Abstract
Despite significant structural diversity, present evidence suggests that EWS/ETS fusion proteins promote oncogenesis by transcriptionally modulating a common set of target genes. In order to identify these genes, microarray expression analyses were performed on NIH 3T3 polyclonal populations expressing one of three EWS/ETS fusion genes. The majority of these genes can be grouped into seven functional categories, including cellular metabolism and signal transduction. The biologic significance of these target genes was pursued. The effects of modulating genes involved in metabolism were assessed by flux studies and demonstrated shifts in glucose utilization and lactate production as a result of EWS/FLI1 expression. The proto-oncogene coding for serine/threonine kinase PIM3 was found to one of several genes encoding signal transduction proteins that were up-regulated by EWS/ETS fusions. PIM3 was found to be expressed in a panel of human Ewing's family tumor cell lines. Forced expression of PIM3 promoted anchorage-independent growth. Coexpression of a kinase-deficient PIM3 mutant attenuated EWS/FLI1-mediated NIH 3T3 tumorigenesis in immunodeficent mice.
The EWS/ETS fusions comprise a group of structurally related oncoproteins that are found specifically in Ewing's family tumors (EFTs). The oncogenes coding for these oncoproteins are the result of chromosomal translocations that fuse a portion of the amino-terminal region of EWS to one of five members of the ETS family of transcription factors: FLI1, ERG, FEV, ETV1, and E1AF (for review, see reference 1). While ETS proteins have been extensively studied, the normal functions of EWS are still coming to light. EWS is part of a family of putative RNA binding proteins that also includes TLS and TAFII68. A growing body of evidence suggests that members of this family (the TET family) may be involved in multiple levels of mRNA metabolism, from roles as transport chaperones to splicing factors (6, 9, 32, 34, 35). Interestingly, each of these three TET members is involved in tumor-associated chromosomal translocations associated with various human sarcomas, including desmoplastic small round cell tumor and clear cell sarcoma (3, 11, 17, 21, 25, 26). In all cases, these translocations lead to the expression of chimeric proteins analogous to EWS/ETS fusions by incorporating DNA-binding domains (DBDs) from different transcription factors. Tumor histologies appear to correlate with the specific DBDs rather than the particular TET component in these fusions.
All members of the ets-coded family of transcription factors contain a conserved 85-amino-acid domain that mediates site-specific DNA binding at target gene promoters and enhancers (for review, see reference 16). Based on the primary amino acid sequences of their respective ets DBDs, the five EWS/ETS fusions can be partitioned into two groups. Group 1 contains EWS/FLI1, EWS/ERG, and EWS/FEV, and its members display up to 98% amino acid identity between their DBDs (FLI1 versus ERG). Similarly, in group 2, the ETV1 and E1AF DBDs are identical to each other except at 2 amino acid residues but are only 60% identical to the FLI1 DBD. In spite of these structural differences, members of both groups are capable of promoting oncogenesis in a characteristic fashion. NIH 3T3 cells expressing either group 1 or group 2 EWS/ETS fusions form tumors in immunodeficient mice that display a round cell histology that is very similar to that seen in human EFTs (30).
While differences in translocation breakpoints can lead to considerable variability in the precise EWS and ETS exons incorporated into individual fusions, all EWS/ETS fusions found in tumors retain an intact ets DBD. This consistent presence of the DBD suggests that EWS/ETS proteins utilize this domain to bind DNA in a site-specific manner and regulate gene expression. This finding and other biochemical features found in EWS/ETS fusions have led to the current hypothesis that these chimeric proteins function at least in part as aberrant transcription factors that promote cellular transformation by trancriptionally modulating a repertoire of target genes. While there are several potential strategies to identify these target genes, a major hurdle lies in determining the biologic significance of individual targets. EWS/ETS proteins like other aberrant transcription factors are unlikely to modulate only genes that play active roles in cellular transformation. There are bound to be regulated genes that are incidental targets and are not directly involved in promoting abnormal cell growth or differentiation.
To address this central issue, we utilized a microarray-based strategy to identify a set of genes that are similarly regulated by structurally divergent EWS/ETS genes in a common cellular background. Our hypothesis was based on the observation that despite their structural differences, given their highly similar and characteristic tumorigenic effects on cells, group 1 and 2 EWS/ETS fusions were likely to transform cells via common genetic pathways. By expressing EWS/FLI1, EWS/ERG, and EWS/ETV1 at similar levels in NIH 3T3 cells, we have identified a core set of EWS/ETS target genes. To demonstrate the potential biologic relevance of this cohort, we singled out one gene, that coding for serine/threonine kinase PIM3, for further study. We found that forced expression of PIM3 promotes anchorage-independent growth and that a dominant-negative mutant PIM3 antagonizes EWS/FLI1-mediated tumorigenesis.
MATERIALS AND METHODS
Plasmid constructs and tissue culture.
N-terminal Flag-tagged EWS/FLI1, EWS/ERG, and EWS/ETV1 were cloned into the replication-deficient retroviral construct pSRα-MSVtkneo and have been described previously (22, 30). The cDNA for the rat PIM3 gene was a generous gift of H. Herschman and was also described previously (13). The gene was then cloned with EcoRI-HindIII fragments into the retroviral construct behind a green fluorescent protein (GFP)-internal ribosomal entry site (IRES) cassette, and a Flag tag was added to the N terminus. PIM3 K69M was generated by a two-step PCR process with specific primer pairs containing the desired nucleotide changes and also contains a N-terminal Flag tag. NIH 3T3 cells were passed under standard conditions in Dulbecco's modified Eagle's medium (DMEM) plus 5% calf serum as well as high levels of glucose and glutamine. The growth curves were determined by plating 1.5 × 103 cells in quadruplicate in five 24-well plates. Each day, the cells from an individual plate were treated with 400 μl of a 1-mg/ml solution of 3-(4,5dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) (Sigma)-RPMI (Invitrogen). The cells were incubated for 3 h at 37°C, at which time a solubility solution (1% Triton X-100, 0.04 M HCl, 95% isopropanol) was added to the MTT solution. The A540 of the resulting solution was measured on a microplate reader. A standard curve was established by plating known quantities of cells, treating the cells with the MTT solubility solution, and measuring the A540. The number of cells in the experimental samples was determined by comparing their respective A540 with the standard curve.
Transfections and infections.
To make the retroviral stocks, 293T cells were transfected with the retroviral vector and a Ψ-packaging plasmid as previously described (2). Briefly, 15 μg of the pSRα constructs was CaPO4 transfected with 15 μg of ecotrophic Ψ-packaging plasmid into 293T cells. After 2 days, viral collections were made for 24 h, filtered, and applied to cells. Following infections, cells were selected with 450 μg of G418 per ml for a period of 7 to 10 days. GFP-positive cells were sorted by fluorescence-activated cell sorter (FACS) analysis.
GeneChip probe preparation and array analysis.
