Abstract
β-Catenin signaling plays an important role in the development of many organisms and has a key part in driving the malignant transformation of epithelial cells comprising a variety of cancers. β-Catenin can activate gene expression through its association with transcription factors of the lymphoid enhancer factor 1 (LEF-1)/T-cell factor (TCF) family. We designed a screen in human cells to identify novel genes that activate a β-catenin-LEF/TCF-responsive promoter and isolated the high-mobility group box transcription factor, UBF2. UBF1 and UBF2 are splice variants of a common precursor RNA. Although UBF1 has been shown to activate RNA polymerase I-regulated genes, the function of UBF2 has remained obscure. Here, we show for the first time that both UBF1 and UBF2 activate RNA polymerase II-regulated promoters. UBF2 associates with LEF-1, as shown by coimmunoprecipitation experiments, and potentiates transcriptional activation stimulated by LEF-1/β-catenin from a synthetic promoter with multimerized LEF/TCF binding sites and a natural cyclin D1 promoter with consensus LEF/TCF binding sites. Downregulation of endogenous UBF expression using an RNA interference approach reduces transcriptional activation of a β-catenin-LEF/TCF-responsive promoter by means of overexpressed β-catenin, further implicating UBF as a transcriptional enhancer of the β-catenin pathway.
The Wnt/Wingless signaling pathway plays a key role in development, cellular differentiation, and oncogenesis and uses similar signaling components to drive these diverse biological processes (59). A pivotal regulator of the pathway is the β-catenin protein, whose stability and subcellular localization determine whether the pathway is active (83). The β-catenin protein has been localized to three subcellular compartments, including (i) the plasma membrane, complexed with E-cadherin and α-catenin to promote cell adhesion; (ii) the cytoplasm, as part of a multiprotein complex including glycogen synthase kinase (GSK) 3β, adenomatous polyposis coli tumor suppressor (APC), axin, and protein phosphatase 2A, which regulates β-catenin phosphorylation and ubiquitin-mediated degradation; and (iii) the nucleus, where direct binding to lymphoid enhancer factor (LEF)/T-cell factor (TCF) transcription factors activates downstream target genes (49, 84).
In nontransformed cells and in the absence of growth factor signals, β-catenin levels are tightly regulated by phosphorylation events that bring about its ubiquitin-mediated degradation (1). The β-catenin degradation process can be inhibited by growth factor signaling or by mutations in β-catenin and APC that have been later on detected in a variety of tumors (9, 51, 58, 60).
In either case, stabilized β-catenin complexes with LEF/TCF proteins and activates transcription from a variety of genes, including those for cyclin D1, c-Myc, and matrix metalloproteinase 7 (7, 10, 20, 25, 28, 48, 70, 76). Constitutive activation of β-catenin signaling and downstream target genes is thought to potentiate malignant processes, such as enhanced cell proliferation, inhibition of apoptosis, stimulation of cell motility, and tumor formation (3, 51, 56, 82).
The vertebrate LEF-1 and TCF (TCF-1, TCF-3, and TCF-4) proteins belong to the high-mobility group (HMG) family of transcription factors (64). These transcription factors serve as contexts repressors in some and as transcriptional activators in others. For example, LEF/TCF proteins associate with the Groucho family proteins to repress Wnt/β-catenin target genes, whereas in association with β-catenin they can serve as activators of the same genes (7, 14, 34, 42, 48, 65).
Since members of the β-catenin pathway and its effector genes likely represent important targets for therapeutic intervention in cancers, we established a functional screen in human cells to identify novel genes that activate the β-catenin pathway. We first constructed a stable reporter cell line containing a LEF/TCF-responsive green fluorescent protein (GFP) reporter gene, along with a LEF-1 expression vector to enhance the detection of active cDNAs. These cells were used to screen a cDNA library derived from human tumor RNA, and the β-catenin signaling events were detected by sorting GFP-positive cells. After several rounds of enrichment for active cDNA clones, the HMG box transcription factor, UBF2, was isolated. Mammalian cells contain two UBF isoforms with molecular masses of 97 and 94 kDa, which are generated by alternative splicing of a common RNA precursor (8, 33, 53). The 97-kDa isoform is referred to as UBF1, and the 94-kDa isoform, which contains a 37-amino-acid deletion in HMG box 2, is referred to as UBF2. The UBF1 transcription factor is a component of the RNA polymerase I preinitiation complex and activates rRNA genes by recruiting human SL1 (hSL1) (murine TIF-1B [mTIF-IB]) to the UCE and CORE promoter elements (8, 30, 38). Like UBF1, UBF2 interacts with SLI; however, it activates rRNA genes very poorly (39, 41).
Here, we show by using overexpressed proteins that UBF2 coimmunoprecipitates with LEF-1 and potentiates the β-catenin/LEF-1 signal from a synthetic promoter with multimerized LEF/TCF binding sites and a natural cyclin D1 promoter with consensus LEF/TCF binding sites. UBF1 showed a similar ability to activate transcription. Endogenous UBF mRNA was targeted for degradation by a retroviral vector expressing a short hairpin RNA (shRNA). In the presence of the UBF-targeted shRNA, reporter gene expression induced by activated β-catenin was diminished. As shown by immunofluorescence staining, UBF2 colocalized with LEF-1 in the nuclei of cells, a site for mRNA synthesis by RNA polymerase II. Our results show that UBF1 and UBF2 are potentiators of the β-catenin signal transduction pathway and can direct RNA polymerase II transcription.
MATERIALS AND METHODS
Vectors.
The mammalian expression vector, pCGN (74), was used to express cDNAs encoding wild-type (wt) β-catenin, ΔN89 β-catenin, Wnt-1, LEF-1, TCF-4, wt UBF1, wt UBF2, fusion (fu) UBF2, and C-terminal deletion (cd) UBF2 proteins in transfected cells. pCGN vector utilizes a cytomegalovirus promoter and allows the synthesis of proteins with an amino-terminal influenza virus hemagglutinin (HA) epitope tag. For expression of the cDNA library, the pCGN vector was modified by removing the thymidine kinase leader with initiating methionine and the HA epitope tag, allowing translation from AUGs internal to the cDNA. In addition, a new polylinker containing EcoRI, XbaI, KpnI, SmaI, XhoI, and BamHI sites was added. The cDNA library was cloned into this modified vector using the EcoRI-XhoI restriction sites. The cDNA library was prepared from a mixture of RNAs obtained from human breast, colon, and prostate tumor tissues. The library was size selected, and the 1.6- to 3.0-kb fraction used for the screen contained 5 × 104 independent inserts. The reporter plasmids consisted of six tandem copies of the wt or mutant LEF/TCF binding site, similar to the sequences in the pTOPFLASH and pFOPFLASH plasmids (40), inserted upstream of a minimal interleukin 2 (IL-2) promoter driving luciferase or enhanced GFP expression (62). The 6×TCF binding sites were transferred to the pLH-Z12-I-PL retroviral vector (62), which contained the hygromycin resistance gene for stable integration and selection in the HT1080 cells. The cyclin D1 reporter (a gift from Frank McCormick) consisted of the cyclin D1 promoter region from +134 to −962 driving luciferase expression and a mutant derivative with a mutation in the primary LEF/TCF site at position −75 (76). The pMSCVhygro vector (Clontech) was used for retroviral delivery of shRNA. The human U6 promoter was cloned into pMSCVhygro using the BglII and HpaI restriction sites to generate MSCV-U6. A small polylinker region was inserted into the HpaI site to generate additional cloning sites, including BamHI, NotI, and MluI. The shRNA against UBF was cloned into the BamHI and MluI restriction sites of MSCV-U6 as a double-stranded oligonucleotide. The sequence of the top strand was 5′-GATCCGTACATTGACAGAATTGATCTTCAAGAGAGATCAATTCTGTCAATGTACTTTTTGGAAA-3′.
Transfections.
HT1080 cells were grown in Dulbecco modified Eagle's medium supplemented with 10% fetal bovine serum and 1% nonessential amino acids. COS7 cells were grown in Dulbecco modified Eagle's medium supplemented with 10% fetal bovine serum. All transfections were performed using FuGENE6 (Boehringer). With the GFP reporter gene, fluorescence-activated cell sorting (FACS) was used to measure the mean GFP fluorescence of all cells (Fig. 1B and D, 2A, and 3; see also Fig. 10A) or the number of GFP-positive cells (see Fig. 5A). Luciferase activity was measured with the dual-luciferase reporter assay system (Promega) (see Fig. 5C, 6, and 9A). To control for the transfection efficiency of the luciferase reporter gene, 10 ng of pRL-TK Renilla luciferase expression vector (Promega) was cotransfected with each sample. All experiments were performed in duplicate or triplicate. The HT1080, REP, and REP-LEF cells were seeded on 12-well plates and transfected at 30% confluence using 0.5 μg of each reporter plasmid, 0.25 μg of each expression vector, or 0.05 μg of each expression vector for cooperation experiments. The total amount of DNA was adjusted to 1.5 μg with pUC119. For the coimmunoprecipitation experiment, COS7 cells were seeded on 10-cm2 plates and transfected at 50% confluence using 10 μg of each expression vector plus 10 μg of pUC119 DNA. For the RNA interference (RNAi) experiment, REP-LEF cells were seeded on six-well plates and transfected at 30% confluence using 0.5 μg of β-catenin expression plasmid and 2.5 μg of MSCV-U6 or MSCV-U6-UBF.
FIG. 1.
