectivity of tRNA import in our in vitro system is not the result of an interaction with receptors specific to the naturally imported tRNAs versus the cytosol-specific ones. However, quite interestingly, an oligodeoxyribonucleotide corresponding to the complete sequence of the cytosolic tRNAMet-e or pBluescript vector digested with AluI appeared to be a very poor competitor for tRNA binding. As shown in Fig. 9E, these single- or double-stranded DNA molecules were unable to inhibit more than 25% of tRNAAla binding to mitochondria when used in a 100-fold molar excess, thus demonstrating that the binding of tRNAs on the outer mitochondrial membrane is RNA specific.
When competition experiments where performed in import assays (Fig. 9D), the amount of labeled RNAAla protected against RNases was decreased by about 85% (as an average of three experiments) when a 100-fold molar excess of unlabeled tRNAAla transcript over the labeled input tRNA was added to the assay. By contrast, the tRNAPhe and tRNAMet-e unlabeled transcripts did not efficiently compete out the internalization of the labeled tRNAAla (Fig. 9D, lines 4 and 5). As an average of three experiments, only 50 and 30% of tRNAAla import inhibition were obtained when tRNAPhe and tRNAMet-e were used as competitors, respectively. These results strongly suggest that the outer membrane and/or the inter membrane space might function as a selective barrier to exclude non-naturally imported tRNAs. However, we cannot exclude that in vivo, cytosolic factors might act in synergy to enhance the specificity of the process.
DISCUSSION
In this paper, we report for the first time the establishment of an in vitro assay for the import of nuclearly encoded tRNAs into isolated plant mitochondria. The potato mitochondria used in this system were previously shown to be fully intact and functional, in particular for protein import (15, 18, 32, 38). Incubation of such isolated mitochondria with labeled tRNAAla led to protection of imported tRNA transcripts from added RNase A and RNase T1. We have shown that this protection is ATP dependent and requires a membrane potential. The use of mitoplasts, obtained by hypotonic disruption of the outer membrane and purification on discontinuous sucrose gradient, demonstrated that the protected tRNA molecules were mostly found in the matrix, where mitochondrial translation occurs. These data demonstrate that the RNA molecules were able to cross the double mitochondrial membrane, thus reflecting the in vivo situation. In addition, this in vitro RNA import does not require soluble cytosolic protein factors and is dependent upon at least one proteinaceous factor on the surface of mitochondria.
Treatment of isolated mitochondria with increasing amounts of trypsin resulted in a loss of their ability to import the labeled tRNAAla transcripts. The same results were obtained with proteinase K-treated mitochondria (data not shown). These data show that tRNA import into plant mitochondria depends on surface-accessible membrane protein(s). Similar results were also observed for S. cerevisiae, L. tropica, L. tarentolae, and T. brucei (for a review, see reference 17), suggesting that the requirement of protease-sensitive outer membrane receptor(s) is quite essential for tRNA import whatever the organism considered.
Another interesting feature of the tRNA import system that we have developed is its requirement for ATP. In this respect, it resembles all the in vitro tRNA import systems described so far for S. cerevisiae and for trypanosomatids (for a review, see reference 17). However, as for tRNA import into L. tarentolae mitochondria (41), the ATP concentration required for an efficient translocation of RNAs (5 to 10 mM) is higher than the ATP concentration (usually 1 mM) used for efficient in vitro protein import. For plants, for instance, it has been reported that an ATP concentration of 0.05 mM is sufficient for protein translocation into mitochondria (10). These contrasting results might reflect fundamental differences between tRNA and protein import pathways, but we cannot exclude that the in vitro system is not optimized and requires more ATP than necessary in vivo. We clearly demonstrated the ATP dependence of the in vitro import system, but further experiments will be necessary to define the individual step(s) requiring ATP for tRNA internalization. For instance, several receptors and/or channels could be involved in an ATP-dependent manner, as already suggested for the L. tropica in vitro import system (36), and elucidation of the energy requirement for tRNA translocation through the outer and inner mitochondrial membranes in plants will necessitate further studies with mitochondrial subfractions.