NIH 3T3 cells were transduced with retroviral stocks for EWS/FLI1, EWS/ERG, EWS/ETV1, and the control, Tk Neo. This procedure was repeated to generate a total of three independent transduced populations for each condition. Cells were harvested, and the resultant total RNAs were quantitated and pooled in equal proportions to create normalized starting populations. An additional Tk Neo population consisting of a single empty vector retroviral transduction was also created. Biotin-labeled cRNA was prepared according to Affymetrix protocols. Briefly, total RNA was isolated according to the RNeasy total RNA isolation protocol (Qiagen catalog no. 75142). Twenty micrograms of the total RNA was used to synthesize first-strand cDNA (Gibco BRL). The T7-(dT)24 oligomer [5′GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3′] (AmbionMEGAScript T7 kit, catalog no. 1334) was used for priming first-strand cDNA synthesis instead of oligo(dT). Second-strand cDNA was synthesized according to standard protocols. Biotin-11-cytidine-5′-triphosphate (Bio-11-CTP; ENZO catalog no. 42818) and biotin-16-uridine-5′-triphosphate (Bio-16-UTP; ENZO catalog no. 42814) were then incorporated into an in vitro transcription reaction with T7 polymerase (Ambion MEGAScript T7 kit catalog no. 1334). Affymetrix Mu11kSubA, Mu11kSubB, Mu19SubA, Mu19kSubB, and Mu19kSubC GeneChips were hybridized with biotin-labeled cRNA probes from each sample at the University of California, San Diego, GeneChip Core facility. The biotin-labeled probes were bound to R-phycoerythrin-steptavidin (Molecular Probes P/N S-866), and scanned on a Hewlett-Packard GeneArray scanner. Chips were scaled to a median average difference intensity of 100. Gene expression results were analyzed with GeneSpring software (Silicon Genetics, Inc.). We defined a threshold of 20 (one-fifth the median intensity) for the negative and low values (10, 29). Using these parameters, we searched for genes that were modulated at least twofold in all three EWS/ETS populations over the averaged value of the two Tk Neo populations. Genes were then culled individually. Borderline genes with raw intensities significantly greater than median intensity (>150), but which were modulated slightly less than twofold, were generally allowed. Alternatively, genes with low raw intensity scores (<100), the Tk Neo values of which were highly variable (more than two- to threefold), were treated with suspicion. In these cases, only those genes with EWS/ETS values more than twofold different from both Tk Neo values were retained.
Western and Northern analysis.
Western and Northern blots were performed by standard procedures. For protein analysis, cell lysates were made with NP-40 lysis buffer (50 mM Tris, 300 mM NaCl, 10% glycerol, 1 mM EDTA). Eighty micrograms of total protein lysates was run on denaturing sodium dodecyl sulfate gels, transferred to nitrocellulose membranes on a Bio-Rad semidry transfer apparatus, and incubated with anti-KID1 (PIM3) rabbit polyclonal antibody (a generous gift from H. Herschman) or anti-Flag (Sigma, Inc.) mouse monoclonal antibody in a mixture containing 5% milk and 1× Tris-buffered saline-Tween (TBS-T). Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibody (Transduction Labs; catalog no. R14745) and Western blot chemiluminescent reagent (NEN; catalog no. NEL101) were used for detection. Coomassie staining of the gel and Ponceau staining of the membrane were used to ensure equal loading. For Northern blots, RNA lysates were made with STAT-60 (TEL-TEST, Inc.). Approximately 8 μg was run under denaturing conditions on a 1% agarose-formaldehyde gel, transferred to nitrocellulose membrane by capillary methods, and hybridized and washed by standard methods. Ethidium staining of the gel was used to ensure equal loading.
In vitro kinase assay.
293T cells were CaPO4 transfected with Srα/GFP-IRES-PIM3, Srα/GFP-IRES-PIM3 K69M, or empty vector, and protein lysates were harvested in radioimmunoprecipitation assay buffer 48 h later. Five hundred micrograms of total lysate was immunoprecipitated with the anti-KID1 (PIM3) antibody and protein A-Sepharose beads. The immunoprecipitated product was subjected to immunoblot analysis to confirm the presence of PIM3 in the immunoprecipitated product. The immunoprecipitated product was incubated in kinase buffer (40 μM Tris-HCl [pH 7.5], 80 μM HEPES, 8 μM MgCl2, 2 μM ATP, 2 μM dithiothreitol, 10 μCi of [γ-32P]dATP). In the histone H1 assay, 10 μg of histone H1was added to the reaction mixture described above. The reaction mixture was incubated for 30 min at 37°C. The reactions were run on a 10% polyacrylamide gel, dried, and imaged.
Transformation assays and tumor assays.
NIH 3T3 fibroblasts (1 × 106) expressing EWS/FLI1, PIM3, EWS/FLI1, and PIM3 K69M or empty vector were subcutaneously injected into the nape region of the neck of CB17 SCID mice. All mice were female and about 6 to 8 weeks of age at the time of injection. Mice were observed until a grossly visible tumor 1.5 cm in diameter was present, at which time, the animals were sacrificed and the tumors were dissected. Tumor cell lines were created by culturing minced tumors until cell lines were established and expanded. For statistical analysis, only the mice that formed tumors were considered. Agar assays were performed as described previously (33). Briefly, 5 × 104 NIH 3T3 cells expressing PIM3, EWS/FLI1, or empty vector were seeded in semisolid agar under high-serum conditions (20%) and incubated for a period of 4 weeks. Photographs were taken at days 21 and 28; the day 21 images are shown.
Glucose utilization assays.
The requisite cell lines were generated as described above. Cells were incubated in standard medium containing 500 mg of glucose per liter. Cells (7.5 × 105) were plated and maintained in normal medium under standard incubation conditions. A medium-only control was incubated under the same conditions in parallel. Twenty-four hours later, the medium was collected, and the cells were enumerated. Concentrations of glucose and lactate in plasma were determined with a COBAS MIRA analyzer (Roche Diagnostic Systems, Inc., Montclair, N.J.) by using reagents provided by Raichem (San Diego, Calif.). Glucose UV reagent (catalog no. 80017) was used for glucose determination, and the Stat-Pack rapid lactate test (catalog no. 869218) was used for lactate determination.
RESULTS
EWS/FLI1, EWS/ERG, and EWS/ETV1 share a common set of target genes.
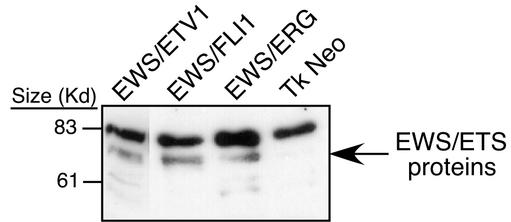
Microarray expression profiling was used to identify genes that are transcriptionally regulated by EWS/FLI1, EWS/ERG, and EWS/ETV1. NIH 3T3 cells were transduced with replication-deficient retroviruses containing epitope-tagged constructs of the genes coding for these three oncoproteins or empty vector as a negative control. After brief antibiotic selection and expansion, polyclonal cell populations were harvested for total RNA and protein. Relatively equivalent levels of EWS/ETS proteins across samples were documented by immunoblotting (Fig. 1). Pooled RNAs consisting of three independent transductions were used to make biotinylated cRNA probes. These probes were then hybridized to murine Mu11k and Mu19k oligonucleotide microarrays (Affymetrix, Inc.) that consisted of a total of 30,000 probe sets corresponding to an estimated 25,000 unique genes. After filtering out genes expressed at subthreshold levels (see Materials and Methods), genes were identified that were activated or repressed by an EWS/ETS fusion by at least twofold compared to those by an empty vector.
FIG. 1.
Immunoblot analysis of NIH 3T3 cells expressing N-terminal Flag-tagged EWS/FLI1, EWS/ERG, EWS/ETV, or empty vector. Cell lysates represented on this blot were derived from the same cell lines utilized in the array experiments for the EWS/ETV, EWS/ERG, and empty vector samples. The EWS/FLI1 sample lysate was derived from an NIH 3T3 cell line that was independent from the array studies but was generated in the same manner with comparable retroviral stocks. Previous studies have shown little variation in the level of protein expression between these fusion proteins generated in this manner (30).
