Introduction
Photoisomerization triggers mammalian vision, converting the chromophore of rhodopsin and cone visual pigments, 11-cis-retinal, to all-trans-retinal. Regeneration of 11-cis-retinal for continued vision occurs in neighboring retinal pigment epithelial (RPE) cells and involves several enzymatic reactions and transcellular diffusion of the retinoids. The overall process is called the visual cycle.1,2 Developments in our understanding of these reactions have been reviewed.3–6 At least two different retinoid dehydrogenases play important roles. In photoreceptor cells, an all-trans-specific retinol dehydrogenase catalyzes the reduction of all-trans-retinal by NADPH.7–9 This reaction is the ultimate step in quenching the reactivity of photoactivated rhodopsin10 and its rate in mouse retina is equal to the rate of appearance of 11-cis-retinal in rhodopsin.11,12 In RPE cells a cis-specific retinol dehydrogenase catalyzes the oxidation of 11-cis-retinol to 11-cis-retinal by NAD/NADP.8,13,14 Both dehydrogenases are members of the short-chain dehydrogenase/reductase superfamily of oxidoreductases.7,14 Additional cis- and trans-specific dehydrogenases may be involved in retinoid metabolism in RPE cells15 (see discussion below).
Spectroscopic methods for assaying pyridine nucleotide-dependent dehydrogenases rely on the change in absorbance or fluorescence of the coenzyme. These methods have not been useful with retinoid dehydrogenases because of the relatively low extinction coefficient of the reduced pyridine nucleotides, the turbidity of membrane-associated enzymes, and the fluorescence of retinols and retinyl esters. We developed a phase partition assay for retinoid dehydrogenases that is rapid, accurate, and sensitive.16 The assay relies on the transfer of tritium from water-soluble [3H]NADPH or [3H]NADH to water-insoluble retinoids. Thus, the activity can be detected by measuring the amount of tritium appearing in a hexane extract of the reaction mixture. Alternatively, the reaction can be followed with [15-3H]retinoid substrate by measuring the amount of tritium that appears in the aqueous phase as [3H]NADPH or [3H]NADH after hexane extraction. These methods are suitable for dehydrogenase reactions involving water-insoluble reactants and products, such as retinoid, steroid, and eicosanoid dehydrogenases.
In this chapter we present methods for the assay of retinoid dehydrogenases by phase partition and high-performance liquid chromatography (HPLC). Examples of the utility and limitation of these assays are provided from studies of retinoid metabolism in the visual cycle. In principle, dehydrogenases that interconvert alcohols and carbonyls can be assayed either in the direction of reduction or oxidation. Pyridine nucleotide-dependent dehydrogenase reactions are pH dependent (see reaction below): low pH favors reduction and high pH favors oxidation.
In general, we have used pH 5.5 for phase partition assays that depend on the transfer of tritium to the hydrophobic substrate (reduction). However, we also give examples of conditions at higher pH for the phase partition assay in which tritium is transferred from the hydrophobic substrate to the pyridine nucleotide (oxidation).
Methods
Chemical Procedures
General
All procedures involving retinoids are conducted under dim, red illumination or in aluminum foil-covered vials to minimize photoisomerization and photodecomposition. Chemical reactions are carried out in screwcap vials fitted with Teflon cap liners. Buffers are bubbled with argon and stored under argon.
Materials
9-cis-, 13-cis-, and all-trans-Retinal are purchased from Sigma (St. Louis, MO) and purified by HPLC. 11-cis-Retinal is obtained from R. Crouch (Medical University of South Carolina, Charleston, SC) and the National Eye Institute (Bethesda, MD).
Preparation of [3H]NADH and [3H]NADPH
NaB3H4 (5 mCi; Du Pont New England Nuclear, Boston, MA) is dissolved in 0.1 ml of H2O and immediately added to 5 mg of NADP or NAD (Sigma) in 0.5 ml of ice-cold 80 mM Tris buffer, pH 8.5. After 10 min of reaction at 0°, the reaction products are separated by HPLC with a column of Nucleosil SB 10 (4.6 × 250 mm) (Macherey-Nagel, Dueren, Germany) equilibrated in 10 mM KH2PO4, pH 7.
