Abstract
Early B-cell factor (EBF) is a DNA binding protein required for early B-cell development. It activates transcription of several B-cell-specific genes, including the λ5 gene, which encodes a protein necessary for signaling by the pre-B-cell receptor. In an effort to understand the mechanism by which EBF activates transcription, we examined its interaction with the coactivator protein p300/CBP. We found that two domains of EBF each bind the histone acetyltransferase (HAT)/CH3 domain of p300/CBP both in vitro and in vivo. Surprisingly, transcriptional activation by EBF was not sensitive to E1A, a potent p300/CBP inhibitor. In fact, overexpressed EBF mimicked E1A by severely repressing the activity of several other transcription factors, including E47, a protein that acts cooperatively with EBF to promote transcription of the λ5 gene. This broad inhibitory profile correlated with EBF's ability to repress the HAT activity of p300/CBP in vivo and in vitro. However, such a repressed complex is not likely to form at the λ5 promoter in vivo since (i) EBF could not bind p300/CBP and DNA simultaneously and (ii) the cooperativity imparted by E47 was sensitive to E1A. Our data reveal an intriguing inhibitory property of EBF—a property shared only by E1A, Twist, Pu.1, and the Hox family of homeodomain proteins—and suggest that E47 and EBF play distinct roles during λ5 promoter activation.
Early B-cell factor (EBF) belongs to an evolutionarily conserved family of DNA binding proteins (9). EBF was first identified in B lymphocytes as a transcriptional regulator of the mb-1 gene (11, 17). In separate studies it was identified as Olf-1, a regulator of genes in olfactory neurons (23, 51). Thus far, three EBF homologues have been identified, and they share a high degree of sequence identity in the DNA binding and dimerization domains (52). EBF binds DNA as a homodimer or a heterodimer to variants of the palindromic sequence ATTCCCNNGGGAAT (48). A dimerization domain facilitates DNA binding, which is mediated by a unique zinc coordination motif (16). One transcriptional activation domain has been mapped to the C-terminal region of the protein, while a second context-dependent activation domain appears to reside within the DNA binding domain (16). In hematopoietic cells expression of EBF is limited to B lymphocytes. Expression is high throughout the various stages of B-cell ontogeny with the exception of terminally differentiated plasma cells, where EBF expression is extinguished (15, 17). The most profound phenotype exhibited by mice homozygous for a targeted deletion of the EBF gene is the complete lack of B cells, indicating a critical role for EBF in B-cell specification (25). EBF target genes include mb-1 (17), λ5 and V-preB (46), and B29 (2), all of which are critically important for early B-cell development (6, 37, 41).
Although EBF has been characterized as a transcriptional activator, the mechanisms by which it activates transcription are not fully understood. Studies of the λ5 and V-preB promoters have shown that EBF collaborates with E47 (14, 45, 46), another transcription factor necessary for early B-cell development (4, 55). Genetic studies also point to a collaborative relationship between EBF and E47. Mice carrying single copies of both the EBF and E47 genes (compound heterozygotes) show a more profound B-cell phenotype than do mice carrying single copies of either gene alone (36). While E47 is known to recruit p300/CBP to DNA (10), the role played by EBF in promoter activation has not been determined.
Transcriptional coactivators can alter chromatin in two ways: by covalently modifying histones and by remodeling chromatin (12, 31, 49). Histone-modifying enzymes include histone acetyltransferases (HATs), kinases, and methylases (5, 20). Such modifications can exert either positive or negative effects on transcription. Histone acetylation is normally associated with transcriptional activation because acetylated histones cannot readily pack DNA into higher-ordered chromatin and because acetylated histones may directly recruit transcriptional activators (20). Several HATs have been identified, including p300, CBP (34), and PCAF (53), and typically exist in multiprotein complexes. Histone deacetylases (HDACs) are enzymes that deacetylate histones and therefore antagonize HATs (21). A dynamic and regulated balance between HAT and HDAC recruitment can be critical to the activity of a given gene. The chromatin remodelers comprise several multiprotein complexes, exemplified by the SWI/SNF complex. In vitro assays for chromatin remodeling typically measure the repositioning of nucleosomes on DNA and require the hydrolysis of ATP. The manner in which chromatin remodelers work in vivo is less clear, but it is widely assumed that they generate access to both transcriptional regulatory proteins, including in some cases histone-modifying enzymes, and components of the basal transcription apparatus (1, 8, 22, 47). Unlike HATs and HDACs, regulatory pathways that specifically target chromatin remodelers have not been described.
Although HDACs likely represent the most common means of antagonizing HAT-mediated transcription, proteins that inhibit HATs directly have also been described. E1A (12S), for example, binds to and inhibits the HAT activity of both p300 and PCAF (7, 18, 39). Interestingly, similar inhibition has also been described for the DNA binding proteins Twist (18) and Pu.1 (19) and for the Hox family of homeodomain proteins (44). A multiprotein complex (INHAT) has been isolated from HeLa cells that also inhibits p300 HAT activity but does so by binding histones and blocking access of the enzyme to nucleosomal templates (42). Forced expression of Pu.1 in MEL cells blocks both dimethyl sulfoxide-induced differentiation and the corresponding increase in CBP acetyltransferase activity, supporting a role for Pu.1 in inhibiting CBP activity during normal erythropoiesis (19). Although provocative, the in vivo significance of the other inhibitory activities has not been addressed.
In the present study, we sought to examine the manner in which EBF stimulates transcription. Although EBF binds p300/CBP, we found that it represses HAT activity when not bound to DNA. Accordingly, EBF is not likely to activate transcription by recruiting p300/CBP per se, although p300/CBP recruitment may contribute to the cooperativity imparted by other DNA binding proteins at EBF target genes. Our data reveal an intriguing property of EBF and suggest that it plays a role functionally distinct from that of E47 at the λ5 promoter.
