Abstract
Approximately 800 transcripts in Saccharomyces cerevisiae are cell cycle regulated. The oscillation of ∼40% of these genes, including a prominent subclass involved in nutrient acquisition, is not understood. To address this problem, we focus on the mitosis-specific activation of the phosphate-responsive promoter, PHO5. We show that the unexpected mitotic induction of the PHO5 acid phosphatase in rich medium requires the transcriptional activators Pho4 and Pho2, the cyclin-dependent kinase inhibitor Pho81, and the chromatin-associated enzymes Gcn5 and Snf2/Swi2. PHO5 mitotic activation is repressed by addition of orthophosphate, which significantly increases cellular polyphosphate. Polyphosphate levels also fluctuate inversely with PHO5 mRNA during the cell cycle, further substantiating an antagonistic link between this phosphate polymer and PHO5 mitotic regulation. Moreover, deletion of PHM3, required for polyphosphate accumulation, leads to premature onset of PHO5 expression, as well as an increased rate, magnitude, and duration of PHO5 activation. Orthophosphate addition, however, represses mitotic PHO5 expression in a phm3Δ strain. Thus, polyphosphate per se is not necessary to repress PHO transcription but, when present, replenishes cellular phosphate during nutrient depletion. These results demonstrate a dynamic mechanism of mitotic transcriptional regulation that operates mostly independently of factors that drive progression through the cell cycle.
Coordination of cell growth and division is essential to all living organisms and is innately tied to the cell division cycle. Periodic increases in certain transcripts at distinct cell cycle phases can meet specific, one-time requirements. For instance, nucleotide biosynthetic and histone genes are activated prior to and during S phase, respectively, to ensure adequate substrate concentrations for chromosomal duplication (9, 16). In addition, the sequential activation and proteolytic destruction of cyclins that partner with cyclin-dependent kinase (CDK) activities drives progression through the cell cycle (29). In contrast to this posttranslational mode of cell cycle control, it is clear that much cell cycle regulation occurs at the level of initiation of transcription by RNA polymerase II. In Saccharomyces cerevisiae, three major classes of transcriptional activators, MBF and SBF, Swi5/Ace2, and Mcm1-associated factors, predominantly regulate various gene clusters at the G1/S transition; M and the M/G1 boundary; and G1, M, or M/G1, respectively (42).
Spellman et al. (42) identified ca. 800 genes exhibiting cell cycle oscillation. While the regulation of ∼500 of these genes can be ascribed to the three known classes of cell cycle transactivators, the mode of regulation of the rest is not understood. Many of these ∼300 genes participate in nutrient acquisition, and their transcripts show peak expression in M or M/G1. For instance, mitotic expression has been observed for genes involved in phosphate metabolism (28, 35) encoding low-affinity (PHO89) and high-affinity (PHO84) transporters of inorganic orthophosphate (Pi), as well as repressible acid phosphatases (rAPases; PHO5, PHO11, and PHO12) and constitutive APase (PHO3). Based on the timing of their expression, the mitotically induced rAPase genes are grouped with 34 other genes comprising the MCM cluster (42). However, there is no evidence for a direct or indirect role of Mcm1, or another known DNA binding cell cycle regulator, in their transcriptional activation. Moreover, as all the experiments of Spellman et al. (42) were conducted in rich medium that contained Pi, it is unclear why these PHO genes, which ordinarily are induced upon Pi deprivation, are activated during specific cell cycle phases. Thus, PHO5, PHO11, and PHO12, as well as other cell cycle-regulated PHO genes, fall into a large group of genes involved in maintaining nutritional homeostasis for which it is not understood how cell cycle periodicity is accomplished or why it is needed.
To date, studies of the mechanisms of yeast PHO gene activation have focused on regulatory events that occur in asynchronous cultures as a function of limiting Pi. By an as-yet-unknown mechanism, Pi limitation initiates a signal transduction cascade that activates Pho81, a CDK inhibitor. Pho81 inhibits the phosphorylation activity of the cyclin-CDK Pho80-Pho85 (39), leading to nuclear retention of the otherwise cytoplasmic helix-loop-helix factor Pho4, the primary PHO transactivator (22, 34). In the nucleus, Pho4, either by itself or as a heterodimer with Pho2, a nuclear homeodomain protein, activates >20 different genes in the PHO system (19, 33). These include several phosphate metabolism (PHM) genes, PHM1 (VTC2), PHM2 (VTC3), PHM3 (VTC4), PHM4 (VTC1), and PHM5 (PPN1). Phm1 to -4 form a biochemical complex in vivo, the integrity of which is disrupted by phm3 or phm4 null mutations (6, 31). As polyphosphate (polyP), a linear polymer of up to hundreds of Pi residues linked by high-energy phosphoanhydride bonds, is not detectable in such strains, the Phm/Vtc complex is proposed to be a polyP synthetase (33). However, this complex is also needed for the proper morphology of vacuoles (31), where 99% of the total cellular polyP is stored (44).
PolyP is present in all organisms examined to date and, in S. cerevisiae, can reach concentrations as high as 120 mM in the vacuole, as much as 60% of the total phosphate content (23). It has been suggested that polyP functions as a phosphate reservoir, since its levels drop when cells need Pi, e.g., during log-phase growth or in Pi-limiting growth media (11, 41). It has also been suggested that polyP serves a cellular protective function in that it chelates cations, such as Ca2+, in the vacuole (23). However, yeast strains deficient in polyP accumulation are not sensitive to high concentrations of calcium (33). Finally, polyP levels increase as yeast cultures approach stationary phase for reasons and by mechanisms that are not clear (40, 47). Thus, while conditions and enzymes involved in polyP synthesis and hydrolysis in eukaryotes have been identified, its metabolic, regulatory, and physiological function(s) remain obscure.
Relative to our understanding of the connection between nutrient sensing and the decision to execute Start (36), little is known about how nutrient levels influence regulation in other stages of the cell cycle. To gain insight into this problem, we have focused on elucidating both the molecular mechanisms and the physiological basis for M phase activation of PHO5. We report that the unexpected mitotic induction of PHO5 in rich medium is under the control of Pho4, Pho2, and Pho81. Increasing the metabolic pools of polyP represses mitotic expression of PHO5. Conversely, elimination of polyP by deletion of PHM3 leads to PHO5 activation. Moreover, we demonstrate that polyP levels are influenced by progression of the cell cycle, declining prior to and being replenished after M phase. Our results suggest that polyP is a dynamic phosphate reserve, and they define a regulatory influence of polyP on PHO mitotic gene expression. Further, our studies demonstrate that Pho4/Pho2 mediates a novel, cell cycle stage-specific mode of regulation that is fine tuned in response to nutrient availability and operates mostly independently of activities that drive cell cycle progression. More generally, our findings suggest that nutrient deficiencies may underlie the transcriptional periodicity of a large class of nutrient transporters that are cell cycle regulated.
MATERIALS AND METHODS
Yeast strains and methods.
All S. cerevisiae strains were constructed by standard genetic methods (37) from CCY694, MATa/MATα leu2Δ0/leu2Δ0 lys2Δ0/lys2Δ0 ura3Δ0/ura3Δ0 pho3Δ::R/pho3Δ::R (S288C background) (2), where R is a Zygosaccharomyces rouxii recombinase site that remains after intramolecular recombination (21). The strains and their relevant (all are MATa leu2Δ0 lys2Δ0 ura3Δ0 pho3Δ::R) genotypes are DNY742 (wild type; MATa bar1Δ::R-URA3-R), THY868 (MATa bar1Δ::R-URA3-R pho4Δ::kanMX4), DNY989 (MATa bar1Δ:: R-URA3-R pho2Δ::kanMX4), DNY925 (MATa bar1Δ::R-URA3-R pho81Δ::kan MX4), DNY986 (MATa bar1Δ::R-URA3-R gcn5Δ::kanMX4), DNY1309 (MATa snf2/swi2Δ::kanMX4), DNY1673 (MATa bar1Δ::R-URA3-R phm3Δ::kanMX4), DNY2464 (MATa his3Δ1 phm1Δ::kanMX4), DNY2465 (MATa bar1Δ::R-URA3-R met15Δ0 phm2Δ::kanMX4), and DNY2467 (MATa bar1Δ::R-URA3-R his3Δ1 phm4Δ::kanMX4).