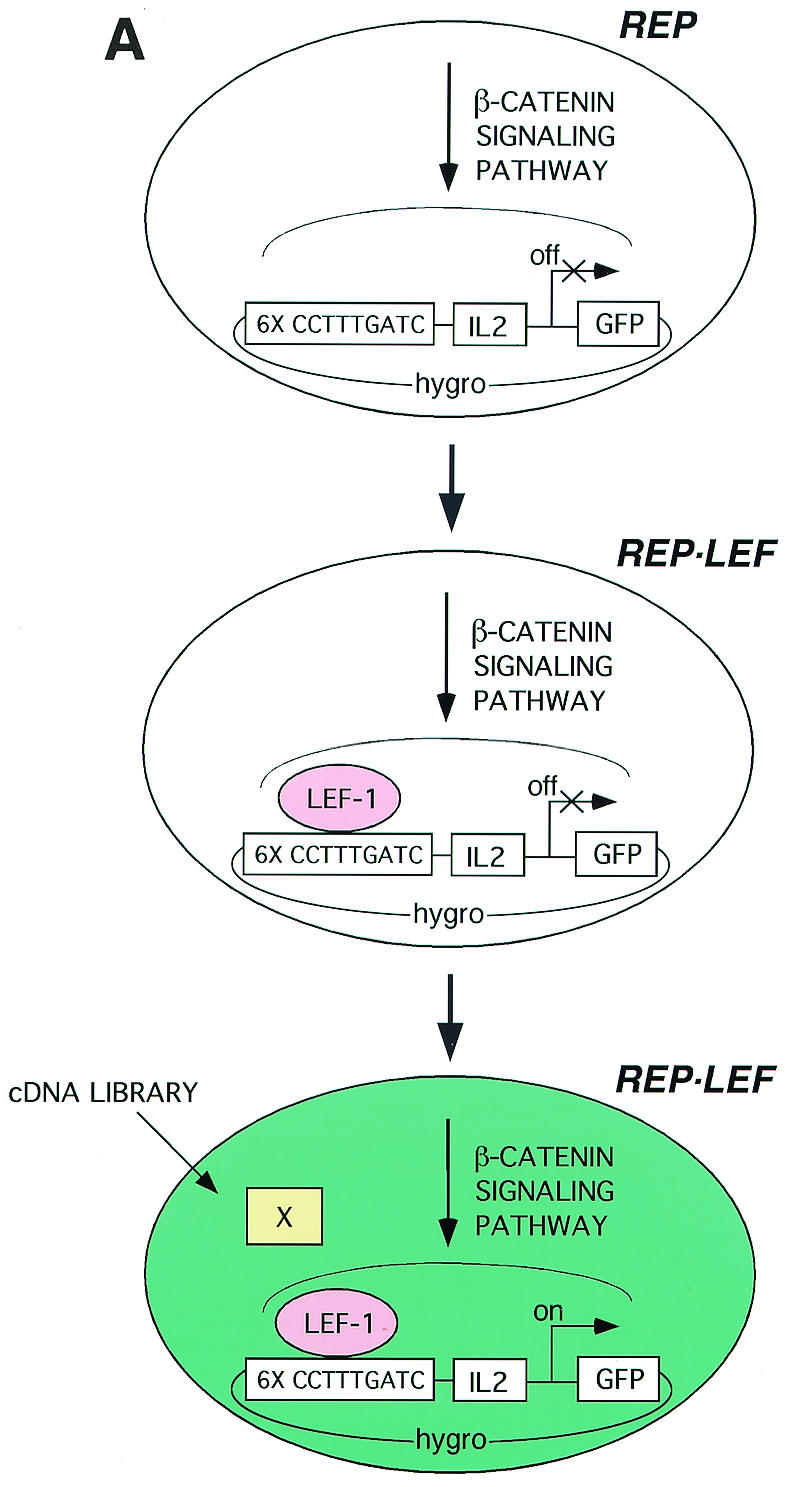
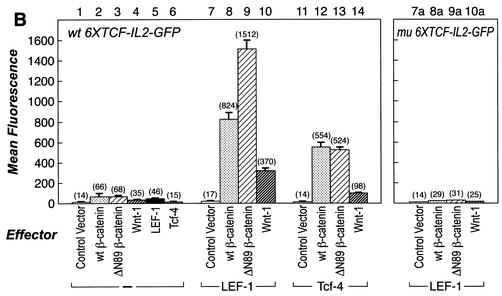
(A) Overview of strategy and process for a mammalian functional screen. Six tandem copies of a LEF/TCF response element (6× CCTTTGATC) were positioned upstream of an IL-2 minimal promoter to drive expression of the reporter GFP gene. The reporter construct was transfected into HT1080 human fibrosarcoma cells, and a stable cell line (REP) was selected. The REP cell lines were further modified by transfection with an expression vector encoding LEF-1 followed by selection of a stable cell line (REP-LEF). The reporter gene is inactive in REP and REP-LEF cells. The REP-LEF cells were used to screen a plasmid-based cDNA library prepared from human tumor cells, GFP-expressing cells were selected by FACS. (B) Reporter gene evaluation by transient transfection with test plasmids. HT1080 cells were cotransfected with the wt reporter gene (wt 6×TCF-IL-2-GFP) (bars 1 to 14) or a reporter gene with mutations in the LEF/TCF binding site (mu 6×TCF-IL-2-GFP) (bars 7a to 10a) and either an empty expression vector (Control Vector) (bars 1, 7, 11, and 7a) or an expression vector encoding β-catenin (wt β-catenin) (bars 2, 8, 12, and 8a), an activated mutant form of β-catenin (ΔN89β-catenin) (bars 3, 9, 13, and 9a), Wnt-1 (bars 4, 10, 14, and 10a), LEF-1 (bar 5), or TCF-4 (bar 6). Bars 1 to 6, cells transfected with the indicated individual expression vectors; bars 7 to 10 and 7a to 10a, cells transfected with the indicated expression vectors along with a LEF-1 expression vector; bars 11 to 14, cells transfected with the indicated expression vectors along with a TCF-4 expression vector. Forty-eight hours after transfection, the fluorescence level of GFP-expressing cells was determined by FACS. The error bars indicate standard deviations. (C) Confirmation of protein expression. Protein extracts were prepared from the transfected cells evaluated in panel B and analyzed by Western immunoblotting with an antibody directed against the HA epitope tag encoded at the amino terminus of each protein. LEF-1 comigrates with a background band. (D) Evaluation of the REP cell line, containing a stable integration of the reporter gene, with test vectors. REP cells were transfected with either an empty expression vector (Control Vector) (bars 1, 7, and 11) or expression vectors encoding wt β-catenin (bars 2, 8, and 12), an activated mutant form of β-catenin (ΔN89 β-catenin) (bars 3, 9, and 13), Wnt-1 (bars 4, 10, and 14), LEF-1 (bar 5), or TCF-4 (bar 6). Bars 1 to 6, cells transfected with the indicated individual expression vectors; bars 7 to 10, cells transfected with the indicated expression vectors along with a LEF-1 expression vector; bars 11 to 14, cells transfected with the indicated expression vectors along with a TCF-4 expression vector. Forty-eight hours after transfection, the fluorescence level of GFP-expressing cells was determined by FACS.
FIG. 2.
Stable integration of a LEF-1 expression plasmid in REP cells to generate REP-LEF cells for a functional screen. (A) Evaluation of the REP-LEF cell line, containing a stable integration of the reporter gene as well as an expression vector encoding LEF-1, with test vectors. REP-LEF cells were transfected with either an empty expression vector (Control Vector) (bar 1) or expression vectors encoding wt β-catenin (bar 2), an activated mutant form of β-catenin (ΔN89 β-catenin) (bar 3), Wnt-1 (bar 4), or LEF-1 (bar 5). Forty-eight hours after transfection, the fluorescence level of GFP-expressing cells was determined by FACS. The error bars indicate standard deviations. (B) Confirmation of LEF-1 protein expression in REP-LEF cells. High-salt nuclear extracts were prepared from REP (lanes 1 and 4), REP-LEF (lanes 2 and 5), and COS7 (lane 3 and 6) cells and analyzed by Western immunoblotting with an antibody directed against the HA epitope tag encoded at the amino terminus of LEF-1 (lanes 1 to 3) or with an antibody directed against LEF-1 (lanes 4 to 6). Markers on the left are in kilodaltons.
FIG. 3.
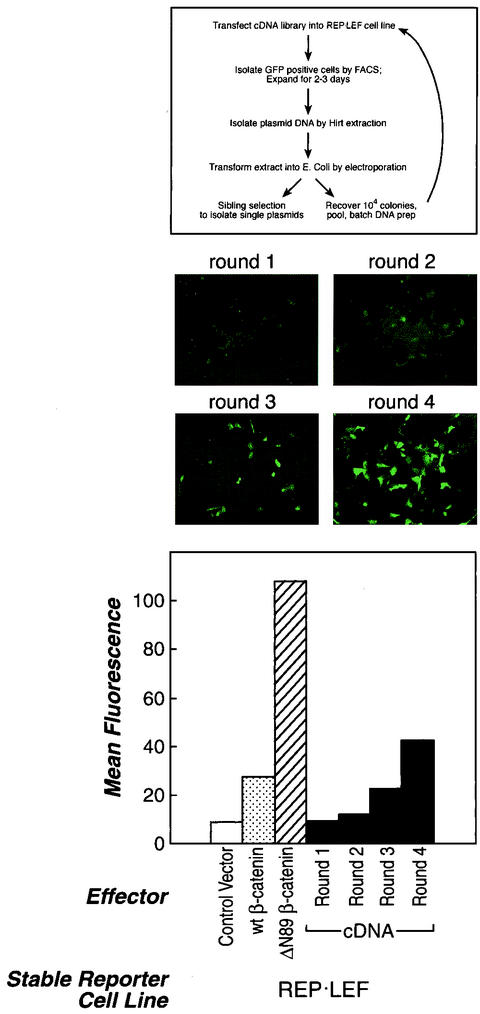
Selection of genes from a tumor cDNA library using the REP-LEF reporter system. (Top) Outline of protocol for the functional screen with REP-LEF cells. (Middle) GFP-positive REP-LEF cell cultures after initial transfection with the cDNA library (round 1) or after each subsequent round of transfection and enrichment (rounds 2 to 4). The cells were visualized with a fluorescence microscope 48 h after transfection with pools of cDNA expression vectors. (Bottom) FACS analysis of transfected REP-LEF cells after initial transfection (round 1) and after each subsequent round of transfection and enrichment (rounds 2 to 4). The mean GFP fluorescence intensity was measured by FACS in pools of cells 48 h after transfection. For comparison, REP-LEF cells were also transfected with a control vector or expression vectors encoding wt β-catenin or ΔN89 β-catenin.
FIG. 10.