Preparation of pro-S-[3H]NADH and pro-S-[3H]NADPH
l-Glutamate dehydrogenase (400 μl; Sigma) is extensively dialyzed against 0.1 M NaCl and 10 mM Bis–Tris propane {1,3-bis[tris(hydroxymethyl)methylamino]-propane, BTP, pH 7.5 at 4°} and mixed with 0.25 mCi of l-[2,3-3H]glutamic acid (24 Ci/mmol; Du Pont New–England Nuclear), 50 μl of 0.1 M l-glutamic acid, 50 μl of 27 mM ADP, and 75 μl of 0.1 M β-NAD, dissolved in H2O; Sigma) or 200 μl of 0.1 M β-NADP (sodium salt, dissolved in H2O; Sigma), in a final volume of 1 ml in a 1.5-ml polypropylene tube. The pH of the reaction mixture is checked with pH paper and adjusted to pH 7–8 with 1 M Tris-HCl, pH 8.4, if necessary. The tube is wrapped in aluminum foil and held at room temperature overnight with occasional mixing. Insoluble material is pelleted by centrifugation, and the pellet is washed twice with 1 ml of 10 mM BTP, pH 7.4. The supernatant is combined in a 15-ml tube and diluted with 10 mM BTP, pH 7.4, to a final volume of 9.2 ml. Half of the solution is applied to a Mono Q HR 5/5 column (Pharmacia, Piscataway, NJ) equilibrated with 10 mM BTP, pH 7.4. The column is developed with a linear gradient from 0 M NaCl in 10 mM BTP to 0.5 M NaCl over 60 min at a flow rate of 1 ml/min. The absorbance is monitored at 340 nm. The elution of radiolabeled NADH and NADPH and nonlabeled NAD and NADP is shown in Fig. 1.
Fig. 1.
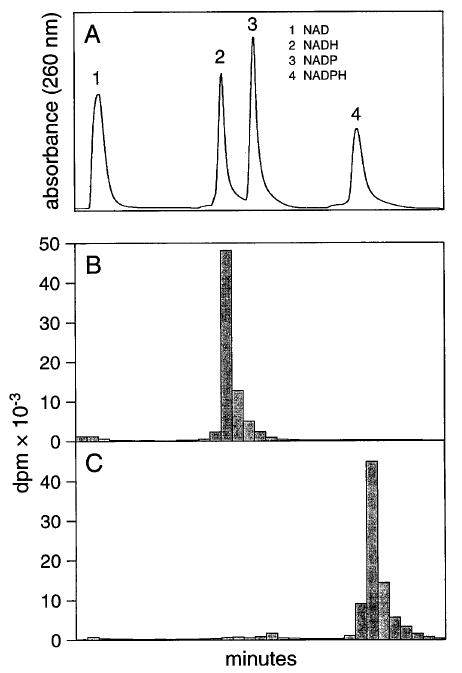
Separation of pyridine nucleotides by Mono Q Sepharose ion-exchange chromatography. (A) Profile obtained by measuring the absorbance of the eluate. (B) Preparation of [3H]NADH. (C) Preparation of [3H]NADPH. NADP and NAD were reduced with l-[2,3-3H]glutamate and glutamate dehydrogenase as described in text. The profile was obtained by following the radioactivity of the fractions.
Preparation of all-trans-[15-3H]Retinal
all-trans-[15-3H]Retinal is prepared by oxidizing all-trans-[15-3H]retinol (Du Pont–New England Nuclear) to all-trans-[15-3H]retinal with MnO2 as described in [19] in this volume.17 The specific radioactivity is adjusted to ~40,000 dpm/nmol by the addition of all-trans-retinal.
Preparation of Membranes and Enzymes
Photoreceptor Membranes
Photoreceptor membranes are prepared as described previously18 in the dark or under red illumination for studies of all-trans-retinol dehydrogenase with endogenous substrate. The membranes are bleached in the presence of NH2OH and extracted with petroleum ether when exogenous substrate is employed.
Retinal Pigment Epithelium Microsomes
Microsomes are isolated as described previously.19
Insect Cell Membranes
Insect cells (Sf9, Spodoptera frugiperda ovary cells) are transfected with a baculovirus shuttle vector (bacmid) carrying the cloned DNA of interest. The empty vector is used as a control. Insect cells are cultured at 27° in Sf-900 IISFM (Life Technologies, Gaithersburg, MD) and harvested 72–96 hr after infection by centrifugation for 15 min at 1200g. The cells are homogenized in water containing 2 mM benzamidine, 0.1 mM NADP, 0.5 mM dithiothreitol (DTT) and centrifuged at 100,000g for 30 min at 4°. The membrane fraction is washed in the same solution, pelleted by centrifugation at 100,000g for 5 min at 4°, and resuspended in five times the volume of the pellet.