Media and growth conditions.
Cells were grown at 30°C on plates or in liquid cultures of complete synthetic medium- 2% glucose (Bio 101), yeast extract (Difco)- peptone (Difco)- 2% glucose (YPD), or YPD supplemented with 13.4 mM KH2PO4 (YPD + Pi) as appropriate. The defined Pi-free medium used in one step of the phosphate overplus experiments (see Fig. 5B), as well as for Pi starvation (see Fig. 2E and 9), contained 0.7 g of yeast nitrogen base without (NH4)2SO4, phosphate, or amino acids (Bio 101), 2 g of glutamine, 20 g of glucose, and 3.9 g of MES (2-N-morpholino ethanesulfonic acid), pH 5.5, per liter.
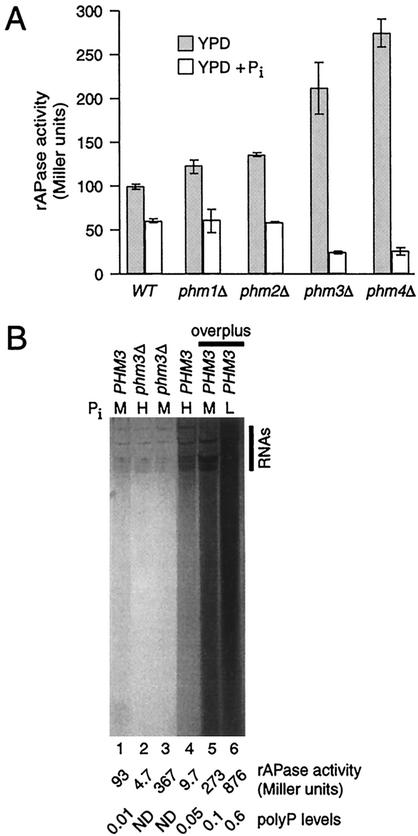
FIG. 5.
Loss of polyP leads to derepression of PHO5. (A) Total rAPase activities of wild-type (WT), phm1Δ, phm2Δ, phm3Δ, and phm4Δ strains. Cells were grown in YPD and diluted in YPD or YPD + Pi medium for 6 h prior to assay of rAPase activity (n = 3; mean ± 1 standard deviation). The severity of loss of polyP accumulation increases from left to right and parallels the extent of disruption of the Phm/Vtc complex, with phm3Δ and phm4Δ strains having no detectable polyP or Phm/Vtc complex (31, 33). (B) Analysis of polyP levels. Wild-type (PHM3) or phm3Δ strains were grown for 2 days on plates and then for 24 h in liquid YPD-low Pi, YPD, or YPD + Pi medium containing a low (L; lane 6), moderate (M; lanes 1, 3, and 5), or high (H; lanes 2 and 4) concentration of Pi, respectively. For lanes 1 to 4, after the 24-h incubation period, internal aliquots of each culture were assayed for rAPase activity and polyP levels by the metachromatic absorbance shift method (as indicated at the bottom; ND, nondetectable). The overplus samples (lanes 5 to 6) received additional treatments of Pi starvation followed by Pi addition (see Materials and Methods) before rAPase and polyP levels were determined. PolyP was also visualized in the gel after electrophoresis of equal amounts (10 μg) of total RNA and staining with toluidine blue O dye, a basic dye that binds polyanions. RNA species at the top of the gel are indicated on the right.
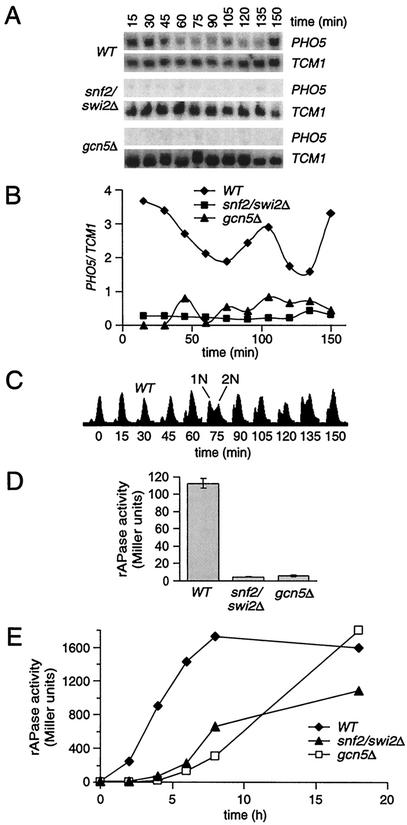
FIG. 2.
PHO5 activation is SNF/SWI and Gcn5 dependent. (A) Northern analysis of synchronous cultures of wild-type (WT), snf2/swi2Δ, and gcn5Δ strains released from nocodazole arrest in YPD medium. (B) Normalized PHO5 transcript levels from panel A. (C) Flow cytometric analysis of wild-type cells in panel A. (D) Total rAPase activities of asynchronous YPD cultures (n = 3; mean ± 1 standard deviation). (E) Time course of PHO5 activation. Asynchronous cultures grown on defined Pi-free medium with 13.4 mM Pi added back were starved for Pi and assayed for total rAPase activity at the indicated times.
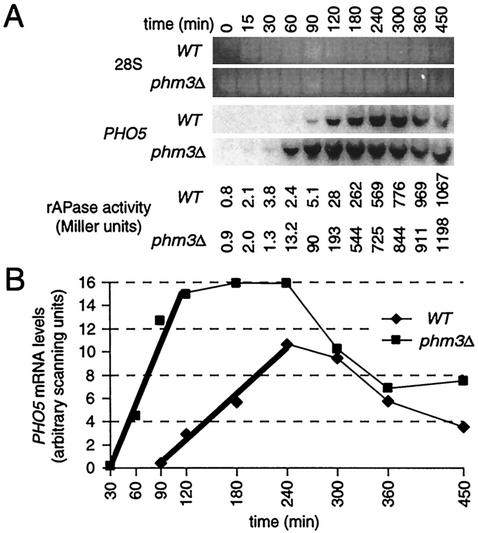
FIG. 9.
Deletion of PHM3 increases the rate of PHO5 activation. (A) Northern analysis of RNA isolated during a time course of PHO5 activation. Wild-type (WT) and phm3Δ strains grown on defined Pi-free medium with 13.4 mM Pi added back (to build up stores of polyP in the wild-type strain) were then starved for Pi by being washed and resuspended in Pi-free medium, and RNA was isolated at the indicated times. The total rAPase activities obtained from an internal aliquot of each culture are indicated below each lane. (B) Absolute PHO5 transcript levels from panel A. The thick trend line indicates the initial, linear period of transcript accumulation.