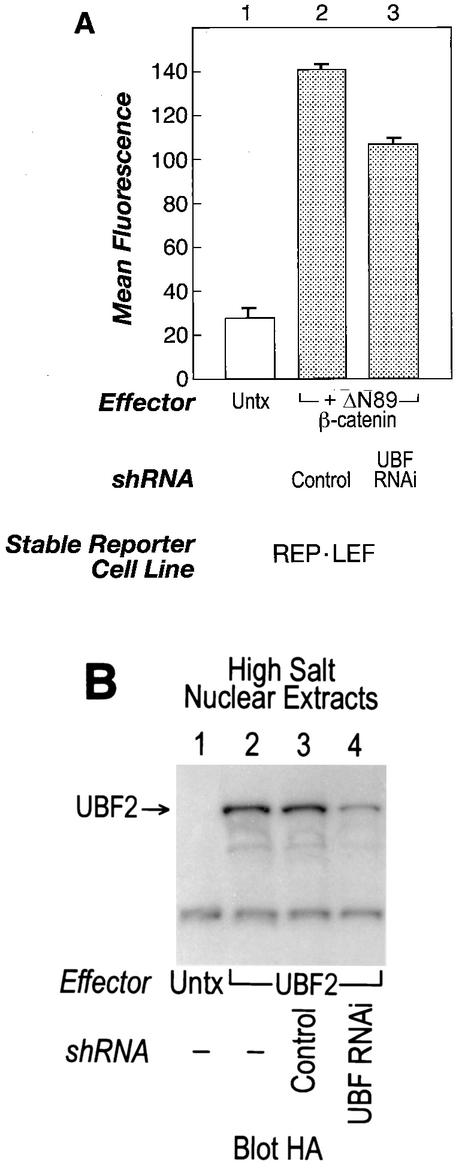
Downregulation of endogenous UBF by shRNA reduces the ability of ΔN89 β-catenin to activate a LEF/TCF-responsive reporter. (A) REP-LEF cells were cotransfected with an expression vector encoding ΔN89 β-catenin along with either MSCV-U6 empty vector (bar 2) or MSCV-U6-UBF (bar 3). Forty-eight hours after transfection, the fluorescence level of GFP-expressing cells was determined by FACS. Untx, untransfected. (B) REP-LEF cells were cotransfected with an expression vector encoding HA-UBF2, along with either MSCV-U6 empty vector (lane 3) or MSCV-U6-UBF (lane 4). High-salt nuclear extracts were prepared from the transfected cells and analyzed by Western immunoblotting with an antibody directed against the HA epitope tag encoded at the amino terminus of UBF2. −, absent.
FIG. 5.
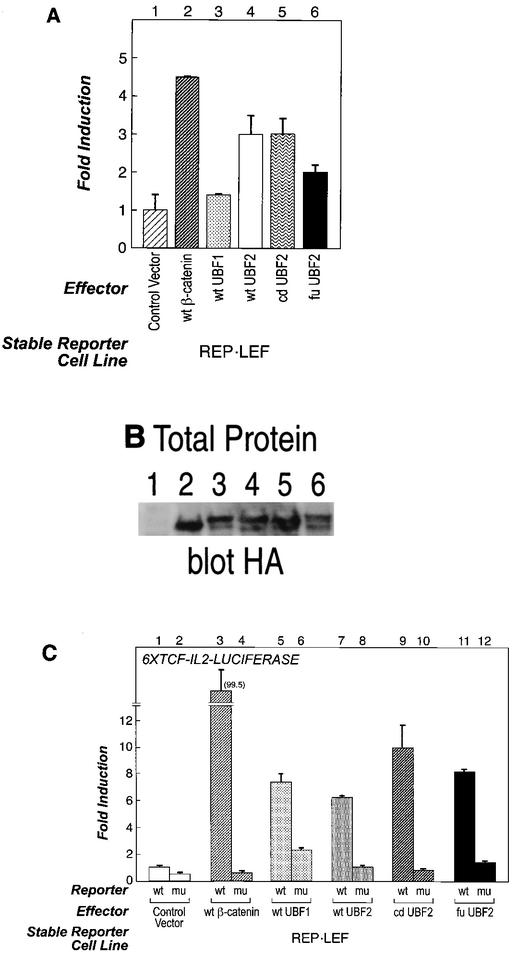
Activities of UBF proteins in the REP-LEF reporter cell line. (A) REP-LEF cells were transfected with a control vector or expression vectors encoding wt β-catenin, wt UBF1, wt UBF2, cd UBF2, or fu UBF2, and GFP induction was analyzed by FACS 48 h after transfection. The error bars indicate standard deviations. (B) Protein extracts were prepared from the cells evaluated in panel A and analyzed by Western immunoblotting with an antibody directed against the HA epitope tag encoded at the amino terminus of each protein. (C) REP-LEF cells were cotransfected with either a wt luciferase reporter gene (6×TCF-IL-2-LUCIFERASE) or a similar reporter gene with mutated LEF/TCF binding sites (mu) and expression vectors encoding yellow fluorescent protein (control), wt β-catenin, wt UBF1, wtUBF2, cdUBF2, or fuUBF2. The transfections were performed in triplicate, and 48 h after transfection, the induced luciferase activity was measured and normalized to the Renilla activity expressed from a cotransfected internal control vector. The data are presented as n-fold induction, where activated transcription is divided by the basal transcription from the control expression plasmid.
FIG. 6.
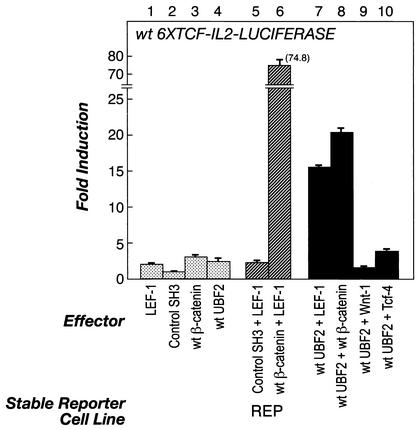
UBF2 cooperates with LEF-1 and β-catenin to potentiate a transcriptional response. REP cells were cotransfected with the wt 6×TCF-IL-2-LUCIFERASE reporter gene along with individual expression plasmids encoding the test genes for LEF-1 (bar 1), control Src-SH3 domain (bar 2), wt β-catenin (bar 3), and wt UBF2 (bar 4). LEF-1 was also tested in combination with control Src-SH3 domain (bar 5), wt β-catenin (bar 6), or wt UBF2 (bar 7), and wt UBF2 was tested in combination with wt β-catenin (bar 8), Wnt-1 (bar 9), and TCF-4 (bar 10). The transfections were performed in triplicate, and 48 h after transfection, the induced luciferase activity was measured and normalized to the Renilla activity expressed from a cotransfected internal control vector. The data are presented as n-fold induction where activated transcription is divided by the basal transcription from the control expression plasmid. The error bars indicate standard deviations.
FIG. 9.
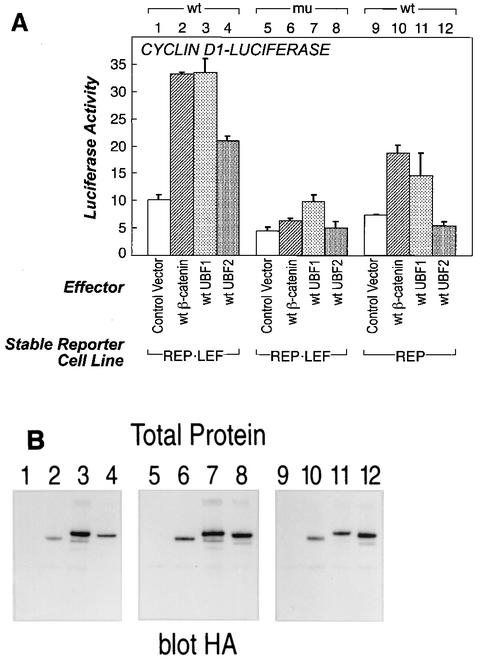
UBF1 and UBF2 proteins activate a cyclin D1 promoter-reporter gene by cooperative interactions with LEF-1 protein. (A) REP-LEF cells were cotransfected with either a promoter-luciferase reporter construct consisting of the wt cyclin D1 promoter, including a LEF/TCF binding site (bars 1 to 4), or a similar reporter gene with a mutation in the LEF/TCF binding site (mu) (bars 5 to 8), along with individual expression vectors encoding a control yellow fluorescent protein gene (bars 1 and 5), wt β-catenin (bars 2 and 6), wt UBF1 (bars 3 and 7), or wt UBF2 (bars 4 and 8). The wt cyclin D1 reporter was also cotransfected into the REP cell line, along with the same test vectors (bars 9 to 12). The transfections were performed in triplicate, and the induced luciferase activity was measured and normalized to the Renilla activity from a constitutive expression vector cotransfected as an internal control. The final numbers represent n-fold induction, where activated transcription is divided by the basal transcription from the control expression plasmid. The error bars indicate standard deviations. (B) Protein extracts were prepared from the cells evaluated in panel A and analyzed by Western immunoblotting with an antibody directed against the HA epitope tag encoded at the amino terminus of each protein.
Establishment and implementation of the functional screen.
The REP stable reporter cell line was constructed by transfecting the retrovirus vector, consisting of the 6×TCF- IL-2-GFP reporter gene and the hygromycin resistance gene, into HT1080 cells, followed by Zeocin selection of transfected clones. The REP-LEF stable cell line was constructed by cotransfecting pCGN-LEF-1 and pBABE-NEO into REP cells, followed by G418 and Zeocin selection of transfected cell clones. For both cell types, transfected clones that exhibited low basal reporter gene expression and optimal induced expression when evaluated by transfection of test vectors expressing LEF-1 or β-catenin were selected. For the functional screen, 5 × 105 REP-LEF cells were transfected with a cocktail containing 2.5 μg of a cDNA library and 2.5 μg of pCR-Blunt carrier DNA (Invitrogen). After 72 h, FACS was used to isolate the population of GFP-expressing cells, which were cultured for 2 to 3 days, followed by Hirt extraction to isolate plasmid DNA (32). The Hirt extract was transformed into DH5α bacterial cells to isolate DNA for subsequent rounds. Approximately 104 bacterial colonies were recovered at each round, and plasmids were isolated and retransfected into REP-LEF cells either as batch pools or by sibling selection, starting with pools of plasmid DNA obtained from five colonies. For batch analysis at rounds 1 and 2 of selection, the ratio of cDNA library to carrier DNA was 1:1, and for rounds 3 and 4, the ratio of cDNA library to carrier DNA was 1:50.
Nuclear extracts, immunoprecipitations, and Western immunoblot analysis.