The accuracy of microarray data was confirmed at several levels. First, the levels of previously identified EWS/FLI1 target genes that were also represented on microarrays were assessed. Known up-regulated genes, including the E2-C and stromelysin-1 genes, were also shown to be up-regulated in these microarray data (2, 7). In addition, Northern blot experiments were performed with 30 genes that by microarray profiling were either up- or down-regulated. There was a high degree (93%) of concordance between the microarray results and Northern blots (data not shown). Together these assays indicated that our microarray data accurately reflected target gene expression levels in our transduced NIH 3T3 populations.
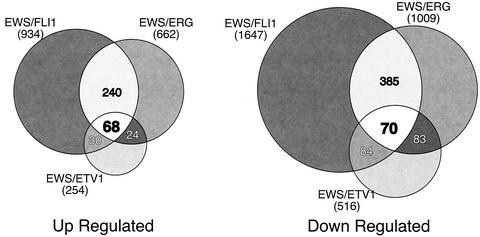
In comparing target gene patterns generated by the three EWS/ETS fusions, there were some notable trends. First, in comparison to either EWS/FLI1 or EWS/ERG, EWS/ETV1 appeared to be a generally weaker transcription factor, altering the expression of smaller numbers of genes and to quantitatively lesser degrees. Considering that EWS/ETV1 is expressed at approximately the same level as the other two EWS/ETS fusions in our transduced cells, this global difference in target gene modulation is likely due to biologic activity and is not simply an artifact of our model system. Second, despite their high degree of structural similarity, there was only a 34% overlap in up-regulated genes between EWS/FLI1 and EWS/ERG (Fig. 2). A similar trend is seen with repressed genes, among which 29% are shared. By comparing the expression profiles of all three EWS/ETS fusions, a core set of 68 up-regulated and 70 down-regulated genes were found. From these gene sets, approximately 70% of the up-regulated and 60% of the down-regulated genes have known functional identities. These were subdivided into one of seven functional categories (Table 1).
FIG. 2.
Venn diagrams of genes up- and down-regulated by EWS/ETS proteins in NIH 3T3 cells. RNA from NIH 3T3 cells stably expressing EWS/FLI1, EWS/ERG, or EWS/ETV1 were used to analyze gene expression patterns on the Affymetrix Mu11k and Mu19k series GeneChips as compared to RNA from parental control cells as described in Materials and Methods. Genes were considered up- or down-regulated when their levels of expression differed from that of the control by a factor of 2 or greater. The sizes of the Venn circles are proportional to the total number of modulated genes by each EWS/ETS fusion. Venn numbers correspond to regions of overlap between EWS/ETS fusion cohorts.
TABLE 1.
EWS/ETS up- and down-regulated genesa
| Gene accession no. and category (n) | Product and function |
|---|---|
| Up-regulated | |
| Cell cycle regulation (6) | |
| AA268341_s_at | Ube2C (ubiquitin-conjugating enzyme E2-C) |
| Msa.15090.0_s_at | p55 CDC (cell division control protein) |
| TC27100_f_at | Plk (polo-like kinase homolog; Drosophila) |
| TC27987_at | EMK2 (Mus musculus ELKL motif kinase 2 long form) |
| U15562_s_at | Cdc25c (cell division cycle 25 homolog C) |
| W08016_s_at | Ccnd1 (cyclin D1) |
| Cell metabolism (9) | |
| D44464_s_at | Upp (uridine phosphorylase) |
| M32240_s_at | Pmp22 (peripheral myelin protein, 22 kDa) |
| Msa.1148.0_f_at | Rpo2-1 (RNA polymerase II 1) |
| Msa.27354.0_r_at | Elongation factor TU, mitochondrial precursor |
| Msa.42771.0_s_at | MCM3, DNA replication licensing factor |
| Msa.6010.0_f_at | Na/K-transporting ATPase, α1 chain |
| TC35975_at | Ube2i (ubiquitin-conjugating enzyme E2I) |
| TC37350_at | Pyrroline-5-carboxylate reductase |
| TC41913_s_at | Pgam1 (phosphoglycerate mutase 1) |
| Cytoskeleton matrix (8) | |
| AA616243_s_at | Similar to rat cytokeratin 21, human cytokeratin 20 |
| TC27833_at | drebrin 1 |
| TC31950_at | XT-II (xylosyltransferase 2) |
| TC37486_at | M. musculus mRNA for mouse smoothelin, large isoform |
| U74079_s_at | Solute carrier family 9 (Na/H exchanger), isoform 3 regulator 1 |
| X52046_s_at | Col3a1 (procollagen, type III, α1) |
| X66402_s_at | Mmp3 (matrix metalloproteinase 3) |
| X69902_s_at | Itga6 (integrin α6) |
| Immune related (6) | |
| AA516966_s_at | RS21-C6 (Tdrg-TL1) |
| M18466_f_at | Ly6c (lymphocyte antigen 6 complex, locus C) |
| Msa.27769.0_s_at | CD97 antigen |
| TC29142_at | BLC (B-lymphocyte chemoattractant) |
| TC31829_at | Ifrg28 (28-kDa alpha interferon-responsive protein) |
| U02298_s_at | Scya5 (small inducible cytokine A5) |
| Signal transduction (9) | |
| AA104254_s_at | PIM-3 (proto-oncogene serine/threonine protein kinase) |
| AA408555_at | Rap2B |
| AF000236_at | Cmkorl (chemokine orphan receptor 1) |
| L75822_s_at | Insulin-like growth factor binding protein 7 |
| M81477_s_at | Ptpn2 (protein tyrosine phosphatase, nonreceptor type 2) |
| Msa.2879.0_s_at | Tob1 (transducer of ErbB-2.1) |
| Msa.907.0_at | Tumor necrosis factor (ligand) superfamily, member 9 |
| TC27106_at | PP2A (protein phosphatase, 55-kDa regulatory subunit, α) |
| TC35879_s_at | ARIP1 (M. musculus mRNA activin receptor-interacting protein 1) |
| Transcriptional regulation (4) | |
| Msa.788.0_s_at | Id1B (inhibitor of DNA binding 1) |
| TC35425_at | M. musculus SIK similar protein mRNA |
| TC37014_at | Very similar to Lbh (limb bud heart) |
| U63648_s_at | P160 (M. musculus p160 myb-binding protein mRNA, complete cds) |
| Other (5) | |
| C77409_rc_f_at | Hba-a1 (hemoglobin α, adult chain 1) |
| TC27925_i_at | M. musculus PAF actylhydrolase mRNA, complete cds |
| TC31637_at | Wiz (widely interspaced zinc finger motifs) |
| TC37528_at | M. musculus mRNA for α-1,6-fucosyltransferase, complete cds |
| X16133_s_at | Prg (proteoglycan, secretory granule) |
| Unidentifiable sequences (21) | |
| Total (68) | |
| Down-regulated | |
| Cell metabolism (15) | |
| AA059528_s_at | Diphosphomevalonate decarboxylase |
| AA122717_at | Dpp7 (dipeptidyl peptidase 7) |
| AA230405_s_at | Highly similar to CRTR (Na-dependent creatinine transporter [human]) |
| TC20149_s_at | Lta4h (leukotriene A4 hydrolase) |
| TC20760_at | Stard5 (StAR-related lipid transfer [START] domain-containing 5) |
| TC24414_at | RNase 4 |
| TC24927_at | Ppp3ca (protein phosphatase 3, catalytic subunit, alpha isoform) |
| TC28512_at | eIF-4B, translation initiation factor homolog |
| TC28860_at | ATPase class 1, member h |
| TC31137_at | ATPase membrane sector-associated protein M8-9 |
| TC31838_s_at | Cryab (crystallin, alpha B) |
| TC37832_at | Gyg1 (glycogenin 1) |
| TC40519_at | Nedd4 (WW domain-binding protein 4) |
| U49351_s_at | Gaa (glucosidase, alpha, acid) |
| X97755_s_at | Ebp (phenylalkylamine Ca2+ antagonist [emopamil] binding protein) |
| Cytoskeleton matrix (5) | |