Retinal Dehydrogenase
A supernatant prepared by homogenization of bovine retina is used as the source of retinal dehydrogenase.20
Phase Partition Assays
all-trans-Retinol Dehydrogenase with [3H]NADPH and Endogenous Substrate
In this assay,18 the conditions described employ a pH close to neutrality to mimic more closely the in vivo milieu of the photoreceptor outer segment. More rapid reaction rates are obtained at pH 5.5. The reaction mixture contains 10 mM HEPES (pH 7.5) and 20 μM [3H]NADPH, in a final volume of 60 μl. Reactions are initiated by the addition of rod outer segment (ROS) membranes (0.5 to 1.0 mg/ml, unbleached) under red illumination and are either incubated in the dark (control) or flashed to produce from 0.1 to 30% bleach of the visual pigment (Auto 322 Thyrister; SunPak, Newark, NJ). After the flash, reaction mixtures are incubated for 10 min at 37°. Reactions are stopped and extracted as described below.
all-trans-Retinol Dehydrogenase with [3H]NADPH and Exogenous Substrate
The reaction mixture contains 40–50 mM 2-(N-morpholino)ethanesulfonic acid (MES, pH 5.5), 6–10 μM [3H]NADPH (20,000 dpm/nmol), and membranes from insect cells (final concentration of 2.5 to 3 mg/ml) or bleached rod outer segments (final concentration of 0.1 mg/ml) in a final volume of 100–150 μl. all-trans-Retinal in dimethylformamide (DMF) is added to give a final concentration of 100–140 μM (final concentration of DMF, <0.1%). After 5 to 10 min at 30°, the reaction is quenched with 400 μl of methanol, 50 μl of 100 mM NH2OH (pH 7), and 50 μl of 1 M NaCl. After 2–3 min of vortexing, the radioactive product is extracted with 500 μl of hexane. Radioactivity is determined in 250–300 μl of the organic phase.
11-cis-Retinol Dehydrogenase with [3H]NADPH and Exogenous Substrate
Reaction and extraction conditions employed are similar to those described for all-trans-retinol dehydrogenase with exogenous substrate (above). 11-cis-Retinal, or an isomer of interest, is added at 20 μM final concentration. The reaction is initiated by addition of RPE microsomal membranes (100 μg/ml) or insect cell membranes (2.5 to 3 mg/ml).
11-cis-Retinol Dehydrogenase with 11-cis-[15-3H]Retinol
In this assay,16 the reaction conditions employ a pH close to neutrality. More rapid oxidation is observed at pH 8.5. The reaction (200-μl final volume) is performed in plastic, 1.5-ml centrifuge tubes at 37° for 20 to 30 min. Conditions include 50 mM Tris (pH 7.2), 30 μM bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), 60 μM NAD, and 10 μM 11-cis-[15-3H]retinol (added as a solution in ethanol; final ethanol concentration <1%). The reaction is initiated by the addition of 5 μl of RPE microsomal suspension (~20 μg of protein). The reaction is stopped by the addition of 400 μl of cold methanol and 20 μl of neutralized NH2OH, followed by incubation for 5 min at 37° to allow retinal oximes to form. After addition of 180 μl of 1 M NaCl, the tubes are placed on ice and 400 μl of dichloromethane is added with vortexing. A brief centrifugation is used to separate the phases. After removal of the lower (organic) phase with a Hamilton syringe, the upper phase is extracted twice more with 400 μl of dichloromethane. Finally, radioactivity is determined in 400 μl of the upper phase.
Retinaldehyde Dehydrogenase
all-trans-[15-3H]Retinal (10 μM final concentration) is dissolved in 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS, pH 7), containing 150 mM KCl, 20 μM BSA, 2 mM DTT, and 60 μM NAD, in a final reaction volume of 200 μl.16,20 The reaction is initiated by the addition of retinal supernatant (0.1 to 0.2 mg of protein per milliliter). After 30 min at 37°, 920 μl of dichloromethane–methanol (1:1, v/v) is added, followed by 22 μl of freshly neutralized 1 M NH2OH (final concentration 40 mM). After an incubation of 15 min to allow formation of retinal oximes, 46 μl of 1 M NaCl is added. After vortexing, the phases are separated by centrifugation (1000g, 1–2 min, 4°) and the upper (aqueous) phase is removed. The remaining aqueous phase is extracted again with another addition of 460 μl each of methanol and 1 M NaCl and radioactivity is determined in the combined upper phases.