For the Western immunoblots in Fig. 2B (also see Fig. 10A) and the coimmunoprecipitation and Western immunoblots (see Fig. 7), cells were harvested and resuspended in 200 μl of hypotonic buffer consisting of 20 mM HEPES (pH 7.9), 20 mM NaF, 1 mM Na3VO4, 1 mM Na4P2O7, 0.125 μM okadaic acid, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 1 μg each of leupeptin, aprotinin, and pepstatin/ml (67). The cells were incubated on ice for 15 min and lysed by five passages through a 25-gauge needle. The nuclei were collected by centrifugation and resuspended in 100 μl of the buffer described above containing 420 mM NaCl and 10% glycerol (high-salt buffer) for the extraction of nuclear proteins. For immunoprecipitations, the nuclear extracts were precleared in a reaction mixture containing 50 μl of nuclear extract, 350 μl of hypotonic buffer, and 50 μl of protein A-Sepharose. Following centrifugation to remove the protein A-Sepharose, 5 μl of an antibody directed against LEF-1 (mouse monoclonal; Oncogene Research Products) or against β-catenin (mouse monoclonal; Transduction Laboratories) was added to the reaction mixture, and the mixture was incubated overnight at 4°C. The immune complexes were recovered by the addition of 50 μl of protein A-Sepharose and washed five times in hypotonic buffer containing 57 mM NaCl and 0.1% NP-40. After the final wash, 2× sodium dodecyl sulfate sample buffer was added, and the beads were heated to 90°C for 5 min and analyzed by NuPAGE (Invitrogen). Western immunoblots were incubated in phosphate-buffered saline (PBS) containing 5% nonfat dry milk. The HA (monoclonal antibody; Covance) and LEF-1 (mouse monoclonal; Exalpha [Fig. 2B]) antibodies were diluted 1:1,000, followed by detection with goat anti-mouse immunoglobulin G-horseradish peroxidase conjugate (Bio-Rad Laboratories) and chemiluminescence (Amersham). For Western immunoblot analysis of total protein, transfected cells were harvested and resuspended in 100 μl of high-salt buffer with 0.5% NP-40. The cells were incubated on ice for 15 min, and the DNA was sheared by five passages through a 25-gauge needle. Sodium dodecyl sulfate sample buffer was added, followed by heating to 90°C for 5 min.
FIG. 7.
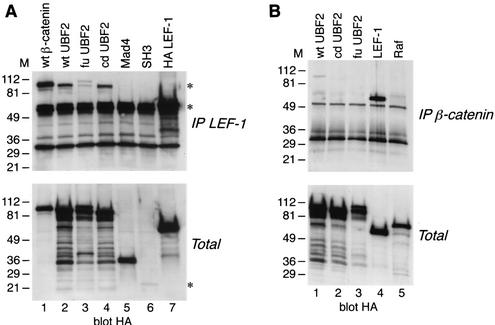
Coimmunoprecipitation of UBF proteins with LEF-1. (A) COS7 cells were cotransfected with an expression vector encoding LEF-1 and expression vectors encoding either wt β-catenin as a positive control (lanes 1), wt UBF2 (lanes 2), fu UBF2 (lanes 3), cd UBF2 (lanes 4), Mad4 as a negative control (lanes 5), or Src-SH3 domain as an additional negative control (lanes 6). HA-tagged LEF-1 (lanes 7) was transfected alone as a positive control for LEF-1 expression and HA epitope tag detection. Extracts from transfected cells were immunoprecipitated with an antibody directed against LEF-1, and immunoprecipitates (IP) (top), along with aliquots of total cell extract (Total) (bottom), were analyzed by Western immunoblotting with an antibody against the HA epitope tag encoded at the amino terminus of each protein. The top asterisk indicates the position of the coimmunoprecipitated proteins, the middle asterisk indicates the HA-tagged LEF-1 protein, and the bottom asterisk indicates the HA-tagged Src-SHC domain. (B) COS7 cells were cotransfected with an expression vector encoding β-catenin and expression vectors encoding either wt UBF2 (lanes 1), cd UBF2 (lanes 2), fu UBF2 (lanes 3), LEF-1 as a positive control (lanes 4), or Raf as a negative control (lanes 5). Extracts from transfected cells were immunoprecipitated with an antibody directed against β-catenin, and immunoprecipitates (top), along with aliquots of total cell extract (bottom), were analyzed by Western immunoblotting with an antibody against the HA epitope tag encoded at the amino terminus of each protein. M, markers.
Immunofluorescence.
REP cells were seeded on coverslips in 12-well plates and transfected at 40% confluence with 0.25 μg of each test plasmid adjusted to 1.5 μg of total DNA with pUC119 DNA. The cells were fixed with 3.7% formaldehyde in PBS for 20 min and then permeabilized for 20 min with 0.1% Triton X-100 in PBS. All antibodies were diluted 1:100 in PBS, including antisera directed against the HA tag (monoclonal antibody; Covance), against LEF-1 (mouse monoclonal; Oncogene Research Products), and against β-catenin (mouse monoclonal; Transduction Laboratories), followed by detection with rhodamine (tetramethyl rhodamine isocyanate)-conjugated or fluorescein (fluorescein isothiocyanate)-conjugated anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories). Images were obtained by fluorescence microscopy (Zeiss).
RESULTS
Strategy for a functional screen in human cells.
A functional screen was established in human cells to identify regulators of the β-catenin signaling pathway using promoter-reporter gene activation as a readout for β-catenin-TCF/LEF-dependent signal transduction. The overall scheme for the genetic screen is depicted in Fig. 1A and is summarized as follows. A consensus LEF/TCF response element was multimerized six times (6×TCF) and inserted upstream of an IL-2 minimal promoter to drive expression of a GFP reporter gene (40, 62). A reporter cell line (REP cells) was established by stable integration of the promoter-reporter gene into the HT1080 human fibrosarcoma cell line. To sensitize the transcriptional response to β-catenin, a LEF-1 expression plasmid was also stably integrated into the REP cells (REP-LEF cells). A plasmid-based cDNA library was transfected into REP-LEF cells, and FACS was used to isolate GFP-positive cells. Pools of cDNA clones were isolated from GFP-expressing cells and reintroduced into REP-LEF cells. Following several rounds of enrichment, individual plasmids were isolated for the ability to activate the reporter gene.
Establishment and testing of a β-catenin-LEF/TCF-responsive reporter cell line.
It was previously reported that the β-catenin-LEF/TCF-responsive reporter, pTOPFLASH, has low basal activity when transfected into a B-cell line lacking LEF/TCF proteins and that exogenous β-catenin and TCF-4 cooperate to induce the reporter gene (40). We observed similar results with HT1080 cells. The reporter construct exhibited very low basal activity when transfected into HT1080 cells in conjunction with a control vector (Fig. 1B, lane 1). Positive control genes encoding wt or activated mutant β-catenin (ΔN89 β-catenin), LEF-1, TCF-4, or Wnt-1 led to a small induction of GFP reporter activity when expressed individually (Fig. 1B, bars 2 to 6). However, wt β-catenin, ΔN89 β-catenin, or Wnt-1 in combination with either LEF-1 or TCF-4 led to significant induction of the reporter, as expected (Fig. 1B, bars 7 to 14) (40). Interestingly, LEF-1 yielded more cooperative activity than TCF-4 when combined with β-catenin and Wnt-1 (Fig. 1B, compare bars 7 to 10 with bars 11 to 14). ΔN89 β-catenin was more active than the wt β-catenin in combination with LEF-1 (Fig. 1B, compare bars 8 and 9) (52). To verify that transcriptional activation was dependent on the LEF/TCF binding sites, a reporter gene with six mutant LEF/TCF binding sites was constructed, similar to pFOPFLASH (40). The positive control genes could no longer activate the mutant reporter, indicating that the β-catenin-LEF/TCF response element was necessary for the transcriptional response (Fig. 1B, bars 7a to 10a). These data demonstrate by transient transfection that the reporter gene was strongly and specifically activated by either β-catenin or Wnt-1 when combined with a LEF-1 or TCF-4 expression plasmid. The LEF-1 expression plasmid provided more cooperative activity than TCF-4, and ΔN89 β-catenin was more active than either wt β-catenin or Wnt-1.
To confirm protein expression, extracts were prepared from each transfection and analyzed by Western immunoblotting using antibodies directed against the HA epitope tag located at the amino terminus of each protein. Figure 1C shows that all of the test proteins were expressed. The cooperation experiments used one-fifth the amounts of LEF-1 and TCF-4 expression plasmids (lanes 8 to 10, 12 to 14, and 8a to 10a), and consequently, those lanes reflect lower protein levels.
To generate a reporter cell line for the functional screen, the wt 6×TCF reporter construct was transfected into HT1080 cells, and stable clones were selected by utilizing the Zeocin resistance gene linked to the reporter gene. A single clonal isolate with low basal GFP expression and strong induction in the presence of β-catenin and LEF-1 expression plasmids (REP cells) was chosen. While absolute levels of GFP induction were lower in the stable reporter cell line than in the transient transfections, β-catenin and Wnt-1 expression plasmids in combination with LEF-1 or TCF-4 again significantly induced the reporter activity (Fig. 1D, lanes 7 to 14). None of the individual genes elicited significant GFP induction in the REP cells, with the exception of LEF-1 (Fig. 1D, lanes 1 to 6). Similar to results obtained with the transient-reporter assay, the LEF-1 gene provided significantly more cooperative activity than TCF-4, and ΔN89 β-catenin was more active than either wt β-catenin or Wnt-1 (Fig, 1D, compare bars 7 to 10 with bars 11 to 14). These results demonstrate that the integrated reporter maintains a pattern of activation similar to that of the transiently transfected reporter, albeit with reduced levels of activity.
Stable integration of a LEF-1 expression plasmid in REP cells to implement a functional screen.