MATERIALS AND METHODS
Plasmids.
The in vivo EBF expression plasmid pCMV-EBF has been described previously (15). For in vitro EBF expression, the EBF cDNA was subcloned into pcDNA3 (Invitrogen). Constructs for the expression of six-Myc-tagged EBF and six-Myc-tagged EBF fragments have been described previously (45). pCMX-Gal4-CBP was a gift from Debu Chakravarti (University of Pennsylvania). pCMV-E47 and pCMV-Gal4-E47(Act) have been described previously (35, 43). The various CBP fragments used for in vitro transcription and translation were cloned into pBSKII and were provided by Gerd Blobel (Children's Hospital of Philadelphia). pGEX-EBF was made by isolating the EBF cDNA from pCMV-EBF by XhoI digestion and subcloning it into the XhoI site of pCMV-Tag3B (Clontech), followed by EcoRI digestion and subcloning the EBF cDNA into the EcoRI site of pGEX4T-1 (Pharmacia). Detailed methods for the construction of glutathione S-transferase (GST) expression plasmids carrying the truncated GST-EBF proteins (amino acids [aa] 1 to 310, aa 108 to 591, and aa 311 to 591) are available upon request. Plasmids expressing the Gal4-CBPHAT and Gal4-CBPHATΔ fusion proteins have been described previously (29).
Reporter constructs Gal4-E1b-Luc and [μE5 + μE2]6-TATA-Luc were made by replacing the chloramphenicol acetyltransferase gene in Gal4-E1b-CAT and [μE5 + μE2]6-TATA-CAT (35) with the luciferase gene. λ5-Luc and Gal4-AdML-TATA-CAT have been described previously (29, 46). Luciferase reporters under the control of the Fos promoter have been described previously (46) with or without the mouse immunoglobulin heavy chain (IgH) enhancer inserted 3′ to the luciferase gene (at the BamHI site).
Protein-protein interaction assays. (i) In vitro.
GST fusion proteins were purified from Escherichia coli BL21 cells. In vitro transcription and translation of the various CBP and EBF fragments were carried out with the TNT coupled transcription-translation assay system (Promega) in the presence of [14C]leucine. The synthesized proteins were then incubated overnight in binding buffer (75 mM NaCl, 25 mM Tris-Cl [pH 7.8], 6% glycerol, 1% bovine serum albumin [BSA], 0.1% NP-40, 1 mM EDTA, 1 mM dithiothreitol, and protease inhibitor cocktail [Roche]) with appropriate GST fusion proteins bound to glutathione-Sepharose 4B beads. The beads were then washed five times with wash buffer (75 mM NaCl, 25 mM Tris, 0.15% Triton X-100). The bound proteins were resolved by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and visualized with a phosphorimager.
(ii) In vivo.
Expression vectors for six-Myc-tagged EBF or six-Myc-tagged EBF fragments were transfected into 293T cells, and after 48 h the cells were harvested and lysed in NP-40 buffer (1% NP-40, 75 mM NaCl, 25 mM Tris-Cl [pH 7.8], 6% glycerol, 1% BSA, 1 mM EDTA,1 mM dithiothreitol, and protease inhibitor cocktail) on ice for 1 h. After centrifugation, the lysates were precleared with protein A/G plus agarose beads (Santa Cruz) in NP-40 buffer and then incubated with myc9e10 antibody-conjugated agarose beads (Santa Cruz) at 4°C overnight. The beads were then washed five times with wash buffer (75 mM NaCl, 25 mM Tris, 0.15% Triton X-100). The precipitated proteins were resolved by SDS-PAGE, followed by Western blotting with anti-CBP antibody (Santa Cruz) as the primary antibody and horseradish peroxidase-conjugated anti-rabbit antibody (Amersham) as secondary antibody.
Transient-transfection and reporter assays.
NIH 3T3 and 293T cells were cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 2 mM glutamine, and 1× penicillin-streptomycin (Gibco). U2OS cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 1× penicillin-streptomycin, and 2 mM sodium pyruvate. NIH 3T3 and U2OS cells were transfected by using Fugene (Roche). Cells were harvested 48 h after transfection. S194 plasmacytoma and 230-238 pre-B cells were transfected as previously described (3, 46). Luciferase assays were carried out with the dual luciferase assay system (Promega). Chloramphenicol acetyltransferase assays were carried out as described previously (35).
RT-PCR assays.
Reverse transcription-PCR (RT-PCR) assays of transfected NIH 3T3 cells were carried out as previously described with minor modifications (54). Briefly, total RNA was prepared with the RNeasy purification kit (Qiagen). For reverse transcription, 5 μg of total RNA was used, and the reaction was carried out with Moloney murine leukemia virus reverse transcriptase (Promega). One-tenth of the reverse transcription reaction mixture was used in the subsequent PCR, in which AmpliTaq Gold polymerase was used (Perkin-Elmer). PCRs were carried out by first heating the samples at 95°C for 10 min and then subjecting them to 25 to 40 cycles of PCR: 30 s at 95°C, 1 min at 60°C, and 1 min at 72°C. Iμ primers were 5′-GGTGGCTTTGAAGGAACAATTCCAC and 5′-TCTGAACCTTCAAGGATGCTCTTG. λ5 primers were 5′-CTTGAGGGTCAATGAAGCTCAGAAGA and 5′-CTTGGGCTGACCTAGGTTG. Primers for β-actin were from Stratagene.
HAT assays.