The requirement for an electrochemical membrane potential has been shown with an ionophore (valinomycin) or protonophores (FCCP and CCCP). Such a requirement has also been observed by several laboratories for plant mitochondrial protein import (50), as well as for yeast tRNA import (47). In trypanosomatids, the results are quite contradictory. Depending on the trypanosomatid considered and the method used to purify the mitochondria, an electrochemical membrane potential is or is not required (for a review, see reference 17). It is also quite clear that in potato, the presence of oligomycin, which inhibits the phosphorylation of ADP into ATP, or KCN, which blocks the electron transport, strongly reduces tRNA import, again demonstrating the importance of mitochondrial activity. Since oligomycin is an inhibitor of ATP synthesis on the matrix side of mitochondria, the strong reduction of tRNA import seen in the presence of this phosphorylation uncoupler could also be explained by the requirement of ATP inside mitochondria.
Binding of tRNAAla onto the surface of mitochondria is a rapid and efficient process occurring within a few minutes after coincubation of the labeled transcript with isolated mitochondria. This rapid binding is followed by a fast internalization of the transcripts into mitochondria: most of the protected RNA is detected within 5 min of incubation of tRNAAla transcripts with isolated mitochondria. A rapid import of the tandemly linked tRNAThr and tRNALeu into T. brucei isolated mitochondria (52), of mature tRNAIle or tRNAGln into isolated L. tarentolae mitochondria (41), or of tRNALys (CUU) into S. cerevisiae mitochondria (49; I. Tarassov, personal communication) has also been reported. In all of these cases, the maximum level of nuclease protection is reached within 10 to 15 min of incubation with isolated mitochondria. So far the only exception to this time course has been described for the import of synthetic antisense transcripts (from the 5′-upstream region of the β-tubulin gene) into L. tropica mitochondria, which requires a lag of 30 min before detection of nuclease-protected transcripts (31). The latter result might be explained by differences in the protocols used to isolate mitochondria as already suggested (52), but we cannot exclude that it might reflect important differences in the import pathways.
Interestingly, the time course observed here for import of tRNA into potato mitochondria is much shorter than the time course reported for protein import into plant mitochondria (plant mitochondrial protein import seems to be linear up to 30 min [E. Glaser and J. Whelan, personal communication]). Such differences in the time course of import between tRNAs and proteins might reflect the use of different channels to direct these two types of macromolecules into plant mitochondria. Another explanation could be that the same channel is used (i.e., the protein import channel), but tRNAs are not engaged in this import system in the same way and their transport is facilitated compared to protein import, at least in vitro. However, considering tRNAs as large, hydrophilic and negatively charged macromolecules, they are unlikely to be good substrates for protein transporters through mitochondrial membranes.
Results presented in this paper show the preference for the cytosolic tRNAAla (localized both in the cytosol and in the mitochondria in vivo) as an import substrate over cytosolic tRNAPhe or tRNAMet-e substrates (localized in the cytosol in vivo in most species), thus reflecting the selectivity of our in vitro import assay. The specificity seen when studying tRNA internalization is not transposable to the binding experiments, where the three tRNA substrates behave similarly. The latter result is strengthened by the fact that both tRNAPhe and tRNAMet-e transcripts are able to compete out labeled tRNAAla transcript for binding as efficiently as does unlabeled tRNAAla. Interestingly, however, the interactions between tRNAs and outer membrane receptors seem to involve specific RNA-binding proteins that are not able to efficiently bind DNA. By contrast, in import studies, only the unlabeled tRNAAla transcript was able to almost fully compete out the internalization of the labeled tRNAAla. Taken together, these data strongly suggest, first, that the binding of the tRNAs on receptor(s) present at the surface of the outer membrane is not responsible for the specificity occurring in vivo, and second, that the passage through the outer membrane is at least partially responsible for this selectivity. Furthermore, when mitoplasts were used in the import assays, the uptake of tRNA transcripts was far more efficient, whatever the substrate (naturally imported or not), suggesting that in plants, the inner membrane constitutes neither a strong nor a specific barrier for tRNA internalization into the mitochondrial matrix.