| L04262_s_at | Lox (lysyl oxidase) |
| L07803_s_at | Thbs2 (thrombospondin 2) |
| Msa.2222.0_s_at | Co14a5 (procollagen, type IV, alpha 5) |
| TC14683_at | Adam17 (a disintegrin and metalloproteinase domain 17) Cetn2 (centrin 2) |
| TC15442_at | Cetn2 (centrin 2) |
| Signal transduction (9) | |
| D86177_s_at | Pipk5b (phosphatidylinositol-4-phosphate 5-kinase, type 1 beta) |
| Msa.2867.0_s_at | Pea15 (phosphoprotein enriched in astrocytes 15) |
| TC22639_at | Capn6 (calpain 6) |
| TC25579_at | Osmr (oncostatin M-specific receptor) |
| TC27402_s_at | Ap3s1 (adapter-related protein complex AP-3, sigma 1 subunit) |
| TC37308_at | Dcamkl1 (double-cortin and calcium/calmodulin-dependent protein kinase-like 1) |
| TC37337_at | CDK-related protein kinase PNQLARE |
| U88909_s_at | Api2 (apoptosis inhibitor 2) |
| Z22649_s_at | Mp1 (myeloproliferative leukemia virus oncogene) |
| Transcriptional regulation (3) | |
| TC29760_at | PLU1 (putative zinc finger DNA binding protein) |
| TC31578_at | ETS-2 |
| TC36768_at | LZTR-1 (leucine zipper-like transcriptional regulator 1) |
| Other (11) | |
| C81270_rc_s_at | Sdfr2 (stromal cell-derived factor receptor 2) |
| Msa.3025.0_s_at | Cp (ceruloplasmin) |
| TC15088_at | Mapbpip (Golgi-associated MP1 adapter protein) |
| TC15484_at | VPS41 (vacuolar assembly protein homolog [human]) |
| TC16885_at | Rnf13 (ring finger protein 13) |
| TC21239_g_at | AP3B1 (adapter-related protein complex AP-3, beta 1 subunit) |
| TC23611_at | SEP5 (Septin 5) |
| TC27740_at | Pepf (pepsinogen F) |
| TC27759_at | Ttc3 (tetratricopeptide repeat domain) |
| TC37400_at | Bicaudal C homolog 1 (Drosophila) |
| X52129_s_at | Tcp11 (t-complex protein 11) |
| Unidentifiable sequences (27) | |
| Total (70) |
Genes with expression levels activated by at least a factor of 2 in NIH 3T3 cells by all three EWS/ETS proteins compared with levels in control (Tk Neo) cells and that have associated identities are displayed (see Materials and Methods). The genes with identities are categorized into six functional groups: cell cycle regulation, cell metabolism, cytoskeleton/matrix, immune related, signal transduction, and transcriptional regulation. Genes without an identity that fell within one of these categories are classified as “other.”
EWS/FLI1 alters cellular metabolism.
Genes involved in cellular metabolism accounted for approximately one-quarter (24 of 90) of all target genes and represented the largest single proportion of the core EWS/ETS-modulated genes with functions that could be categorized. Within this category, there is a cohort of genes with functions associated with carbohydrate metabolism. The genes coding for Glycogenin 1 and glucosidase (Gyg 1, EC 2.4.2.186; and Gaa, EC 3.2.1.3, respectively) are down-regulated by EWS/ETS proteins and function in the biosynthesis of glycogen and the metabolism of glycogen to glucose, respectively (31). Genes up-regulated by EWS/ETS proteins in this category include those coding for phosphoglycerate mutase and uridine phosphorylase. Phosphoglycerate mutase (PGM, EC 5.4.2.4) functions in the oxidation phase of glycolysis, while uridine phosphorylase (Upase, EC 2.3, 2.4) is involved in nucleotide recycling and is linked to carbohydrate metabolism through ribose production involving the pentose phosphate pathway (18). Given the interdependency of carbohydrate metabolic pathways, it seemed likely that transcriptional modulation of genes associated with these various pathways would alter glucose metabolism. To examine this directly, glucose utilization and lactate production were measured in NIH 3T3 populations with and without EWS/FLI1.
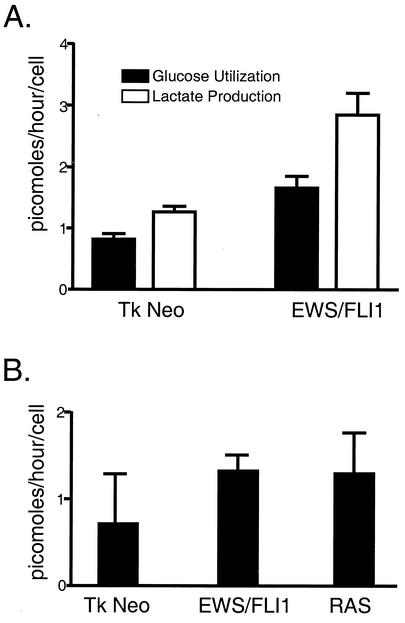
NIH 3T3 cells were transduced with retrovirus containing EWS/FLI1 or empty vector and selected with G418. Expression of EWS/FLI1 was confirmed through immunoblot analysis (data not shown). In order to assess the degree of glucose flux, transduced cell lines were incubated in normal medium containing glucose for 24 h. The medium was then collected, and the cells were harvested and enumerated. Plasma glucose levels were determined by COBAS MIRA analysis. Glucose and lactate levels were determined by comparing the amount of glucose present in the media collected from the cell lines with the amount from a medium-only control that was incubated under the same conditions without cells. Differences in the amounts of glucose present between the medium-only control and the cell lines represent the degree of glucose utilized by the respective cell line. NIH 3T3 cells expressing EWS/FLI1 displayed a twofold increase in glucose utilization and lactate production compared to the level expressed with the empty vector control (Fig. 3A).
FIG. 3.
Analysis of glucose utilization in response to EWS/FLI (A) and in response to EWS/FLI1 and RAS (B). The amount of glucose and lactate utilized is represented as picomoles (of substrate produced or utilized) per hour per cell. These experiments were performed in quadruplicate and are representative of three independent experiments.
To assess the relative specificity of these shifts in metabolism observed in the EWS/FLI-expressing cell lines, glucose utilization experiments were repeated with RAS-transformed NIH 3T3 cells as a positive control. Independent NIH 3T3 populations were transduced with replication-deficient retroviruses containing EWS/FLI1, activated RAS, or empty vector. After antibiotic selection, polyclonal NIH 3T3 cell lines were subjected to glucose utilization studies as described above. Again, expression of EWS/FLI1 boosted glucose utilization twofold (Fig. 3B). In addition, ectopic expression of activated RAS also resulted in a quantitatively similar increase in glucose uptake. Such data suggest that both oncoproteins promote overlapping metabolic effects on cells (see Discussion).
Murine PIM3 is up-regulated by EWS/ETS fusions and expressed in human EFT cell lines.
The second largest category of EWS/ETS target genes consisted of genes the products of which modulate intra- or intercellular signal transduction (19 of 91). Lacking a precise way to globally assess signaling within cells, the significance of our expression data was validated by choosing one candidate gene from within this category and investigating its effects on cellular growth in our model systems. The gene upon which we focused was the gene coding for PIM3, a member of a family of at least three serine/threonine protein kinases in mammals that have orthologs in several species. PIM genes were initially discovered through proviral insertional activation by murine leukemia virus and have been shown to cooperate with c-MYC in murine leukemia (5, 8).