HAT assays were performed as previously described (7). Flag-tagged p300 protein was purified from baculovirus-infected Sf9 cells. GST-EBF was purified from bacteria, and approximately 200 ng of full-length protein was added to each reaction mixture (most of the protein sample consisted of degradation products). The reaction was carried out at 30°C for 10 min. The reaction products were resolved by SDS-14% PAGE and quantified with a phosphorimager.
Electrophoretic mobility shift assays (EMSAs).
EMSAs were carried out as previously described (45). Briefly, oligonucleotides were annealed and end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Invitrogen). In vitro-transcribed and -translated EBF or the EBF fragment consisting of the DNA binding and dimerization domains was incubated for 30 min at room temperature with labeled probe in the absence or presence of in vitro-transcribed and -translated CBP fragment IV. The reaction mixtures were then subjected to electrophoresis on a 6% polyacrylamide-Tris-borate-EDTA gel and analyzed with a phosphorimager.
RESULTS
Interaction of EBF and CBP in vitro and in vivo.
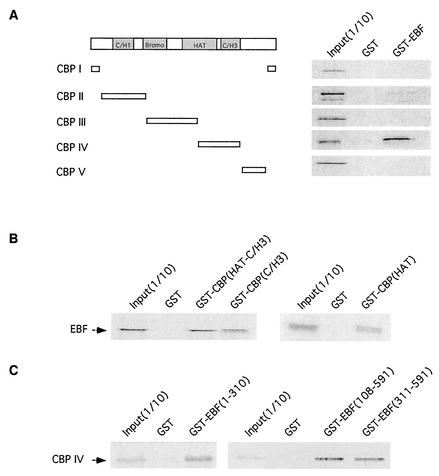
The vast majority of transcription factors appear to utilize the transcriptional coactivator p300/CBP. To determine if this is the case for EBF, we first asked if a GST-EBF fusion protein could bind in vitro-translated CBP. We assayed various segments of CBP and found that fragment IV, encompassing aa 1626 to 2260 of CBP, interacted well with GST-EBF in vitro (Fig. 1A). We also detected a weak interaction between GST-EBF and CBP fragment II, which includes aa 117 to 737. CBP fragment IV contains a portion of the C-terminal region of the HAT domain and the cysteine-histidine-rich C/H3 domain (28). The C/H3 domain of p300/CBP is involved in the interaction with many p300/CBP-binding proteins such as E1A, E47, and GATA-1. In our in vitro binding analyses, we found that both the C/H3 domain and the C-terminal region of the HAT domain could interact with full-length EBF (Fig. 1B). Employing GST fusion proteins containing various fragments of EBF, we observed that CBP fragment IV could interact with both the N-terminal half of EBF (aa 1 to 310), containing the DNA binding domain, and the C-terminal half of the protein (aa 311 to 591), comprising the dimerization domain and the major transactivation domain (Fig. 1C).
FIG. 1.
CBP and EBF interact in vitro. (A) GST and GST-EBF were assessed for their abilities to interact with the indicated series of in vitro-translated CBP fragments: CBP I (aa 1 to 117 plus 2394 to 2442), CBP II (aa 117 to 737), CBP III (aa 737 to 1626), CBP IV (aa 1626 to 2260), and CBP V (aa 2260 to 2389). The input lane represents 1/10 of the total labeled CBP used in the binding reaction. (B) GST, GST-CBP(HAT-C/H3) (aa 1196 to 1896), GST-CBP(C/H3) (aa 1718 to 1896), and GST-CBP(HAT) (aa 1196 to 1718) were assessed for their abilities to interact with in vitro-translated full-length EBF. (C) GST, GST-EBF(1-310), GST-EBF(108-591), and GST-EBF(311-591) were assessed for their abilities to interact with in vitro-translated CBP fragment IV.
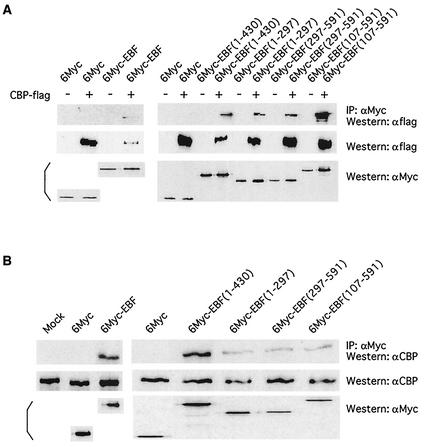
To assess the ability of EBF and CBP to interact in vivo, we employed coimmunoprecipitation assays. We transfected 293T cells with expression vectors for Myc-tagged full-length EBF, or various EBF fragments, along with Flag-tagged CBP. Cell lysates were treated with an anti-Myc antibody, and precipitated proteins were analyzed by Western blotting with an anti-Flag antibody. We observed Flag-tagged CBP in the precipitates, but only in the presence of Myc-tagged EBF (Fig. 2A). Consistent with our in vitro experiments, we also found that both the N-terminal region and the C-terminal region of EBF could interact with CBP. We also looked for interactions between EBF and endogenous CBP. 293T cells were transfected with only the EBF expression plasmid, and an anti-CBP antibody was used in the Western analysis of precipitated proteins. Again, confirming the interaction, endogenous CBP was observed in the anti-Myc precipitates only in the presence of Myc-tagged EBF (Fig. 2B).
FIG. 2.
