Abstract
Several homeobox transcription factors, such as HOXB3 and HOXB4, have been implicated in regulation of hematopoiesis. In support of this, studies show that overexpression of HOXB4 strongly enhances hematopoietic stem cell regeneration. Here we find that mice deficient in both Hoxb3 and Hoxb4 have defects in endogenous hematopoiesis with reduced cellularity in hematopoietic organs and diminished number of hematopoietic progenitors without perturbing lineage commitment. Analysis of embryonic day 14.5 fetal livers revealed a significant reduction in the hematopoietic stem cell pool, suggesting that the reduction in cellularity observed postnatally is due to insufficient expansion during fetal development. Primitive Lin− ScaI+ c-kit+ hematopoietic progenitors lacking Hoxb3 and Hoxb4 displayed impaired proliferative capacity in vitro. Similarly, in vivo repopulating studies of Hoxb3/Hoxb4-deficient hematopoietic cells resulted in lower repopulating capability compared to normal littermates. Since no defects in homing were observed, these results suggest a slower regeneration of mutant HSC. Furthermore, treatment with cytostatic drugs demonstrated slower cell cycle kinetics of hematopoietic stem cells deficient in Hoxb3 and Hoxb4, resulting in increased tolerance to antimitotic drugs. Collectively, these data suggest a direct physiological role of Hoxb4 and Hoxb3 in regulating stem cell regeneration and that these genes are required for maximal proliferative response.
Class I Homeobox (Hox) genes encode a family of 39 transcription factors sharing a highly conserved DNA-binding domain. In mammals they play a major role in specifying position and tissue fate in the embryo, as has been demonstrated by several lack-of-function Hox gene mutants that exhibit various developmental abnormalities (see, for example, references 6, 29, 30, 38, 41, 49, and 58). Hox genes are also expressed postnatally, and several of them are expressed in primitive hematopoietic cells and committed progenitors but downregulated upon differentiation to mature cells (44).
Murine models have been generated where enforced expression of Hox genes is used to determine the effect of overexpression on self-renewal, differentiation, and other cell fate decisions during hematopoiesis (for reviews, see references 10 and 55). Such models include overexpression of HOXA10, as well as HOXA9, which both affected myelo- and lymphopoiesis and ultimately lead to myeloid leukemia (5, 9, 23, 52, 53). Expression of HOXB3 and HOXB4 is found in the primitive CD34+ population that is highly enriched for human hematopoietic stem cells (HSCs) but is rapidly downregulated as the cells differentiate into committed progenitors (44). Despite very similar expression pattern of HOXB3 and HOXB4, suggesting a common role or collaboration between these factors, the consequences from overexpressing these genes are very different. Although enforced expression of HOXB3 blocks both T- and B-cell development and causes a myeloproliferative disorder (46), overexpression of HOXB4 greatly enhances the regenerative capacity of HSCs in serial transplantation models and results in selective expansion of HOXB4-transduced cells without causing altered lineage decisions or malignant transformations (1, 45, 54). It is noteworthy that this expansion continues until the stem cell pool is normal in size (without overriding it), differing significantly from transplantation of untreated bone marrow (BM) cells, which can only regenerate up to 10% of the number of HSCs found in normal mice. Furthermore, a recent report demonstrates that an ∼40-fold net expansion of murine repopulating HSCs can be achieved by enforced expression of HOXB4 ex vivo for 10 to 14 days (2). This finding is in sharp contrast to ordinary cytokine induced cultures which can support maintenance of HSC numbers or at most expand by a factor of 2 to 4 (4, 12, 28). Less is known about the feasibility of using HOXB4 for human HSC expansion. Recent findings indicate that HOXB4 overexpression in human hematopoietic progenitors affects fate decisions in a concentration-dependent manner to determine whether self-renewal, differentiation, or a differentiation block ensues (7a, 8, 46a). These findings emphasize the importance of understanding the physiological effects of HOXB4 in HSCs in vivo. Interestingly, overexpression studies of HOXC4 also result in expansion of primitive human hematopoietic progenitors, suggesting a common role for paralog 4 genes on these progenitors (13).
Since HOXB3 and HOXB4 are expressed in the stem cell compartment and gain-of-function studies result in enhanced HSC regeneration, we wanted to further analyze the physiological role of these genes in controlling stem cell fate in a lack-of-function mouse model. Lack-of-function mouse models have generated important insight into the role of various transcription factors in hematopoiesis. These include targeting of genes such as GATA-2, SCL/tal-1, Rbtn2/Lmo2, AML1, PU.1/Spi1, Ikaros, Hoxb6, and Hoxa9 (reviewed in references 34 and 51). Hoxa9-deficient mice exhibit prominent defects in erythroid, myeloid, and lymphoid development, including early T-cell development, as well as apparent defects in HSC function, although these have not been fully described (20, 27, 27a). Hoxb6 deficiency mainly affects the erythroid development, increasing the numbers of erythroid progenitors (22). Here we describe a mouse model that is deficient in the contiguous Hoxb3 and Hoxb4 genes. All exons and intermediate sequences of these genes were excised by utilizing the Cre/LoxP technique (18, 43). Homozygous mice deficient in Hoxb3 and Hoxb4 (Hoxb3/b4−/−) were born at normal Mendelian ratios, showed no major abnormalities in skeletal structure, and remained healthy. However, the hematopoietic organs of Hoxb3/b4-deficient mice exhibited a significant reduction in cellularity and reduced numbers of primitive hematopoietic progenitors, in particular of the HSC pool in fetal livers (FLs) from 14.5-day-old embryos. The proliferative capacity of primitive hematopoietic progenitors from mutant mice was diminished in vitro and the regenerative capacity of Hoxb3/b4−/− HSCs was reduced after primary and secondary transplantation. The defects in repopulating abilities were not caused by aberrations in homing but were more likely due to diminished proliferation of HSCs. This is supported by studies after hematopoietic stress, which demonstrated that repopulating Hoxb3/b4−/− HSCs exhibited slower cell cycle kinetics and a larger proportion of resting stem cells. In summary, these findings show that Hoxb3/b4-deficient HSCs harbor a functional defect, impairing the proliferation capacity when rapid regeneration is required.
MATERIALS AND METHODS
Cloning of Hoxb4 and Hoxb3 and generation of targeting constructs.
A 390-bp (SalI/FspI) cDNA fragment from exon 1 of Hoxb4 was used to screen a 129/SvJ mouse genomic BAC (bacteria artificial chromosome) library (Stratagene). A clone containing more than 100 kb of the Hoxb locus was identified and isolated. For generating the Hoxb4 targeting construct, two overlapping subclones were used, a 9-kb EcoRI clone (pBS-ERI; Hoxb4 and 3′ sequence) and a 6-kb EagI clone (pBS-EagI; Hoxb4 and 5′ sequence). For generating the Hoxb3 targeting construct, a 7.5-kb NheI fragment (pSL-NheI; Hoxb3 exons III and IV), as well as an overlapping 3′ ApaI fragment (pBS-ApaI; 3′ sequence) were used. Briefly, for the generation of the Hoxb4 targeting vector, the 6-kb EagI fragment was ligated into a modified pBluescript (pBS-ΔHindIII) and then opened with HindIII and blunted. Into this site the loxP flanked (floxed) neomycin expression cassette, a 1.3-kb XbaI/SalI fragment isolated from pl2neo, was ligated. To the 3′ end of this subclone a 1-kb EagI/HindIII fragment (from pBS-ERI) was ligated, and this construct was then digested with PshAI/ClaI, resulting in a 7.2-kb targeting fragment (with 1.6-kb homologous arm upstream and 4.4-kb arm downstream of the neomycin cassette). The targeting fragment was ligated into a SmaI/ClaI-cut pBS, resulting in pBS-B4KO. The herpes simplex virus thymidine kinase (tk) gene driven by the PGK promoter was isolated from pPNT by EcoRI and HindIII digestion. The fragment was blunt ligated into the ClaI site in the 3′ polylinker of pBS-B4KO, resulting in the final targeting construct, pBS-B4KOtk.
For generation of the Hoxb3 targeting construct, a 1.7-kb XbaI fragment, located 3′ of exon IV, was isolated from pSL-NheI and ligated into pBS. This subclone was digested with MscI and BamHI and blunted, and into this site the floxed neomycin cassette was ligated, generating pBS-b3lnl3′. pBS-b3lnl3′ was then digested with SmaI, and a blunted 1.6-kb MscI fragment from pBS-ApaI was ligated in that site, resulting in pBS-b3lnl3′+ with a 3′ homologous short arm of 1.6 kb. pBS-b3lnl3′+ was opened in the 5′ polylinker with XbaI and NotI, and into this a 4.5-kb XbaI/NotI (where the NotI has replaced the endogenous Tth111I site) fragment from pSL-NheI was ligated, resulting in pBS-B3KO with a 5′homologous long arm of 4.5 kb. The tk fragment was blunt ligated into the ClaI site in the 3′ polylinker of pBS-B3KO, resulting in the final targeting construct, pBS-B3KOtk.
Gene targeting in ES cells.