All cultures, including overnight cultures, were maintained in early to mid-logarithmic phase growth. When Pi starvation was employed, the cells grew for only two to three additional generations after being washed and resuspended in Pi-free medium. Phosphate overplus experiments (see Fig. 5B) were performed as previously described (33); pregrowth in YPD or YPD-low Pi (YPD from which Pi was removed by precipitation) (15) followed by 2 h of Pi starvation in defined Pi-free medium and, finally, addition of KH2PO4 to 13.4 mM for 2 h. Note that while YPD is limiting for Pi (see Fig. 3 to 5), the designation YPD-low Pi indicates that the Pi concentration of the medium is decreased further but it is not Pi free. For measuring rAPase activities in asynchronous cultures (see Fig. 1D, 2D, 4A, and 5A), overnight cultures (10 ml) were grown and diluted the next day to an optical density at 600 nm (OD600) of 0.03 in the appropriate fresh medium and incubated for another 6 h. For the Pi starvations (see Fig. 2E and 9), strains were plated on defined Pi-free medium with 13.4 mM KH2PO4 added back for 3 days. After overnight growth of starter cultures (10 ml) in the same medium, the cells were diluted in defined Pi-free medium at an OD600 of 0.2 and assayed at the appropriate times for rAPase activity and/or PHO5 mRNA.
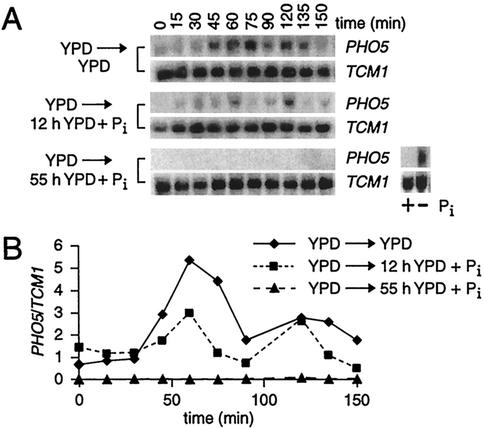
FIG. 3.
PHO5 mitotic activation is repressed by addition of orthophosphate (Pi). (A) Northern analysis of α-factor-synchronized cultures. Single colonies from a YPD plate were inoculated into YPD overnight cultures and then diluted in YPD or YPD + Pi (13.4 mM) for 12 or 55 h (as indicated on the left) prior to α-factor arrest. Cells grown for 6 h in defined Pi-free medium with (+) or without (−) Pi were included as a positive control in the Northern analysis of the samples grown for 55 h in YPD + Pi. (B) Normalized PHO5 transcript levels from panel A.
FIG. 1.
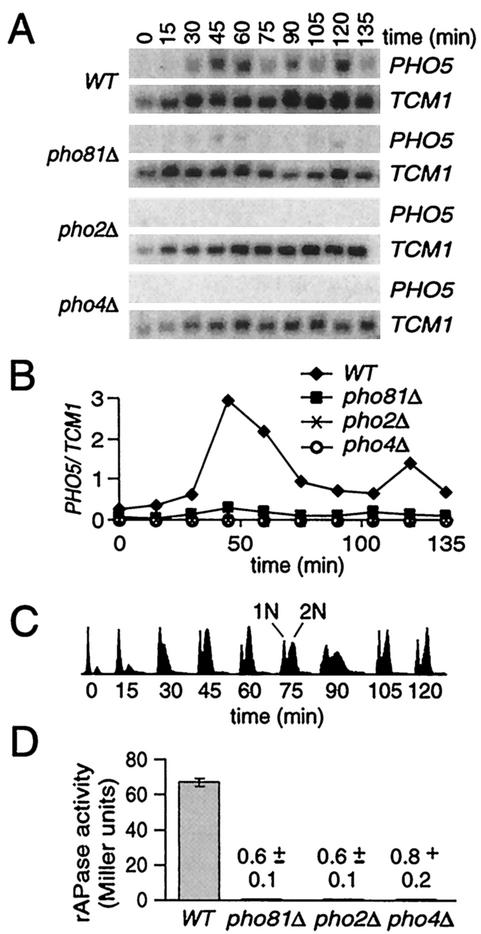
Mitotic induction of PHO5 requires PHO2, PHO4, and PHO81. (A) Northern analysis of wild-type (WT), pho81Δ, pho2Δ, and pho4Δ cultures at various times following release from synchronization with α-factor. The wild-type strain was analyzed in parallel with each null strain as a positive control. (B) Transcript levels of PHO5 (normalized to TCM1) from panel A. (C) Flow cytometric analysis of Sytox Green-stained cells. The x and y axes indicate the fluorescence intensity and numbers of cells analyzed (which was identical in all panels), respectively. 1N and 2N refer to haploid and diploid DNA contents, respectively. (D) Total rAPase activities of asynchronous YPD cultures. The means ± 1 standard deviation from three independent experiments are shown.
FIG. 4.
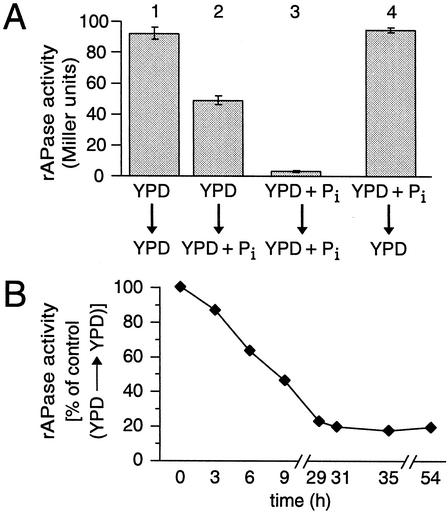
Repression of PHO5 mitotic expression by added Pi is time dependent. (A) Total rAPase activities of asynchronous cultures. Cultures were grown overnight in either YPD (bars 1 and 2) or YPD + Pi (bars 3 and 4) and then washed and resuspended in YPD (bars 1 and 4) or YPD + Pi (bars 2 and 3) for an additional 6 h, as indicated (n = 3; mean ± 1 standard deviation). (B) Time course of decrease in total rAPase activity following addition of Pi. Cells were plated and pregrown on YPD and transferred to YPD (control) or YPD + Pi. The percentage of activity of the control (growth in parallel in YPD) is plotted as a function of time.
Cell cycle synchronizations, fluorescence-activated cell sorter analysis, and RNA-polyP isolation.
Yeast cultures in early logarithmic growth in YPD were arrested at late G1 and G2/M by the addition of α-factor and nocodazole to final concentrations of 12 ng/ml and 17 μg/ml, respectively. After 2 h, the cells were released from cell cycle arrest by being filter washed three times and subsequently resuspended with YPD containing 0.1 mg of pronase E/ml (α-factor arrest) or YPD without pronase E (nocodazole arrest).
For flow cytometric analysis (FACSCalibur; Becton Dickinson), cells (1 ml) were removed every 15 min after release from cell cycle arrest and fixed overnight in 70% ethanol at 4°C. RNA was degraded by overnight incubation at 37°C in 50 mM sodium citrate, pH 7.1, containing 0.25 mg of RNase A/ml. The cells were then washed, and the DNA was stained by overnight incubation at 4°C in 50 mM sodium citrate, pH 7.1, containing 1 μM Sytox Green (Molecular Probes).
Total RNA isolation, which also recovers polyP, was performed as described by Cross and Tinkelenberg (7), except that the cells were resuspended in 350 μl of 1× LETS buffer (0.1 M LiCl, 10 mM EDTA, 10 mM Tris [pH 8.0], 0.5% [wt/vol] sodium dodecyl sulfate [SDS]) and 350 μl of acid phenol-chloroform (1:1) and lysed with glass beads by being vortexed at 4°C for 15 min. Following centrifugation of the mixture at 14,000 × g for 15 min, RNA and polyP were recovered from the aqueous phase by ethanol precipitation. The pellet consisting of RNA and polyP was resuspended in 0.1% SDS, quantified by absorbance at 260 nm, and stored at −80°C.
Northern hybridization and polyP analysis.