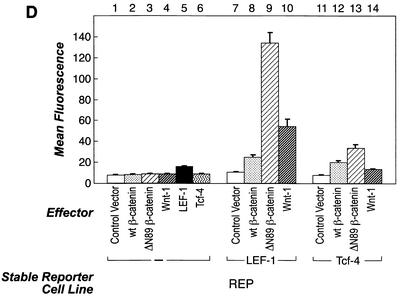
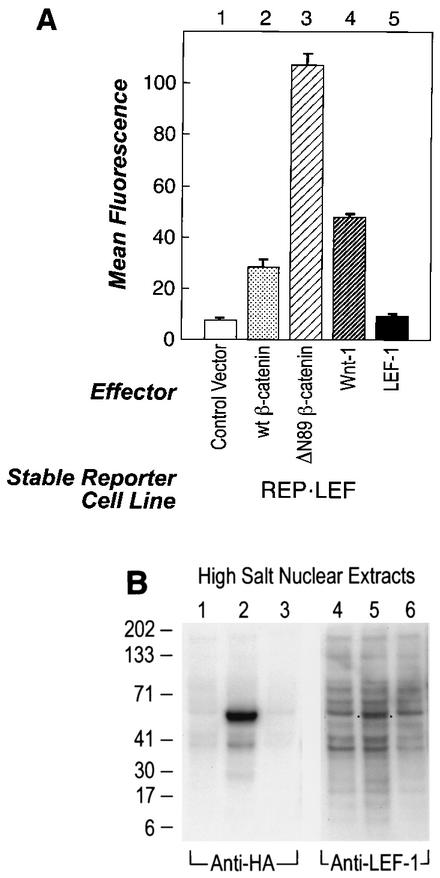
Since overexpression of either β-catenin or Wnt-1 protein alone could only weakly activate the reporter gene in REP cells, it appeared that the endogenous level of LEF/TCF in HT1080 cells was limiting for transcriptional activation. To perform a functional screen using a cDNA library, it was necessary to establish a system in which a single transfected cDNA plasmid could induce reporter gene activity. In order to sensitize the reporter cell system to a single, overexpressed gene that could stimulate β-catenin-LEF/TCF-dependent transcription, a pCGN-LEF-1 expression plasmid was transfected and stably integrated into the REP cell line by cotransfection with a neomycin resistance gene. Single transfected cell clones were evaluated to identify one with low basal GFP reporter activity and strong induction upon transfection of a β-catenin plasmid. Figure 2A shows that the cell line chosen for the functional screen, called REP-LEF, exhibited a 15-fold induction of reporter activity by ΔN89 β-catenin (bar 3), 4-fold induction by wt β-catenin (bar 2), and 6-fold induction by Wnt-1 (bar 4). Transfection of a LEF-1 expression plasmid did not further induce the reporter gene (bar 5). These results showed that the REP-LEF cell line, containing both the reporter gene and overexpressed LEF-1 protein, enabled a single gene to induce the reporter that was essential for library screening.
Our data suggested that LEF/TCF protein levels in REP cells were insufficient to support β-catenin-mediated activation of the reporter gene, and this was confirmed upon stable integration of a LEF-1 expression plasmid (REP-LEF cells), which enabled transcriptional activity. To directly compare LEF-1 protein levels in the REP and REP-LEF cell lines, protein extracts were prepared from REP, REP-LEF, and COS7 cells (used for coimmunoprecipitation experiments) and analyzed by Western immunoblotting using antibodies directed against LEF-1. For comparison, the same extracts were analyzed by Western immunoblotting with an antibody directed against the HA epitope tag located at the amino terminus of the overexpressed LEF-1 protein. Figure 2B shows that the LEF-1 protein was readily detectable in REP-LEF cells using the HA antibody (lane 2) and, as expected, was not detectable in REP cells (lane 1) or COS7 cells (lane 3). Western immunoblotting of the same extracts using antibodies against LEF-1 also showed expression of LEF-1 protein in REP-LEF cells (lane 5), but none was detectable in REP cells (lane 4) and COS7 cells (lane 6). These data demonstrate that LEF-1 protein was overexpressed in REP-LEF cells and that endogenous LEF-1 was either absent or undetectable with the LEF-1 antibody.
Selection of genes from a tumor cDNA library using the REP-LEF reporter cell system.
Our protocol for selecting and enriching cDNA clones that activate the REP-LEF reporter cell line is outlined in Fig. 3, top panel. A cDNA library was prepared using pooled human tumor tissues from breast, colon, and prostate. The library was transfected into REP-LEF cells, and GFP-expressing cells were isolated by FACS. The GFP-positive cells were expanded for several days, and plasmid DNA was isolated by a Hirt extraction protocol (32). The recovered plasmids were transformed into Escherichia coli by electroporation, ∼104 bacterial colonies were pooled, and plasmid DNA was prepared. The plasmid DNA was again transfected into REP-LEF cells, and the procedure was repeated several times to enrich for active cDNA clones. The middle and bottom panels show GFP expression in pools of REP-LEF cells after each round of transfection. There was a progressive increase in fluorescence intensity from rounds 1 to 4, and the signal in round 4 was comparable to that obtained by transfecting a wt β-catenin expression plasmid. Even though the fluorescence intensity was the greatest by round 4, active plasmids were identified in the second round by sibling selection. The enriched cDNA populations from rounds 2 to 4 were subjected to sibling selection in which pools of five plasmids were tested for activation of the REP-LEF reporter cell line, followed by testing of individual plasmids from the positive pools. If the cDNA clones were able to activate the REP-LEF reporter cell line on a single-gene basis, then they were sequenced.
Isolation of a gene encoding UBF2 as a ribosomal fusion protein.
All 24 positive clones selected in the screen yielded the same cDNA, encoding a fusion protein comprised of the transcription factor UBF2 and a small sequence derived from the ribosomal protein L31 gene (Fig. 4, fu UBF2). No cDNA clones encoding wt UBF2 were selected. Nucleotide sequencing of the fu UBF2 clones revealed that the encoded UBF2 protein was missing its carboxyl-terminal 16 amino acids, which were replaced by 41 novel amino acids derived from out-of-frame translation of the ribosomal protein L31 sequences (Fig. 4, bottom). UBF2 belongs to a family of transcription factors that contain multiple DNA-binding domains homologous to HMG proteins 1 and 2 (2, 33, 38, 47). The UBF gene is differentially spliced, giving rise to two protein isoforms referred to as UBF1 and UBF2, the form with HMG box 2 deleted (Fig. 4, top) (8, 33, 44, 53). The UBF1 transcription factor activates rRNA genes, whereas UBF2 is essentially inactive. Here, we show that fu UBF2 activates an mRNA promoter corresponding to the LEF/TCF reporter gene.
FIG. 4.
Enrichment and selection of a cDNA encoding a UBF2 fusion protein. (Top) Comparison of amino acid sequences from HMG box 2 of UBF1 and UBF2 isoforms. UBF1 and UBF2 are derived from the same gene by differential splicing; UBF2 contains a 37-amino-acid deletion in HMG box 2. The dashes represent missing amino acids. (Bottom) Amino acid sequence of the carboxyl terminus of fu UBF2, selected in the screen, compared to the carboxyl-terminal sequence of wt UBF1 and wt UBF2. fu UBF2 encodes a fusion between the UBF2 protein, with a carboxyl-terminal truncation of 16 amino acids, and 41 novel carboxyl-terminal amino acids (underlined) derived from out-of-frame translation of ribosomal protein L31. cd UBF2 was constructed to represent the truncated UBF2 protein without the 41-amino-acid carboxyl-terminal extension. The wt UBF1 and wt UBF2 clones were isolated from a human placenta cDNA library.
UBF1 and UBF2 activate integrated and transient reporter genes in REP-LEF cells.
As defined by the functional screen, the fu UBF2 expression plasmid activates the integrated reporter two- to threefold in REP-LEF cells (Fig. 5A). Since the fu UBF2 cDNA was isolated many times from the screen with no evidence of the wt proteins, it was important to establish whether reporter activation by fu UBF2 required the novel 41 amino acids or if the truncated UBF2 protein was sufficient. Therefore, we tested full-length UBF2 (wt UBF2) and a UBF2 carboxyl-terminal truncation missing the last 16 amino acids (cd UBF2) for the ability to activate transcription. Both wt UBF2 and cd UBF2 activated the REP-LEF reporter by approximately threefold and were slightly more active than fu UBF2 (Fig. 5A, compare bars 4 to 6), indicating that neither the novel 41 carboxyl-terminal amino acids nor truncation of the UBF2 protein contributed to the transcriptional activity of fu UBF2. The UBF1 and UBF2 isoforms have very different functions. A role for UBF1 in RNA polymerase I transcription has been clearly defined, whereas UBF2 is not active in this context (39, 41). To evaluate the role of UBF1 in RNA polymerase II transcription, UBF1 was cloned from a human placenta cDNA library and tested for its ability to activate the REP-LEF cell line. Figure 5A demonstrates that an expression plasmid producing wt UBF1 also activated the reporter gene (bar 3). The various UBF proteins were expressed at similar levels, as determined by Western immunoblot analysis using an antibody directed against the HA tag at the amino terminus of each protein (Fig. 5B).
To further test the abilities of the UBF1 and UBF2 proteins to activate transcription and to determine whether activation was dependent on LEF/TCF enhancer elements, we employed a sensitive and quantitative luciferase reporter assay. Two additional reporters were constructed that contain either wt or mutant 6×TCF sites inserted upstream of the minimal IL-2 promoter to drive luciferase reporter gene expression. Transient transfections were performed in REP-LEF cells to take advantage of the additional LEF-1 protein. Similar to results with the GFP reporter, wt UBF1, wt UBF2, cd UBF2, and fu UBF2 were all capable of significantly inducing the wt luciferase reporter (Fig. 5C, bars 5, 7, 9, and 11) compared to the negative control (bar 1). These results show that all of the UBF proteins can activate transient and integrated β-catenin- LEF/TCF-dependent reporter genes.
Mutation of the LEF/TCF enhancer elements completely abolished all reporter activity induced by the different UBF proteins, with the exception of wt UBF1, which showed some residual activity on the mutant reporter (Fig. 5C, bars 4, 6, 8, 10, and 12). These results show that, like β-catenin, UBF requires a functional LEF/TCF binding site for transcriptional activation.
UBF2 requires LEF-1 or β-catenin for transcriptional activation.
As described above, UBF2 activates a LEF/TCF response element in LEF-1-overexpressing cells. However, these results do not distinguish between UBF2 binding alone to activate transcription and UBF2 interacting with LEF-1 and/or β-catenin to activate transcription. To distinguish between these possibilities, a luciferase reporter assay was performed in REP cells without exogenous LEF-1 protein by cotransfecting the wt 6×TCF- IL-2-LUCIFERASE reporter gene and expression plasmids producing LEF-1, control Src-SH3 domain, wt β-catenin, wt UBF2, Wnt-1, and TCF-4 proteins, alone or in combination. Figure 6 shows that together, UBF2 and LEF-1 expression plasmids strongly cooperated to activate the reporter gene (bar 7) compared to the individual plasmids (bars 1 and 4). A similar cooperative effect was observed between UBF2 and β-catenin (bar 8). As expected, the positive control of wt β-catenin plus LEF-1 showed a dramatic cooperative induction (bar 6), whereas the negative control showed no activity (bar 5). To test if UBF2 cooperates with other members of the Wnt signaling pathway, we assayed for cooperative interactions between UBF2 and either Wnt-1 or TCF-4. UBF2 did not cooperate with Wnt-1 (bar 9) or TCF-4 (bar 10) under the same conditions in which it cooperates with LEF-1 or β-catenin. Our results suggest that UBF has a novel function in β-catenin signaling, which is to potentiate a transcriptional response through cooperative interactions among the UBF, LEF-1, and β-catenin proteins.