The targeting constructs were linearized with NotI and purified. A total of 25 to 30 μg of DNA was electroporated (Bio-Rad; 0.26 kV, 500 μF) into ca. 107 RI embryonic stem (ES) cells that were grown according to standard procedure (56). The cells were then cultured in selective medium (300 to 500 μg of G418/ml and 4 μM ganciclovir [when appropriate]) for 7 to 9 days. To verify homologous recombination (HR) the surviving colonies were screened by PCR with the external primers B3L-ext (GCAGCATGGGCACTTCCACAAG) and B4U-ext (GAGGACAACATTGCCATGCCTAGAT) and the internal primers NeoU (TTGGCTGCAGGTCGCTTCGGTGTT) and NeoL (CTTCTTGACGAGTTCTTCTGAGGGGAT). Positive clones were further analyzed by Southern blot with external probes (data not shown). Southern blotting was also used to verify single integration of the targeting vectors. Genomic DNA was digested with either ApaLI or KpnI to screen Hoxb3 and Hoxb4 targeting, respectively, and then probed with a neomycin-specific probe (0.9-kb EcoRI fragment), resulting in a 8.5-kb band from the ApaLI digest and a 5.5-kb band from the KpnI digest (Fig. 1). Targeted clones were expanded and ca. 2 × 106 cells were electroporated with 15 μg of the plasmid pIC-Cre for excision of the neomycin gene. Resulting neomycin sensitive (Neos) clones were screened by PCR with the primers P2 (GAGTGTCACCAATGCCCTCCTGCT) and P6 (GCTTGCCCATTCTCCAGTCTCTCA) or the primers P1 (GTTGACATAAACACTCCGCTCATA) and P5 (ATGGCAACCTTATGTTTTCAGGGC) for the loxP sites at the Hoxb3 and Hoxb4 alleles, respectively (Fig. 1). For analysis of Cre-mediated total deletion of both the Hoxb3 and Hoxb4 genes, primers P1 and P2 were used (Fig. 1). Independently targeted clones of both the Hoxb3/b4-floxed and the Hoxb3/b4-deleted versions were injected into 3.5-day-old C57BL/6 blastocysts and transferred into pseudopregnant (C57BL/6 × CBA)F1 fostermothers by standard techniques (56). Chimeric males were mated to C57BL/6J females, resulting in offspring with a 129Sv/C57BL/6J genetic background. For screening of germ line offspring, DNA was isolated from tail biopsies and analyzed by PCR. Primers P3 (GGAAGCAAGAAAAGGAGGAAGAAAGGA) and P4 (CAAAGTGGGTACAGACAGGGAGGAAAG) were used to distinguish between homozygous and heterozygous offspring of Hoxb3/b4-deleted mice. Total RNA was isolated from the peripheral blood (PB) and BM (RNeasy; Qiagen) of Hoxb3/b4 mice and used for reverse transcription-PCR (RT-PCR) analysis to verify the presence or absence of the RNA transcripts.
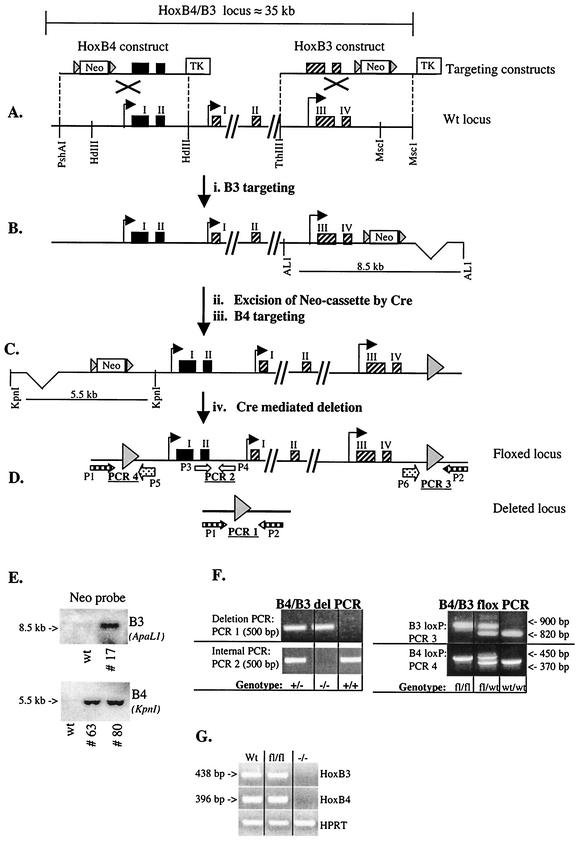
FIG. 1.
Schematic overview of the Hoxb3/b4 locus and the targeting strategy. The gene targeting strategy successfully removes the Hoxb3 and Hoxb4 genes and generates mice without expression of Hoxb3 and Hoxb4 mRNA. (A) The Hoxb4 (exons shown as black boxes) and Hoxb3 (exons shown as hatched boxes) targeting constructs and the wild-type locus. The targeting constructs were generated by inserting a loxP flanked neomycin (Neo) cassette into the HindIII (HdIII) site upstream of Hoxb4 and in the MscI site downstream of Hoxb3. A thymidine kinase (TK) gene was included in the constructs for negative selection against random integration. Gray arrowheads depict loxP sites. (B) Recombinant allele after Hoxb3 targeting. To verify HR, Southern blot analysis with a 5′ external probe was performed, as well as a PCR strategy with one 3′ external and one 5′ internal primer (data not shown). (C) Recombinant allele after transient Cre expression, resulting in excision of the neomycin cassette and after the Hoxb4 targeting. HR was verified by PCR, with a 5′ external primer and a vector-specific primer within the floxed neomycin cassette (data not shown). (D) Recombinant allele after the second transient Cre expression. In the floxed version the Hoxb4 and Hoxb3 genes are flanked by loxP sites, but in the deleted version both of the genes have been excised by the Cre enzyme. (E) Southern blot analysis verifying single homologous integration of both Hoxb4 and Hoxb3 constructs separately, prior to Cre excision of the neomycin cassette. Genomic DNA was digested with ApaLI (AL1) or KpnI for screening of Hoxb3 and Hoxb4 targeting, respectively (see panels B and C) and probed with a neomycin-specific probe. Hoxb3 clone 17 was used for further targeting with the Hoxb4 construct, and the resulting independently doubly targeted clones 63 and 80 were used for experiments described in the text. (F) Examples of PCR results from genotyping of mice carrying the floxed (B4/B3-flox) or the deleted (B4/B3 del) version described in D. fl, floxed allele. (G) RT-PCR, verifying expression or lack thereof, of Hoxb3 and Hoxb4 in BM cells from a wild-type (Wt) mouse, a homozygous floxed mouse (fl/fl), and a knockout mouse (−/−). Hypoxanthine guanine phosphoribosyltransferase (HPRT) RT-PCR was done to verify presence of cDNA in all samples.
Cell harvest.
Hoxb3/b4 knockout and wild-type littermate mice were sacrificed at different ages (mainly 10 to 16 weeks), and cells were harvested from the PB, BM, spleen, thymus, and lymph nodes. Various other organs were also removed for pathological analysis. An aliquot of PB was used (Sysmex K1000; TOA Medical Elektronics Co., Ltd.) to measure some of the hematological parameters described in Table 1. Femurs and tibias were crushed in a mortar in the presence of phosphate-buffered saline (Gibco-BRL) containing 2% fetal calf serum (Gibco-BRL) and then filtered through a 70-μm (pore-size) cell strainer. Cells were isolated from the spleen, thymus, and lymph nodes by meshing the organs through a 70-μm-pore-size cell strainer. FL cells were harvested from embryos at 14.5 days postcoitus for further analysis.
TABLE 1.
Hoxb3/b4-deficient mice are born in normal Mendelian ratios
| Mouse genotypea | No. of pups (% frequency) | Avg wt (g) ± SD atb:
|
|
|---|---|---|---|
| 3 wk | 12 wk | ||
| Hoxb3/b4+/+ | 45 (23.3) | 16.5 ± 0.5∗ | 21.9 ± 1.9† |
| Hoxb3/b4+/− | 99 (51.3) | 16.3 ± 0.5 | 21.6 ± 1.8 |
| Hoxb3/b4−/− | 49 (25.4) | 15.2 ± 0.5∗ | 20.9 ± 1.7† |
Ten pups per genotype.
∗, P < 0.05; †, P = 0.2.
Clonogenic assays.
For myeloid clonogenic progenitor assays, BM cells were cultured in 35-mm petri dishes. For the cell CFU (CFU-C) assay, rich methylcellulose (M3534 containing stem cell factor [SCF; 50 ng/ml], interleukin-3 [IL-3; 10 ng/ml], and IL-6 [10 ng/ml]; Stem Cell Technologies), with the addition of 5 U of human erythropoietin (hEpo; Janssen-Cilag AB)/ml, was used. For the erythroid cell burst-forming unit (BFU-E) assay, serum-free methylcellulose (M3236; Stem Cell Technologies) supplemented with 50 ng of SCF (Amgen)/ml, 50 ng of thrombopoietin (TPO; Kirin Brewery Co., Ltd.)/ml, and 5 U of hEpo/ml was used. Colonies were scored on days 7 to 12 of incubation. For the spleen cell CFU (CFU-S) assay, 50,000 to 75,000 fresh BM cells were injected into lethally irradiated recipients (950 cGy at 110 cGy/min, 137Cs gamma rays); 12 days later, the spleens were harvested and the macroscopic colonies were enumerated.