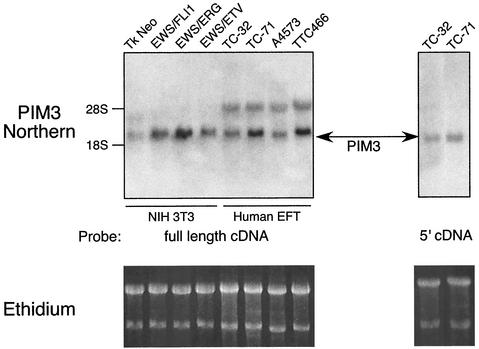
Northern blot experiments were performed to confirm that PIM3 was indeed up-regulated as indicated in our microarray data. Size-fractionated total RNA from polyclonal NIH 3T3 cells transduced with one of the three EWS/ETS fusions was hybridized to a full-length rat PIM3 cDNA probe (95% nucleotide, 93% amino acid identity between murine and rat PIM3) (13). PIM3 was found to be up-regulated by all three EWS/ETS fusions in approximately the same relative proportions found in the microarray data (EWS/ERG > EWS/FLI1 > EWS/ETV) (Fig. 4).
FIG. 4.
Northern blot demonstrating that PIM3 is up-regulated by all three of the EWS/ETS proteins in NIH 3T3 cells and expressed in Ewing's tumor cell lines, including three EWS/FLI1-derived tumors (TC32, TC71, and A4573) and one EWS/ERG tumor (TTC466). In the left panel, full-length cDNA to PIM3 hybridizes to two transcripts in the human tumor samples. Hybridization of a probe created from the 5′ portion of PIM3 (a sequence unique to this PIM family member) to RNA from TC32 and TC71 cells displays specific binding to the smaller transcript. Ethidium staining of the RNA prior to transfer is shown as loading controls.
In a similar fashion, we assessed whether PIM3 was expressed in human tumor-derived EFT cell lines. Three of these cell lines were derived from EFTs harboring an EWS/FLI1 fusion (TC32, TC71, and A4573), and one carried an EWS/ERG fusion (TTC466). A PIM3 transcript of the same size and level as that seen in EWS/ETS-transduced NIH 3T3 cell lines was detected in all four human EFT cell lines (Fig. 4). In addition, a larger PIM transcript was also detected in human cells. Nucleotide sequence comparison between murine and human PIM genes revealed that the region of greatest divergence among PIMs mapped to the 5′ 120 bp. Northern blots probed with this 5′-PIM3 cDNA segment demonstrated only the smaller PIM3 transcript, which was comparable to that seen in the murine NIH 3T3 model system. These results indicate that the smaller transcript corresponds to human PIM3 and suggest that additional PIM genes may also be expressed in human EFT cell lines.
Ectopic expression of PIM3 promotes anchorage-independent growth.
The observation that PIM genes could promote oncogenesis in other model systems suggested that PIM3 could potentially be playing similar roles in EWS/ETS-mediated cellular transformation. Loss of anchorage dependence for cell growth is a hallmark of the transformed phenotype in many cell systems. As an initial estimate of its oncogenic potential, the effects of isolated forced expression of PIM3 on NIH 3T3 anchorage-independent cell growth was assessed by plating cells in soft agar.
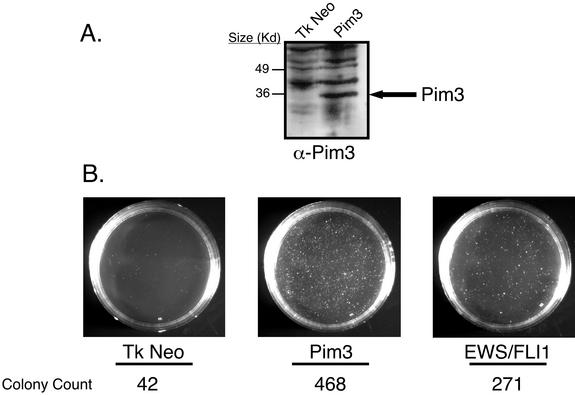
We cloned full-length rat PIM3 cDNA into retroviral vectors and transduced them into NIH 3T3 cells by using replication-deficient retroviruses (13). As positive and negative controls, respectively, separate populations of NIH 3T3 cells transduced with either EWS/FLI1 or empty vector were also prepared. After a short period of antibiotic selection, polyclonal populations were plated in soft agar. Immunoblots using rabbit antisera raised to full-length PIM3 confirmed stable expression of PIM3 in transduced cells (Fig. 5A). Northern analysis revealed an approximately 20-fold increase in PIM3 transcript levels in ectopic retroviral expression, when compared to the level of endogenous induction by EWS/FLI1 (data not shown). Forced expression of PIM3 led to a dramatic increase in macroscopic colony formation on the order of that seen with EWS/FLI1 (Fig. 5B). This entire experiment was repeated two additional times, and in all cases, the results were the same, indicating that enforced expression of PIM3 renders NIH 3T3 cells anchorage independent.
FIG. 5.
(A) Immunoblot analysis displaying ectopic expression of PIM3 in NIH 3T3 cells. This blot is representative of the levels of PIM3 expression in the cells used in the agar and tumorigenesis assays. (B) Agar assays of NIH 3T3 cells expressing PIM3, EWS/FLI1, and empty vector. Images were recorded at day 21 and are representative of three independent experiments. Colony numbers represent the number of macroscopic colonies present on each plate at day 21. EWS/FLI1 and the empty vector serve as controls to measure the capacity of PIM3 to support anchorage-independent growth. EWS/FLI1 is able to promote colony formation, whereas the empty vector does not.
Anchorage-independent growth reflects only one aspect of the oncogenic phenotype. Tumorigenesis assays assess oncogenic potency in a more physiologic in vivo setting. Therefore, experiments were performed to determine the ability of PIM3 to accelerate NIH 3T3 tumorigenesis. The same polyclonal NIH 3T3 cell populations that had been transduced with PIM3, EWS/FLI1, or empty vector and used for agar assays were injected subcutaneously into SCID mice, and the time to formation of 1.5-cm-diameter tumors was measured. In two independent experiments, PIM3-expressing NIH 3T3 cells failed to accelerate NIH 3T3 tumorigenesis over the background rate seen with empty vector controls (data not shown).
A kinase-deficient PIM3 mutant inhibits EWS/FLI1-mediated tumorigenesis.
Although forced expression of PIM3 was able to promote some elements of cellular transformation, it was insufficient to recapitulate the tumorigenic effects seen with expression of EWS/FLI1. This prompted the question of whether PIM3 was necessary for EWS/FLI1-mediated tumorigenesis. In order to address this question, a kinase-deficient PIM3 mutant was constructed as a potential inhibitor of normal PIM3. Other groups had previously shown that similar kinase-defective PIM1 mutants behave in a dominant-negative fashion to antagonize normal PIM1 (24). Based on primary amino acid comparison of PIM1 versus PIM3, we made an analogous lysine-to-methionine mutation at position 69 of PIM3 by PCR-based site-directed mutagenesis.