Sterol Dehydrogenases
In this assay,7,21 sterol substrates dissolved in ethanol are added to a reaction mixture containing 25 mM MES (pH 5.5) and 20 μM [3H]NADPH to give a final concentration of 0.5 mM. Membranes (typically bacterial or insect cell membranes) are added to give a final concentration of 1 mg/ml. After 10 min at 30°, steroids are extracted with a 10-fold volume of dichloromethane and radioactivity is determined. The reaction product can be further identified by fluorography and thin-layer chromatography on silica with chloroform–ethyl acetate (3:1, v/v). As a positive control, the reduction of 4-androstene-3,17-dione to testosterone by rat liver microsomes is used. The following substrates are used for assays of 13α-, 11β-, or 17β -hydroxysteroid dehydrogenase activities: androsterone (5α-androstan-3α-ol-17-one), 5α-androstan-17β-ol-3-one, dihydroandrosterone (5α-androstane-3α,17β-diol), 11-dehydrocorticosterone (4-pregnen-21-ol-3,11,20-trione), corticosterone (4-pregnene-11β,21diol-3,20-dione), esterone (l,3,5[10]-estratrien-3-ol-17-one), β-estradiol (l,3,5[10]-estratrien-3,17β-diol), testosterone (4-androsten-17β-ol-3-one), 4-androstene-3,17-dione, and 5α-androstane-3,17-dione. The steroids are dissolved in ethanol to give 15–50 mM stock solutions.
High-Performance Liquid Chromatography Assays
all-trans-Retinal Dehydrogenase with Exogenous Substrate
In this assay,18 reaction conditions are as described for the phase partition assay (see above). Reactions are stopped by the addition of 1 ml of cold ethanol and 0.1 ml of neutralized O-ethylhydroxylamine (Fluka, Milwaukee, WI) (final concentration 40 mM). After 10 min at room temperature, an additional 1 ml of ethanol and 5 ml of hexane are added and mixed with a Pasteur pipette. Two milliliters of H2O is added, and the phases are separated by centrifugation. The upper (hexane) phase is removed, dried in a stream of argon, and taken up in acetonitrile, and retinoids are separated by isocratic reversed-phase HPLC (see [19] in this volume17). The addition of O-ethyl-hydroxylamine results in conversion of the residual retinal substrate to O-ethyl oximes, which are separated from the retinols on reversed-phase HPLC.22
11-cis-Retinol Dehydrogenase
In this assay,16 11-cis-retinal (or another isomer), dissolved in ethanol, is added to a reaction mixture containing 50 mM MOPS, (pH 5.5), 30 μM BSA, 60 μM NADPH, and 1 mM DTT (final concentration of ethanol, <0.1%). After a brief incubation at 37°, a suspension of RPE microsomes is added to give a final microsomal protein concentration of 30 μg/ml. After an appropriate time (usually 10 min) a 1-ml portion is removed and the reaction quenched by addition of 1 ml of ice-cold ethanol. After addition of 4 ml of hexane, the upper phase is repeatedly introduced into the lower phase with a Pasteur pipette, taking care not to introduce air into the sample. The upper hexane phase is removed, an additional 4 ml of hexane is introduced, and the extraction is repeated. The combined upper phases from three extractions are back-extracted with H2O to remove dissolved ethanol, and the hexane is evaporated to dryness with flowing argon, taken up in a small volume of hexane (<200 μl), and introduced into an HPLC column. Retinoids are resolved by normal-phase HPLC as described in [19] in this volume.17
Discussion
The methods reported here for assay of visual cycle and other dehydrogenases can be employed advantageously for a variety of specialized purposes, or in combination to obtain further information.
Phase-Partition Assay
Improvements in the separation of pyridine nucleotides on ion-exchange resins has simplified the preparation of [3H]NADPH and [3H]NADH for use with the phase partition assay (Fig. 1). In addition, the use of stereospecific enzymes for the production of pro-4S and pro-4R forms of the pyridine nucleotides further extends the utility of the method. As an example, we used the phase partition assay to determine that all-trans-retinol dehydrogenase (RDH) from photoreceptor outer segments catalyzed transfer of the pro-4S proton of NADPH to retinol.7 The phase partition assay is particularly useful when multiple determinations are required; for instance, during the development of purification procedures20 and for kinetic analysis.18 Reaction products should always be verified by HPLC analysis before routine use of the phase partition assay, especially if the substrate is readily isomerized or if additional, competing enzymes are present.