For S. cerevisiae, only one mature nuclearly encoded tRNALys is imported into mitochondria. For trypanosomatids, a wide variety of mitochondrial import substrates has been successfully used (e.g., mature tRNAs, cotranscribed tRNAs, and various heterologous RNA substrates), and the functional significance of these data remains to be clarified. In higher plants, imported tRNAs represent one-third to one-half of the tRNA population (19, 26). Here, we show that at least mature tRNAs can be imported into plant mitochondria, but this does not exclude the possibility that other substrates could be used. It will be necessary in the future to study the extent of this specificity using other naturally imported or nonimported tRNAs, as well as heterologous RNAs, as substrates. We can then expect that further experiments will help in elucidating the sequence and/or structural features characteristic for the imported tRNAs. The characterization of the mitochondrial proteins responsible on the one hand for the binding of tRNAs on the surface of mitochondria and on the other hand for the selectivity of the translocation through the outer membrane will now be crucial for further understanding of this specific import process.
It should be noted that a low level of incorporation was observed in the case of tRNAPhe and tRNAMet-e, but in contrast to tRNAAla import, this residual nuclease protection seems to be independent of the presence of ATP and of the membrane potential (data not shown). In the case of tRNAAla, a background incorporation was also observed in the absence of ATP or when mitochondria were treated with agents dissipating the electrochemical membrane potential. This residual amount of protected tRNA transcripts, which could be efficiently removed by a mild proteinase K treatment of mitochondria prior to RNase digestion, is thus due mostly to nonspecific association of labeled tRNA transcripts with the outer membrane.
Quite surprisingly, it seems that our in vitro system does not require addition of a cytosolic protein extract. Previous data have shown that the enzymatic extract used in these experiments is able (i) to correctly aminoacylate plant cytosolic tRNAAla transcript (9) and (ii) to efficiently replace the yeast cytosolic S-100 supernatant required for import of tRNALys into isolated S. cerevisiae mitochondria (I. Tarassov, N. Entelis, A. Dietrich, and L. Maréchal-Drouard, unpublished data), suggesting that mature and precursor forms of mitochondrial aaRSs are present in our protein fraction. Furthermore, the conditions we used for in vitro tRNA import are very similar to those described for in vitro protein import into isolated potato mitochondria (51), and as a matter of fact, in vitro protein import was efficient under these tRNA import conditions (data not shown), in case coimport would have to occur. It should also be noted that preaminoacylation of tRNAAla did not improve its import efficiency in our in vitro system (data not shown), whereas aminoacylation is an essential requirement for import of tRNALys into S. cerevisiae mitochondria (48). These results appear somehow in contradiction with the in vivo situation, since previous work has shown that the aaRSs probably play an important role in the tRNA import process. In particular, a striking correlation between alanylation and tRNAAla import has been previously reported with transgenic tobacco plants (11). Similarly, we obtained recent evidence that when aminoacylation of normally imported cytosolic tRNAVal is switched from valine to methionine (cytosolic tRNAMets are not imported into plant mitochondria), its import into mitochondria of transgenic plants is completely abolished (10a).
Several hypotheses can be proposed to explain these discrepancies between the in vitro and in vivo situations. AaRSs might play an important role in determining the specificity of the tRNA import process, but not by participating directly in the translocation of the tRNA through the double mitochondrial membranes. Specificity could be given by the aminoacylation of the tRNAs (hence the necessity for the aaRS), but the in vitro data presented above argue against this hypothesis. Also, it does not explain the selective import of only some tRNAGly isoacceptors observed in potato and bean mitochondria (7). AaRSs might be involved in carrying the imported tRNAs to the surface of the outer membrane of mitochondria, and/or they might be used to stabilize tRNAs once they have entered mitochondria. If one of these two hypotheses is correct, external addition of these enzymes would not be necessary for import of mitochondrial tRNA in vitro, and the transport of tRNAs through the mitochondrial membranes could use a specific but still unknown RNA import channel, rather than the protein import channel.