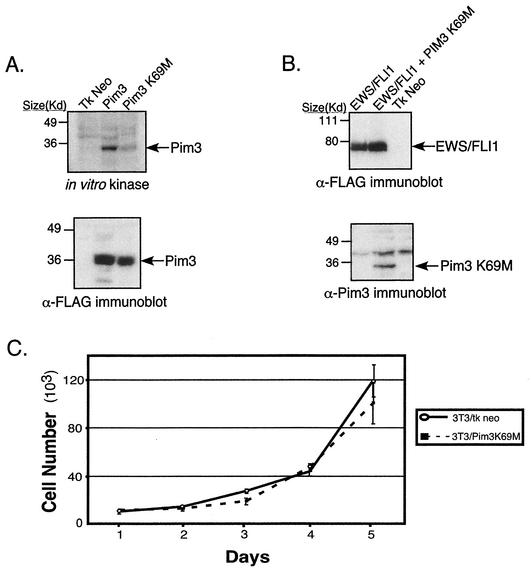
In vitro kinase assays were performed in order to assess the ability of PIM3 K69M to phosphorylate substrate relative to wild-type PIM3. Expression constructs containing either of these two genes were transiently transfected into 293T cells. After 48 h, cells were harvested, and total protein lysates were immunoprecipitated with anti-PIM3 antisera. Kinase assays were performed with immunoprecipitated lysates and demonstrated that the ability of the PIM3 K69M mutant to phosphorylate itself (Fig. 6A) or histone H1 (data not shown) was significantly attenuated, although not entirely absent compared to that of normal PIM3. As an initial test of the effects of this mutant, the growth rates in tissue culture of NIH 3T3 cells expressing PIM3 K69M and empty vector were compared. In two independent experiments, the growth rates of these two populations were not significantly different (Fig. 6C), indicating this dominant-negative PIM3 mutant does not have a direct effect on cellular proliferation.
FIG. 6.
(A) In vitro kinase assay demonstrating that the PIM3 K69M mutant has attenuated intrinsic kinase activity. The lysates used on the immunoblot represent an equal proportion of product immunoprecipitated from 293T cells transfected with either wild-type PIM3 or PIM3 K69M. The amounts of PIM3 protein present in the immunoprecipitated product are equal between the wild-type and K69M mutant samples. (B) Immunoblot analysis demonstrating expression of EWS/FLI1 and PIM3 K69M in the NIH 3T3 cell lines utilized in the tumorigenesis assay. (C) Growth curves of NIH 3T3 cells expressing PIM3 K69M or empty vector. The graph is representative of two independent experiments performed in quadruplicate.
Since PIM3 K69M demonstrated a biochemical decrease in kinase, we then tested whether its expression in cells would antagonize EWS/FLI1-mediated tumorigenesis by inhibiting normal PIM3. NIH 3T3 cells were first transduced with recombinant retroviruses containing either epitope-tagged (Flag) EWS/FLI1 or empty vector. After a short period of short antibiotic selection, EWS/FLI1-expressing cells were split, and one-half were transduced with a second retrovirus containing a GFP-IRES-PIM3 K69M expression cassette. Five days postinfection, these cells were sorted for cells displaying the top 25% of GFP fluorescence intensity. Immunoblot analyses were performed on lysates from resultant cell populations to assess protein expression of transduced genes (Fig. 6B). Mutant PIM3 protein was detected in the GFP-IRES-PIM3 K69M-transduced population. Moreover, there were equal levels of EWS/FLI1 expression in cells transduced with both EWS/FLI1 and mutant PIM3 compared to the level in cells that received only EWS/FLI1.
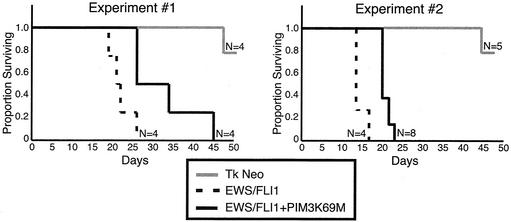
Tumorigenic potentials of transduced NIH 3T3 populations were assessed by a SCID mouse assay. Equal numbers of cells from each polyclonal cell line were injected subcutaneously, and the time to form 1.5-cm tumors was determined. In the first experimental set, the EWS/FLI1 population formed 1.5-cm tumors in an average of 22 days (standard error [SE], 3 days), whereas the EWS/FLI1-PIM3 K69M-expressing cells formed 1.5-cm tumors in an average of 32.75 days (SE, 9 days), representing a 46% (SE, 21%; P < 0.04) difference in the mean time to tumor formation between these cell lines (Fig. 7 and Table 2). In the repeat experiment (experiment 2), cell lines were independently reproduced, and the results showed a similar trend, with the EWS/FLI1-PIM3 K69M cell line showing a 38% (SE, 6%; P < 0.001) increase in tumor generation time compared to the EWS/FLI1 cell line. There is some variability in the mean time to tumor formation between these sets of experiments (i.e., EWS/FLI1 set 1 mean time of 22 days and EWS/FLI1 set 2 mean time of 14.5 days); however, the relative differences within each experimental set are concordant and statistically significant (Table 2).
FIG. 7.
Survival curves of mice injected subcutaneously with NIH 3T3 cells expressing EWS/FLI1, EWS/FLI1-PIM3 K69M, and empty vector. Mice were sacrificed when tumors reached a diameter of 1.5 cm; thus, the proportion of mice surviving on a given day is representative of the number of mice without a 1.5-cm tumor.
TABLE 2.
Summary of results from the mouse tumorigenic assays
| Cell line | No. with tumor induction/ no. injected | Mean (SE) days to inductiona | % Difference (SE)b | P |
|---|---|---|---|---|
| Expt 1 | ||||
| EWS/FLI1-PIM3 K69M | 4/4 | 32.75 (9) | 46 (21) | 0.04 |
| EWS/FLI1 | 4/4 | 22 (3) | ||
| Tk Neo | 2/4 | 55 (7) | ||
| Expt 2 | ||||
| EWS/FLI1-PIM3 K69M | 8/8 | 20 (1.5) | 38 (6) | 0.001 |
| EWS/FLI1 | 4/4 | 14.5 (1) | ||
| Tk Neo | 3/5 | 48 (3) |
The mean time of tumor formation is the average number of days it took a given cell line to form a 1.5-cm-diameter tumor in a SCID mouse. Data from mice that did not form tumors 80 days postinjection are not included in the average rate of tumor formation.
The average percentage of difference represents the difference in the mean time to tumor formation between the EWS/FLI1 and EWS/FLI1-PIM3K69M cell lines; the empty vector control was not included in this analysis. The percentages of increase in the geometric mean time to tumor formation between the EWS/FLI1 and EWS/FLI1-PIM3 K69M cell lines are shown for both experiments. These increases are significantly different from 0 (P < 0.05 by two-tailed test).
DISCUSSION
In an effort to identify target genes commonly regulated by EWS/ETS proteins, we performed microarray analysis with NIH 3T3 cells ectopically expressing EWS/ERG, EWS/ETV1, or EWS/FLI1. Despite their high degree of structural similarity, EWS/ERG and EWS/FLI1 share only approximately one-third of their target genes in common. To a certain extent, part of this difference could be due to variation in experimental conditions and/or microarray hybridizations. In an effort to dampen these effects, each microarray experiment was performed with pooled RNAs from three independent EWS/ETS transductions. Finally, EWS/ETS core target genes were selected from those genes that were transcriptionally modulated in a similar fashion by all three fusions. Our extensive follow-up Northern analysis suggests a low false-positive rate in this target gene set. A caveat to this approach is that due to the limited accessibility of Affymetrix reagents at the time these experiments were performed, we were unable to perform replicate experiments, potentially leading us to underestimate the target gene repertoire. Because we chose to err on the side of stringency, it seems likely that a smaller subset of shared EWS/ETS target genes may have been missed. For example, the gene manic fringe, which has been shown previously to be up-regulated by all three EWS/ETS fusions, did not make our gene list, primarily because of its known weak up-regulation by EWS/ETV. Repeats of these microarray experiments with additional independently derived EWS/ETS-transduced populations and more densely packed microarrays could potentially refine our results further.