Proliferation recruitment.
For single cell cultures, Lin− c-kit+ ScaI+ (LSK) cells were used. Briefly, BM cells were treated with ammonium chloride (NH4Cl; Stem Cell Technologies) and then incubated in a lineage antibody cocktail (CD4, CD8, CD5, Gr1, Mac1, B220, and TER119; all antibodies were from BD Pharmingen unless indicated otherwise). After washing and resuspending steps, sheep anti-rat immunoglobulin G (Fc)-conjugated immunomagnetic beads (Dynal) were added, and lineage-positive cells were removed with a magnetic particle concentrator (MPC-6; Dynal). Lin−/lo cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-ScaI (anti-E13-161.7) and allophycocyanin (APC)-conjugated anti-c-kit (anti-2B8). The cells were washed and stained with 7AAD (Sigma-Aldrich) to exclude dead cells. The LSK cells were sorted on a FACSVantage Cell Sorter (Becton Dickinson [BD]) and seeded into Terasaki plates (Nunc) at a concentration of one cell per well in 20 μl of serum free medium (X-vivo 15; BioWhittaker) supplemented with 1% bovine serum albumin (Stem Cell Technologies), 100 IU of penicillin and 100 μg of streptomycin (Gibco-BRL)/ml, 2 mM l-glutamine (Gibco-BRL), and 10−4 M 2-mercaptoethanol (Sigma). The following cytokines were used in various combinations (see Results): SCF (Amgen), TPO (Kirin), Flt-3 ligand (FL; Immunex), granulocyte-colony stimulating factor (G-CSF; Amgen), and granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-3 (Novartis).
Cell cycle analysis.
Lin− cells were isolated as described above. The cells were then stained with c-kit-APC and ScaI-phycoerythrin (PE) and thereafter preserved in 0.4% formaldehyde (LabKemi) and 0.2% Triton-X (Sigma) overnight. The following day, the cells were labeled with Ki67-FITC and 7AAD (Sigma) and analyzed on a FACSCalibur (BD). When cyclophosphamide (Sendoxan; Asta Medica A.G.) treatment was included, the mice were injected intraperitoneally (200 mg/kg) at 96 h prior to BM harvest.
Transplantation experiments.
For standard competitive transplantation experiments, 2 × 105 fresh BM cells from Hoxb3/b4−/− or control littermates (Ly5.2) were mixed together with 8 × 105 BM cells from B6.SJL (Ly5.1) cells. The mixture was injected into the tail vein of lethally irradiated B6.SJL recipient mice. To measure reconstitution of Ly5.2-derived cells, PB samples were taken at weeks 6 and 12 and when the mice were sacrificed at weeks 17 to 20. In some cases, 106 fresh BM cells from the primary recipients were further transplanted into secondary B6.SJL recipients. When 5-fluorouracil (5-FU; Nycomed AB) treatment was included in the transplantation study, Hoxb3/b4−/− and Hoxb3/b4+/+ littermates were injected with 5-FU (150 mg/kg) at days 1 and 5. On day 6, BM cells were harvested, and a 1/10 femur equivalent was mixed together with fresh 2 × 105 B6.SJL BM cells. This mixture was then injected intravenously (i.v.) into lethally irradiated B6.SJL recipients. For FL transplantation experiments, 2 × 105 Hoxb4−/− or Hoxb4+/+ cells (Ly5.2) derived from 14.5-day-old embryos were used in competition with 3 × 105 B6.SJL cells (Ly5.1) and transplanted into lethally irradiated (B6.SJL × C57BL/6)F1 recipients (express both Ly5.1/Ly5.2).
FACS analysis.
Hematopoietic cell suspensions (PB, BM, spleen, thymus, and lymph nodes) were treated with ammonium chloride (Stem Cell Technologies) prior to fluorescence-activated cell sorting (FACS) analysis. For lineage analysis, the cells were stained with FITC-conjugated anti-Mac1, anti-Gr1, anti-B220, and anti-CD4 antibodies, as well as PE-conjugated anti-CD3, anti-CD8, and anti-TER119 antibodies. For a more detailed analysis of B- and T-cell development, APC-conjugated anti-B220, FITC-anti-CD43, PE-(biotin)-anti-IgM, FITC-αβTCR, and PE-δγTCR were also used. For analysis of reconstitution in transplanted mice, PE-conjugated anti-CD45.1 (Ly5.1) and APC-(biotin-)-anti-CD45.2 (Ly5.2) antibodies were used. For estimation of LSK CD34lo/− (LSK CD34) cells, the cells were incubated in the lineage cocktail described above and then labeled with Tri-Color-conjugated goat F(ab′)2 anti-rat immunoglobulin G (H+L; Caltag Laboratories) and ScaI-FITC, c-kit-PE, and CD34-(biotin)-APC. For evaluation of LSA (Lin− ScaI+ AA4.1+) cells from FL, the antibody against c-kit was replaced by an antibody to AA4.1. Analysis was done on FACSCalibur (BD), and the results were analyzed with CellQuest software (BD).
Homing assays.
Fifteen million whole BM cells from four Ly5.2 Hoxb3/4−/− donors or four littermate control donors were injected into lethally irradiated Ly5.1 recipients. Recipients were sacrificed 24 h later for FACS analysis of viable (7AAD−) donor-derived cells in the BM and spleen by using a Tri-Color labeled lineage cocktail (CD4, CD8, CD5, Gr1, Mac1, B220, and TER119), PE-conjugated anti-Ly5.1, FITC-conjugated anti-Ly5.2, and APC-conjugated anti-c-kit. For analysis of CFU-S at day 12, 1/10 and 1/20 recipient spleen cells were injected into a second cohort of lethally irradiated Ly5.1 recipients (8 recipients per donor spleen), and the numbers of CFU-S were determined 12 days later by counting the visible colonies after fixation in Telleyesniczky's solution. To study the ability of the knockout BM to accept BM grafts, 15 × 106 whole BM cells from five Ly5.1 donors were injected into lethally irradiated Ly5.2 Hoxb3/4−/− or littermate control recipients (one Ly5.1 donor to one Ly5.2 knockout and one Ly5.2 control), and FACS analysis for engrafted Ly5.1 cells was performed 24 h later as described above.
Statistical analysis.
Statistical analysis was performed by using the Student t test (either nonpaired or paired when applicable). A P value of <0.05 was considered significant.
RESULTS
Generation of Hoxb3/b4-deficient mice.
In order to study hematopoiesis in Hoxb3/b4-deficient animals, mice were generated that completely lack expression of Hoxb3 and Hoxb4. The rationale was to generate conditional knockout mice for the contiguous Hoxb3 and Hoxb4 genes to avoid potential developmental abnormalities that could ensue and interfere with examination of postnatal hematopoiesis. LoxP sites were introduced upstream of the Hoxb4 gene and downstream of the Hoxb3 gene by using two different targeting constructs (Fig. 1). After we verified correct targeting in 129Sv ES cells by PCR and Southern blot analysis, the ES cells were used to produce chimeric mice. Mice carrying the floxed (i.e., flanked by loxP sites) Hoxb3/b4 genes were generated in parallel to null mutant mouse lines lacking both genes along with flanking and intermediate regulatory elements (Fig. 1D). Both mouse lines (i.e., the “floxed” and the “deleted”) reproduce normally and are born at normal Mendelian ratios (Table 1). The homozygous Hoxb3/b4 flox/flox mice (Hoxb3/b4fl/fl) appear completely normal, and the presence of the loxP sites does not disturb the expression of the targeted genes (Fig. 1G). However, no Hoxb3 or Hoxb4 mRNA could be detected in the Hoxb3/b4−/− mice. These mice are slightly smaller (8% lower body weight, P < 0.05) than their healthy littermates at the time of weaning (3 weeks), but at 12 weeks of age this difference is usually negligible (Table 1). Therefore, mice used for further analysis were generally 12 to 16 weeks of age and matched with regard to weight. Since the Hoxb3/b4−/− mice were viable and fertile, they were the focus of our studies. It is noteworthy that, in contrast to an earlier report demonstrating severe developmental abnormalities in the thorax structure of Hoxb4 targeted mice (38), no major skeletal alterations could be detected in preliminary analysis of the Hoxb3/b4−/− mice (see Discussion).
Hoxb3/b4−/− mice exhibit reduced cellularity in hematopoietic organs.
Pathological examination of hematopoietic tissues of the Hoxb3/b4−/− mice (BM, spleen, thymus, and lymph nodes) did not reveal any abnormalities. However, the mice displayed significantly reduced cellularity in hematopoietic organs (Table 2). The spleen weight and cellularity was reduced by 30% (P < 0.001) compared to normal littermates, and BM cellularity was reduced by ca. 25% (P < 0.003). A reduction in the red blood cell count and hemoglobin values was also observed, but the white blood cell count was normal. For comparative studies involving the knockout mice, littermate controls were used in all of our experiments. The observed reduction in cellularity led us to ask whether the reduction was restricted to a specific lineage or compartment within the hematopoietic hierarchy. FACS was performed on cells derived from PB, BM, spleen, and thymus with antibodies to Gr1, Mac1, CD4, CD8, B220, CD3, and TER119. The results did not show any significant difference in the lineage distribution of the hematopoietic cells derived from Hoxb3/b4−/− mice compared to Hoxb3/b4+/+ littermates (Fig. 2). In light of previously published data on the blocking effect of overexpression of Hoxb3 on B- and T-cell maturation (46), these populations were analyzed in more detail by FACS analysis. Antibodies to B220 were used in combination with CD43 and IgM for B-cell development, and CD4/CD8 were used in combination with αβTCR or γδTCR for T-cell analysis; however, no difference was observed between Hoxb3/b4−/− cells and control cells (data not shown). Thus, the reduction observed in hematopoietic cell numbers suggested that the phenotype might arise from the primitive progenitor and stem cell compartment.