The in vitro tRNA import system developed here for potato mitochondria was shown to be dependent on ATP, outer membrane protein receptor(s), and a membrane potential, which are requirements similar to those of protein import. Furthermore, the translocation step through the outer membrane seems to play an important role for the specificity of the process. It will be essential now to address the following question: do tRNAs use the protein import machinery to cross the double mitochondrial membrane? The use of antibodies directed against components of the protein import pathway, or the use of covalent conjugates (16) in combination with in vitro tRNA import, will help to solve this problem.
Acknowledgments
We thank Robert Martin, Ian Small, Anne-Marie Duchêne, and Jacques-Henry Weil for helpful discussions and critical reading of the manuscript and/or of the Ph.D. thesis.
This work was supported by the Centre National de la Recherche Scientifique (UPR2357) and by a fellowship from the French Ministère de l'Enseignement Supérieur et de la Recherche to L.D.
REFERENCES
- 1.Adhya, S., T. Ghosh, A. Das, S. K. Bera, and S. Mahapatra. 1997. Role of an RNA-binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem. 272:21396-21402. [DOI] [PubMed] [Google Scholar]
- 2.Akama, K., and M. Kashihara. 1996. Plant nuclear tRNAMet genes are ubiquitously interrupted by introns. Plant Mol. Biol. 32:427-434. [DOI] [PubMed] [Google Scholar]
- 3.Akashi, K., M. Takenaka, S. Yamaoka, Y. Suyama, H. Fukuzawa, and K. Ohyama. 1998. Coexistence of nuclear DNA-encoded tRNAVal (AAC) and mitochondrial DNA-encoded tRNAVal (UAC) in mitochondria of a liverwort Marchantia polymorpha. Nucleic Acids Res. 26:2168-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aphasizhev, R., U. Karmarkar, and L. Simpson. 1998. Are tRNAs imported into the mitochondria of kinetoplastid protozoa as 5′-extended precursors? Mol. Biochem. Parasitol. 93:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, M. F., S. Hofmann, W. Neupert, and M. Brunner. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10:25-31. [DOI] [PubMed] [Google Scholar]
- 6.Bowler, C., L. Slooten, S. Vandenbranden, R. De Rycke, J. Botterman, C. Sybesma, M. Van Montagu, and D. Inzé. 1991. Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. Proc. Natl. Acad. Sci. USA 10:1723-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brubacher-Kauffmann, S., L. Maréchal-Drouard, A. Cosset, A. Dietrich, and A. M. Duchêne. 1999. Differential import of nuclear-encoded tRNAGly isoacceptors into Solanum tuberosum mitochondria. Nucleic Acids Res. 27:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger, G., K. M. Plante, K. M. Lonergan, and M. W. Gray. 1995. The mitochondrial DNA of the amoeboid protozoan, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J. Mol. Biol. 245:522-537. [DOI] [PubMed] [Google Scholar]
- 9.Carneiro, V. T. C., A. Dietrich, L. Maréchal-Drouard, A. Cosset, G. Pelletier, and I. Small. 1994. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol. Biol. 26:1843-1853. [DOI] [PubMed] [Google Scholar]
- 10.Chaumont, F., V. O'Riordan, and M. Boutry. 1990. Protein transport into mitochondria is conserved between plant and yeast species. J. Biol. Chem. 265:16856-16862. [PubMed] [Google Scholar]
- 10a.Delage, L., A. M. Duchêne, and L. Maréchale-Drouard. Plant J., in press.
- 11.Dietrich, A., L. Maréchal-Drouard, V. Carneiro, A. Cosset, and I. Small. 1996. A single base change prevents import of cytosolic tRNAAla into mitochondria in transgenic plants. Plant J. 10:913-918. [DOI] [PubMed] [Google Scholar]
- 12.Dörner, M., M. Altmann, S. Paabo, and M. Mörl. 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol. Biol. Cell 12:2688-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douce, R. 1985. Mitochondria in higher plants. Structure, function and biogenesis. Academic Press Inc., Orlando, Fla.
