Abstract
Members of the Snf1/AMP-activated protein kinase family are activated under conditions of nutrient stress by a distinct upstream kinase. Here we present evidence that the yeast Pak1 kinase functions as a Snf1-activating kinase. Pak1 associates with the Snf1 kinase in vivo, and the association is greatly enhanced under glucose-limiting conditions when Snf1 is active. Snf1 kinase complexes isolated from pak1Δ mutant strains show reduced specific activity in vitro, and affinity-purified Pak1 kinase is able to activate the Snf1-dependent phosphorylation of Mig1 in vitro. Purified Pak1 kinase promotes the phosphorylation of the Snf1 polypeptide on threonine 210 within the activation loop in vitro, and an increased dosage of the PAK1 gene causes increased Snf1 threonine 210 phosphorylation in vivo. Deletion of the PAK1 gene does not produce a Snf phenotype, suggesting that one or more additional protein kinases is able to activate Snf1 in vivo. However, deletion of the PAK1 gene suppresses many of the phenotypes associated with the deletion of the REG1 gene, providing genetic evidence that Pak1 activates Snf1 in vivo. The closest mammalian homologue of yeast Pak1 kinase, calcium-calmodulin-dependent protein kinase kinase beta, may play a similar role in mammalian nutrient stress signaling.
The Snf1 kinase of budding yeast and the mammalian AMP-activated protein kinase (AMPK) play critical roles in signaling nutrient stress (9). When activated, Snf1 and AMPK down-regulate metabolic pathways that consume ATP and stimulate pathways that promote ATP synthesis. Nutrient-sensing pathways, in particular, glucose-sensing pathways, are critical signaling pathways that are deregulated in type 2 diabetes. Recent studies of AMPK underscore its importance as a metabolic switch and suggest that activation of AMPK may provide a therapeutic benefit for patients with type 2 diabetes (38). The budding yeast Snf1 kinase and mammalian AMPK are orthologous proteins, and they play similar roles in controlling cellular metabolism. Snf1 kinase is needed for the proper response to nutrient stress conditions, such as growth on alternative carbon sources and sporulation (2). A full understanding of the Snf1/AMPK signaling pathways will first require identification of all of the components of the pathway. While several downstream targets of Snf1 and AMPK are both known and conserved between species (39), much less is known about components acting upstream of Snf1 and AMPK.
Biochemical and genetic experiments have shown that members of the Snf1/AMP-activated protein kinase family are regulated by phosphorylation of the conserved threonine residue in the kinase activation loop. This mechanism for controlling protein kinase activity is used both by members of the serine threonine protein kinase family and by members of the tyrosine protein kinase family. Biochemical fractionation of rat liver has shown that threonine 172 of the mammalian AMPK enzyme is phosphorylated by a distinct protein kinase called AMPK kinase (AMPKK) (11). In budding yeast, the analogous threonine is located at position 210. Carlson and coworkers first showed that replacement of this residue with alanine inactivates Snf1 (5), suggesting that phosphorylation of Snf1 threonine 210 is required for Snf1 activation. Later, Hardie and colleagues demonstrated that Snf1 is regulated by phosphorylation of its activation loop threonine (37). Despite intensive study, the identity of the Snf1-activating kinase has not been determined. One possible explanation for this is that the Snf1 kinase catalyzes the autophosphorylation of threonine 210. An autophosphorylation event would explain the failure to identify a Snf1-activating kinase in genetic screens. More recently, we developed a phosphopeptide antibody that is specific for the Snf1 protein that has been phosphorylated on threonine 210 (21). By using this antibody, we have shown that phosphorylation of Snf1 threonine 210 correlates with glucose stress and Snf1 kinase activation. Further, phosphorylation of threonine 210 occurs normally in cells lacking a functional Snf1 kinase. Therefore, Snf1 kinase must be activated by an upstream kinase.