TABLE 2.
Cellularity in hematopoietic organs of Hoxb3/b4-deficient mice
| Mouse genotypea | Mean ± SDb
|
|||||
|---|---|---|---|---|---|---|
| Spleen wt (mg) | Spleen cellularity (106) | RBC count (1012/liter) | Hemoglobin level (g/liter) | WBC count (109/liter) | BM cellularity (two femurs [106]) | |
| Hoxb3/b4+/+ | 103 ± 14 | 187 ± 30 | 8.1 ± 0.8 | 151 ± 10 | 4.1 ± 1 | 45 ± 6 |
| Hoxb3/b4−/− | 70 ± 7 | 127 ± 24 | 7.4 ± 0.7 | 139 ± 10 | 3.8 ± 1 | 34 ± 5 |
Ten mice per genotype, each at 12 to 16 weeks of age.
The P values for the hematopoietic parameters were as follows: spleen weight, <0.001; spleen cellularity, 0.001; red blood cell (RBC) count, 0.05; hemoglobin level, 0.03; white blood cell (WBC) count, 0.5; and BM cellularity, 0.003.
FIG. 2.
Hoxb3/Hoxb4-deficient mice exhibit normal distribution of hematopoietic lineages in all hematopoietic organs. Representative data from FACS analysis of hematopoietic cells within BM, PB, spleen, and thymus with the specific antibodies stated in the figure. The frequency (%) of each population is given outside the respective quadrant.
The hematopoietic progenitor cell pool is reduced in Hoxb3/b4-deficient mice.
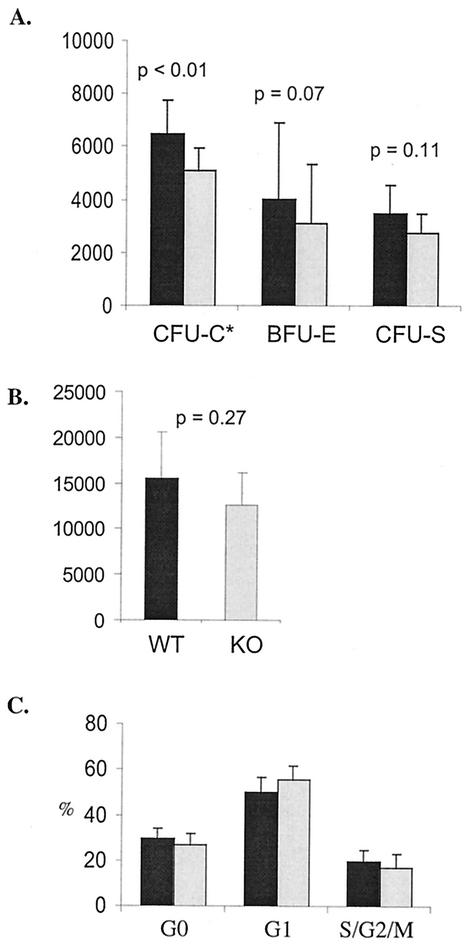
In order to analyze colony-forming abilities of Hoxb3/b4−/−-derived progenitor cells, fresh BM cells were plated out in methylcellulose optimized for either CFU-C or BFU-E assay. BFU-Es and CFU-Cs were enumerated on day 8 and on days 10 and 11, respectively. Hoxb3/b4−/− BM cells showed a nonsignificant reduction in the frequency of hematopoietic progenitor colonies compared to Hoxb3/b4+/+ littermates, both for CFU-C (colonies/10,000 BM cells = 18 ± 4 versus 21 ± 3 [P = 0.2]) and for BFU-E (colonies/100,000 BM cells = 11 ± 3 versus 13 ± 3 [P = 0.3]). However, due to reduced BM cellularity of the Hoxb3/b4−/−, the absolute numbers of CFU-C were significantly reduced (Fig. 3A, P < 0.01) and the numbers of BFU-E were also reduced, although not significantly (Fig. 3A, P < 0.07). The frequency of day 12 CFU-S colonies in Hoxb3/b4−/− mice was also similar to that for control littermates (colonies/50,000 BM cells; 5.2 ± 0.9 for knockout mice versus 5.5 ± 1.4 for wild-type mice [P = 0.8; n = seven mice/group]), whereas the absolute number of CFU-S at day 12 was slightly reduced (Fig. 3A, P = 0.11).
FIG. 3.
Normal cell cycle distribution during endogenous hematopoiesis but reduced numbers of primitive hematopoietic progenitors lacking Hoxb3/Hoxb4. (A) Hoxb3/b4−/− BM cells and BM cells from control littermates were plated out in methylcellulose for measurement of CFU-C and BFU-E ability or injected into lethally irradiated recipients for CFU-S day 12 assay. The findings show a reduction in the absolute numbers of hematopoietic progenitors. The numbers of CFU/femur are shown for BFU-E and CFU-S, but the number of CFU-C is per 1/10 femur. Data from four different experiments are shown (mean ± the standard deviation [SD]; seven Hoxb3/b4−/− and seven Hoxb3/b4+/+ littermates). (B) The absolute numbers of LSK CD34 cells in the BM of both femurs was estimated by FACS analysis. A mild reduction in absolute numbers of LSK CD34− cells was observed in mice deficient for Hoxb3 and Hoxb4. (C) The cell cycle distribution of LSK cells was analyzed by staining with Ki67 and 7AAD, enabling distinction and estimation of G0, G1, and S/G2/M contents. No difference in cell cycle distribution was detected. The results from two experiments are shown (six mice/group). Shaded bars, Hoxb3/b4−/−; solid bars, Hoxb3/b4+/+.
Normal cell cycle distribution but reduced absolute numbers of primitive hematopoietic progenitors in Hoxb3/b4−/−-deficient mice.
Because enforced expression of HOXB4 has been shown to increase expansion and regeneration of HSCs, we analyzed whether there was a reduction in the proportion of cycling HSCs in the Hoxb3/b4−/− mice. The proportion of LSK CD34 cells in the BM of Hoxb3/b4−/− mice and healthy littermates was estimated by FACS analysis. The frequency of these cells was the same in Hoxb3/b4−/− mice as in controls, but the absolute numbers of this primitive hematopoietic population were reduced; this reduction, however, did not reach statistical significance (Fig. 3B; P = 0.27). The cell cycle distribution of freshly isolated LSK progenitors was analyzed by Ki67 and 7AAD staining. The findings demonstrate that in normal endogenous hematopoiesis, the ratio of Hoxb3/b4−/− cells in active cycle versus G0/G1 is not affected compared to controls (Fig. 3C), indicating that reduced cellularity and size of hematopoietic organs is not caused by a lower proportion of proliferating stem cells during endogenous hematopoiesis of adult mice. Rather, this result suggested that already at the fetal stage the expansion of hematopoietic cells is compromised due to the lack of Hoxb3 and Hoxb4. Therefore, we enumerated the FL cells at 14.5 days postcoitus. Although there was no difference in the total number of FL cells from Hoxb3/b4−/− and Hoxb3/4+/+ littermate controls (19.7 ± 6.7 × 106 [n = 24] and 19.2 ± 4.6 × 106 [n = 12], respectively), the absolute number of Lin− ScaI+ AA4.1 cells per FL was significantly reduced in the Hoxb3/b4−/− mice compared to the Hoxb3/b4+/+ littermate controls (29 ± 12 × 103 [n = 12] versus 49 ± 22 × 103 [n = 24]; P = 0.04). These findings support the notion that there may be a reduction in primitive hematopoietic progenitors and stem cells of Hoxb3/b4−/− mice already at the FL stage.
Reduced proliferative capacity of primitive hematopoietic progenitors in vitro.