UBF2 associates with LEF-1 by coimmunoprecipitation.
Since UBF2 was able to cooperate with both β-catenin and LEF-1 to stimulate transcription, we next examined whether the UBF2 protein was also found in physical association with either the β-catenin or LEF-1 protein. To detect an interaction between UBF2 and LEF-1, a coimmunoprecipitation experiment was performed by overexpressing the UBF and LEF-1 proteins in COS7 cells. We were unable to detect interactions between endogenous proteins, since the levels of endogenous UBF and LEF-1 proteins were too low to be detected with available antibodies. COS7 cells were cotransfected with an expression vector encoding LEF-1 plus either of the following HA-containing expression vectors, encoding wt β-catenin, wt UBF2, fu UBF2, cd UBF2, or the negative control proteins Mad4 and Src-SH3. Nuclear extracts were immunoprecipitated with an antibody directed against LEF-1 followed by Western immunoblot analysis with an antibody directed against the HA epitope tag at the amino termini of β-catenin, UBF2 derivatives, and control proteins. Figure 7A shows that, as expected, LEF-1 coimmunoprecipitated with β-catenin (lane 1) but not with the HA-tagged control proteins (lanes 5 and 6). In addition, LEF-1 coimmunoprecipitated with both the wt UBF2 and cd UBF2 proteins (lanes 2 and 4). Interestingly, the fu UBF2/LEF-1 coimmunoprecipitation was weak and was observed only on longer exposures (lane 3). Western immunoblotting of the total protein extracts (Fig. 7A, bottom) showed that all UBF proteins were similarly expressed, suggesting that the weak coimmunoprecipitation of fu UBF2 was probably not due to reduced protein levels. Overall, our results suggest that UBF2 interacts with LEF-1 in the nuclei of cells that overexpress both proteins.
Next, we determined whether UBF2 could also be coimmunoprecipitated with β-catenin. COS7 cells were cotransfected with an expression vector encoding wt β-catenin plus either of the following HA-containing expression vectors, encoding LEF-1, wt UBF2, fu UBF2, cd UBF2, or the negative control Raf. Nuclear extracts were immunoprecipitated with an antibody directed against β-catenin, followed by Western immunoblot analysis with an antibody against the HA epitope tag at the amino terminus of LEF-1, UBF2 derivatives, or Raf. Again, LEF-1 coimmunoprecipitated with β-catenin, but the control protein did not (Fig. 7B, lanes 4 and 5). No fu UBF2 or cd UBF2, and only a trace amount of wt UBF2, was detectable in association with β-catenin (lanes 1 to 3). A Western immunoblot of protein extracts (Fig. 7B, bottom) demonstrated that all recombinant proteins were expressed at similar levels in the transfected cells. These data suggest that UBF2 protein associates with a LEF-1/β-catenin complex in the nuclei of expressing cells. Since β-catenin and LEF-1 form a complex and since these proteins are also endogenous to COS7 cells, the data do not absolutely distinguish whether UBF2 binds directly to LEF-1, β-catenin, or both proteins. However, when expressed by transfection, UBF2 was readily detected in complex with LEF-1, whereas the complex between UBF2 and β-catenin was barely detectable, suggesting that UBF2 binds to LEF-1 rather than to β-catenin.
UBF2 and LEF-1 colocalize in the nuclei of human cells.
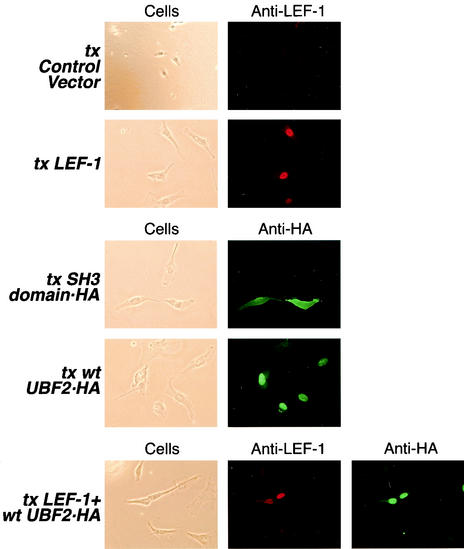
UBF1 was previously reported as an RNA polymerase I transcription factor that predominantly localizes in the nucleoli of cells. Additional staining was also observed in the nucleus, suggesting a distinct but unknown role for UBF (37, 38, 55). The antiserum used in those studies could not distinguish between the UBF1 and UBF2 isoforms. To determine the subcellular localization of UBF2, an expression vector encoding HA-tagged wt UBF2 was transfected into HT1080 cells in the absence or presence of a LEF-1 expression plasmid, and indirect immunofluorescence microscopy was used to locate the proteins. The results shown in Fig. 8 indicate that the subcellular localization of overexpressed UBF2 was both nuclear and nucleolar (tx wt UBF2-HA), whereas overexpressed LEF-1 was nuclear without detectable nucleolar staining (tx LEF-1). The coexpression of wt UBF2 and LEF-1 revealed nuclear colocalization (tx LEF-1+wt UBF2-HA). The staining patterns observed for UBF2 and LEF-1 were specific for those proteins and were not observed in cells transfected with a control vector (tx Control Vector) or a vector producing another epitope-tagged protein (tx SH3 domain-HA). These data indicate that UBF2 and LEF-1 are present in the nuclei of cells, a prerequisite for transactivation of mRNA promoters by RNA polymerase II.
FIG. 8.
UBF2 and LEF-1 colocalize in the nuclei of human cells. HT1080 cells were transfected (tx) with either an empty expression vector or individual vectors encoding LEF-1, HA-tagged Src-SH3 domain, HA-tagged wt UBF2 protein, or a combination of expression vectors encoding LEF-1 plus HA-tagged wt UBF2. The Src-SH3 domain- and wt UBF2-expressing cells were stained with a polyclonal antibody directed against the HA tag, followed by a green fluorescent secondary antibody. Cells transfected with the empty control vector and the expression vector encoding LEF-1 were stained with a monoclonal antibody directed against LEF-1, followed by a red fluorescent secondary antibody.
UBF potentiates LEF-1/β-catenin-dependent activation of a cyclin D1 promoter-reporter gene.
Cyclin D1 is a well-defined cellular target gene for β-catenin transcriptional activation, and sequences within the promoter related to LEF/TCF binding sites are responsive to β-catenin-dependent transcriptional activation (70, 76). To determine whether the UBF proteins could activate a known β-catenin target gene, we tested a reporter construct consisting of the cyclin D1 promoter from sequences +134 to −962, driving expression of a luciferase reporter gene (76). A strong consensus LEF/TCF binding site was located at position −75, surrounded by several weaker consensus sites. The reporter gene was cotransfected into either REP or REP-LEF cells along with expression plasmids that produce wt β-catenin, wt UBF1, or wt UBF2 (Fig. 9A). Consistent with previous reports, β-catenin activated the cyclin D1 promoter (bar 10) (70, 76), and this activity was enhanced in the presence of exogenous LEF-1 (compare bars 2 and 10). The wt UBF1 protein activated the cyclin D1 promoter to levels similar to those obtained with wt β-catenin (compare bars 10 and 11), and again, activation was greater in the presence of exogenous LEF-1 (bar 3). Conversely, the wt UBF2 protein activated the cyclin D1 promoter in the presence, but not in the absence, of exogenous LEF-1 (compare bars 4 and 12). The overall levels of reporter activation by β-catenin, UBF1, and UBF2 were lower in REP cells than in REP-LEF cells, suggesting that LEF-1 overexpression is required for full transcriptional activation (compare bars 2, 3, and 4 with 10, 11, and 12). The ability of β-catenin and wt UBF2 to stimulate reporter expression was abolished upon mutation of the −75 LEF/TCF consensus site in the cyclin D1 promoter; however, some residual activity remained with wt UBF1 (bars 6 to 8). Each transfection was analyzed by Western immunoblotting using antibodies directed against the HA epitope tag located at the amino terminus of each protein (Fig. 9B). These results show that, like β-catenin, the UBF proteins require LEF-1 protein and its binding site to fully activate the cyclin D1 promoter-reporter gene.
Downregulation of endogenous UBF expression by RNAi reduces β-catenin-mediated activation of a LEF/TCF reporter gene.
Thus far, we have established cooperative interactions between UBF, LEF-1, and β-catenin in experiments where each protein was overexpressed. To evaluate the role of endogenous UBF proteins in β-catenin-LEF/TCF-mediated transcriptional activation, RNAi technology was employed to knock down endogenous UBF expression, followed by a reporter gene assay with activated β-catenin to monitor the effects. We have used a previously described retroviral vector (MCSV-U6) to deliver double-stranded inhibitory RNA (shRNA) to cells to induce an RNAi effect (13, 21, 23, 57, 73). The shRNA targeted the amino termini of both UBF1 and UBF2. To examine whether the shRNA could reduce expression of UBF1 and UBF2 in REP-LEF cells and downregulate β-catenin induction of the LEF/TCF-responsive promoter, REP-LEF cells were cotransfected with an expression plasmid encoding ΔN89-activated β-catenin in combination with either an MSCV-U6 empty vector or an MSCV-U6-UBF gene-specific insert. Two days after transfection, the expression of the integrated GFP reporter gene was measured. Figure 10A shows that the ability of ΔN89 β-catenin to activate the integrated GFP reporter was reduced by ∼25% in the presence of MSCV-U6-UBF (bar 3) compared to the MSCV-U6 control (bar 2). Interestingly, MSCV-U6-UBF had no effect on a Wnt-1-induced signal (data not shown), consistent with the data in Fig. 6. Since it was difficult to detect endogenous UBF proteins in REP-LEF cells with available antibodies, we evaluated the ability of the shRNA to downregulate overexpressed HA-tagged UBF protein in REP-LEF cells. REP-LEF cells were cotransfected with expression vectors producing HA-tagged UBF2 protein and the UBF-specific shRNA, and extracts were prepared and analyzed by Western immunoblotting using antibodies directed against the HA epitope tag located at the amino terminus of UBF2. Figure 10B shows that HA-UBF2 protein levels were readily detectable in transfected cells (lane 2), and the levels were diminished upon coexpression of the MSCV-U6-UBF vector (lane 4) but not the control vector (lane 3). UBF1 protein levels were also downregulated by MSCV-U6-UBF (data not shown). These results suggest that endogenous UBF contributes to activated β-catenin-dependent transcription in REP-LEF cells.