Chemical reduction of NADP is not specific to the 4-position of the substituted pyridine ring and the isomers generated are incompletely separated from each other by Nucleosil chromatography. This has not proved to be a problem because of the strict specificity of the dehydrogenases for proton transfer from the 4-position of the pyridine ring. Comparison of the results of the phase partition assay with those of the HPLC assay for all-trans-retinol dehydrogenase of ROS has not revealed major discrepancies.18 Reduction with enzymes and substrates specific for either the pro-R or pro-S forms of the pyridine nucleotide avoids this problem completely and provides further information about the specificity of the dehydrogenase under study. [3H]NADH or [3H]NADPH could be stored over a 3-month period in the dark at −80° with no noticeable decomposition.
The phase partition assay was used to compare the retinoid–dehydrogenase activities of RPE microsomes with those of 11-cis-RDH expressed in insect cells (r11-cis-RDH). The recombinant enzyme showed a marked preference for 11-cis- and 13-cis-isomers of retinal (Fig. 2A and C), which was not apparent with RPE microsomes (Fig. 2B and D). The apparent activity observed with all-trans-retinal resulted from isomerization of the substrate (confirmed by HPLC assay). In addition, r11-cis-RDH was completely inhibited by NADH, whereas the 11-cis-RDH activity of RPE microsomes was only partially inhibited by this nucleotide (Fig. 3). The results suggest that r11-cis-RDH does not account for all the retinoid dehydrogenase activities of RPE microsomes and that other enzymes remain to be characterized.15
Fig. 2.
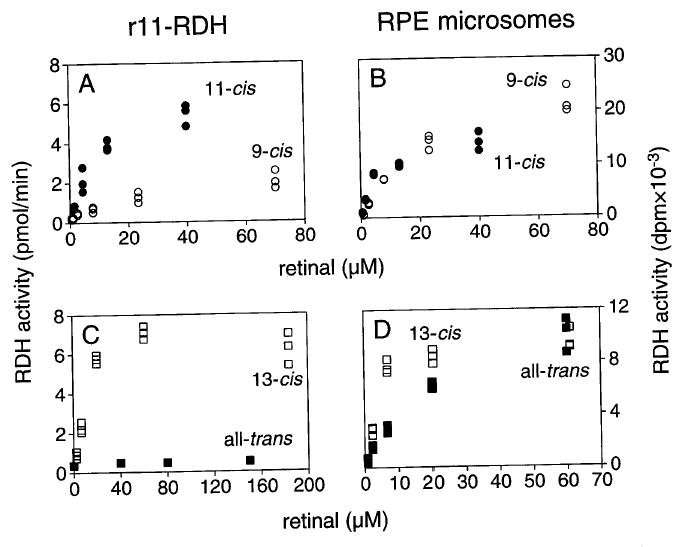
Phase partition assay. Enzymatic activities of recombinant 11-cis-retinol dehydrogenase (r11-cis-RDH) (A and C) and RPE microsomes (B and D). (A)–(D) depict the results obtained when all-trans-, 9-cis-, or 13-cis-retinal and [3H]NADPH were incubated with the indicated source of the enzyme. The large difference in the ratio of 11-cis-retinol to 9-cis-retinol dehydrogenase activities displayed by the two preparations suggests that the activities of RPE microsomes are not accounted for by r11-cis-RDH.
Fig. 3.
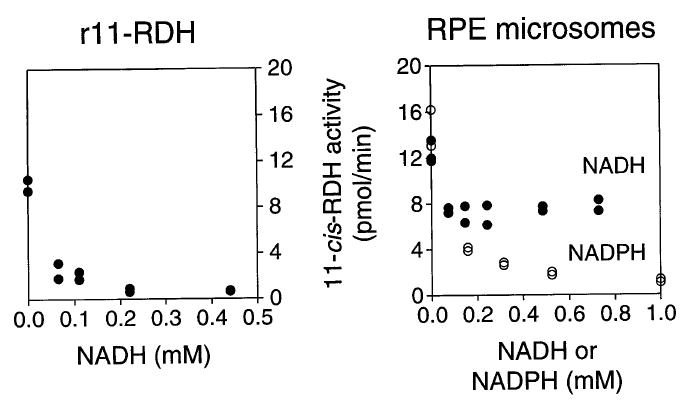
Phase partition assay: inhibition of 11-cis-RDH activity by NADH or NADPH. 11-cis-RDH activity was measured at 37° for 15 min with 11-cis-retinal and [3H]NADPH and with RPE microsomes or recombinant 11-cis-RDH (r11-cis-RDH). Inhibition by varying concentrations of NADH or NADPH was measured. However, NADH only partially inhibits the 11-cis-RDH activity of RPE microsomes whereas r11-cis-RDH activity is inhibited completely.