CBP and EBF interact in vivo. (A) 293T cells grown in 60-mm-diameter plates were transfected with plasmids expressing Flag-tagged CBP (6 μg) and various Myc-tagged versions of EBF (1 μg) as indicted. Extracts were immunoprecipitated with an anti-Myc antibody, and the precipitates were analyzed for the presence of tagged CBP with an anti-Flag antibody (top panels). (B) 293T cells were transfected with the Myc-tagged EBF constructs (1 μg) indicated, and extracts were subjected to immunoprecipitation with an anti-Myc antibody. Precipitates were analyzed for the presence of endogenous CBP by using anti-CBP Western blotting (top panels). Total extracts were assessed for expression of endogenous CBP (middle panels) and tagged EBF proteins (bottom panels) with Western blotting with anti-CBP and anti-Myc antibodies, respectively.
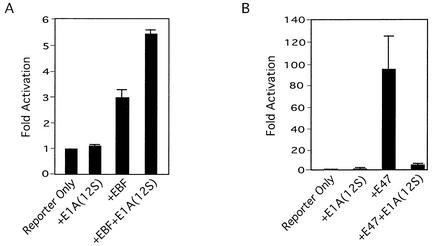
HAT activity of the coactivators p300/CBP and PCAF is sensitive to the adenovirus E1A protein (7, 18, 39). Accordingly, transcription factors requiring p300/CBP or PCAF are sensitive to expression of E1A in vivo. Surprisingly, we found that EBF activity—as measured by its ability to stimulate transcription from the λ5 promoter in transfection assays—was completely insensitive to E1A (Fig. 3A). In fact, E1A reproducibly stimulated EBF activity on both this promoter and the CD19 promoter (data not shown). In contrast, E1A completely blocked the ability of E47 to stimulate transcription from a promoter containing E boxes upstream of a TATA box (Fig. 3B). We conclude that, although EBF and p300/CBP can form a complex, EBF does not require the HAT activity of p300/CBP to stimulate transcription.
FIG. 3.
EBF is not inhibited by E1A. (A) EBF activity was measured in NIH 3T3 cells by using an EBF expression plasmid (100 ng) and a reporter carrying the λ5 promoter driving luciferase (50 ng). (B) E47 activity was measured by using an E47 expression vector (20 ng) and the [E5+E2+E3]6-TATA-Luc reporter (20 ng). A 12S E1A expression vector (20 ng) was added where indicated.
Inhibition of p300/CBP by EBF.
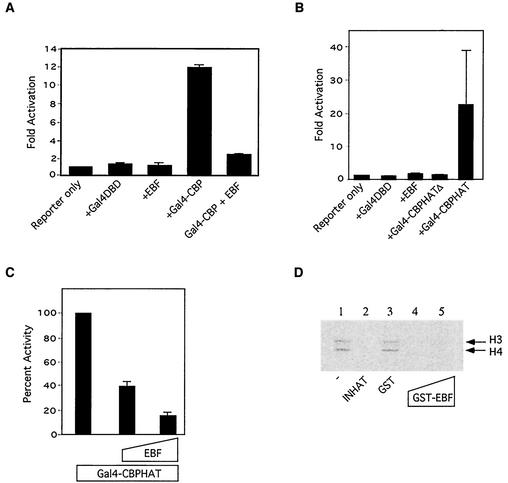
E1A, Twist, Pu.1, and the Hox family of DNA binding proteins bind to, yet inhibit, p300/CBP HAT activity (7, 18, 19, 39, 44). We used several assays to see if EBF might do the same. In the first, we asked if EBF could affect the activity of full-length CBP as a Gal4 fusion protein. Gal4-CBP activated transcription from a promoter bearing Gal4 binding sites upstream of a TATA box, but in the presence of EBF, activity was inhibited (Fig. 4A). Similar results were obtained with Gal4-p300 (data not shown). Previous studies have shown that, when tethered to Gal4, a CBP fragment containing the HAT domain (CBPHAT) can activate transcription from an adenovirus early promoter in U2OS cells. This activity is entirely dependent on the HAT domain of CBP and therefore represents an in vivo assay for HAT function (29). We confirmed this result and showed that a mutant CBP harboring a small, 18-aa deletion in the HAT domain (CBPHATΔ) was unable to activate transcription (Fig. 4B). When we added EBF to the assay mixture, we observed a decrease in transcription (Fig. 4C). We also examined the effect of EBF on HAT activity in vitro. We used purified p300 and measured its ability to acetylate histones H3 and H4 in the presence of radiolabeled acetyl coenzyme A. Activity was assessed by autoradiography and quantitated with a phosphorimager. We confirmed that p300 was able to acetylate both H3 and H4 and that the addition of INHAT inhibited the reaction. The addition of purified GST-EBF also inhibited the reaction while GST alone had no effect (Fig. 4D). We conclude that EBF can inhibit the HAT activity of p300/CBP both in vivo and in vitro.
FIG. 4.
EBF inhibits p300/CBP in vivo and in vitro. (A) Activity of Gal4-CBP (20 ng) was measured in NIH 3T3 cells by using the Gal4-E1b-Luc reporter (100 ng) in the absence or presence of CMV-EBF (20 ng). (B) Activity of the CBPHAT domain as a Gal4 fusion protein (0.5 μg) was assessed in transfected U2OS cells by using the Gal4-AdML-TATA-CAT reporter (500 ng). (C) As for panel B activity of Gal4-CBP was assessed in the presence of an EBF expression vector (0.5 or 1 μg). (D) p300 HAT activity was measured in vitro by using purified histones and radiolabeled acetyl coenzyme A alone or in the presence of the indicated proteins. Acetylated histones were visualized with a phosphorimager.