DISCUSSION
We established a functional screen in mammalian cells to identify genes that activate β-catenin signaling and isolated the HMG domain transcription factor, UBF2. We have demonstrated for the first time that UBF transcription factors can mediate the RNA polymerase II transcriptional activation of promoters by cooperative interactions with LEF-1 and β-catenin. To perform the functional screen, a GFP reporter construct consisting of multimerized LEF/TCF binding sites was integrated into a highly transfectable human cell line, HT1080. As expected, transient expression of the positive control genes encoding wt and ΔN89 β-catenin, Wnt-1, or LEF-1 each activated the reporter gene when tested individually, and combinations of β-catenin plus LEF-1 or TCF-4 induced significantly more expression. Upon stable integration of the reporter construct into HT1080 cells, the individual positive control genes weakly induced reporter gene activity; however, induction was significantly increased by combinations of β-catenin plus LEF-1 and, to a lesser extent, TCF-4. These findings are the likely result of the reduced copy numbers of the reporter gene in stable transfectants compared to the high copy numbers generated in transient transfections. Furthermore, while HT1080 cells express endogenous β-catenin and LEF/TCF proteins, the levels of these proteins, particularly LEF/TCF, are probably limiting for cooperative interactions. To establish a functional screen in which individual genes rather than gene combinations could significantly activate the LEF/TCF reporter, a vector encoding LEF-1 was also integrated into the stable reporter cell line. This modified reporter cell system enabled individual genes, such as those for β-catenin or Wnt-1, to activate the reporter construct, and a cDNA library was then screened to select for new genes that were capable of activating reporter expression.
Many genes were enriched during the rescue and retransfection protocol employed for the functional screen; however, most of these genes did not activate the integrated or transiently expressed reporter gene when transfected individually. Surprisingly, β-catenin was not isolated from the screen, although we were able to detect the presence of β-catenin sequences by PCR in the cDNA library used for the screen. However, the β-catenin cDNA may have been incomplete and/or unable to activate the reporter gene. On the other hand, we were able to repeatedly isolate a β-catenin expression vector from the screen when it was spiked into the cDNA library. As described here, the HMG box transcription factor, UBF2, was enriched from the screen and was able to activate both the integrated and the transiently transfected reporter genes upon individual transfection. The HMG domains of the UBF and LEF/TCF proteins show sequence homology and are referred to as architectural proteins due to their abilities to bend DNA and bring together promoter elements with bound proteins to create higher-order protein-DNA structures (5, 19, 25, 27, 35, 41, 43, 61). Here, we detected an interaction between two HMG domain proteins, LEF-1 and UBF, by coimmunoprecipitation. Based on the experiments presented here, it is likely that UBF specifically associates with LEF-1 rather than β-catenin. This conclusion is inferred from the readily detectable coimmunoprecipitation of LEF-1 with UBF when both proteins are expressed by transfection. In similar experiments, very little or no UBF protein could be detected in complex with β-catenin; the small amount of UBF observed in the β-catenin immunoprecipitate could be due to the endogenous LEF/TCF protein in complex with the overexpressed β-catenin protein.
Coexpression of LEF-1 and UBF proteins leads to the synergistic activation of transient and integrated reporter genes carrying β-catenin-LEF/TCF target sequences. The shRNA experiments allowed us to further validate UBF as a transcriptional enhancer of the β-catenin pathway. Upon depletion of endogenous UBF from the reporter cells, the ability of β-catenin to activate transcription was decreased. Data from others have shown that UBF binds DNA as a dimer, whereas LEF-1 binds as a monomer (24, 46). An intriguing possibility is that UBF and LEF-1 form a heterodimer which functions in concert with β-catenin to potentiate signaling. LEF/TCF proteins are context-dependent transcriptional activators or repressors functioning in conjunction with other nuclear factors (7, 12, 14, 15, 16, 26, 34, 42, 45, 65, 66). LEF/TCF proteins in complex with the corepressor protein Groucho (and related TLE proteins), CtBP, or HBP1 can repress Wnt/β-catenin target genes (11, 14, 65, 68). One mechanism by which β-catenin can activate its target genes is by displacing negative regulators from LEF/TCF while recruiting additional costimulators, such as p300/CBP or Pontin52 (4, 29). Several corepressor proteins belong to the HMG box family of transcription factors, including the LEF/TCF-interacting protein HBP1 and the β-catenin-interacting proteins XSox17A/B and Xsox3 (65, 85). Therefore, it is not surprising that other HMG box proteins, such as UBF1 and UBF2, regulate β-catenin-LEF/TCF complexes. UBF could potentiate LEF-1 transcriptional activation by competing with Groucho proteins or other negative regulators for binding to LEF/TCF, thus abrogating repression and linking β-catenin-activated transcription to the basal transcription machinery.
UBF function is regulated by phosphorylation in a cell cycle-dependent manner, and UBF phosphorylation plays a role in modulating recruitment of SL1 to the rRNA promoter, which influences its transcriptional activity (31, 53, 72, 77, 79, 80, 81). The kinases that phosphorylate UBF during cell cycle progression include cyclin (D1, A, and E)-dependent kinases (79, 80), and these could enhance the ability of UBF to potentiate β-catenin-LEF/TCF signaling. Also of interest are the casein kinase II (CKII) phosphorylation consensus sites within the 95 carboxyl-terminal amino acids of UBF (31, 54, 81). The CKII-mediated phosphorylation of UBF contributes to, but is not sufficient for, transcriptional activation, suggesting that there may be other kinases modifying the carboxyl terminus. It has been reported that phosphorylation by CKII can create substrate recognition sites for GSK-3β, a negative regulator of the β-catenin signaling pathway (22, 54, 63). The data presented here show that a UBF protein with a carboxyl-terminal deletion of 16 amino acids, including several serine phosphorylation sites, is still able to facilitate LEF/β-catenin signaling to the same extent as wt UBF2; however, our data do not rule out the possibility that UBF phosphorylation plays a role in regulating β-catenin-responsive genes. Additional experiments will determine if a UBF deletion mutant that lacks all 95 carboxyl-terminal amino acids along with the potential phosphorylation sites exhibits changes in transcriptional activity and whether wt UBF protein function can be modulated by GSK-3β.
Previous data showed that cooperative interactions between UBF1 and all four subunits of hSL1/mTIF-1B (TBP and TAF48, -63, and -110) can mediate RNA polymerase I transcriptional activation of an rRNA promoter, but not RNA polymerase II transcriptional activation of an mRNA promoter (6, 17, 18, 41). The mechanism of differential activation is under debate, but there is evidence demonstrating that UBF2 binding to the rRNA promoter is weak compared to that of UBF1, which correlates with its inactivity (41). We show that both UBF1 and UBF2 activate mRNA promoters; however, the 37-amino-acid difference between them may contribute to differential promoter activation, depending on the context. Interestingly, the UBF1-TBP-TAF complexes assembled at rRNA promoters are reminiscent of the TBP-TAF complexes assembled at mRNA promoters, but it appears that the specific combination of proteins governs the promoter class and RNA polymerase selectivity (78). We speculate that LEF-1/UBF interactions may contribute to the promoter selectivity of the TBP-TAF complexes, allowing transcriptional activation at mRNA promoters. However, it remains unclear whether LEF-1 can potentiate transcriptional responses from rRNA promoters. Our immunofluorescence experiments argue against an rRNA connection, given that LEF-1 localizes to the nucleus without obvious nucleolar staining (Fig. 8) (66).
From the functional screen, we isolated a mutant form of UBF2 missing the carboxyl-terminal 16 amino acids, with no evidence of wt UBF2. The RNA used to construct the cDNA library was derived from RNA purified from a pool of human tumors, but we were unable to determine whether the sequences encoding mutant UBF2 were present in the original RNA preparation or arose from cloning errors. It will be interesting to determine whether human cancers contain a mutated UBF gene and whether alterations in UBF could lead to changes associated with cellular transformation.
Here, we provide new evidence that UBF transcription factors can activate polymerase II-mediated transcription and serve as a positive effector in the Wnt/β-catenin signaling pathway through cooperation with LEF-1 and β-catenin. Constitutive activation of β-catenin signaling, through mutation or alterations in expression of various pathway components, is important in the genesis of a variety of human tumors (50, 69, 71, 75). A recent study showed that UBF was overexpressed in human hepatocellular carcinoma compared to its expression in normal liver tissue and that UBF overexpression led to increased colony formation along with increased sensitivity to the chemotherapeutic cisplatin (36). Thus, interactions between UBF and β-catenin signaling may potentiate cellular processes involved in malignant transformation.
Acknowledgments
We thank Jacqueline Saleh and Jolene Andrews for flow cytometry, Peter Reavey and M. Draper for several cDNA libraries, M. Lee for sharing reagents and ideas, Mei Xin for establishing the luciferase assay, and the ACGC sequencing facility for HTP sequencing and clustering of genes. We especially thank A. Arnold and R. Pestell for cloning the cyclin D1 promoter and O. Tetsu and F. McCormick for generously sharing their pGL3 cyclin D1 reporter constructs. We thank A. Roy, L. Comai, J. Cherry, and M. Lee for critical reading of the manuscript. A special thank you is due to Lori Rosenthal for generating figures at ARRCO Medical Art & Design.