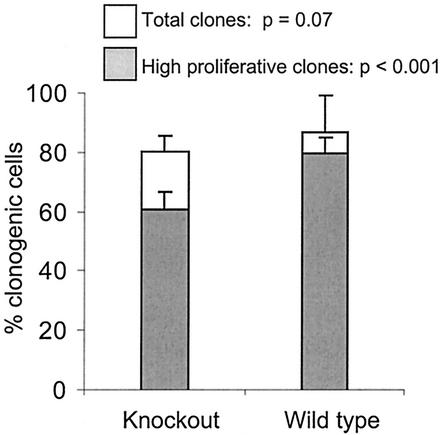
Due to the reduction in hematopoietic cellularity and a mild reduction in the hematopoietic progenitor cell pool caused by the loss of Hoxb3/b4, we sought to determine whether proliferation recruitment of primitive hematopoietic progenitors was affected in Hoxb3/b4−/− mice. LSK cells were sorted out from the BM and plated in serum-free medium in a single cell assay to evaluate survival and proliferation. The cells were plated out in medium containing either TPO or SCF alone and, on day 6, a multicytokine mix (SCF, FL, TPO, G-CSF, IL-3, and GM-CSF) was added to test the viability, which was found to be normal for the mutant cells. Recruitment into proliferation of single LSK cells was tested by supplementing the medium at day 1 with SCF alone, SCF-TPO, SCF-TPO-FL, or the multicytokine mixture described above. Low stimulatory growth conditions (few cytokines) did not give the mutant cells a proliferative disadvantage compared to control cells, but a significant difference in proliferation potential was detected between the Hoxb3/b4−/− and Hoxb3/b4+/+ littermates, when the multicytokine combination was used (Fig. 4 and data not shown). A significantly reduced number of LSK Hoxb3/b4−/− cell clones with high proliferative capacity (progeny cells cover ≥50% of the well) was observed compared to Hoxb3/b4+/+ littermates (P < 0.001). Similarly, there was a reduction in the total number of responding clones from the mutant cells, although this was not statistically significant (Fig. 4, P < 0.07). These results indicate that primitive LSK Hoxb3/b4−/− derived cells proliferate less effectively than their normal counterparts in vitro and that the effects due to lack of Hoxb3 and Hoxb4 are most prominent in settings in which there is extensive pressure for proliferation on this primitive cell population.
FIG. 4.
Reduced proliferation recruitment of single primitive hematopoietic progenitors in vitro. Single LSK cells were seeded directly into serum-free medium containing 25 ng of SCF, FL, TPO, and G-CSF/ml and 10 ng of GM-CSF and IL-3/ml to assay for in vitro expansion. After 10 days of culture, the cloning frequency (open bars) and clone size (shaded bars, representing the percentage of high proliferative clones covering >50% of the well) were evaluated. The results represent the mean ± the SD values of two experiments.
Normal homing of Hoxb3/b4-deficient BM cells.
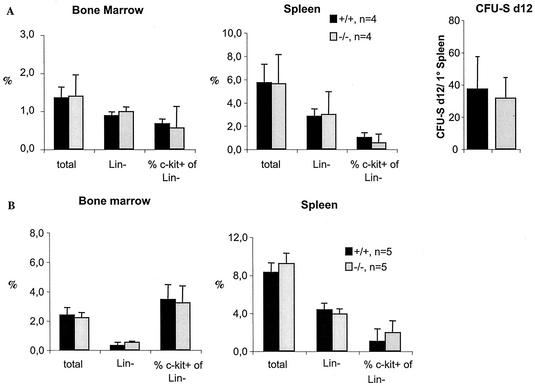
Before evaluating the regeneration capacity of Hoxb3/b4-deficient repopulating HSCs, we sought to determine whether Hoxb3/b4−/− BM cells were defective in homing to hematopoietic sites. Fifteen million Hoxb3/b4−/−and control BM cells were transplanted into irradiated recipients, and homing into BM and spleen was evaluated by FACS. The homing capacity of total, Lin−, and Lin− c-kit+ Hoxb3/b4-deficient donor cells was not affected, and this included primitive, multipotent myeloid (CFU-S at day 12) progenitors (Fig. 5A). There was also no difference in the ability of normal donor cells to home into the BM stroma and spleens of Hoxb3/b4−/− mice compared to control mice, suggesting that the reduced cellularity of hematopoietic organs in Hoxb3/b4−/− mice was not simply due to an impaired ability to retain hematopoietic cells (Fig. 5B).
FIG. 5.
Normal homing of Hoxb3/b4-deficient BM cells. (A) Fifteen million Ly5.2 knockout or wild-type littermate BM cells were transplanted into lethally irradiated wild-type (Ly5.1) recipients. Homing of donor cells in the spleen and BM of recipients was measured as a percentage of engrafted Ly5.2 donor cells (total cells, Lin− cells, and the percentage of c-kit+ cells from the Lin− fraction). Spleens were harvested from the recipients, and cells from 1/10 or 1/20 of the spleens were transplanted into irradiated normal recipients to enumerate the number of CFU-S day 12 multipotent progenitors that had homed into the spleen of the primary recipient. (B) Fifteen million Ly5.1 wild-type BM cells were transplanted into irradiated Ly5.2 Hoxb3/b4−/− and control littermate Hoxb3/b4+/+ mice to evaluate the number of homing sites in the recipient stroma. The percentage of Ly5.1 donor wild-type cells (total cells, Lin− cells, and the percentage of c-kit+ cells in the Lin− fraction) were measured by FACS analysis. Shaded bars, Hoxb3/b4−/−; solid bars, Hoxb3/b4+/+.
Repopulating HSCs from Hoxb3/b4-deficient mice have reduced regenerative capacity.
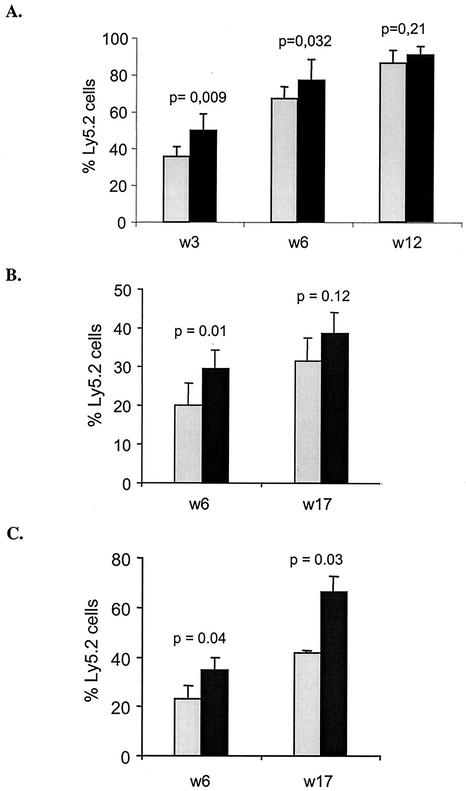
In order to determine whether the lack of Hoxb3/b4 affected the function of repopulating HSCs after transplantation to lethally irradiated mice, a competitive transplantation experiment was performed. Fresh FL cells from Hoxb3/b4−/− mice or normal littermates (expressing the Ly5.2 marker) were transplanted together with B6.SJL competitor cells (expressing Ly5.1) into lethally irradiated B6.SJL recipient mice. PB samples were taken 3, 6, and 12 weeks posttransplant, and the level of reconstitution by the Ly5.2 cells was analyzed. Repopulation by Hoxb3/b4−/− cells lagged behind that of controls at 3 and 6 weeks (Fig. 6A), although by 12 weeks equivalent levels were reached. This finding prompted us to test the regenerative capacity of adult BM HSCs by the same approach. The Hoxb3/b4−/− derived BM cells showed significantly lower regenerative capacity than their Hoxb3/b4+/+ counterparts 6 weeks posttransplantation (P = 0.01), but in long-term reconstituted mice at 17 weeks this difference was less prominent and not significant (P = 0.12; Fig. 6B). The animals were sacrificed at 17 weeks posttransplantation, and the lineage distribution of Ly5.2 cells in BM and PB was analyzed. All transplanted mice showed reconstitution of all hematopoietic lineages, and the lineage distribution was normal in mice transplanted with Hoxb3/b4−/− BM cells (data not shown). Since the Hoxb3/b4−/− cells exhibited lower reconstitution ability in the primary recipients, we sought to determine whether these findings would be more prominent upon further proliferative stress after transplantation to secondary recipients. Reconstitution of Hoxb3/b4−/− cells in the secondary recipients was significantly lower compared to Hoxb3/b4+/+ cells, at 6 and 17 weeks posttransplant (Fig. 6C). These observations strongly suggest that deficiency of Hoxb3/b4 negatively affects the regenerative capacity of adult BM repopulating HSCs.
FIG. 6.
Reduced regenerative capacity of Hoxb3/Hoxb4-deficient repopulating FL and BM HSCs after BM transplantation. The donor cells (Ly5.2) were injected together with competitor B6.SJL cells (Ly5.1) into lethally irradiated recipients. The figure shows in vivo reconstitution of Hoxb3/b4−/− (shaded bars) and Hoxb3/b4+/+ (littermates, solid bars) derived donor cells. (A) Repopulation of FL cells in B6.SJL recipients over time showing slower regeneration at 3 and 6 weeks but full regenerative potential at 12 weeks. (B) Repopulation of BM HSCs in B6.SJL mice. Level of reconstitution in primary transplanted recipients (each bar represents 16 recipients from four donor mice). Shown here is the percentage of reconstitution of Ly5.2 derived cells in the PB at 6 and 17 weeks posttransplant (± the standard error of the mean) from two experiments. (C) Ly5.2 reconstitution in secondary transplant recipients determined by using cells from primary mice (BM recipients) included in the competitive transplantation study depicted in panel B. BM cells (106) from four primary recipients (two Hoxb3/b4−/− and two Hoxb3/b4+/+) were injected into lethally irradiated B6.SJL mice, and the overall reconstitution was analyzed as described in the text. Each bar represents six recipients derived from two primary donor mice. The result (i.e., the percentage of Ly5.2 ± the SD) from one experiment is shown.
Hoxb3/b4−/− HSCs exhibit increased tolerance to 5-FU.