For analysis of PHO5 transcript levels, 10 μg of RNA was electrophoresed at 100 V for 3 h in 1% agarose gels buffered with 1× MOPS (3-N-morpholinopropanesulfonic acid) (20 mM MOPS [pH 7], 5 mM sodium acetate, 0.5 mM EDTA). RNA was blotted by 10× SSC (1.5 M NaCl, 0.15 M sodium citrate) to a positively charged nylon membrane. Membrane prehybridization and hybridization were performed in Church-Gilbert buffer (0.25 M Na2HPO4 [pH 7.4], 7% [wt/vol] SDS, 10 mg of fraction V bovine serum albumin/ml, 1 mM EDTA, and 0.5 mM sodium pyrophosphate). Hybridization probes, generated by PCR amplification using the oligonucleotide primers DNO425 (5′-TCTTTCCCTGGCGA-3′) and DNO426 (5′-GTCATCCAAGTAGGTTGTGT-3′) or DNO429 (5′-GCCAAGAAAGAGAGCTGC-3′) and DNO430 (5′-GAACTTAGAACCTGGTCTGTCC-3′) for PHO5 and TCM1, respectively, were radiolabeled with [α-32P]dCTP by random priming. mRNA levels were quantified by Storm 860 PhosphorImager analysis (Molecular Dynamics).
Gel analysis of polyP was performed by electrophoresis of 10 μg of RNA (containing copurifying polyP [see Fig. 5B] or treated with RNase A [see Fig. 7]) on native 6% polyacrylamide gels, followed by staining with toluidine blue O as described by Ogawa et al. (33). To quantify total levels of polyP, 10 μg of isolated RNA-polyP was treated with RNase A (after the removal of SDS by chloroform extraction, if necessary), and the polyP was ethanol precipitated and resuspended in 50 μl of distilled H2O. Ten microliters of the purified polyP was mixed with 1 ml of toluidine blue solution (6 mg of toluidine blue O dye/liter, 40 mM acetic acid), and the metachromatic shift in the ratio of absorbance at 530 nm to that at 630 nm was measured (5). The A530/A630 ratio of toluidine blue solution alone is a constant of 0.175, which was used as a blank. All samples were diluted to assure assay linearity, and polyP levels are expressed as [(A530/A630) − 0.175]/μg of RNA. As expected, samples from a phm3Δ strain yield a value of zero.
FIG. 7.
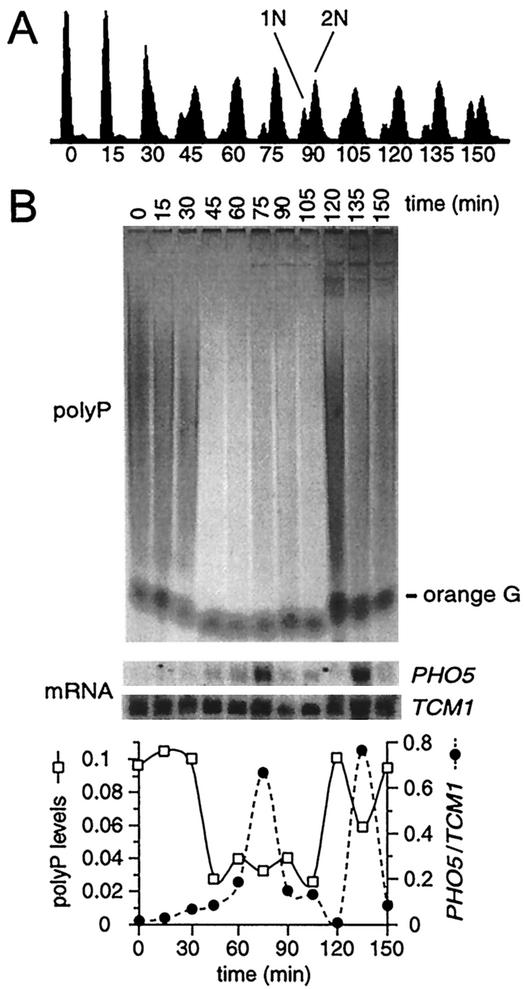
PolyP levels fluctuate during the cell cycle. (A) Flow cytometric analysis of α-factor-synchronized cultures. (B) Analysis of polyP and mRNA levels. Total RNA and polyP were isolated at 15-min intervals following release from α-factor arrest. PolyP was analyzed as for Fig. 5B, except that the samples were treated with RNase A prior to electrophoresis (polyP gel) and metachromatic shift quantification of polyP levels of an internal fraction (graph at bottom). Northern analysis of PHO5 and TCM1 mRNA levels of internal aliquots of RNA-polyP not treated with RNase A (middle) are plotted versus time and compared to polyP levels at the bottom.
Acid phosphatase activity assays.
After the specified growth conditions were achieved, the cells were chilled to 4°C, washed twice, and resuspended in 0.1 M sodium acetate, pH 3.6, at 4°C. After preincubation (10 min) of 500 μl of cell suspension at 30°C, the rAPase activity was assayed by addition of 500 μl of 20 mM p-nitrophenylphosphate (Roche) and incubation at 30°C for 10 min. Enzymatic activity was terminated by the addition of 250 μl of 1 M Na2CO3 and quantified by measuring the absorbance at 420 nm. Activities are reported in Miller units {(A420 × 1,000)/(OD600 × volume [in milliliters] of cells assayed × 10 min)}.
RESULTS
Mitotic induction of PHO5 requires the PHO activators Pho2, Pho4, and Pho81.
Earlier genome-wide determinations of cell cycle-regulated transcripts suggested that PHO5 and other PHO genes are induced during mitosis (42). However, significant cross hybridization of mRNA species can occur above 75% DNA sequence identity (17). Since PHO5 and PHO3 constitute a duplicated gene pair (87% identity over 1,404 bp), we reanalyzed the cell cycle expression profile of PHO5 in a strain with the entire PHO3 coding sequence deleted. Cultures in rich (YPD) medium were arrested at late G1 with the mating pheromone α-factor and released synchronously by being washed with YPD. Northern analysis of RNA samples isolated at 15-min intervals demonstrates that PHO5 mRNA levels oscillate during the cell cycle, with maximal expression occurring at 45 to 60 and 120 min after α-factor removal (Fig. 1A and B). Transcript from the ribosomal protein L3 locus, TCM1, remains constant throughout the cell cycle (4, 42) and serves as a loading control. Flow cytometric analysis confirmed that the cells synchronously traversed two cell cycles and that the points of maximal PHO5 transcript accumulation coincide with M phase (Fig. 1C).
To examine whether the canonical PHO signal transduction pathway is involved in PHO5 mitotic activation in YPD, we deleted three different positive regulators of the low-Pi induction pathway and assayed PHO5 expression during synchronous growth in YPD. Deletions of PHO4 and PHO2, encoding DNA site-specific transactivators of PHO5, abrogate mitotic induction of PHO5 (Fig. 1A and B). A null allele of the CDK inhibitor PHO81 eliminates ∼95% of the mitotic expression. Since PHO5 activation is completely dependent on Pho81 for inhibition of the Pho80-Pho85 cyclin-CDK (39), the residual level of expression in the pho81Δ strain is unexpected and suggests that a weaker, Pho81-independent activation mechanism also functions at PHO5 during mitosis. We also determined the effects of the pho2, pho4, and pho81 null mutations on rAPase activity in asynchronous cultures, which can often be used to observe cell cycle-dependent events (20). Approximately 60 to 110 U of rAPase activity are detected in asynchronous YPD cultures of wild-type strains (Fig. 1D and 2D; also see Fig. 4A and 5). Enzyme activity is reduced by >99% in pho4Δ and pho2Δ strains. Despite the residual level of mitotic PHO5 mRNA, rAPase activity is also eliminated in the pho81Δ strain. We conclude that most of the measurable rAPase activity in asynchronous YPD cultures of strain DNY742 (pho3Δ) can be attributed to expression of PHO5, and presumably that of the minor rAPases PHO11 and PHO12, in M phase (4, 42). Further, mitotic induction of PHO5 is strongly dependent on the positive PHO effectors PHO4, PHO2, and PHO81.