REFERENCES
- 1.Aberle, H., A. Bauer, J. Stappert, A. Kispert, and R. Kemler. 1997. Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachvarov, D., and T. Moss. 1991. The RNA polymerase I transcription factor xUBF contains 5 tandemly repeated HMG homology boxes. Nucleic Acids Res. 19:2331-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, A. I., D. B. Stewart, and W. J. Nelson. 1999. T cell factor-activated transcription is not sufficient to induce anchorage-independent growth of epithelial cells expressing mutant beta-catenin. Proc. Natl. Acad. Sci. USA 96:4947-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, A., S. Chauvet, O. Huber, F. Usseglio, U. Rothbacher, D. Aragnol, R. Kemler, and J. Pradel. 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 19:6121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazett-Jones, D. P., B. Leblanc, M. Herfort, and T. Moss. 1994. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264:1134-1137. [DOI] [PubMed] [Google Scholar]
- 6.Beckmann, H., J. L. Chen, T. O'Brien, and R. Tjian. 1995. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 270:1506-1509. [DOI] [PubMed] [Google Scholar]
- 7.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. P., R. M. Learned, H. M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241:1192-1197. [DOI] [PubMed] [Google Scholar]
- 9.Bienz, M., and H. Clevers. 2000. Linking colorectal cancer to Wnt signaling. Cell 103:311-320. [DOI] [PubMed] [Google Scholar]
- 10.Brabletz, T., A. Jung, S. Dag, F. Hlubek, and T. Kirchner. 1999. Beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am. J. Pathol. 155:1033-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brannon, M., J. D. Brown, R. Bates, D. Kimelman, and R. T. Moon. 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126:3159-3170. [DOI] [PubMed] [Google Scholar]
- 12.Bruhn, L., A. Munnerlyn, and R. Grosschedl. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 11:640-653. [DOI] [PubMed] [Google Scholar]
- 13.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 14.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 15.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, R. T., and J. E. Treisman. 2000. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 14:3140-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comai, L., N. Tanese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965-976. [DOI] [PubMed] [Google Scholar]
- 18.Comai, L., J. C. Zomerdijk, H. Beckmann, S. Zhou, A. Admon, and R. Tjian. 1994. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266:1966-1972. [DOI] [PubMed] [Google Scholar]
- 19.Copenhaver, G. P., C. D. Putnam, M. L. Denton, and C. S. Pikaard. 1994. The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res. 22:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford, H. C., B. M. Fingleton, L. A. Rudolph-Owen, K. J. Goss, B. Rubinfeld, P. Polakis, and L. M. Matrisian. 1999. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene 18:2883-2891. [DOI] [PubMed] [Google Scholar]
- 21.Devroe, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez, I., K. Itoh, and S. Y. Sokol. 1995. Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. USA 92:8498-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudilliere, B., Y. Shi, and A. Bonni. 2002. RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J. Biol. Chem. 277:46442-46446. [DOI] [PubMed] [Google Scholar]
- 24.Giese, K., A. Amsterdam, and R. Grosschedl. 1991. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 5:2567-2578. [DOI] [PubMed] [Google Scholar]
- 25.Giese, K., J. Cox, and R. Grosschedl. 1992. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69:185-195. [DOI] [PubMed] [Google Scholar]
- 26.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 27.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94-100. [DOI] [PubMed] [Google Scholar]
- 28.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 29.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heix, J., J. C. Zomerdijk, A. Ravanpay, R. Tjian, and I. Grummt. 1997. Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc. Natl. Acad. Sci. USA 94:1733-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hershey, J. C., M. Hautmann, M. M. Thompson, L. I. Rothblum, T. A. Haystead, and G. K. Owens. 1995. Angiotensin II-induced hypertrophy of rat vascular smooth muscle is associated with increased 18 S rRNA synthesis and phosphorylation of the rRNA transcription factor, upstream binding factor. J. Biol. Chem. 270:25096-25101. [DOI] [PubMed] [Google Scholar]
- 32.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 33.Hisatake, K., T. Nishimura, Y. Maeda, K. Hanada, C. Z. Song, and M. Muramatsu. 1991. Cloning and structural analysis of cDNA and the gene for mouse transcription factor UBF. Nucleic Acids Res. 19:4631-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu, S. C., J. Galceran, and R. Grosschedl. 1998. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 18:4807-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu, C. H., B. McStay, S. W. Jeong, and R. H. Reeder. 1994. xUBF, an RNA polymerase I transcription factor, binds crossover DNA with low sequence specificity. Mol. Cell. Biol. 14:2871-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, R., T. Wu, L. Xu, A. Liu, Y. Ji, and G. Hu. 2002. Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J. 16:293-301. [DOI] [PubMed] [Google Scholar]
- 37.Imai, H., M. J. Fritzler, R. Neri, S. Bombardieri, E. M. Tan, and E. K. Chan. 1994. Immunocytochemical characterization of human NOR-90 (upstream binding factor) and associated antigens reactive with autoimmune sera. Two MR forms of NOR-90/hUBF autoantigens. Mol. Biol. Rep. 19:115-124. [DOI] [PubMed] [Google Scholar]
- 38.Jantzen, H. M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344:830-836. [DOI] [PubMed] [Google Scholar]
- 39.Jantzen, H. M., A. M. Chow, D. S. King, and R. Tjian. 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6:1950-1963. [DOI] [PubMed] [Google Scholar]
- 40.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn, A., R. Voit, V. Stefanovsky, R. Evers, M. Bianchi, and I. Grummt. 1994. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 13:416-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love, J. J., X. Li, D. A. Case, K. Giese, R. Grosschedl, and P. E. Wright. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376:791-795. [DOI] [PubMed] [Google Scholar]
- 44.Matera, A. G., W. Wu, H. Imai, C. L. O'Keefe, and E. K. Chan. 1997. Molecular cloning of the RNA polymerase I transcription factor hUBF/NOR-90 (UBTF) gene and localization to 17q21.3 by fluorescence in situ hybridization and radiation hybrid mapping. Genomics 41:135-138. [DOI] [PubMed] [Google Scholar]
- 45.Mayall, T. P., P. L. Sheridan, M. R. Montminy, and K. A. Jones. 1997. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 11:887-899. [DOI] [PubMed] [Google Scholar]
- 46.McStay, B., M. W. Frazier, and R. H. Reeder. 1991. xUBF contains a novel dimerization domain essential for RNA polymerase I transcription. Genes Dev. 5:1957-1968. [DOI] [PubMed] [Google Scholar]
- 47.McStay, B., C. H. Hu, C. S. Pikaard, and R. H. Reeder. 1991. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. EMBO J. 10:2297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 49.Morin, P. J. 1999. Beta-catenin signaling and cancer. Bioessays 21:1021-1030. [DOI] [PubMed] [Google Scholar]
- 50.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 51.Muller, T., A. Choidas, E. Reichmann, and A. Ullrich. 1999. Phosphorylation and free pool of beta-catenin are regulated by tyrosine kinases and tyrosine phosphatases during epithelial cell migration. J. Biol. Chem. 274:10173-10183. [DOI] [PubMed] [Google Scholar]
- 52.Munemitsu, S., I. Albert, B. Rubinfeld, and P. Polakis. 1996. Deletion of an amino-terminal sequence beta-catenin in vivo and promotes hyperphosphorylation of the adenomatous polyposis coli tumor suppressor protein. Mol. Cell. Biol. 16:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Mahony, D. J., and L. I. Rothblum. 1991. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc. Natl. Acad. Sci. USA 88:3180-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Mahony, D. J., S. D. Smith, W. Xie, and L. I. Rothblum. 1992. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 20:1301-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Mahony, D. J., W. Xie, S. D. Smith, H. A. Singer, and L. I. Rothblum. 1992. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. J. Biol. Chem. 267:35-38. [PubMed] [Google Scholar]
- 56.Orford, K., C. C. Orford, and S. W. Byers. 1999. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J. Cell Biol. 146:855-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paddison, P., A. Caudy, and G. Hannon. 2002. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 99:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papkoff, J., and M. Aikawa. 1998. WNT-1 and HGF regulate GSK3 beta activity and beta-catenin signaling in mammary epithelial cells. Biochem. Biophys. Res. Commun. 247:851-858. [DOI] [PubMed] [Google Scholar]
- 59.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 60.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 61.Putnam, C. D., G. P. Copenhaver, M. L. Denton, and C. S. Pikaard. 1994. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility-group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol. Cell. Biol. 14:6476-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera, V. M., T. Clackson, S. Natesan, R. Pollock, J. F. Amara, T. Keenan, S. R. Magari, T. Phillips, N. L. Courage, F. Cerasoli, Jr., D. A. Holt, and M. Gilman. 1996. A humanized system for pharmacologic control of gene expression. Nat. Med. 2:1028-1032. [DOI] [PubMed] [Google Scholar]
- 63.Roach, P. J. 1991. Multisite and hierarchal protein phosphorylation. J. Biol. Chem. 266:14139-14142. [PubMed] [Google Scholar]
- 64.Roose, J., and H. Clevers. 1999. TCF transcription factors: molecular switches in carcinogenesis. Biochim. Biophys. Acta 1424:M23-M37. [DOI] [PubMed] [Google Scholar]
- 65.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 66.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sadowski, H. B., and M. Z. Gilman. 1993. Cell-free activation of a DNA-binding protein by epidermal growth factor. Nature 362:79-83. [DOI] [PubMed] [Google Scholar]
- 68.Sampson, E. M., Z. K. Haque, M. C. Ku, S. G. Tevosian, C. Albanese, R. G. Pestell, K. E. Paulson, and A. S. Yee. 2001. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 20:4500-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satoh, S., Y. Daigo, Y. Furukawa, T. Kato, N. Miwa, T. Nishiwaki, T. Kawasoe, H. Ishiguro, M. Fujita, T. Tokino, Y. Sasaki, S. Imaoka, M. Murata, T. Shimano, Y. Yamaoka, and Y. Nakamura. 2000. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 24:245-250. [DOI] [PubMed] [Google Scholar]
- 70.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sparks, A. B., P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 58:1130-1134. [PubMed] [Google Scholar]
- 72.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 73.Sui, G., C. Soohoo, E. Affar, F. Gay, Y. Shi, W. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375-386. [DOI] [PubMed] [Google Scholar]
- 75.Tejpar, S., C. Li, C. Yu, R. Poon, H. Denys, R. Sciot, E. Van Cutsem, J. J. Cassiman, and B. A. Alman. 2001. Tcf-3 expression and beta-catenin mediated transcriptional activation in aggressive fibromatosis (desmoid tumour). Br. J. Cancer 85:98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 77.Tuan, J. C., W. G. Zhai, and L. Comai. 1999. Recruitment of TATA-binding protein-TAF complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19:2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 79.Voit, R., and I. Grummt. 2001. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. USA 98:13631-13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voit, R., M. Hoffmann, and I. Grummt. 1999. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voit, R., A. Schnapp, A. Kuhn, H. Rosenbauer, P. Hirschmann, H. G. Stunnenberg, and I. Grummt. 1992. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 11:2211-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weng, Z., M. Xin, L. Pablo, D. Grueneberg, M. Hagel, G. Bain, T. Muller, and J. Papkoff. 2002. Protection against anoikis and down-regulation of cadherin expression by a regulatable beta-catenin protein. J. Biol. Chem. 277:18677-18686. [DOI] [PubMed] [Google Scholar]
- 83.Wong, N. A., and M. Pignatelli. 2002. Beta-catenin—a linchpin in colorectal carcinogenesis? Am. J. Pathol. 160:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yap, A. S., W. M. Brieher, and B. M. Gumbiner. 1997. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 13:119-146. [DOI] [PubMed] [Google Scholar]
- 85.Zorn, A. M., G. D. Barish, B. O. Williams, P. Lavender, M. W. Klymkowsky, and H. E. Varmus. 1999. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell 4:487-498. [DOI] [PubMed] [Google Scholar]