EWS/ETV was notably weaker in transcriptional modulation of target genes than either EWS/FLI1 or EWS/ERG. This would suggest that the EWS/ETS fusions in group 2 might be biologically less potent than those in group 1. In fact, EWS/ETV is unable to promote anchorage-independent growth and accelerates tumorigenesis in NIH 3T3 cells less effectively than EWS/FLI1 (30). However, from a clinical standpoint, there is no apparent difference in survival in EFT patients expressing different EWS/ETS fusions (15). It should be noted, however, that these comparisons primarily involve EWS/FLI1 and EWS/ERG. Group 2 fusions occur so infrequently that derivation of accurate patient survival statistics is difficult given the small number of patients.
The net result of our strategy was identification of a core set of shared EWS/ETS response genes that is considerably smaller than the repertoires of target genes from any individual EWS/ETS gene. For example, in NIH 3T3 cells, EWS/FLI1 was found to up-regulate 934 of the approximately 25,000 genes cumulatively represented on the five microarray types used in our experiments. Given the increase in the number of genes interrogated, this estimate is concordant with recent microarray expression findings of other investigators using a TET-regulated EWS/FLI1 construct in primary human fibroblasts (20). Cross-referencing with two other EWS/ETS fusions reduces the number of target genes dramatically to less than 140 and permits more focused experiments to assess the biologic significance of specific target gene networks.
Comparison of our findings with those of other groups has proven difficult for both biological and technical reasons. The primary human fibroblast system developed by Lessnick et al. (20) is subjected to p53-dependent arrest upon EWS/FLI1induction, whereas our NIH 3T3 system supports EWS/FLI1 oncogenesis. Such differences in the biological response to EWS/FLI1 expression likely lead to differences in the target gene modulation. Moreover, there are limited gene annotation resources between the Affymetrix human U95A2 arrays utilized by these investigators and the murine Affymetrix 11K and 19K arrays we employed in these studies. Comparison with our previous RDA findings has proven more useful in validating our findings. Of the genes present on the array sets interrogated, the stromelysin, E2-C, and manic fringe genes are all concordantly modulated in these microarray studies, whereas the EAT-2 gene is not present on our arrays.
Within the core set of EWS/ETS-modulated genes, subgroups begin to emerge that suggest functional patterns. For example, consistent with previous findings, EWS/ETS fusions up-regulate cyclin D1, which could facilitate passage through the G1/S cell cycle checkpoint. Our data also demonstrate coordinate up-regulation of polo-like kinase 1 (Plk1), cdc25C phosphatase, the ubiquitin conjugase E2-C, and p55cdc, which are all involved in regulating the G2/M transition (4, 19, 27). These results suggest that EWS/ETS fusions can accelerate passage through cell cycle by deregulating both major cell cycle checkpoints.
Although subsets of EWS/ETS target genes can be clustered by putative function, the net result of transcriptional modulation of these genes on the cell can be hard to predict. For example, down-regulation of glycogenin 1 and glucosidase would suggest a shift away from gluconeogenesis. Up-regulation of phosphoglycerate mutase would favor an increase in glycolysis and aerobic glucose utilization. The effects of up-regulation of uridine phosphorylase are hard to anticipate, since this enzyme is involved in both nucleotide scavenging and glucose metabolism through the pentose phosphate pathway. Direct measurement of glucose flux was necessary to show that the end result of these genetic modulations was a twofold increase in glucose utilization and lactate production in EWS/FLI1-expressing cells as compared to parental NIH 3T3 cells. This suggests that even having a core set of commonly modulated EWS/ETS target genes in hand, the physiologic relevance of individual genes and pathways will still need to be validated by independent experiments.
The increase in glucose uptake likely coincides with the increased proliferation rate of EWS/FLI1- and RAS-expressing cells. It may be that these shifts in glucose metabolism are a mechanism utilized by the oncogenes coding for these oncoproteins to facilitate cellular transformation. Such a model is supported by the fact that RAS upregulates both uridine phosphorylase and phosphoglycerate mutase (data not shown). Alternatively, it is possible that such shifts are a cellular response to the mitogenic signals propagated by these oncogenes. Future studies are aimed at delineating the relationships between oncogene expression and alterations in global cellular metabolic profiles.
PIM3 is an EWS/ETS target gene that appears to play an active role in EWS/ETS-mediated oncogenesis. PIM3 is the third member of a family of mammalian protein kinases that were initially discovered through their ability to potentiate the oncogenic effects of c-MYC in a murine lymphoma model (5, 8). Like PIM1 and -2, retroviral integration 5′ to PIM3 leads to transcriptional up-regulation of PIM3 and a decrease in tumor latency in mice expressing a c-MYC transgene. PIM3 was independently discovered in a screen for genes transcriptionally up-regulated by ionic depolarization in the rat neuronal PC12 cell line (13). Subsequent to this, changes in both PIM1 and PIM3 have been linked to brain synaptic activity in rodents (12).
While the biology of PIM3 remains largely undefined, there is evidence suggesting involvement of PIM1 in cell cycle regulation. PIM1 is thought to be a mediator in gp130-mediated cell cycle progression in murine pro B cells (14, 28). Furthermore, the G1/S regulator cdc25A appears to be activated through phosphorylation by PIM1 both in vitro and in vivo (23). Our initial experiments have failed to show a similar interaction between PIM3 and cdc25A (data not shown). The roles that PIM3 may be playing in EWS/ETS-transformed cells will be a subject of future study.
Although forced expression of PIM3 promotes anchorage-independent growth, it does not accelerate tumorigenesis as do all EWS/ETS fusions. In addition, levels of ectopic PIM3 expression are likely to mediate biological effects that would be less apparent at significantly lower levels. However, when these data are coupled with the antitumorigenic effects of dominant-negative PIM3, it seems likely that PIM3 is actively contributing to EWS/FLI1 oncogenesis. Specifically, these data suggest that this protein kinase promotes but is not sufficient for EWS/ETS-mediated tumorigenesis. The partial inhibition of EWS/FLI1 tumorigenesis by PIM3 K69M could be due to the fact that this mutant's kinase activity is reduced, although not absent. It also suggests that while PIM3 is a likely mediator of EWS/ETS tumorigenesis, it cannot account for the full effect. These findings coupled with those described for the proteins Manic Fringe and PDGF-C further reinforce the concept that individual target genes are only able to partially recapitulate EWS/ETS biology (22, 36). It appears more and more likely that the final cellular phenotype is dictated by the summed effects of a network of modulated genes.
Acknowledgments
We thank Harvey Herschman for providing the anti-KID1 (PIM3) antibodies and cDNA, Elliot Landaw for insight and assistance with statistical analysis, and Iris Williams for assistance with the FACS analysis and sorting.
This work was supported by NCI grant CA87771 from the National Institutes of Health.
B.D. and S.M.W. contributed equally to this work.