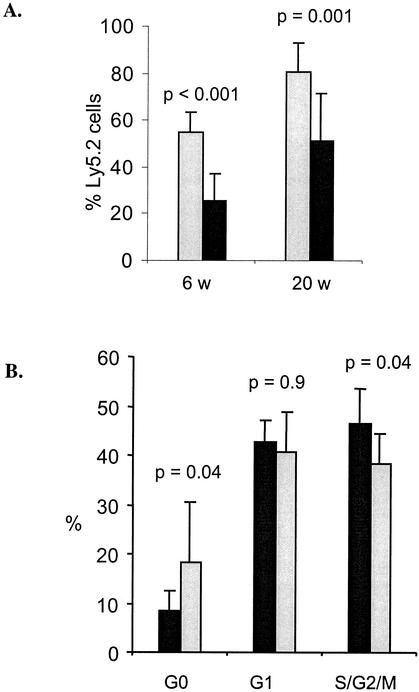
To further investigate the effects of Hoxb3/b4 deficiency on the repopulating HSC compartment in adult mice, we sought to determine whether mutant HSCs exhibited abnormal proliferation kinetics. Therefore, the mice were treated with 5-FU twice (on day 1 and on day 5) to test the hypothesis that slower proliferation kinetics result from Hoxb3/b4 deficiency. The first treatment of 5-FU eliminated actively cycling cells, thereby forcing the primitive resting population into cycle. A second treatment of 5-FU then killed the primitive cells that had been recruited into proliferation as a response to the previous hit. Hoxb3/b4−/− and Hoxb3/b4+/+ littermates were injected i.v. with 5-FU (150 mg/kg) on day 1 and on day 5 (96 h apart), BM was harvested the following day and transplanted together with fresh B6.SJL cells into lethally irradiated B6.SJL recipients. FACS analysis showed no difference in lineage distribution between the knockout donor cells and their normal counterparts (data not shown). However, there was a clear and significant difference in overall reconstitution of the recipient mice when the Hoxb3/b4−/− cells were compared to the Hoxb3/b4+/+ cells. After two hits with 5-FU, the Hoxb3/b4−/− cells now had a clear advantage over the Hoxb3/b4+/+ derived cells, showing two- to threefold-higher levels of reconstitution at 6 weeks posttransplantation (P < 0.001) and, on average, 40% higher reconstitution after 20 weeks (P = 0.001; Fig. 7A). These data indicate that the primitive compartment is less activated after the first 5-FU treatment in Hoxb3/b4−/− mice than in the normal littermates, resulting in a higher proportion of noncycling, protected stem and progenitor cells that are not affected by the second 5-FU hit.
FIG. 7.
Hoxb3/Hoxb4-deficient HSCs exhibit slower proliferation kinetics and delayed activation into active cell cycle after hematopoietic stress induced by cytotoxic agents. (A) Effects of two hits of 5-FU (150 mg/kg, given i.v.) on the reconstitution of Hoxb3/b4−/− (shaded bars) and Hoxb3/b4+/+ (littermates, solid bars) derived cells. Donor mice (Ly5.2) were given 5-FU on day 1 and on day 5. On day 6, the BM was harvested and 1/10 femur equivalent was transplanted, along with fresh competitor B6.SJL cells (Ly5.1), into lethally irradiated recipients. The level of reconstitution was measured in PB cells at weeks 6 and 20 posttransplantation. Data from two independent experiments were pooled. Each bar represents 20 recipient mice, originating from five Ly5.2 donor mice (i.e., the percentage of Ly5.2 ± the standard error of the mean). (B) Hoxb3/b4−/− mice and littermates were treated with cyclophosphamide; 96 h later the BM was harvested, and the cell cycle distribution of LSK cells was analyzed with Ki67 and 7AAD. A significantly smaller proportion of primitive Hoxb3/b4−/− LSK cells were in active cell cycle after treatment with cyclophosphamide. The data (means ± the SD) from two experiments are shown (seven mice/group).
Activation of Hoxb3/b4-deficient HSCs from G0 into cell division is delayed after hematopoietic stress.
In order to support the hypothesis that primitive Hoxb3/b4−/− cells are less activated than control cells after treatment with cytotoxic drugs such as 5-FU, we performed cell cycle analysis after cyclophosphamide treatment. Cyclophosphamide acts similarly to 5-FU; however, it does not alter the expression of cellular markers such as c-kit and Mac1 posttreatment, as has been observed after 5-FU treatment (39, 50). Therefore, the primitive hematopoietic LSK cells have an unaltered immunophenotype after cyclophosphamide treatment. Hoxb3/b4-deficient mice and normal littermates were treated with cyclophosphamide (200 mg/kg, given intraperitoneally); 96 h later, the BM cells were harvested and the LSK population was stained with Ki67 and 7AAD for an analysis of the cell cycle distribution. A significantly higher proportion of Hoxb3/b4−/− LSK cells were found in the noncycling G0 phase (P = 0.04), and a significantly lower proportion was in active (S/G2/M) cycle compared to control cells (P = 0.04; Fig. 7B). The frequencies of cells in G1 were the same in both groups. Collectively, these findings demonstrate that there is delayed activation and recruitment into proliferation of Hoxb3/b4-deficient HSCs after hematopoietic stress that is induced by cytotoxic drugs.
DISCUSSION
Accumulating data show that Hox transcription factors play an important role in hematopoiesis. Here we report that deficiency of Hoxb4 and Hoxb3 decreases proliferation capacity of hematopoietic cells with repopulating ability and causes significantly reduced cellularity in hematopoietic organs. The reduction in cellularity is most pronounced in the spleen (30%) and in the BM (25%), with a more marginal reduction observed in the PB. The reduction is not caused by alterations in lineage commitment of hematopoietic cells, but rather the defect seems to be at a more primitive level, within the multipotent progenitor and stem cell compartment. The colony-forming ability of clonogenic progenitors seems not to be significantly affected by the deficiency of Hoxb3 and Hoxb4; however, their absolute numbers were reduced. Interestingly, the cell cycle distribution of endogenous LSK CD34 Hoxb3/b4−/− cells was normal; however, absolute numbers of this primitive population were mildly reduced. These findings suggest that the numbers of primitive hematopoietic progenitors may be reduced during embryogenesis. During fetal hematopoiesis a robust expansion of stem cells and progenitors occurs in the FL at days 11 to 15 (32). We therefore analyzed the FL of 14.5-day-old old embryos and observed a significant reduction in the absolute number of the repopulating Lin− ScaI+ AA4.1+ cells (21) in Hoxb3/b4−/− embryos compared to controls and, moreover, a competitive transplantation assay demonstrated a slower regeneration capacity of FL HSCs. Furthermore, competitive repopulation assays with BM-derived cells demonstrated an impaired regeneration rate of Hoxb3/4−/− HCSs in primary recipients, and this effect is amplified somewhat in secondary recipients, possibly in part due to a mild reduction of HSCs in the primary recipient. Collectively, these findings support the notion that deficiency of Hoxb3 and Hoxb4 negatively affects cycling of multipotent progenitors and stem cells undergoing rapid proliferation. However, other important parameters such as survival, differentiation, and homing of these primitive cells seem not to be affected. We have also generated mice with a deficiency in Hoxb4 alone (complete deletion), where the impact of the deficiency is qualitatively similar but is less pronounced quantitatively (5a). Therefore, the additional deletion of Hoxb3 seems only to enhance the effects seen without causing a new phenotype, a dose-dependent phenomena that has been observed in other compound Hox knockout models (see discussion below).
It is of interest to compare the findings presented here with those of previous reports that have used gain-of-function models to define the role of Hoxb4 and Hoxb3 in hematopoiesis. The loss of Hoxb3 expression did not affect B- or T-cell maturation and had no aberrant effects on the myeloid lineages, as reported by Sauvageau et al. in a retrovirally engineered overexpression study (46). This would indicate that Hoxb3 expression is redundant but that downregulation of this gene is important for normal differentiation. The loss of Hoxb3 and Hoxb4 does not seem to affect the hematopoietic lineage commitment pathways but rather reduces the proliferation capacity of stem cells, although the effects of deficiency are clearly not as dramatic as enforced HOXB4 expression resulting in 50-fold expansion of long-term repopulating HSCs (1, 45, 54). In a recent, elegant overexpression study, a new additional function of HOXB4 is suggested in promoting primitive HSCs, derived from yolk sac as well as from ES cells, to become definitive (26, 40). However, this promotion cannot entirely depend on Hoxb4 since the lack of this gene does not block the onset of definitive hematopoiesis. These effects could also be dose dependent, where exceeding a certain threshold-level changes the functional effect of the transcription factor, as has been seen for GATA-1, where the lineage outcome is correlated with the level of GATA-1 expression (25).