The phase partition assay is particularly useful for determining the enzymatic activities of expressed cDNAs encoding short-chain dehydrogenase/reductases (SDRs) thought to be involved in retinoid metabolism. Some of these enzymes also accept steroid substrates,21 and the phase partition assay with appropriate steroid substrates can be used to assess this parameter.7 Expression in insect cells proved to be superior to expression in bacteria and addition of NADP to the insect cell homogenate increased the stability of the dehydrogenase.
High-Performance Liquid Chromatography Assay
The HPLC assay, while less amenable to multiple samples, provides a positive identification of the product and is useful when further metabolism of the product occurs or when isomerization of the substrate is likely. These two factors are illustrated by the metabolism of various isomers of retinal by RPE microsomes in the presence of NADH (Fig. 4). Addition of each of the cis-isomers of retinal resulted in the appearance of the corresponding alcohol and varying amounts of retinyl ester (Fig. 4A–C). Thus, the retinol produced by the dehydrogenase reaction was esterified by the powerful lecithin: retinol acyltransferase (LRAT, EC 2.3.1.135) activity known to be present in RPE microsomes.23,24 In contrast, addition of all-trans-retinal (Fig. 4) did not result in the appearance of all-trans-retinol, illustrating the stereospecificity of the reaction. Some substrate is converted to 13-cis-retinol, probably because of isomerization to 13-cis-retinal followed by reduction.
Fig. 4.
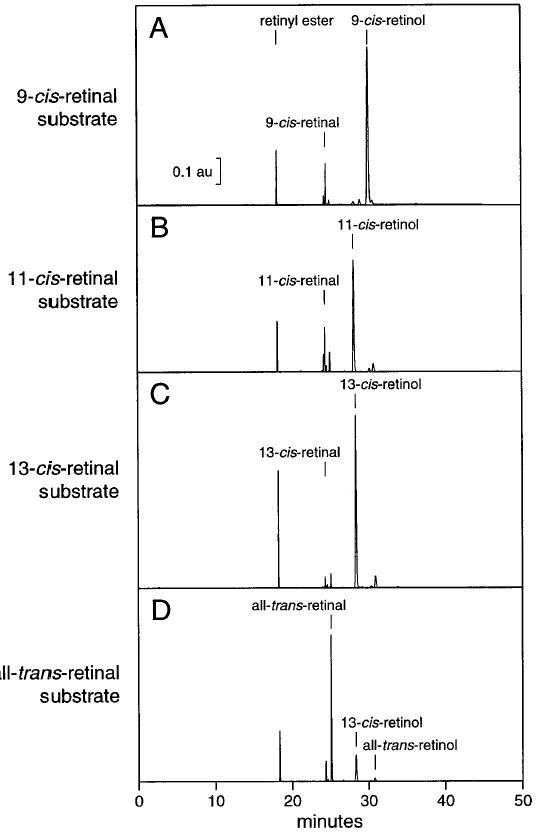
HPLC assay: activity of RPE microsomes with isomers of retinal. The retinoids were incubated at 10 μM with RPE microsomes and 200 μM NADH as described in the text. After 10 min of reaction, the retinoids were extracted and analyzed by normal-phase HPLC as described. (A–C) Reduction of each of the cis-isomers to the corresponding retinol is evident. The retinyl ester peak indicates that the product retinol was further esterified. (D) No activity was observed with all-trans-retinal as the substrate. The 13-cis-retinol and retinyl ester observed are likely to have resulted from reduction of 13-cis-retinal, formed by nonenzymatic isomerization during the incubation.
The HPLC assay is also useful when pairs of competing substrates are presented to an enzyme. For instance, a cis-specific RDH activity of RPE microsomes preferentially metabolizes 11-cis- or 13-cis-retinals when the substrates are presented paired with 9-cis-retinal (Fig. 5A). Others have suggested that this 11-cis-RDH of RPE microsomes catalyzes the first step in the biosynthesis of 9-cis-retinoic acid.25,26 In RPE, where 11-cis-retinoids are relatively abundant, it appears unlikely that this enzyme could play a major role in the production of 9-cis-retinoic acid unless the reaction occurs in a defined intracellular compartment.