Recently, genomic studies of yeast protein complexes have used mass spectrometry to identify proteins in hundreds of affinity-purified complexes (6, 14). Affinity purification of protein complexes with tagged versions of the Snf1 and Snf4 proteins led to the identification of several other proteins that may be present in a complex with Snf1 kinase. Not surprisingly, the beta and gamma subunits of Snf1 kinase (Snf4, Sip1, Sip2, and Gal83) were identified as members of the Snf1 complex. More interesting was the finding that two protein kinases, Pak1 and Cka1, are also associated with either Snf1 or Snf4 (6). PAK1 (polymerase alpha kinase 1) was isolated in a screen for multicopy suppressors of a temperature-sensitive mutation in CDC17, the gene encoding DNA polymerase alpha (15), however, a direct connection between Pak1 and DNA polymerase alpha was not demonstrated. Pak1 is likely to be a Snf1 kinase complex component since it was found independently in complexes isolated with both Snf1 and Snf4 as the tagged proteins (6) and since Pak1 was not found in association with any other proteins. The gene for protein kinase Cka1 is one of two yeast genes that encode orthologues of mammalian casein kinase II. The significance of the purported interaction between Cka1 and the Snf1 kinase complex is questionable since Cka1 was found to be present in several complexes of disparate functions (6, 14). Finally, analysis of proteins associated with protein kinase Tos3, a close homologue of Pak1, identified several proteins, including Snf4, the gamma subunit of the Snf1 kinase (14). The significance of this interaction is also questionable since no other Snf1 kinase components were detected in association with Tos3. In this study, we investigated the possibility that Pak1, Tos3, or Cka1 directly activates Snf1 by phosphorylation of the threonine residue in the Snf1 activation loop.
We focused on the Pak1 protein kinase since it was found in the Snf1 complex with both Snf1 and Snf4 as the tagged proteins. Furthermore, the closest mammalian homologue of Pak1 is calcium/calmodulin-dependent protein kinase kinase beta (CaMKK-β), a kinase known to phosphorylate and activate other protein kinases (1). The closest mammalian homologue of Tos3 is CaMKK-α. Pak1 and Tos3 are members of the calcium/calmodulin-dependent kinase family, which includes the Cmk1 and Snf1 kinases (16). The name PAK1 can be somewhat confusing since a distinct and much larger body of literature uses the term PAK to refer to p21-activated kinases. Several mammalian kinases in the p21-activated kinase family go by the name PAK1, as does a PAK-encoding gene in Schizosaccharomyces pombe (17, 34). To make matters even more confusing, another yeast kinase gene, PRK1 (YIL095w), was once annotated as PAK1 (32). The net result of the presence of multiple names for the same genes and different genes with the same name is that the many databases contain incorrect annotations for these genes. Lest there be any confusion, this report concerns the product of Saccharomyces cerevisiae gene PAK1 encoded by open reading frame YER129w.
MATERIALS AND METHODS
Strains and growth conditions.
Growth of yeast was done with standard medium at 30°C (26). Glucose was present at 2 or 0.05% (grams per 100 ml), as indicated. Medium containing 2-deoxyglucose contained yeast extract (10 g/liter), peptone (20 g/liter), sucrose (20 g/liter), and 2-deoxyglucose (0.2 g/liter). The S. cerevisiae strains used in this study were MSY182 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200), FY1193 (MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 snf1Δ10), MSY608 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 kin1::KAN), MSY678 (MATα ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 pak1::KAN), MSY809 [MATa ura3(52/Δ0) leu2(Δ1/Δ0) his3(Δ1/Δ200) trp1Δ63 met15Δ0 snf1Δ10 pak1::KAN], MSY839 [MATα ura3(52Δ0) leu2Δ0 lys2Δ0 trp1Δ63 reg1Δ::URA3 pak1Δ::KAN], PY102 (MATa his9-917Δ lys2 leu2Δ1 ura3-52 trp1Δ63 reg1Δ::URA3), and PY450 (MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 reg1Δ::URA3 snf1Δ10).
Plasmids.
The PAK1 gene was cloned from a plasmid library of yeast genomic DNA by screening pools of bacterial clones by PCR (28). A 5.7-kb fragment (MunI-EagI) encompassing the entire PAK1 gene was subcloned into pRS424 (29) and designated pPAK1-424. The tandem affinity purification tag (25) was amplified by PCR and introduced into the 3′ end of the PAK1 gene in pPAK1-424 by homologous recombination. Pak1 was tagged at the C terminus with three copies of the myc epitope with plasmid pYM3 (36), PCR amplification, and homologous recombination. Clones of yeast cells expressing GST-Cka1 and GST-Tos3 (19) were isolated from pools of transformants by PCR prior to use. A plasmid expressing glutathione S-transferase (GST)-Mig1 in Escherichia coli has been described previously (22).