Mitotic activation of PHO5 requires the chromatin remodelers Gcn5 and SNF/SWI.
Next, we investigated the requirements, in mitotic activation of PHO5, for chromatin-remodeling enzymes, including Gcn5, the catalytic subunit of several histone H3 acetyltransferase complexes (3, 12), and the Snf2/Swi2 ATPase subunit of the SNF/SWI chromatin-remodeling complex (27). Since gcn5Δ cells could not be arrested with α-factor, presumably because Gcn5 is required for expression of the CDK inhibitor FAR1 (18), we synchronized cells in G2/M with nocodazole, an inhibitor of microtubule polymerization. After release from G2/M, wild-type cultures progressed through three cell cycles and showed maximal PHO5 induction at 15, 105, and 150 min (Fig. 2A and B), each peak corresponding to M phase (Fig. 2C). In contrast, no induction was observed in the snf2/swi2Δ and gcn5Δ strains, indicating that mitotic induction of PHO5 is highly dependent on these transcriptional coactivators. Greater than 90% of the rAPase activity of asynchronous cultures is eliminated in each null strain, demonstrating that synchrony is not required to observe the requirement for SNF/SWI and Gcn5 (Fig. 2D). We conclude that the previously reported Gcn5- (18) and Snf2/Swi2-dependent (43) transcription of PHO5 in asynchronous YPD cultures is explained by the pronounced need for these enzymes in PHO5 mitotic expression.
Previous studies using asynchronous cell populations demonstrated that SNF/SWI and Gcn5 are not required for activation of PHO5 following overnight starvation for Pi (10, 14). More recently, it has been shown that deletion of GCN5 decreases PHO5 activation at early times after Pi starvation, but not 8 h postactivation (1). Figure 2E demonstrates that asynchronous cultures of gcn5Δ and snf2/swi2Δ strains both exhibit a decreased initial rate of PHO5 transactivation following transfer to Pi-free medium. Thus, we have defined novel requirements for transcriptional coactivators at PHO5, including PHO5 among other genes for which activation in M phase is strongly Gcn5 and SNF/SWI dependent (24). Our data also confirm a role for Gcn5 in the rate of PHO5 activation and extend this observation to the SNF/SWI complex.
Mitotic activation of PHO5 occurs under conditions of limiting Pi.
Activation of PHO5 by Pho4 in asynchronous cultures is dependent on Pi deprivation and, ordinarily, is rapidly repressed by the addition of exogenous Pi (38). Since increased PHO5 transcription during mitosis is Pho4 dependent (Fig. 1), we tested if this cell cycle-specific activation in YPD is responsive to Pi levels. Surprisingly, despite 12 h of growth in YPD + Pi prior to α-factor arrest and release, mitotic activation of PHO5 is still apparent (Fig. 3). However, when the incubation in YPD + Pi is extended to 55 h before α-factor synchronization, PHO5 transcript is not detectable. This result indicates that exogenous Pi attenuates mitotic induction of PHO5 in a time-dependent manner.
Next, we further investigated the time dependence of repression of mitotic PHO5 induction. Relative to cells growing continuously on YPD, rAPase activity decreased by 50% when the cultures were shifted from YPD to YPD + Pi (Fig. 4A, compare bars 1 and 2). Full repression of PHO5 required longer incubation in YPD + Pi (compare bars 1 and 3). In the opposite experiment, pregrowth on YPD + Pi and shift to YPD, full derepression is restored (compare bars 1 and 4). This result suggests that repression of mitotic expression takes longer than its induction. The time required to achieve full repression of PHO5 was assessed by pregrowth of cells in YPD followed by transfer to YPD + Pi. As in Fig. 4A, rAPase activity was reduced to ∼60% of that of the control YPD culture at 6 h (Fig. 4B). Maximal PHO5 repression was achieved after ∼29 h and remained at this level for the rest of the time course. The time required to reach full repression is not likely due to rAPase stability, since the half-life of rAPase activity is 2 to 2.5 h, as determined by the phosphate overplus procedure (Pi starvation followed by Pi addition; see Materials and Methods and data not shown). The remaining activity may be due to basal levels of PHO5 expression and that of the more minor rAPases, PHO11 and PHO12, which vary somewhat between experiments. These findings demonstrate that YPD can be limiting for Pi. The lag in full repression may be due to active growth in YPD that leads to rapid metabolism of a PHO-repressive signal.
PolyP reserves increase, but are not essential for, repression of PHO5.
PolyP is the most prevalent form of phosphate in yeast, and it is believed to serve as a phosphate reservoir (11, 41, 44). Such a phosphate reservoir might sustain intracellular Pi levels more effectively during Pi deprivation and hence repress M phase expression of PHO5. To test this hypothesis, we asked if strains singly null for four PHM genes had increased levels of total rAPase activity. Strains with PHM3 and PHM4 deleted are severely deficient in the Phm/Vtc complex and thus lack detectable polyP, but they do not exhibit growth defects. The phenotypic defects of phm1Δ and phm2Δ strains are much less pronounced (31, 33). In the experiment shown in Fig. 5A, each strain was grown in YPD and then diluted in YPD or YPD + Pi for 6 h. Relative to wild-type cells, rAPase activity during continuous growth in YPD increases 2- to 2.5-fold in phm3Δ or phm4Δ strains and only modestly in phm1Δ or phm2Δ cells (Fig. 5A). Thus, the magnitude of PHO5 derepression under Pi-limiting conditions (YPD) correlates well with the severity of the polyP accumulation defect and extent of disruption of Phm/Vtc complex integrity: phm1Δ ≈ phm2Δ < phm3Δ ≈ phm4Δ (31, 33). Interestingly, incubation in YPD + Pi for 6 h reverses the PHO5 derepression, and lower levels of expression are established in the absence of Phm3 and Phm4.
To explore the relationship between PHO5 mitotic activation and polyP levels further, we tested if high-Pi conditions increase cellular polyP and, if so, its effects on total rAPase activity. Consistent with our finding that YPD is limiting for Pi (Fig. 3 to 5A), a wild-type (PHM3) strain grown in YPD has low but significant levels of polyP (Fig. 5B, lane 1). The isogenic phm3Δ strain has no detectable polyP (33) (Fig. 5B, compare lanes 1 and 3). In this experiment, rAPase activity is increased fourfold in the phm3Δ strain relative to the wild-type strain, again demonstrating that loss of polyP leads to increased PHO5 activation in YPD. When wild-type (PHM3) cells are grown in high-Pi medium (YPD + Pi), polyP amounts increase 5-fold (Fig. 5B, compare lanes 1 and 4), which is accompanied by a 10-fold reduction in rAPase activity and undetectable PHO5 transcript levels in M phase (Fig. 3). During growth in complete synthetic medium (7.3 mM Pi), wild-type strains accumulate levels of polyP [0.07 (A530/A630)/μg of RNA] and rAPase (13 U) activities similar to those in YPD + Pi (13.4 mM). As expected, the phm3Δ strain had no detectable polyP in YPD + Pi (33) but, as in Fig. 5A, had repressed levels of rAPase activity (Fig. 5B, lane 2). This indicates that polyP is not required to repress PHO5 when Pi is plentiful. We conclude that, under conditions of limiting Pi, accumulated polyP acts as a cellular phosphate reserve that can contribute to the repression of PHO5 expression. However, as PHO5 is completely repressed by excess Pi in strains that are unable to accumulate polyP, the polymer is not necessary for PHO5 promoter inactivation.