Fig. 5.
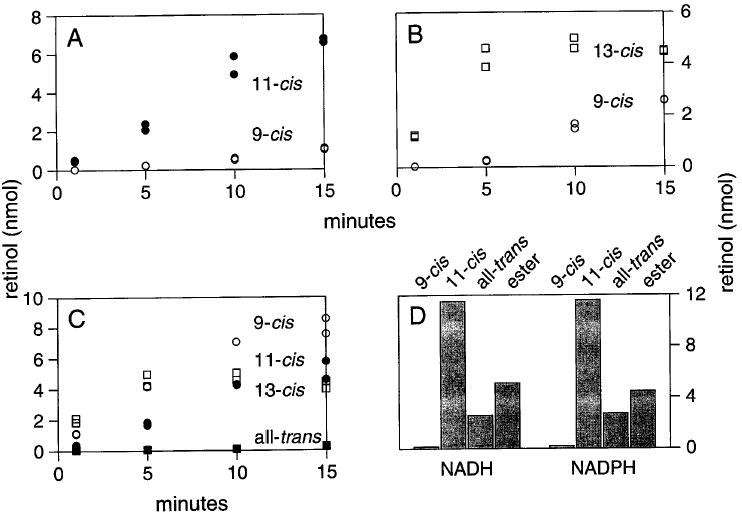
HPLC assay: activity of RPE microsomes with pairs of retinal substrates. RDH activity with pairs of retinal substrates was measured at 37° with 10 μM retinoids and 200 μM NADH. (A) 11-cis-Retinal is reduced in preference to 9-cis-retinal. (B) Reduction of 9-cis-retinal is delayed until reduction of 13-cis-retinal plateaus. (C) Reduction of retinal substrates presented individually. 9-cis-Retinal is readily reduced when it is the only substrate. (D) Comparable amounts of reduction of 11-cis-retinal are obtained with NADH or NADPH. (D) also depicts the amount of nonenzymatic isomerization of 11-cis-retinal with this preparation and the amount of retinyl ester produced by subsequent esterification of the retinol.
Perspective
Study of the physiologic roles of SDRs is complicated by the general lack of substrate specificity exhibited by these enzymes. For instance, all-trans-retinol dehydrogenases from liver27 have been shown to catalyze the efficient oxidation of hydroxysteroids, raising the question of the identity of their physiological substrates.21 A similar problem exists with defining the physiologic role of 11-cis-retinol dehydrogenase (11-cis-RDH), which has been suggested to catalyze the oxidation of 9-cis-retinol to 9-cis-retinal during the synthesis of 9-cis-retinoic acid.25 Rigorous identification of a physiological substrate must include the presence of both the enzyme and its substrate in the same cellular environment. The genome of Caenorhabditis elegans contains ~80 genes encoding proteins with sequences containing SDR motifs (http://www.sanger.ac.uk/Projects/C_elegans/). The mammalian genome is likely to contain many more SDR-related sequences and it seems certain that assigning the physiological roles of these enzymes will be complicated. Furthermore, the presence of several enzymes with overlapping substrate specificities makes it likely that knockout animals will have a subtle phenotype.
Acknowledgments
The methods reported here were developed or utilized in the authors’ laboratories with support from the National Eye Institute (EY02317, 01730, 09339), and by an unrestricted grant to the Department of Ophthalmology Research from Research to Prevent Blindness, Inc. (RBP). J.C.S. is a Senior Scientific Investigator of RBP and K.P. is a Jules and Doris Stein Professor of RBP.
References
- 1.Dowling JE. Nature (London) 1960;168:114. doi: 10.1038/188114a0. [DOI] [PubMed] [Google Scholar]
- 2.Wald G. Science. 1968;162:230. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 3.Bok D. J Cell Sci Suppl. 1993;17:189. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- 4.Crouch RK, Chader GJ, Wiggert B, Pepperberg DR. Photochem Photobiol. 1996;64:613. doi: 10.1111/j.1751-1097.1996.tb03114.x. [DOI] [PubMed] [Google Scholar]
- 5.Rando RR. Chem Biol. 1996;3:255. doi: 10.1016/s1074-5521(96)90105-2. [DOI] [PubMed] [Google Scholar]
- 6.J. C. Saari, in “The Retinoids: Biology, Chemistry, and Medicine,” 2nd Ed. (M. B. Sporn, A. B. Roberts, and D. S. Goodman, eds.), p. 351. Raven Press, New York, 1994.