Western blotting.
Tagged proteins were detected by Western blotting under conditions described previously (21). Mouse monoclonal antibodies against the hemagglutinin (HA; F7) and myc (9E10) epitopes were purchased from Santa Cruz Biotechnology. Antibodies against Snf1 protein phosphorylated on threonine 210 have been described previously (21). Antibodies were incubated with membranes for 2 h at a dilution of 1/1,000, except for the myc antibody, which was used at a dilution of 1/250 and incubated with membranes overnight.
Immunoprecipitation.
Protein extracts were prepared from snf1Δ10 pak1Δ (MSY809) cells transformed with empty plasmid vectors or with plasmids expressing Snf1-3HA or Pak1-3myc. Snf1 complexes were collected from 800 μg of total protein by incubation with 2.5 μl of undiluted monoclonal antibodies against HA for 1.5 h at 4°C and then incubated with 20 μl of protein A beads (Sigma). The beads were washed three times with radioimmunoprecipitation assay buffer, and bound proteins were eluted by incubation in sodium dodecyl sulfate (SDS) sample buffer at 95°C.
In vitro kinase assays.
Protein kinase assay mixtures contained 0.2 mM [γ-32P]ATP (1,000 cpm/pmol) in 20 μl of kinase buffer (20 mM HEPES [pH 7.0], 0.5 mM EDTA, 0.5 mM dithiothreitol, 5 mM Mg acetate) and were incubated at 30°C for 20 min. Substrate proteins (GST-Mig1) were present at approximately 10 μg/ml. Snf1 kinase was added to autophosphorylation reaction mixtures at a final concentration of approximately 25 ng/ml. When GST-Mig1 was present as the phosphate acceptor, Snf1 kinase was added to a final concentration of 2.5 ng/ml. Proteins were precipitated on ice with 200 μl of 10% trichloroacetic acid, washed in ice-cold acetone, and resolved by SDS-polyacrylamide gel electrophoresis. Incorporation of 32P was detected by autoradiography of dried gels.
Protein purification.
Snf1 kinase complexes were isolated from cultures grown on sucrose medium by tandem affinity purification (25), followed by Mono Q chromatography as previously described (22). Pak1 kinase was purified by tandem affinity purification (25). GST-Cka1 and GST-Tos3 were purified by glutathione agarose chromatography (24).
RESULTS
Pak1-dependent phosphorylation of Snf1protein.
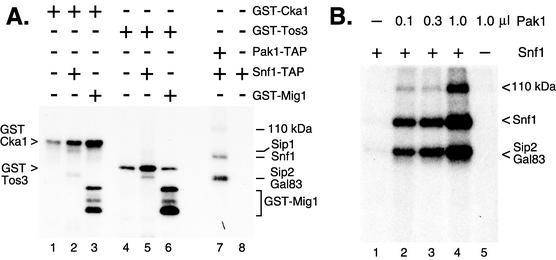
Three protein kinases, Pak1, Cka1, and Tos3, were identified as potential members of the Snf1 kinase complex (6, 14). To test whether these kinases might also have the ability to phosphorylate and activate Snf1, we used affinity purification methods to isolate Pak1-TAP, GST-Cka1, and GST-Tos3 from yeast and assayed their ability to phosphorylate the Snf1 protein in vitro (Fig. 1A). Purified GST-Cka1 phosphorylated itself (lane 1) and GST-Mig1 (lane 3). When incubated with limiting quantities of purified Snf1, GST-Cka1 caused incorporation of 32P into two additional proteins. On the basis of results reported in an earlier study (22), we have identified these bands as Sip2/Gal83 and Sip1. GST-Cka1 failed to promote any incorporation of 32P into the Snf1 polypeptide. Similarly, the GST-Tos3 kinase phosphorylated itself (lane 4), GST-Mig1 (lane 6), and the beta subunits Sip2 and Gal83 (lane 5) but failed to phosphorylate the Snf1 polypeptide. With the same limiting quantities of purified Snf1 enzyme, purified Pak1 did promote phosphorylation of the Snf1 protein, as well as the Sip2 and Gal83 proteins and a 110-kDa polypeptide (lane 7). The ability of purified Pak1 to promote Snf1 phosphorylation was examined in more detail over a 10-fold range of Pak1 kinase concentration (Fig. 1B). Addition of increasing quantities of purified Pak1 kinase resulted in increased incorporation of 32P into the Snf1 polypeptide. Three prominent phosphorylated species were detected in this reaction mixture. The Snf1 protein and the Sip2 and Gal83 proteins, subunits of the Snf1 kinase complex, are efficiently labeled with 32P. Since a constant level of Snf1 kinase was present in these reaction mixtures (approximately 2.5 ng/ml) and the level of phosphorylation increased with the addition of Pak1 kinase, we concluded that the Pak1 protein kinase promotes phosphorylation of Snf1 kinase in vitro. A 110-kDa protein and the Sip2/Gal83 proteins are also phosphorylated in a Pak1-dependent and dose-responsive manner.