When S. cerevisiae encounters a period of Pi starvation followed by a high-Pi environment, a phenomenon called “overplus” or “overcompensation” occurs in which large amounts of polyP accumulate (23). This method was previously used to evaluate the role of PHM genes in polyP synthesis (33). In parallel with the samples in Fig. 5B, lanes 1 to 4, wild-type cells subjected to the overplus procedure (see Materials and Methods) accumulate 60-fold more polyP than during continuous growth in YPD (Fig. 5B, compare lanes 5 and 1). Interestingly, overplus polyP levels surpass those of cultures grown continuously in YPD + Pi (compare lanes 4 and 5). The highest level of polyP is observed upon growth in YPD-low Pi before the addition of Pi (compare lanes 6 and lanes 1 to 5). Therefore, the amount of polyP that accumulates during the overplus procedure is proportional to the severity of prior Pi starvation, consistent with increased transcription of PHM and/or other genes that may play direct or indirect roles in polyP biosynthesis (19, 33). In contrast, as relatively high levels of polyP are synthesized in wild-type cells when excess Pi is present, polyP synthesis does not require a preceding period of Pi starvation.
Accumulation of polyP under limiting Pi conditions requires SNF/SWI and Gcn5.
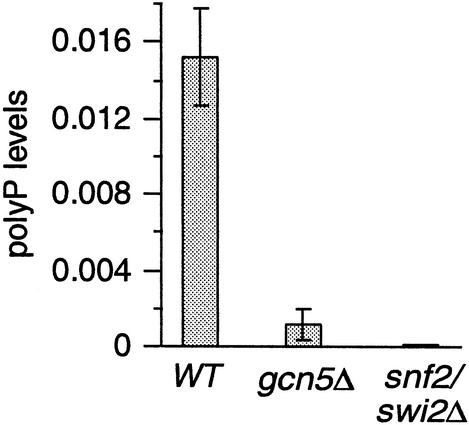
The levels of polyP and PHO5 expression are inversely correlated (Fig. 5B, lanes 1 and 4 or 3 and 4). Thus, the loss of mitotic PHO5 activation in snf/swi and gcn5 mutants (Fig. 2) might be due to a pleiotropic hyperaccumulation of polyP. We determined, however, that polyP levels were decreased >10-fold and near the detection limit in gcn5Δ and snf2/swi2Δ strains, respectively, during continuous growth in YPD (Fig. 6). This result suggests that SNF/SWI and Gcn5 are needed for M phase activation of PHM genes, as well as PHO5, when Pi is limited.
FIG. 6.
SNF/SWI and Gcn5 are required for polyP accumulation. Steady-state levels of polyP from wild-type (WT), gcn5Δ, and snf2/swi2Δ strains were measured (n = 3; mean ± 1 standard deviation) by the metachromatic shift method after asynchronous growth in YPD (the same conditions used for rAPase activity measurements in Fig. 2D).
PolyP levels fluctuate during the cell cycle.
Because PHO5 expression is inversely related to polyP reserves (Fig. 5), we investigated if mitotic activation of PHO5 is correlated with a decrease in polyP levels before M phase in α-factor-synchronized cultures (Fig. 7). Maximal levels of polyP are observed at G1 (0 to 15 min) and reach a minimum when cells are well into S phase, 45 to 60 min after α-factor removal. The 6- to 10-fold increase in polyP levels compared to Fig. 5B and 6 is reproducibly observed following synchronization. Nevertheless, a four- to fivefold drop in polyP levels precedes peak accumulation of PHO5 mRNA in mitosis, which occurs at 75 min. PolyP amounts remain at the minimum through 105 min and then increase fivefold at 120 min. Presumably, this is because enhanced mitotic expression of PHO5 (Fig. 7B), other rAPase genes, and PHO84, the high-affinity Pi transporter, lead to increased scavenging of Pi and hence polyP synthesis. These data indicate that, in Pi-limiting medium (e.g., YPD), polyP levels fluctuate with the cell cycle. Importantly, this result also suggests that the physiological basis for mitotic cycling of PHO5 is due to significant, but not complete, depletion of polyP reserves prior to M phase.
Loss of polyP increases the rate, magnitude, and duration of PHO5 activation.
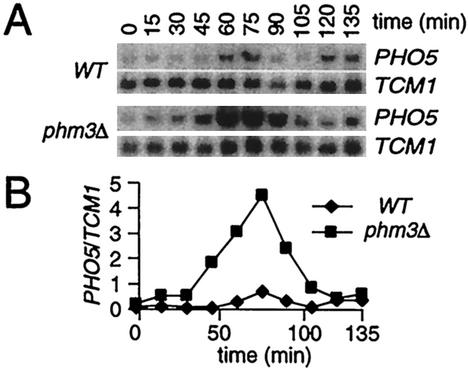
We also assessed the effects of the absence of polyP deficiency on PHO5 activation during synchronous cell cycle progression. Following α-factor synchronization and release in YPD medium, induction of PHO5 in the phm3Δ strain, which lacks detectable polyP (33), increases dramatically in comparison to that in wild-type cells (Fig. 8). Furthermore, PHO5 mRNA was evident earlier in the phm3Δ strain, indicating that the null cells lose the repressive signal inactivating PHO5 transcription before their wild-type counterparts. Flow cytometric analysis demonstrated that the length and timing of each cell cycle phase in the phm3 strain was indistinguishable from those in the wild type (data not shown), consistent with its wild-type growth phenotype (6). Therefore, the absence of Phm3 causes increased levels of PHO5 transcription that are not due to a higher proportion of cells in M phase.
FIG. 8.
Mitotic activation of PHO5 is increased in phm3Δ strains. (A) Northern analysis of α-factor-synchronized cultures of wild-type (WT) and phm3Δ cells. (B) Normalized PHO5 transcript levels from panel A.
Strains lacking the Phm/Vtc complex are defective in sustaining transport of Pi that is added during the overplus procedure (33), making it possible that PHO5 would be derepressed (26). However, a low level of PHO5 transcript that precedes and follows the mitotic peak (Fig. 8) strongly suggests that PHO5 transcription is not constitutively derepressed in phm strains. Alternatively, it was hypothesized (33) that compromised Pi transport could indirectly result from the inability to synthesize polyP; the failure to divert added Pi to vacuoles leads to increased cytosolic Pi concentrations that signal degradation of Pho84, the high-affinity Pi transporter (48). Nevertheless, employing the overplus treatment with asynchronous wild-type and phm3Δ cultures, an identical precipitous drop in PHO5 transcript levels occurs from 20 to 30 min after Pi addition (data not shown). We conclude that the initial 5-min period of Pi transport observed in phm3Δ cells during the phosphate overplus (33) is sufficient to signal PHO5 repression.