Given the functional complexity of the Hox gene clusters with regard to redundancy, as well as shared internal regulation, the deletion of a single gene might not give an accurate picture of its role since a neighboring or a paralog gene(s) might rescue the phenotype. Indeed, compound knockouts of paralog genes can display dose-dependent degrees of synergism in the homeotic transformations observed, based on the mutant combinations analyzed (19, 59). Surprisingly, in our double-knockout model, homozygous Hoxb3/b4−/− mice were born at a normal Mendelian ratios without signs of any life-threatening phenotype as reported for Hoxb4 mutants, which died at or around the time of birth due to a split sternum (38). The penetrance of this lethal phenotype was reported to be stronger (100%) in a pure 129SvEv genetic background but is still quite significant (50%) in the mixed C57BL/6J × 129SvEv background, which is the one that we have mainly used. The absence of this phenotype in our model is intriguing. A possible explanation could lie within the very different targeting strategies used since in the study by Ramirez-Solis et al., Hoxb4 mutants were generated either by disrupting the first exon by insertion of a double selection cassette and stop codons or by inserting a stop codon in the second exon. The split sternum phenotype was only observed in the first model (38). This might indicate that aberrant splice variants or the presence of a selection cassette driven by strong promoters within the complex regulated Hox cluster could affect the observed phenotype (i.e., see reference 6). In the present study, all exons and introns of Hoxb4 and Hoxb3 are completely removed, along with intermediate and flanking sequences containing specific and shared regulatory elements (i.e., see references 15, 16, 17, 31, and 47). This leaves no possibility for expression of abnormal splice variants or truncated proteins with aberrant function. However, the deletion brings Hoxb2 and Hoxb5 together, thereby possibly altering the sequential expression pattern of the Hoxb cluster during embryogenesis (as well as in hematopoiesis).
The mechanism by which Hox genes affect transcription remain largely unknown, as well as their target genes. In addition to classical transcriptional activities, including DNA binding or direct involvement in transcription complexes, recent studies suggest that these genes might also be involved in chromatin modulation by affecting the acetylation of histones, thereby either functioning as repressors or activators (14a, 42, 48). With regard to Hoxb4, cellular proliferation induced by Hoxb4 has been reported to cause increased activity of the AP-1 complex and higher levels of cyclin D1, both directly involved in cell cycle regulation (24). Hoxb4 has also been reported to bind to and participate in downregulation of c-myc, resulting in differentiation of a promyelocytic leukemic cell line (35, 36).
Regulating maintenance and expansion of the stem cell pool involves a very complex mixture of internal and external signals. In addition to Hoxb4, a number of other molecules have also been reported to have an important role in this scheme. Among these is the cyclin-dependent kinase inhibitor, p21cip1/waf1, which is necessary for maintaining stem cells in a quiescent state, and deficiency of this molecule leads to stem cell exhaustion (11). Interestingly, p21cip1/waf1 has been suggested to be a transcriptional target of another Hox transcription factor, namely, HOXA10 (7). Other factors important for maintenance of hematopoietic activity include molecules such as Pbx1 (14) and Rae28 (33), both with strong connections to Hox genes. A few candidate genes with the ability to expand stem cells have been reported. An example of this is HOXA9 (52); however, long-term overexpression of this gene leads to AML (23). Other examples include Hedgehog and Sonic hedgehog, which enhance proliferation of primitive human hematopoietic cells via BMP regulation (3), and Notch1 (57) and the homeobox gene Lhx2 (37), which have been successfully used to generate immortalized HSCs.
In summary, the data presented here suggest that proliferation, but not commitment of true stem cells, is negatively affected by Hoxb3/b4 deficiency. Furthermore, it appears that Hoxb3 and Hoxb4 are mainly important under conditions that call for a rapid proliferation response and are dispensable for normal, steady-state hematopoiesis. These findings are important for understanding the regulatory mechanisms that control fate, particularly self-renewal, of HSCs. Further studies are required to elucidate fully the mechanism of Hoxb4 action in HSCs in order to determine whether enforced expression of Hoxb4 can be used safely to generate or expand stem cells ex vivo for cell or gene therapy.
Acknowledgments
These studies were generously supported by grants from Cancerfonden, Sweden, and Barncancerfonden, Sweden; Astra Draco AB (now AstraZeneca); and Donation Funds, Lund University Hospital, to S.K.; and by a clinical research award (ALF) from Lund University Hospital to S.K. and N.L. J.M.B. was supported by a Graduate Student Award from the Medical Faculty, Lund University.
We thank C. Largman for providing the Hoxb4 probe, H. Gu for the pl2neo plasmid, R. Jaenish for the pNT, and R. Fässler for providing the RI ES cells. We also thank Per Levéen and Mary-Ann Sällström for help with blastocyst injections, Kristina Sundgren and Eva Gynnstam for expert animal care, Lilian Wittman for help with animal experiments, and Sten Eirik Jacobsen and members of The Department of Stem Cell Biology, Lund University, for helpful advice and discussions.
J.M.B. and N.L. contributed equally to this work.
REFERENCES
- 1.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2001. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp. Hematol. 29:1125-1134. [DOI] [PubMed] [Google Scholar]
- 2.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. [DOI] [PubMed] [Google Scholar]
- 3.Bhardwaj, G., B. Murdoch, D. Wu, D. P. Baker, K. P. Williams, K. Chadwick, L. E. Ling, F. N. Karanu, and M. Bhatia. 2001. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat. Immunol. 2:172-180. [DOI] [PubMed] [Google Scholar]
- 4.Bhatia, M., D. Bonnet, U. Kapp, J. C. Wang, B. Murdoch, and J. E. Dick. 1997. Quantitative analysis reveals expansion of human hematopoietic repopulating cells after short-term ex vivo culture. J. Exp. Med. 186:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjornsson, J. M., E. Andersson, P. Lundstrom, N. Larsson, X. Xu, E. Repetowska, R. K. Humphries, and S. Karlsson. 2001. Proliferation of primitive myeloid progenitors can be reversibly induced by HOXA10. Blood 98:3301-3308. [DOI] [PubMed] [Google Scholar]
- 5a.Björnsson, J. M., A. Brun, N. Larsson, E. Repetowska, and S. Karlsson. 2001. HoxB4 deficiency causes reduced cellularity in hematopoietic organs and diminished proliferative capacity response of primitive hematopoietic progenitors. Blood 98:69a. (Abstract.) [Google Scholar]
- 6.Boulet, A. M., and M. R. Capecchi. 1996. Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev. Biol. 177:232-249. [DOI] [PubMed] [Google Scholar]
- 7.Bromleigh, V. C., and L. P. Freedman. 2000. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 14:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Brun, A., X. Fan, K. Humphries, and S. Karlsson. 2001. The effects of HoxB4 in human primitive hematopoietic progenitors are concentration dependent: high levels of HoxB4 lead to increased myeloid differentiation and a reduction in proliferative capacity. Blood 98:66a. (Abstract.) [Google Scholar]
- 8.Buske, C., M. Feuring-Buske, C. Abramovich, K. Spiekermann, C. J. Eaves, L. Coulombel, G. Sauvageau, D. E. Hogge, and R. K. Humphries. 2002. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood 100:862-868. [DOI] [PubMed] [Google Scholar]
- 9.Buske, C., M. Feuring-Buske, J. Antonchuk, P. Rosten, D. E. Hogge, C. J. Eaves, and R. K. Humphries. 2001. Overexpression of HOXA10 perturbs human lymphomyelopoiesis in vitro and in vivo. Blood 97:2286-2292. [DOI] [PubMed] [Google Scholar]
- 10.Buske, C., and R. K. Humphries. 2000. Homeobox genes in leukemogenesis. Int. J. Hematol. 71:301-308. [PubMed] [Google Scholar]
- 11.Cheng, T., N. Rodrigues, H. Shen, Y. Yang, D. Dombkowski, M. Sykes, and D. T. Scadden. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287:1804-1808. [DOI] [PubMed] [Google Scholar]
- 12.Conneally, E., J. Cashman, A. Petzer, and C. Eaves. 1997. Expansion in vitro of transplantable human cord blood stem cells demonstrated using a quantitative assay of their lympho-myeloid repopulating activity in nonobese diabetic-scid/scid mice. Proc. Natl. Acad. Sci. USA 94:9836-9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daga, A., M. Podesta, M. C. Capra, G. Piaggio, F. Frassoni, and G. Corte. 2000. The retroviral transduction of HOXC4 into human CD34+ cells induces an in vitro expansion of clonogenic and early progenitors. Exp. Hematol. 28:569-574. [DOI] [PubMed] [Google Scholar]
- 14.DiMartino, J. F., L. Selleri, D. Traver, M. T. Firpo, J. Rhee, R. Warnke, S. O'Gorman, I. L. Weissman, and M. L. Cleary. 2001. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98:618-626. [DOI] [PubMed] [Google Scholar]
- 14a.Eklund, E. A., R. Kakar, and A. Jalava. 2000. HoxA10 recruits transcriptional co-repressors to the CYBB and NCF2 genes. Blood 96:286a. (Abstract.) [Google Scholar]