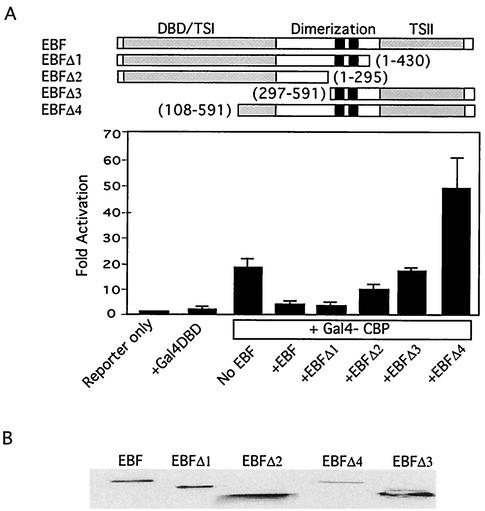
A series of EBF deletion mutants were used to map the region on EBF required for inhibition of Gal4-CBP in vivo. We found that the DNA binding domain of EBF (aa 1 to 297) was sufficient to inhibit Gal4-CBP and that addition of the dimerization domain (aa 1 to 430) led to more effective inhibition (Fig. 5A). Expression levels of the various deletions revealed that EBF(108-591) was expressed at relatively low levels (Fig. 5B). To ensure that the lack of inhibitory activity of this mutant was not a function of low protein levels, we carried out a titration with increasing amounts of EBF(108-591) and confirmed that it lacked activity at higher expression levels (data not shown). We conclude that EBF can target p300/CBP and that inhibition requires the DNA binding domain of EBF.
FIG. 5.
The DNA binding domain of EBF inhibits CBP in vivo. (A) Full-length EBF and the various EBF deletion mutants indicated (all carrying Myc tags at their N termini) were assessed for their abilities to inhibit Gal4-CBP as described for Fig. 4A. (B) Western blot showing relative expression levels of the various EBF fragments with use of an anti-Myc antibody.
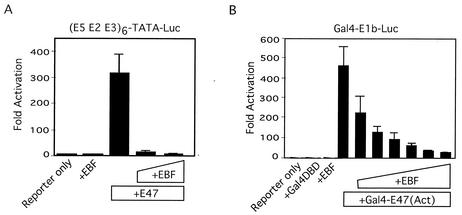
The ability of EBF to inhibit p300/CBP prompted us to examine EBF's effect on transcriptional activation by other transcription factors. As shown above, E47 is sensitive to E1A, a well-established inhibitor of p300/CBP. We found that EBF also inhibited transcriptional activation by E47 and by the E47 activation domain fused to Gal4 (Fig. 6). EBF also inhibited transcriptional activation by Gal4-VP16, TFE3, and Notch (data not shown), all of which utilize p300/CBP (10, 13, 38, 50).
FIG. 6.
EBF can inhibit E47 activity. (A) E47 activity was measured by using an E47 expression vector (20 ng) and the [E5+E2+E3]6-TATA-Luc reporter (20 ng). An EBF expression vector (20 or 50 ng) was included in the transfections where indicated. (B) Activity of the E47 activation domain was measured with a Gal4-E47 fusion protein (containing roughly the N-terminal half of the E47 protein) and a reporter containing five Gal4 binding sites linked to the adenovirus E1b TATA box (100 ng). Assays were carried out with increasing amounts of an EBF expression vector (20, 50, 100, 200, 500, or 1,000 ng).
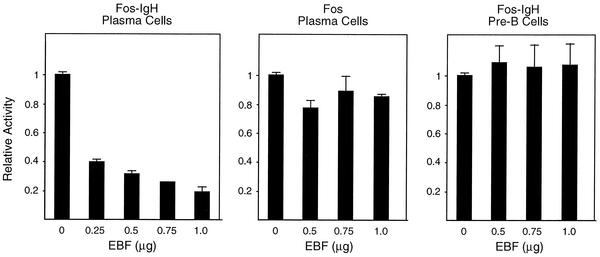
These data raised the possibility that EBF may be able to antagonize the activity of E47 in B lymphocytes, despite genetic data that indicate that the two proteins collaborate to promote B-cell development. To address this, we first attempted to generate S194 and J558 plasmacytoma cells that express retrovirally transduced EBF. Plasma cells do not normally express EBF, so we reasoned that such cells would let us evaluate the effect of EBF on the expression of E47 target genes (e.g., immunoglobulin genes) and EBF target genes (e.g., the V-preB and λ5 genes). However, of over 70 virus-positive clones, none expressed active EBF as judged by EMSA, whereas expression was readily detected in transduced BaF3 cells (data not shown). These data argue that plasma cells cannot tolerate EBF. Therefore, in the absence of stably transduced clones, we transiently transfected EBF into B lymphocytes and assessed the effect on a reporter carrying a Fos promoter linked to the IgH enhancer, a target of E47. We found that in S194 cells EBF decreased activity of the reporter in a dose-dependent manner (Fig. 7, left panel), while having no effect on a reporter driven by the Fos promoter alone (center panel). Interestingly, EBF did not repress activity of the IgH enhancer in transfections of the pre-B-cell line 230-238 (right panel). Although we will need to analyze many additional cell types to get a more complete picture, these results suggest that the ability of EBF to repress p300/CBP is cell type dependent and that EBF does not antagonize E47 in pre-B cells. The nature of this apparent cell type specificity is currently being investigated.
FIG. 7.
EBF inhibits IgH enhancer activity in plasmacytoma cells but not a pre-B-cell line. Either S194 plasmacytoma cells (left and center panels) or 230-238 pre-B cells (right panel) were transfected with the indicated reporters along with increasing amounts of an EBF expression plasmid. Luciferase values are expressed relative to those obtained with the reporter alone (100%) and are the averages of at least four transfections.
Distinct properties of free and DNA-bound EBF.