The rapid onset of accumulation of PHO5 transcript in the phm3Δ strain (Fig. 8), even prior to G2/M, where all cells have achieved a 2N DNA content, suggests an increased rate of PHO5 activation in the absence of polyP reserves. To evaluate this possibility, we tested how polyP stores influence the initial rate of PHO5 mRNA accumulation in asynchronous cultures in response to Pi starvation (Fig. 9). Wild-type and phm3Δ strains were first grown in defined Pi-free medium with 13.4 mM Pi added back so that substantial cellular polyP would accumulate. Subsequently, Pi was removed by washing, both cultures were shifted to Pi-free medium, and the levels of PHO5 mRNA, as well as rAPase activity, were monitored over time. PHO5 transcript accumulates linearly in both the wild-type (R2 = 0.96) and phm3Δ (R2 = 0.99) strains for 150 and 60 min, respectively. Strikingly, the initial, rapid response to Pi starvation occurs 60 min earlier and at a threefold-higher rate in the phm3Δ strain. Moreover, PHO5 transcript levels in the phm3Δ strain are significantly higher (120-fold more at 60 min) at every time point up to and including 4 h, achieve maximum 2 h earlier, and are maintained at this higher level longer than in wild-type cells. Subsequently, PHO5 mRNA levels plateau and then, interestingly, decline to similar levels in both cultures around 6 h after Pi starvation. As expected, the levels of internally assayed rAPase activity lag behind and qualitatively support these observations (Fig. 9A). The absence of a decrease in rAPase activity at the later time points likely reflects the increased stability of the protein relative to PHO5, PHO11, and PHO12 transcripts (46). We conclude that the initial rates of accumulation of PHO5 transcript and rAPase activity, and presumably the rates of transcriptional initiation, are enhanced in cells lacking polyP. Further, the repressive influence of polyP reserves on PHO5 transcription was observed in a defined medium, which complements our results obtained using YPD medium.
DISCUSSION
We have investigated the molecular mechanisms and physiological basis for activation of PHO promoters, which are in a prominent class of mitotically induced genes with nutritional roles (42). In contrast to previous findings using Pi-starved, asynchronous cultures, we find that PHO5 activation in M phase in synchronized cultures grown in rich medium is strongly dependent on the SNF/SWI complex, an ATP-dependent chromatin remodeler, and the histone acetyltransferase Gcn5 (Fig. 2). Additionally, our work shows that M phase induction of PHO5 requires the transactivators Pho2 and Pho4, as well as the CDK inhibitor Pho81, suggesting that PHO5 mitotic induction occurs through the PHO signal transduction pathway (Fig. 1). In accord with this, addition of exogenous Pi suppresses mitotic activation of PHO5 (Fig. 3) and increases polyP levels, which are inversely correlated with PHO5 expression in wild-type cells (Fig. 5B). Further, in synchronized cultures, polyP amounts decrease between G1 and M phases, demonstrating that metabolic pools of polyP fluctuate during the cell cycle (Fig. 7). Beyond these correlations, we show that strains with undetectable levels of polyP (phm3Δ) exhibit a dramatically enhanced PHO response in both synchronous and asynchronous cultures (Fig. 8 and 9). These results establish that polyP causally represses PHO5 expression. Moreover, the physiological stimulus for activation of PHO5 in M phase is Pi depletion that ensues after reduction of vacuolar polyP reserves. Thus, our results expand the role of the PHO signaling system to include coordination of the mitotic activation with Pi availability during cell cycle progression. Our data also highlight a novel function for polyP as a negative regulator of PHO activation in mitosis.
Our findings support the model that a central, physiological role of polyP is that of a phosphate reservoir (11, 41). Under conditions of limiting Pi, inasmuch as degradation of polyP increases intracellular Pi pools, the polymer antagonizes PHO5 activation. It is likely that our findings apply generally to other PHO (and PHM) genes, since the rate of PHO84 transcript accumulation is also enhanced upon Pi starvation in a phm3Δ strain (D. W. Neef and M. P. Kladde, unpublished data). It must be emphasized that Phm/Vtc complex activity, and hence polyP, is not required to maintain (Fig. 9, zero point) or establish (Fig. 5) repression of PHO5 when Pi is abundant. Further, a lack of detectable polyP does not impair the ability to repress PHO5 transcription before and after the mitotic peak in synchronized populations (Fig. 8). Thus, polyP is a modulator rather than an essential cell cycle regulator of PHO expression, replenishing intracellular levels of Pi when metabolic demand for it is high.
The inability to make polyP in the absence of the Phm/Vtc complex allowed us to address the interplay between polyP and PHO5 regulation during M phase. Extensive Pi starvation leads to G1 arrest (30), primarily due to controls that operate at Start (36). Although the mechanisms are unclear, it is widely accepted that these controls involve attaining sufficient cell size and nutrient acquisition for cell division. In view of this, while cells have already gauged sufficient phosphate resources to execute Start, why do they rapidly mobilize their polyP reserves with synchronous progression through S phase (Fig. 7)? It is possible that polyP reserves are directly sensed as a Pi source prior to Start. However, if this were the case, constitutive activation of PHO5 outside of M phase would be expected in the absence of polyP (phm3Δ cells), which is not observed (Fig. 8). Instead, since phm3Δ cells lack reserve Pi afforded by polyP, they exaggerate mitotic induction of PHO genes (Fig. 8). Thus, while the cell division commitment decision is made at G1, our data suggest that the sensing of and response to nutrient imbalances is not restricted to G1 (45). This view is supported further by the fact that PHO5 is strongly activated by Pi starvation during sustained mitotic arrest (Neef and Kladde, unpublished). Since the cells could not anticipate the post-Start starvation, we conclude that they are able to sense Pi at cell cycle stages other than G1.
We propose the following model for PHO5 mitotic activation under conditions of limiting Pi (e.g., YPD). A yeast cell maintains cytosolic Pi at levels sufficient to satisfy metabolic needs and stores any excess as polyP. Upon reaching critical size and nutrient status, the cell will initiate Start and probably decrease cytosolic Pi rapidly while synthesizing nucleotides for DNA replication and transcription, as well as phospholipids for the enlarging bud. PolyP reduction also correlates with marked increases in sugar phosphates during synchronous entry into S phase (11, 32). As the intracellular Pi concentration declines and is unable to be replenished by preferred extracellular Pi (11), it may first be more expedient to degrade polyP to Pi, presumably via endopolyPases (Ppn1) and exopolyPases (25, 40). Subsequently, when polyP reserves and hence cytosolic Pi fall critically low, transcription of the PHO and PHM genes is activated. During M phase, and possibly into G1, rAPase synthesis leads to hydrolysis of phosphoesters in the medium, which supplies Pi for import via the high-affinity transporter Pho84, which is also expressed in mitosis (42). Increased cytosolic Pi (or some anabolite of Pi) leads to attenuation of the PHO mitotic induction. After metabolic growth requirements are met, excess Pi is assimilated into polyP, thus reconciling the seeming paradox that cells synthesize polyP in response to Pi deprivation.
The primary reason for PHO mitotic expression may be to sequester as much Pi as is feasible as vacuolar polyP. This view is supported by the fact that pho4Δ cells exhibit a wild-type growth rate in YPD, despite the absence of detectable polyP (33) (Fig. 5B) and inability to activate PHO genes in mitosis (Fig. 1). Hence, the amount of Pi (or phosphate esters) and basal levels of PHO gene expression are adequate for pho4Δ cells to meet their Pi growth requirements. Therefore, mitotic PHO activation may simply confer an adaptive advantage through hoarding Pi as polyP or significant conservation of transcriptional resources, antagonizing excessive activation of PHO5 (Fig. 8 and 9) and ∼20 additional PHO, PHM/VTC, and other genes (19, 33).
We find that activation of PHO5 during mitosis in YPD and the initial period of Pi starvation are strongly dependent on SNF/SWI and Gcn5 (Fig. 2). In contrast, activation of PHO5 in asynchronous cultures extensively starved for Pi is refractory to the loss of both chromatin-remodeling activities (10, 13). This apparent discrepancy is potentially explained by a global requirement for Gcn5 and SNF/SWI in mitotic gene expression (24). The relatively short period (∼15 to 30 min) of mitotic activation may also contribute to the pronounced Gcn5 and SNF/SWI dependence, since these coactivators are needed for wild-type rates of PHO5 activation (1) (Fig. 2E). Because snf/swi and gcn5 mutants have very low levels of polyP (Fig. 6), it is unlikely that they exhibit lower rates of PHO5 activation due to impaired growth and hence slower depletion of cellular Pi. Moreover, the rates of transactivation of double snf/swi phm3 and gcn5 phm3 mutants with undetectable polyP are also lower than those of phm3 cells (data not shown).