- 15.Giannola, D. M., W. D. Shlomchik, M. Jegathesan, D. Liebowitz, C. S. Abrams, T. Kadesch, A. Dancis, and S. G. Emerson. 2000. Hematopoietic expression of HOXB4 is regulated in normal and leukemic stem cells through transcriptional activation of the HOXB4 promoter by upstream stimulating factor (USF)-1 and USF-2. J. Exp. Med. 192:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould, A., A. Morrison, G. Sproat, R. A. White, and R. Krumlauf. 1997. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11:900-913. [DOI] [PubMed] [Google Scholar]
- 17.Graham, A., N. Papalopulu, J. Lorimer, J. H. McVey, E. G. Tuddenham, and R. Krumlauf. 1988. Characterization of a murine homeo box gene, Hox-2.6, related to the Drosophila Deformed gene. Genes Dev. 2:1424-1438. [DOI] [PubMed] [Google Scholar]
- 18.Gu, H., Y. R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155-1164. [DOI] [PubMed] [Google Scholar]
- 19.Horan, G. S., R. Ramirez-Solis, M. S. Featherstone, D. J. Wolgemuth, A. Bradley, and R. R. Behringer. 1995. Compound mutants for the paralogous hoxa-4, hoxb-4, and hoxd-4 genes show more complete homeotic transformations and a dose-dependent increase in the number of vertebrae transformed. Genes Dev. 9:1667-1677. [DOI] [PubMed] [Google Scholar]
- 20.Izon, D. J., S. Rozenfeld, S. T. Fong, L. Komuves, C. Largman, and H. J. Lawrence. 1998. Loss of function of the homeobox gene Hoxa-9 perturbs early T-cell development and induces apoptosis in primitive thymocytes. Blood 92:383-393. [PubMed] [Google Scholar]
- 21.Jordan, C. T., C. M. Astle, J. Zawadzki, K. Mackarehtschian, I. R. Lemischka, and D. E. Harrison. 1995. Long-term repopulating abilities of enriched fetal liver stem cells measured by competitive repopulation. Exp. Hematol. 23:1011-1015. [PubMed] [Google Scholar]
- 22.Kappen, C. 2000. Disruption of the homeobox gene Hoxb-6 in mice results in increased numbers of early erythrocyte progenitors. Am. J. Hematol. 65:111-118. [DOI] [PubMed] [Google Scholar]
- 23.Kroon, E., J. Krosl, U. Thorsteinsdottir, S. Baban, A. M. Buchberg, and G. Sauvageau. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krosl, J., and G. Sauvageau. 2000. AP-1 complex is effector of Hox-induced cellular proliferation and transformation. Oncogene 19:5134-5141. [DOI] [PubMed] [Google Scholar]
- 25.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250-1262. [DOI] [PubMed] [Google Scholar]
- 26.Kyba, M., R. C. Perlingeiro, and G. Q. Daley. 2002. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109:29-37. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, H. J., C. D. Helgason, G. Sauvageau, S. Fong, D. J. Izon, R. K. Humphries, and C. Largman. 1997. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89:1922-1930. [PubMed] [Google Scholar]
- 27a.Lawrence, H. J., S. T. Fong, Y. H. Hsiang, G. Sauvageau, and R. K. Humphries. 1998. Evidence for a stem cell defect in mice with targeted interruption of the Hoxa9 homoebox gene. Blood 92:55a. (Abstract.) [Google Scholar]
- 28.Luens, K. M., M. A. Travis, B. P. Chen, B. L. Hill, R. Scollay, and L. J. Murray. 1998. Thrombopoietin, kit ligand, and flk2/flt3 ligand together induce increased numbers of primitive hematopoietic progenitors from human CD34+ Thy-1+ Lin− cells with preserved ability to engraft SCID-hu bone. Blood 91:1206-1215. [PubMed] [Google Scholar]
- 29.Manley, N. R., and M. R. Capecchi. 1997. Hox group 3 paralogous genes act synergistically in the formation of somitic and neural crest-derived structures. Dev. Biol. 192:274-288. [DOI] [PubMed] [Google Scholar]
- 30.Medina-Martinez, O., A. Bradley, and R. Ramirez-Solis. 2000. A large targeted deletion of Hoxb1-Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev. Biol. 222:71-83. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, A., L. Ariza-McNaughton, A. Gould, M. Featherstone, and R. Krumlauf. 1997. HOXD4 and regulation of the group 4 paralog genes. Development 124:3135-3146. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, S. J., H. D. Hemmati, A. M. Wandycz, and I. L. Weissman. 1995. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 92:10302-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta, H., A. Sawada, J. Y. Kim, S. Tokimasa, S. Nishiguchi, R. K. Humphries, J. Hara, and Y. Takihara. 2002. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 195:759-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orkin, S. H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 35.Pan, Q., and R. U. Simpson. 2001. Antisense knockout of HOXB4 blocks 1,25-dihydroxyvitamin D3 inhibition of c-myc expression. J. Endocrinol. 169:153-159. [DOI] [PubMed] [Google Scholar]
- 36.Pan, Q., and R. U. Simpson. 1999. c-myc intron element-binding proteins are required for 1,25-dihydroxyvitamin D3 regulation of c-myc during HL-60 cell differentiation and the involvement of HOXB4. J. Biol. Chem. 274:8437-8444. [DOI] [PubMed] [Google Scholar]
- 37.Pinto Do, O. P., K. Richter, and L. Carlsson. 2002. Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood 99:3939-3946. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Solis, R., H. Zheng, J. Whiting, R. Krumlauf, and A. Bradley. 1993. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell 73:279-294. [DOI] [PubMed] [Google Scholar]
- 39.Randall, T. D., and I. L. Weissman. 1997. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood 89:3596-3606. [PubMed] [Google Scholar]
- 40.Rideout, W. M., K. Hochedlinger, M. Kyba, G. Q. Daley, and R. Jaenisch. 2002. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell 109:17-27. [DOI] [PubMed] [Google Scholar]
- 41.Rossel, M., and M. R. Capecchi. 1999. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126:5027-5040. [DOI] [PubMed] [Google Scholar]
- 42.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer, B. 1996. Multiplex Cre/lox recombination permits selective site-specific DNA targeting to both a natural and an engineered site in the yeast genome. Nucleic Acids Res. 24:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauvageau, G., P. M. Lansdorp, C. J. Eaves, D. E. Hogge, W. H. Dragowska, D. S. Reid, C. Largman, H. J. Lawrence, and R. K. Humphries. 1994. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 91:12223-12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauvageau, G., U. Thorsteinsdottir, C. J. Eaves, H. J. Lawrence, C. Largman, P. M. Lansdorp, and R. K. Humphries. 1995. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 9:1753-1765. [DOI] [PubMed] [Google Scholar]
- 46.Sauvageau, G., U. Thorsteinsdottir, M. R. Hough, P. Hugo, H. J. Lawrence, C. Largman, and R. K. Humphries. 1997. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity 6:13-22. [DOI] [PubMed] [Google Scholar]
- 46a.Schiedlmeier, B., H. Klump, E. Will, G. Arman-Kalcek, Z. Li, Z. Wang, A. Rimek, J. Friel, C. Baum, and W. Ostertag. 2003. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood 101:1759-1768. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe, J., S. Nonchev, A. Gould, J. Whiting, and R. Krumlauf. 1998. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 17:1788-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen, W. F., K. Krishnan, H. J. Lawrence, and C. Largman. 2001. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 21:7509-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suemori, H., and S. Noguchi. 2000. Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev. Biol. 220:333-342. [DOI] [PubMed] [Google Scholar]
- 50.Szilvassy, S. J., and S. Cory. 1993. Phenotypic and functional characterization of competitive long-term repopulating hematopoietic stem cells enriched from 5-fluorouracil-treated murine marrow. Blood 81:2310-2320. [PubMed] [Google Scholar]
- 51.Tenen, D. G., R. Hromas, J. D. Licht, and D. E. Zhang. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90:489-519. [PubMed] [Google Scholar]
- 52.Thorsteinsdottir, U., A. Mamo, E. Kroon, L. Jerome, J. Bijl, H. J. Lawrence, K. Humphries, and G. Sauvageau. 2002. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood 99:121-129. [DOI] [PubMed] [Google Scholar]
- 53.Thorsteinsdottir, U., G. Sauvageau, M. R. Hough, W. Dragowska, P. M. Lansdorp, H. J. Lawrence, C. Largman, and R. K. Humphries. 1997. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell. Biol. 17:495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thorsteinsdottir, U., G. Sauvageau, and R. K. Humphries. 1999. Enhanced in vivo regenerative potential of HOXB4-transduced hematopoietic stem cells with regulation of their pool size. Blood 94:2605-2612. [PubMed] [Google Scholar]
- 55.Thorsteinsdottir, U., G. Sauvageau, and R. K. Humphries. 1997. Hox homeobox genes as regulators of normal and leukemic hematopoiesis. Hematol. Oncol. Clin. N. Am. 11:1221-1237. [DOI] [PubMed] [Google Scholar]
- 56.Torres, R. M., and R. Kühn. 1997. Laboratory protocols for conditional gene targeting. Oxford University Press, Oxford, United Kingdom.
- 57.Varnum-Finney, B., L. Xu, C. Brashem-Stein, C. Nourigat, D. Flowers, S. Bakkour, W. S. Pear, and I. D. Bernstein. 2000. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nat. Med. 6:1278-1281. [DOI] [PubMed] [Google Scholar]
- 58.Zakany, J., and D. Duboule. 1996. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384:69-71. [DOI] [PubMed] [Google Scholar]
- 59.Zakany, J., C. Fromental-Ramain, X. Warot, and D. Duboule. 1997. Regulation of number and size of digits by posterior Hox genes: a dose-dependent mechanism with potential evolutionary implications. Proc. Natl. Acad. Sci. USA 94:13695-13700. [DOI] [PMC free article] [PubMed] [Google Scholar]