- 7.Haeseleer F, Huang J, Lebioda L, Saari JC, Palczewski K. J Biol Chem. 1998;273:21790. doi: 10.1074/jbc.273.34.21790. [DOI] [PubMed] [Google Scholar]
- 8.Lion F, Rotmans JP, Daemen FJM, Bonting SL. Biochim Biophys Acta. 1975;384:283. doi: 10.1016/0005-2744(75)90030-3. [DOI] [PubMed] [Google Scholar]
- 9.Wald G, Hubbard R. J Gen Physiol. 1949;32:367. doi: 10.1085/jgp.32.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann KP, Pulvermüller A, Buczylko J, Van Hooser P, Palczewski K. J Biol Chem. 1992;267:15701. [PubMed] [Google Scholar]
- 11.Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC. Biochemistry. 1999;37:12012. doi: 10.1021/bi990504d. [DOI] [PubMed] [Google Scholar]
- 12.Saari JC, Garwin GG, Van Hooser JP, Palczewski K. Vision Res. 1998;38:1325. doi: 10.1016/s0042-6989(97)00198-3. [DOI] [PubMed] [Google Scholar]
- 13.Driessen CAGG, Janssen BPM, Winkens HJ, van Vugt AHM, de Leeuw TLM, Janssen JJM. Invest Ophthalmol Visual Sci. 1995;36:1988. [PubMed] [Google Scholar]
- 14.Simons A, Hellman U, Wernstedt C, Eriksson U. J Biol Chem. 1995;270:1107. [PubMed] [Google Scholar]
- 15.Driessen CAGG, Winkens HJ, Kuhlmann ED, Janssen APM, van Vugt AHM, Deutman AF, Janssen JJM. FEBS Lett. 1998;428:135. doi: 10.1016/s0014-5793(98)00473-6. [DOI] [PubMed] [Google Scholar]
- 16.Saari JC, Bredberg DL, Garwin GG, Buczylko J, Wheeler T, Palczewski K. Anal Biochem. 1993;213:128. doi: 10.1006/abio.1993.1395. [DOI] [PubMed] [Google Scholar]
- 17.Garwin GG, Saari JC. Methods Enzymol. 1999;316(19) doi: 10.1016/s0076-6879(00)16731-x. this volume. [DOI] [PubMed] [Google Scholar]
- 18.Palczewski K, Jager S, Buczylko J, Crouch RK, Bredberg DL, Hofmann KP, Asson-Batres MA, Saari JC. Biochemistry. 1994;33:13741. doi: 10.1021/bi00250a027. [DOI] [PubMed] [Google Scholar]
- 19.Saari JC, Bredberg DL. Methods Enzymol. 1990;190:156. doi: 10.1016/0076-6879(90)90020-2. [DOI] [PubMed] [Google Scholar]
- 20.Saari JC, Champer RJ, Asson-Batres MA, Garwin GG, Huang J, Crabb JW, Milam AH. Visual Neurosci. 1995;12:263. doi: 10.1017/s095252380000794x. [DOI] [PubMed] [Google Scholar]
- 21.Biswas MG, Russell DW. J Biol Chem. 1997;272:15959. doi: 10.1074/jbc.272.25.15959. [DOI] [PubMed] [Google Scholar]
- 22.Van Kuijk FJGM, Handelman GJ, Dratz EA. J Chromatogr. 1985;348:241. doi: 10.1016/s0021-9673(01)92458-6. [DOI] [PubMed] [Google Scholar]
- 23.Barry RJ, Cañada FJ, Rando RR. J Biol Chem. 1989;264:9231. [PubMed] [Google Scholar]
- 24.Saari JC, Bredberg DL. J Biol Chem. 1989;264:8636. [PubMed] [Google Scholar]
- 25.Mertz JR, Shang E, Piantedosi R, Wei S, Wolgemuth DJ, Blaner WS. J Biol Chem. 1997;272:11744. doi: 10.1074/jbc.272.18.11744. [DOI] [PubMed] [Google Scholar]
- 26.Romert A, Tuvendal P, Simon A, Dencker L, Uriksson U. Proc Natl Acad Sci USA. 1998;95:4404. doi: 10.1073/pnas.95.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boerman MHEM, Napoli JL. Biochemistry. 1995;34:1027. doi: 10.1021/bi00021a014. [DOI] [PubMed] [Google Scholar]