- 14.Douce, R., E. L. Christensen, and W. D. Bonner. 1972. Preparation of intact plant mitochondria. Biochem. Biophys. Acta 275:148-160. [DOI] [PubMed] [Google Scholar]
- 15.Duchêne, A. M., N. Peeters, A. Dietrich, A. Cosset, I. D. Small, and H. Wintz. 2001. Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J. Biol. Chem. 276:15275-15283. [DOI] [PubMed] [Google Scholar]
- 16.Entelis, N. S., O. A. Kolesnikova, S. Dogan, R. P. Martin, and I. A. Tarassov. 2001. 5S rRNA and tRNA import into human mitochondria: comparison of in vitro requirements. J. Biol. Chem. 276:45642-45653. [DOI] [PubMed] [Google Scholar]
- 17.Entelis, N. S., O. A. Kolesnikova, R. P. Martin, and I. A. Tarassov. 2001. RNA delivery into mitochondria. Adv. Drug. Deliv. Rev. 49:199-215. [DOI] [PubMed] [Google Scholar]
- 18.Fey, J., M. Vermel, J. Grienenberger, L. Maréchal-Drouard, and J. M. Gualberto. 1999. Characterization of a plant mitochondrial active chromosome. FEBS Lett. 458:124-128. [DOI] [PubMed] [Google Scholar]
- 19.Glover, K. E., D. F. Spencer, and M. W. Gray. 2001. Identification and structural characterization of nucleus-encoded transfer RNAs imported into wheat mitochondria. J. Biol. Chem. 276:639-648. [DOI] [PubMed] [Google Scholar]
- 20.Hancock, K., and S. L. Hajduk. 1990. The mitochondrial tRNAs of Trypanosoma brucei are nuclear-encoded. J. Biol. Chem. 265:19208-19215. [PubMed] [Google Scholar]
- 21.Hauser, R., and A. Schneider. 1995. tRNAs are imported into mitochondria of Trypanosoma brucei independently of their genomic context and genetic origin. EMBO J. 14:4212-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heins, L., H. Mentzel, A. Schmid, R. Benz, and U. K. Schmitz. 1994. Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J. Biol. Chem. 269:26402-26410. [PubMed] [Google Scholar]
- 23.Hermann, J. M., and W. Neupert. 2000. Protein transport into mitochondria. Curr. Opin. Microbiol. 3:210-214. [DOI] [PubMed] [Google Scholar]
- 24.Hoogenraad, N. J., and M. T. Ryan. 2001. Translocation of protein into mitochondria. IUBMB Life 51:345-350. [DOI] [PubMed] [Google Scholar]
- 25.Kapushoc, S. T., J. D. Alfonzo, M. A. Rubio, and L. Simpson. 2000. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J. Biol. Chem. 275:37907-37914. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, R., L. Maréchal-Drouard, K. Akama, and I. Small. 1996. Striking differences in mitochondrial tRNA import between different plant species. Mol. Gen. Genet. 252:404-411. [DOI] [PubMed] [Google Scholar]
- 27.Lamattina, L., D. Gonzalez, J. M. Gualberto, and J. M. Grienenberger. 1993. Higher plant mitochondria encode an homologue of the nuclear-coded 30 kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217:831-838. [DOI] [PubMed] [Google Scholar]
- 28.LeBlanc, A. J., A. E. Yermovsky-Kammerer, and S. L. Hajduk. 1999. A nuclear encoded and mitochondrial imported dicistronic tRNA precursor in Trypanosoma brucei. J. Biol. Chem. 274:21071-21077. [DOI] [PubMed] [Google Scholar]
- 29.Lima, B. D., and L. Simpson. 1996. Sequence-dependent in vivo import of tRNAs into the mitochondrion of Leishmania tarentolae. RNA 2:429-440. [PMC free article] [PubMed] [Google Scholar]
- 30.Mahapatra, S., S. Ghosh, S. K. Bera, T. Ghosh, A. Das, and S. Adhya. 1998. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 26:2037-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahapatra, S., T. Ghosh, and S. Adhya. 1994. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 22:3381-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maréchal-Drouard, L., A. Cosset, C. Remacle, D. Ramamonjisoa, and A. Dietrich. 1996. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell. Biol. 16:3504-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maréchal-Drouard, L., P. Guillemaut, A. Cosset, M. Arbogast, F. Weber, J. H. Weil, and A. Dietrich. 1990. Transfer RNAs of potato (Solanum tuberosum) mitochondria have different genetic origins. Nucleic Acids Res. 18:3689-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maréchal-Drouard, L., I. Small, J. H. Weil, and A. Dietrich. 1995. Transfer RNA import into plant mitochondria. Methods Enzymol. 260:310-327. [DOI] [PubMed] [Google Scholar]