REFERENCES
- 1.Arvand, A., and C. T. Denny. 2001. Biology of EWS/ETS fusions in Ewings family tumors. Oncogene 20:5747-5754. [DOI] [PubMed] [Google Scholar]
- 2.Arvand, A., H. Bastians, S. M. Welford, A. D. Thompson, J. V. Ruderman, and C. T. Denny. 1998. EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 17:2039-2045. [DOI] [PubMed] [Google Scholar]
- 3.Attwooll, C., M. Tariq, M. Harris, J. D. Coyne, N. Telford, and J. M. Varley. 1999. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene 18:7599-7601. [DOI] [PubMed] [Google Scholar]
- 4.Bastians, H., L. M. Topper, G. L. Gorbsky, and J. V. Ruderman. 1999. Cell cycle-regulated proteolysis of mitotic target proteins. Mol. Biol. Cell 10:3927-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns, A., H. Mikkers, P. Krimpenfort, J. Allen, B. Scheijen, and J. Jonkers. 1999. Identification and characterization of collaborating oncogenes in compound mutant mice. Cancer Res. 59:1773-1777. [PubMed] [Google Scholar]
- 6.Bertolotti, A., Y. Lutz, D. J. Heard, P. Chambon, and L. Tora. 1996. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15:5022-5031. [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, B. S., R. Frieden, S. L. Lessnick, W. A. May, and C. T. Denny. 1995. Identification of target genes for the Ewing's sarcoma EWS/FLI fusion protein by representational difference analysis. Mol. Cell. Biol. 15:4623-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breuer, M., R. Slebos, S. Verbeek, M. van Lohuizen, E. Weintjens, and A. Berns. 1989. Very high frequency of lymphoma induction by a chemical carcinogen in pim-1 transgenic mice. Nature 340:61-63. [DOI] [PubMed] [Google Scholar]
- 9.Calvio, C., G. Neubauer, M. Mann, and A. I. Lamond. 1995. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA 1:724-733. [PMC free article] [PubMed] [Google Scholar]
- 10.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Alava, E., and W. L. Gerald. 2000. Molecular biology of the Ewing's sarcoma/primitive neuroectodermal tumor family. J. Clin. Oncol. 18:204-213. [DOI] [PubMed] [Google Scholar]
- 12.Feldman, J. D., L. Vician, M. Crispino, G. Tocco, M. Baudry, and H. R. Herschman. 1998. Seizure activity induces PIM-1 expression in brain. J. Neurosci. Res. 53:502-509. [DOI] [PubMed] [Google Scholar]
- 13.Feldman, J. D., L. Vician, M. Crispino, G. Tocco, V. L. Marcheselli, N. G. Bazan, M. Baudry, and H. R. Herschman. 1998. KID-1, a protein kinase induced by depolarization in brain. J. Biol. Chem. 273:16535-16543. [DOI] [PubMed] [Google Scholar]
- 14.Fukada, T., Y. Yoshida, K. Nishida, T. Ohtani, T. Shirogane, M. Hibi, and T. Hirano. 1999. Signaling through Gp130: toward a general scenario of cytokine action. Growth Factors 17:81-91. [DOI] [PubMed] [Google Scholar]
- 15.Ginsberg, J. P., E. de Alava, M. Ladanyi, L. H. Wexler, H. Kovar, M. Paulussen, A. Zoubek, B. Dockhorn-Dworniczak, H. Juergens, J. S. Wunder, I. L. Andrulis, R. Malik, P. H. Sorensen, R. B. Womer, and F. G. Barr. 1999. EWS-FLI1 and EWS-ERG gene fusions are associated with similar clinical phenotypes in Ewing's sarcoma. J. Clin. Oncol. 17:112-123. [DOI] [PubMed] [Google Scholar]
- 16.Graves, B. J., and J. M. Petersen. 1998. Specificity within the ets family of transcription factors. Adv. Cancer Res. 75:1-55. [DOI] [PubMed] [Google Scholar]
- 17.Ichikawa, H., K. Shimizu, Y. Hayashi, and M. Ohki. 1994. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 54:2865-2868. [PubMed] [Google Scholar]
- 18.Ishitsuka, H., M. M. Takemoto, K. Kukouka, A. Itoga, and H. B. Marruyama. 1980. Role of uridine phosphorylase for antitumor activity of 5′-deoxy-5-fluorouridine. Jpn. J. Cancer Res. 71:112-123. [PubMed] [Google Scholar]
- 19.Kallio, M., J. Weinstein, J. R. Daum, D. J. Burke, and G. J. Gorbsky. 1998. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 141:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessnick, S., C. S. Dacwag, and T. R. Golub. 2002. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 4:393-401. [DOI] [PubMed] [Google Scholar]
- 21.Mastrangelo, T., P. Modena, S. Tornielli, F. Bullrich, M. A. Testi, A. Mezzelani, P. Radice, A. Azzarelli, S. Pilotti, C. M. Croce, M. A. Pierotti, and G. Sozzi. 2000. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene 19:3799-3804. [DOI] [PubMed] [Google Scholar]
- 22.May, W. A., A. Arvand, A. D. Thompson, B. S. Braun, M. Wright, and C. T. Denny. 1997. EWS/FLI1-induced manic fringe renders NIH 3T3 cells tumorigenic. Nat. Genet. 17:495-497. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki, T., C. Kitanaka, K. Noguchi, T. Muramatsu, A. Asai, and Y. Kuchino. 1999. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J. Biol. Chem. 274:18659-18666. [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki, T., K. C. Noguchi, A. Sugiyama, S. Kagaya, S. Chi, A. Asai, and Y. Kuchino. 1997. Pim-1 kinase stimulates c-Myc mediated death signaling upstream of caspase 3 (CPP32)-like protease activation. Oncogene 15:1471-1480. [DOI] [PubMed] [Google Scholar]
- 25.Panagopoulos, I., M. Hoglund, F. Mertens, N. Mandahl, F. Mitelman, and P. Aman. 1996. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene 12:489-494. [PubMed] [Google Scholar]
- 26.Panagopoulos, I., M. Mencinger, C. U. Dietrich, B. Bjerkehagen, G. Saeter, F. Mertens, N. Mandahl, and S. Heim. 1999. Fusion of the RBP56 and CHN genes in extraskeletal myxoid chondrosarcomas with translocation. Oncogene 18:7594-7598. [DOI] [PubMed] [Google Scholar]
- 27.Roshak, A. K., E. A. Capper, C. Imburgia, J. Fornwald, G. Scott, and L. A. Marshall. 2000. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell Signal. 12:405-411. [DOI] [PubMed] [Google Scholar]
- 28.Shirogane, T., T. Fukada, J. M. Muller, D. T. Shima, M. Hibi, and T. Hirano. 1999. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11:709-719. [DOI] [PubMed] [Google Scholar]
- 29.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, S. Kitareewan, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, A. D., M. A. Teitell, A. Arvand, and C. T. Denny. 1999. Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumorigenic pheno-type on NIH3T3 cells. Oncogene 18:5506-5513. [DOI] [PubMed] [Google Scholar]
- 31.Van Maanen, M., P. A. Fournier, T. N. Palmer, and L. J. Abraham. 1999. Characterization of mouse glycogenin-1 cDNA and promoter region. Biochim. Biophys. Acta 1447:284-290. [DOI] [PubMed] [Google Scholar]
- 32.Weighardt, F., G. Biamonti, and S. Riva. 1996. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. BioEssays 18:747-756. [DOI] [PubMed] [Google Scholar]
- 33.Welford, S. M., S. P. Hebert, B. Deneen, A. Arvand, and C. T. Denny. 2001. DNA binding domain-independent pathways are involved in EWS/FLI1 mediated oncogenesis. J. Biol. Chem. 276:41977-41984. [DOI] [PubMed] [Google Scholar]
- 34.Yang, L., L. J. Embree, S. Tsai, and D. D. Hickstein. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 273:27761-27764. [DOI] [PubMed] [Google Scholar]
- 35.Zinszner, H., J. Sok, D. Immanuel, Y. Yin, and D. Ron. 1997. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell Sci. 110:1741-1750. [DOI] [PubMed] [Google Scholar]
- 36.Zwerner, J. A., and W. A. May. 2001. PDGF-C is an EWS/FLI induced transforming growth factor in Ewing's family of tumors. Oncogene 20:626-633. [DOI] [PMC free article] [PubMed] [Google Scholar]