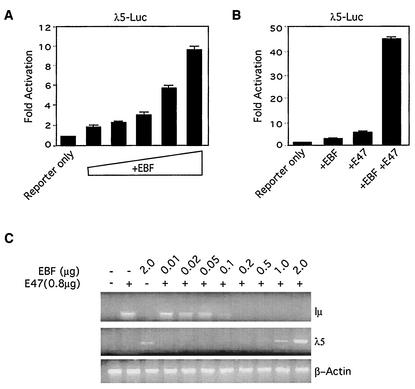
Our data raised an interesting question: how can EBF inhibit E47 in reporter assays in fibroblasts and yet in the same assays work collaboratively with E47 to stimulate transcription from the λ5 promoter? We first confirmed that EBF and E47 synergize at the λ5 promoter by using transient transfections of NIH 3T3 cells. As reported previously, EBF was able to activate the λ5 promoter on its own (Fig. 8A), and yet the combination of EBF and E47 was much more effective than was either protein alone (Fig. 8B). We also measured the ability of EBF and E47 to activate transcription from the endogenous λ5 gene in transfected NIH 3T3 cells. Again, as reported previously, EBF was sufficient to activate transcription from the endogenous λ5 gene and transcription was enhanced in the presence of transfected E47 (Fig. 8C, top panel). When we simultaneously measured the ability of E47 to activate transcription from the endogenous IgH locus, by using Iμ sterile transcripts as a readout, EBF effectively inhibited transcription (Fig. 8C, middle panel). Thus, the ability of EBF to activate or inhibit transcription is also promoter dependent. Interestingly, EBF inhibited transcription at much lower concentrations than those needed to detect transcriptional activation of the λ5 gene. However, we cannot conclude from this that widespread inhibition by EBF occurs at concentrations that are normally associated with transcriptional activation. It is just as likely that activation of the endogenous λ5 gene in NIH 3T3 cells requires abnormally high levels of EBF.
FIG. 8.
EBF can both activate and repress transcription. (A) Activation of the λ5 promoter. Increasing amounts of an EBF expression vector (20, 50, 100, 200, or 500 ng) were added to a reporter carrying the λ5 promoter driving luciferase (50 ng). (B) Activity of the λ5 promoter was measured with EBF alone (50 ng of expression plasmid), E47 alone (50 ng of expression plasmid), or EBF plus E47 (50 ng each). (C) Effect of EBF and E47 on transcription of the endogenous λ5 and IgH loci in NIH 3T3 cells. Cells were transfected with the indicated plasmids, and total RNA was subjected to RT-PCR analyses with primer pairs corresponding to Iμ, λ5, or β-actin.
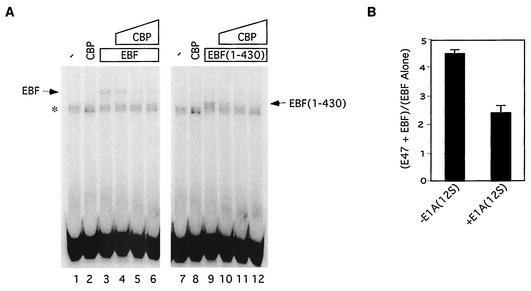
The λ5 promoter has binding sites for both EBF and E47, whereas the heavy chain gene is not known to bind EBF (3). We reasoned, therefore, that EBF might generally activate transcription of genes containing EBF binding sites and might repress transcription of those without EBF sites. However, given the effect of EBF on p300/CBP, we wanted to know if DNA-bound EBF was also capable of inhibiting p300/CBP. Since the inhibitory domain of EBF mapped to its DNA binding domain, we asked if EBF could bind DNA and CBP at the same time. EMSAs were used to analyze the DNA binding of EBF, in the absence or the presence of increasing amounts of CBP. Using either in vitro-translated full-length EBF (Fig. 9A, left panel) or the EBF DNA binding and dimerization domains (Fig. 9A, right panel), we found that in vitro-translated CBP inhibited DNA binding by EBF. We conclude that EBF cannot bind DNA and p300/CBP at the same time and, therefore, that DNA-bound EBF does not inhibit p300/CBP. Consistent with this, we found that transcriptional synergy imparted by E47 at the λ5 promoter was partially inhibited by E1A (Fig. 9B), suggesting that p300/CBP contributes to transcriptional activation at the λ5 promoter in the presence of DNA-bound EBF.
FIG. 9.
DNA-bound EBF does not inhibit p300/CBP. (A) CBP inhibits DNA binding by EBF. EMSAs were carried out with a probe corresponding to the EBF binding site in the mb-1 promoter. EBF and CBP fragment IV proteins were generated by in vitro transcription and translation and employed in the binding reactions as indicated. The bands marked by * were present in unprogrammed reticulocyte lysates (lanes 1 and 7). (B) Cooperativity at the λ5 promoter is sensitive to E1A. The contribution of E47 to activation of the λ5 promoter in the presence of both E47 and EBF was calculated as the ratio of fold stimulation by E47 plus EBF to fold stimulation by EBF alone. This value was determined in the presence or absence of 12S E1A.