- 35.Martin, R. P., J. M. Schneller, A. J. Stahl, and G. Dirheimer. 1979. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 18:4600-4605. [DOI] [PubMed] [Google Scholar]
- 36.Mukherjee, S., S. N. Bhattacharyya, and S. Adhya. 1999. Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J. Biol. Chem. 274:31249-31255. [DOI] [PubMed] [Google Scholar]
- 37.Nabholz, C. E., E. K. Horn, and A. Schneider. 1999. tRNAs and proteins are imported into mitochondria of Trypanosoma brucei by two distinct mechanisms. Mol. Biol. Cell 10:2547-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuburger, M., E. P. Journet, R. Bligny, J. P. Carde, and R. Douce. 1982. Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys. 217:312-323. [DOI] [PubMed] [Google Scholar]
- 39.Perret, V., A. Garcia, H. Grosjean, J. P. Ebel, C. Florentz, and R. Giege. 1990. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature 344:787-789. [DOI] [PubMed] [Google Scholar]
- 40.Ramamonjisoa, D., S. Kauffmann, N. Choisne, L. Maréchal-Drouard, G. Green, H. Wintz, I. Small, and A. Dietrich. 1998. Structure and expression of several bean (Phaseolus vulgaris) nuclear transfer RNA genes: relevance to the process of tRNA import into plant mitochondria. Plant Mol. Biol. 36:613-625. [DOI] [PubMed] [Google Scholar]
- 41.Rubio, M. A., X. Liu, H. Yuzawa, J. D. Alfonzo, and L. Simpson. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA 6:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schneider, A., and L. Maréchal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol. 10:509-513. [DOI] [PubMed] [Google Scholar]
- 44.Simpson, A. M., Y. Suyama, H. Dewes, D. A. Campbell, and L. Simpson. 1989. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 17:5427-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Small, I., L. Maréchal-Drouard, J. Masson, G. Pelletier, A. Cosset, J. H. Weil, and A. Dietrich. 1992. In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants: towards novel ways of influencing mitochondrial gene expression? EMBO J. 11:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, T. H. P., R. Pach, A. Crausaz, A. Ivens, and A. Schneider. 2002. tRNAs in Trypanosoma brucei: Genomic organization, expression and mitochondrial import. Mol. Cell. Biol. 22:3707-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarassov, I., N. Entelis, and R. P. Martin. 1995. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol. 245:315-323. [DOI] [PubMed] [Google Scholar]
- 48.Tarassov, I., N. Entelis, and R. P. Martin. 1995. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 14:3461-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarassov, I. A., and N. S. Entelis. 1992. Mitochondrially-imported cytoplasmic tRNA(Lys) (CUU) of Saccharomyces cerevisiae: in vivo and in vitro targetting systems. Nucleic Acids Res. 20:1277-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan, J., and E. Glaser. 1997. Protein import into plant mitochondria. Plant Mol. Biol. 33:771-789. [DOI] [PubMed] [Google Scholar]
- 51.Wischmann, C., and W. Schuster. 1995. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA mediated transfer and exon shuffling at the integration site. FEBS Lett. 374:152-156. [DOI] [PubMed] [Google Scholar]
- 52.Yermovsky-Kammerer, A. E., and S. L. Hajduk. 1999. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol. Cell. Biol. 19:6253-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]