PHO5 transcription is also Gcn5 and SNF/SWI dependent in asynchronous cultures grown in YPD (18, 43) but does not require SNF/SWI in synthetic minimal medium (43). We propose that this medium-specific difference for SNF/SWI can now be explained in terms of polyP. Mitotic expression in YPD is significant due to its low levels of Pi and hence vacuolar polyP. In contrast, the high Pi concentration (7.3 mM) of minimal medium leads to substantial increases in polyP that repress M phase expression (Fig. 5).
The antagonism between polyP/Pi levels and the magnitude of PHO activation suggests that nutrient sensing may underlie the transcriptional periodicity of other cell cycle-regulated genes. In particular, transcripts for genes involved in the transport of a variety of nutrients in addition to Pi, including carbohydrates, heavy metals, ions, and amino acids, fluctuate during the cell cycle (42). Genes involved in glycogen (GSY1) and fatty acid (FAA1 and FAS1) synthesis are also cell cycle regulated in rich medium. Depending on the particular sensing system, the presence or absence of nutrients may lead to periodic increases or decreases in cell cycle transcription. Alternatively, the low levels of Pi in YPD may contribute indirectly to the periodic expression of some genes. For example, mitotic induction of the plasma membrane ATPase, PMA1, may generate the proton gradient that is requisite for Pho84-mediated symport of Pi (and many other nutrients). Finally, since polyP chelates cations (8), it is plausible that their uptake is coordinated with polyP synthesis. It remains to be seen if the loss of polyP affects the transcription of any of these additional cell cycle-regulated genes.
Acknowledgments
We thank Christopher Carvin for strains and Mary Bryk, Michael Polymenis, and members of our laboratory for critical reading of the manuscript.
This work was supported in part by a grant from the Texas Higher Education Coordinating Board.
REFERENCES
- 1.Barbaric, S., J. Walker, A. Schmid, J. Q. Svejstrup, and W. Hörz. 2001. Increasing the rate of chromatin remodeling and gene activation—a novel role for the histone acetyltransferase Gcn5. EMBO J. 20:4944-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 3.Brownell, J. E., J. X. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 4.Cho, R. J., M. J. Campbell, E. A. Winzeler, L. Steinmetz, A. Conway, L. Wodicka, T. G. Wolfsberg, A. E. Gabrielian, D. Landsman, D. J. Lockhart, and R. W. Davis. 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2:65-73. [DOI] [PubMed] [Google Scholar]
- 5.Clark, J. E., H. Beegen, and H. G. Wood. 1986. Isolation of intact chains of polyphosphate from “Propionibacterium shermanii” grown on glucose or lactate. J. Bacteriol. 168:1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, A., N. Perzov, H. Nelson, and N. Nelson. 1999. A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 274:26885-26893. [DOI] [PubMed] [Google Scholar]
- 7.Cross, F. R., and A. H. Tinkelenberg. 1991. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65:875-883. [DOI] [PubMed] [Google Scholar]
- 8.Dunn, T., K. Gable, and T. Beeler. 1994. Regulation of cellular Ca2+ by yeast vacuoles. J. Biol. Chem. 269:7273-7278. [PubMed] [Google Scholar]
- 9.Elledge, S. J., and R. W. Davis. 1989. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 9:4932-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudreau, L., A. Schmid, D. Blaschke, M. Ptashne, and W. Hörz. 1997. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89:55-62. [DOI] [PubMed] [Google Scholar]
- 11.Gillies, R. J., K. Ugurbil, J. A. den Hollander, and R. G. Shulman. 1981. 31P NMR studies of intracellular pH and phosphate metabolism during cell division cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2125-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, P. D., A. Schmid, M. Zavari, L. Lui, S. L. Berger, and W. Hörz. 1998. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol. Cell 1:495-505. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Hörz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, M., U. J. Kim, P. Kayne, and M. Grunstein. 1988. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 7:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hereford, L. M., M. A. Osley, T. R. Ludwig, and C. S. McLaughlin. 1981. Cell-cycle regulation of yeast histone mRNA. Cell 24:367-375. [DOI] [PubMed] [Google Scholar]
- 17.Hess, K. R., W. Zhang, K. A. Baggerly, D. N. Stivers, and K. R. Coombes. 2001. Microarrays: handling the deluge of data and extracting reliable information. Trends Biotechnol. 19:463-468. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Huang, D., J. Moffat, and B. Andrews. 2002. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 22:5076-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 21.Kladde, M. P., M. Xu, and R. T. Simpson. 1996. Direct study of DNA-protein interactions in repressed and active chromatin in living cells. EMBO J. 15:6290-6300. [PMC free article] [PubMed] [Google Scholar]
- 22.Komeili, A., and E. K. O'Shea. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977-980. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 24.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 25.Kumble, K. D., and A. Kornberg. 1996. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 271:27146-27151. [DOI] [PubMed] [Google Scholar]
- 26.Lau, W. W., K. R. Schneider, and E. K. O'Shea. 1998. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics 150:1349-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurent, B. C., I. Treich, and M. Carlson. 1993. The yeast SNF2/SWI2-protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 7:583-591. [DOI] [PubMed] [Google Scholar]
- 28.Lenburg, M. E., and E. K. O'Shea. 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21:383-387. [PubMed] [Google Scholar]
- 29.Lew, D. J., T. Weinert, and J. R. Pringle. 1997. Cell cycle control in Saccharomyces cerevisiae, p. 607-695. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 30.Lillie, S. H., and J. R. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, O., M. J. Bayer, C. Peters, J. S. Andersen, M. Mann, and A. Mayer. 2002. The Vtc proteins in vacuole fusion: coupling NSF activity to V0 trans-complex formation. EMBO J. 21:259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicolay, K., W. A. Scheffers, P. M. Bruinenberg, and R. Kaptein. 1983. In vivo 31P NMR studies on the role of the vacuole in phosphate metabolism in yeasts. Arch. Microbiol. 134:270-275. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill, E. M., A. Kaffman, E. R. Jolly, and E. K. O'Shea. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209-212. [DOI] [PubMed] [Google Scholar]
- 35.Oshima, Y., N. Ogawa, and S. Harashima. 1996. Regulation of phosphatase synthesis in Saccharomyces cerevisiae—a review. Gene 179:171-177. [DOI] [PubMed] [Google Scholar]
- 36.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle, p. 97-142. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 37.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 38.Schmid, A., K. D. Fascher, and W. Hörz. 1992. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell 71:853-864. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, K. R., R. L. Smith, and E. K. O'Shea. 1994. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266:122-126. [DOI] [PubMed] [Google Scholar]
- 40.Sethuraman, A., N. N. Rao, and A. Kornberg. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirahama, K., Y. Yazaki, K. Sakano, Y. Wada, and Y. Ohsumi. 1996. Vacuolar function in the phosphate homeostasis of the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 37:1090-1093. [DOI] [PubMed] [Google Scholar]
- 42.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urech, K., M. Durr, T. Boller, A. Wiemken, and J. Schwencke. 1978. Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch. Microbiol. 116:275-278. [DOI] [PubMed] [Google Scholar]
- 45.Veinot-Drebot, L. M., G. C. Johnston, and R. A. Singer. 1991. A cyclin protein modulates mitosis in the budding yeast Saccharomyces cerevisiae. Curr. Genet. 19:15-19. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wurst, H., T. Shiba, and A. Kornberg. 1995. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177:898-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wykoff, D. D., and E. K. O'Shea. 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]