DISCUSSION
The results presented here describe an intriguing property of EBF and provide clues as to its mechanism of transcriptional activation. We show that overexpressed EBF can activate transcription but can also function as a potent and general inhibitor of transcription in fibroblasts and plasma cells by targeting the coactivator p300/CBP. Inhibition of p300/CBP correlates with a direct interaction between the two proteins that maps to the HAT and C/H3 domains of p300/CBP. The manner in which inhibition occurs has yet to be investigated, but it is likely that EBF functions as a competitive inhibitor of HAT enzymatic activity. At the present time we can only speculate as to why EBF inhibits p300/CBP. Indeed, the relevance of such an activity for E1A (7, 18), Twist (18), Pu.1 (19), the Hox proteins (44), and the Rb-interacting protein EID-1 (27, 30) is largely conjecture. While it is clear that the levels of p300 and CBP are critical for proper development—mutations in a single copy of the CBP gene in humans lead to Rubinstein-Taybi syndrome (40), and mice heterozygous for a deletion of the CBP gene develop hematopoietic malignancies (24)—it is not known if any of the aforementioned proteins, including EBF, functionally diminish p300/CBP activity within a B cell. Mice carrying only one functional EBF gene do show a mild B-cell phenotype (25, 36), but we cannot say if this is due to elevated p300/CBP activity. In fact, our preliminary data suggest that the inhibition of p300/CBP by EBF may not be relevant to the B-cell types in which it is normally expressed. It remains to be determined if it is relevant to other EBF-expressing cell types such as neurons or adipocytes. Pu.1, which can inhibit p300/CBP HAT activity at physiological levels and whose expression correlates inversely with HAT activity in erythroid cells (19), stimulated rather than inhibited Iμ transcription in the presence of E47 (F. Zhao, unpublished observations). The ability of Pu.1 to activate rather than repress Iμ might be related to the fact that Eμ has a Pu.1 binding site (33). We would propose that the inhibition of p300/CBP would be an important aspect of EBF's function only if there were significant amounts of free EBF in cells. This is not known. Even then, it would likely inhibit only a small set of genes whose activities are particularly sensitive to fluctuations in p300 and/or CBP levels.
Detailed knowledge concerning the individual roles of EBF and E47 in transcriptional activation will require the generation of cell lines that express only EBF or only E47 and the analysis of chromatin and proteins recruited to the endogenous λ5 gene. Nevertheless, our findings clearly indicate that EBF stimulates transcription independently of p300/CBP or PCAF. Thus, it probably serves a purpose at the λ5 promoter other than to recruit HATs. The dose of EBF has been shown elsewhere to affect directly position effect variegation of a λ5 transgene integrated into alpha satellite heterochromatin (26), and so it is likely to play an important role in regulating chromatin structure. We have shown that EBF can interact with BRG1, a component of the mammalian SWI/SNF chromatin remodeling complex (32; Zhao, unpublished observations); however, we do not yet know if EBF actually recruits chromatin remodelers to the λ5 gene in B cells. Given the recently identified role of histone methyltransferases in the regulation of position effect variegation (20), it is possible that EBF interacts with several distinct proteins that affect chromatin dynamics. On the other hand, given its sensitivity to E1A, E47 is likely to recruit HATs to the λ5 promoter.
The interdependent functions of E47 and EBF are supported by genetic studies in mice. O'Riordan and Grosschedl examined lymphopoiesis in mice heterozygous for mutations in both the E2A and EBF genes (36). The compound heterozygotes showed a significant block in pro-B-cell differentiation, while single heterozygotes were much less affected. The block was later than that seen in mice homozygous for mutations in either E2A or EBF. Genes whose expression was diminished included λ5, Pax5/BSAP, mb-1, Rag1, and Rag2. Although we have looked only at the interplay between E47 and EBF at the λ5 promoter, we would assume that they play similarly distinct roles in the transcriptional activation of these other genes as well. In fact, it has been shown elsewhere that E47 cannot activate the endogenous V-preB gene on its own but can synergize with EBF to give a dramatic activation, a scenario much like what we've described for λ5 (46). We propose that the genetic interaction reflects a mutual dependency—each protein making a unique contribution to gene activation—rather than a simple dosage effect.
B-cell development is a complex process, involving a battery of finely tuned transcriptional events that regulate a host of B-cell-specific genes at different times (41). For the case of E47 and EBF, which work together to activate several B-cell-specific genes, we propose that their collaboration arises through their two different but complementary activities. To date, the case for the λ5 promoter is unique. Although there are countless examples of transcription factors that work together to activate gene transcription, for the most part they are all capable of recruiting HATs, and thus it is difficult to know without detailed promoter recruitment studies if they play mechanistically distinct roles. For proteins such as EBF—these would possibly include Twist and the Hox proteins—the case may be simpler because they cannot activate transcription by recruiting p300/CBP. Almost by definition, activation of their target genes would be expected to require some degree of chromatin remodeling. Whether this is a general requirement of the target genes of EBF, Twist, and Hox proteins remains to be determined.
Acknowledgments
We thank Gerd Blobel and Shara Kabak for critical reading of the manuscript and members of the Kadesch lab for valuable input throughout the course of this work. We are also indebted to Debu Chakravarti for supplying reagents and helping with the p300 HAT assays.
This work was supported by funds from the National Institutes of Health (AI 36878 and DK52558, to T.K.).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Akerblad, P., M. Rosberg, T. Leanderson, and M. Sigvardsson. 1999. The B29 (immunoglobulin β-chain) gene is a genetic target for early B-cell factor. Mol. Cell. Biol. 19:392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerblad, P., M. Sigvardsson, and T. Leanderson. 1996. Early B-cell factor (EBF) down-regulates immunoglobulin heavy chain intron enhancer function in a plasmacytoma cell line. Scand. J. Immunol. 44:89-92. [DOI] [PubMed] [Google Scholar]
- 4.Bain, G., E. C. R. Maandag, D. J. Izon, D. Amsen, A. M. Kruisbeek, B. C. Weintraub, I. Krop, M. S. Schlissel, A. J. Feeney, M. van Roon, M. van der Valk, H. P. J. te Riele, A. Berns, and C. Murre. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell 79:885-892. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Busslinger, M., S. L. Nutt, and A. G. Rolink. 2000. Lineage commitment in lymphopoiesis. Curr. Opin. Immunol. 12:151-158. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 8.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 9.Dubois, L., and A. Vincent. 2001. The COE-Collier/Olf1/EBF-transcription factors: structural conservation and diversity of developmental functions. Mech. Dev. 108:3-12. [DOI] [PubMed] [Google Scholar]
- 10.Eckner, R., T. P. Yao, E. Oldread, and D. M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478-2490. [DOI] [PubMed] [Google Scholar]
- 11.Feldhaus, A. L., D. Mbangkollo, K. L. Arvin, C. A. Klug, and H. Singh. 1992. BLyF, a novel cell-type- and stage-specific regulator of the B-lymphocyte gene mb-1. Mol. Cell. Biol. 12:1126-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 13.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gisler, R., and M. Sigvardsson. 2002. The human V-pre-B promoter is a target for coordinated activation by early B cell factor and E47. J. Immunol. 168:5130-5138. [DOI] [PubMed] [Google Scholar]
- 15.Hagman, J., C. Belanger, A. Travis, C. W. Turck, and R. Grosschedl. 1993. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7:760-773. [DOI] [PubMed] [Google Scholar]
- 16.Hagman, J., M. J. Gutch, H. Lin, and R. Grosschedl. 1995. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 14:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman, J., A. Travis, and R. Grosschedl. 1991. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 10:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamamori, Y., V. Sartorelli, V. Ogryzko, P. L. Puri, H. Y. Wu, J. Y. Wang, Y. Nakatani, and L. Kedes. 1999. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96:405-413. [DOI] [PubMed] [Google Scholar]
- 19.Hong, W., A. Y. Kim, S. Ky, C. Rakowski, S. B. Seo, D. Chakravarti, M. Atchison, and G. A. Blobel. 2002. Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Mol. Cell. Biol. 22:3729-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 22.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 13:1412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudrycki, K., C. Stein-Izsak, C. Behn, M. Grillo, R. Akeson, and F. L. Margolis. 1993. Olf-1-binding site: characterization of an olfactory neuron-specific promoter motif. Mol. Cell. Biol. 13:3002-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, H., and R. Grosschedl. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Lundgren, M., C. M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 27.MacLellan, W. R., G. Xiao, M. Abdellatif, and M. D. Schneider. 2000. A novel Rb- and p300-binding protein inhibits transactivation by MyoD. Mol. Cell. Biol. 20:8903-8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmorstein, R. 2001. Structure of histone acetyltransferases. J. Mol. Biol. 311:433-444. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyake, S., W. R. Sellers, M. Safran, X. Li, W. Zhao, S. R. Grossman, J. Gan, J. A. DeCaprio, P. D. Adams, and W. G. Kaelin, Jr. 2000. Cells degrade a novel inhibitor of differentiation with E1A-like properties upon exiting the cell cycle. Mol. Cell. Biol. 20:8889-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 32.Narlikar, G. J., M. L. Phelan, and R. E. Kingston. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 8:1219-1230. [DOI] [PubMed] [Google Scholar]
- 33.Nelsen, B., G. Tian, B. Erman, J. Gregoire, R. Maki, B. Graves, and R. Sen. 1993. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science 261:82-86. [DOI] [PubMed] [Google Scholar]
- 34.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 35.Ordentlich, P., A. Lin, C. P. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 37.O'Riordan, M., and R. Grosschedl. 2000. Transcriptional regulation of early B-lymphocyte differentiation. Immunol. Rev. 175:94-103. [PubMed] [Google Scholar]
- 38.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21:7761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perissi, V., J. S. Dasen, R. Kurokawa, Z. Wang, E. Korzus, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1999. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc. Natl. Acad. Sci. USA 96:3652-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrij, F., R. H. Giles, H. G. Dauwerse, J. J. Saris, R. C. Hennekam, M. Masuno, N. Tommerup, G. J. van Ommen, R. H. Goodman, D. J. Peters, et al. 1995. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 376:348-351. [DOI] [PubMed] [Google Scholar]
- 41.Reya, T., and R. Grosschedl. 1998. Transcriptional regulation of B-cell differentiation. Curr. Opin. Immunol. 10:158-165. [DOI] [PubMed] [Google Scholar]
- 42.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 43.Shen, C. P., and T. Kadesch. 1995. B-cell-specific DNA binding by an E47 homodimer. Mol. Cell. Biol. 15:4518-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, W. F., K. Krishnan, H. J. Lawrence, and C. Largman. 2001. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 21:7509-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigvardsson, M. 2000. Overlapping expression of early B-cell factor and basic helix-loop-helix proteins as a mechanism to dictate B-lineage-specific activity of the λ5 promoter. Mol. Cell. Biol. 20:3640-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigvardsson, M., M. O'Riordan, and R. Grosschedl. 1997. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7:25-36. [DOI] [PubMed] [Google Scholar]
- 47.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 48.Travis, A., J. Hagman, L. Hwang, and R. Grosschedl. 1993. Purification of early-B-cell factor and characterization of its DNA-binding specificity. Mol. Cell. Biol. 13:3392-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, M. M., and R. R. Reed. 1993. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364:121-126. [DOI] [PubMed] [Google Scholar]
- 52.Wang, S. S., R. Y. Tsai, and R. R. Reed. 1997. The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J. Neurosci. 17:4149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, F., A. Vilardi, R. J. Neely, and J. K. Choi. 2001. Promotion of cell cycle progression by basic helix-loop-helix E2A. Mol. Cell. Biol. 21:6346-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell 79:875-884. [DOI] [PubMed] [Google Scholar]