FIG. 1.
Pak1 kinase, but not Cka1 or Tos3, promotes phosphorylation of Snf1 in vitro. (A) Reaction mixtures contained [γ-32P]ATP and additional purified proteins as indicated. Phosphorylated products were detected by autoradiography. (B) Pak1 kinase-dependent phosphorylation of Snf1 kinase components. A limiting concentration (2.5 ng/ml) of Snf1 kinase purified from pak1Δ mutant cells (lanes 1 to 4) was incubated with either no Pak1 or increasing concentrations of Pak1 kinase purified from snf1Δ10 mutant cells. Snf1 kinase was omitted from the reaction mixture in lane 5.
Pak1 associates with the Snf1 kinase complex in vivo.
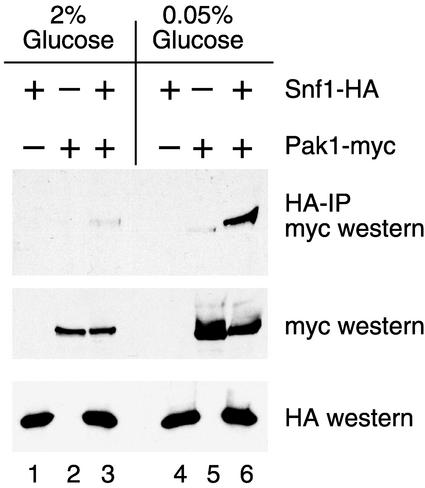
In order to provide independent verification that the Pak1 protein associates with the Snf1 kinase complex in vivo, we coexpressed a Pak1 protein tagged with three copies of the myc epitope with Snf1 protein tagged with three copies of the HA epitope. Snf1 kinase complexes were immunoprecipitated with a monoclonal antibody directed against HA, and the bound proteins were then probed by Western blotting with antibodies directed against the myc epitope (Fig. 2). Since Snf1 activation is regulated in response to the glucose concentration, extracts were prepared from cells grown under high- and low-glucose conditions. Consistent levels of Snf1-HA and Pak1-myc were easily detected in the extracts by Western blotting (lower panels). Following immune precipitation with anti-HA antibodies, the Pak1 protein was detected in a complex with Snf1 from glucose-starved cells (lane 6) but only weakly detected in Snf1 complexes isolated from cells grown in glucose-rich medium (lane 3). We concluded that Pak1 associates with Snf1 in vivo and that the association is greatly enhanced under low-glucose conditions when Snf1 is activated.
FIG. 2.
Pak1 kinase associates with active Snf1 kinase in vivo. Yeast strain MSY809 (snf1Δ pak1Δ) was transformed simultaneously with two plasmids, either an empty vector (−) or a plasmid expressing Snf1-HA or Pak1-myc, as indicated above each lane. Protein extracts were prepared from cells grown in medium containing 2 or 0.05% glucose. Snf1 kinase complexes were immunoprecipitated (IP) with an anti-HA monoclonal antibody and probed by Western blot assay with anti-myc antibodies (top panel). Equivalent aliquots of each protein extract (20 μg) were analyzed directly by Western blotting with anti-myc (middle panel) and anti-HA (bottom panel) antibodies.
Purification of Snf1 kinase from PAK1 and pak1Δ mutant cells.
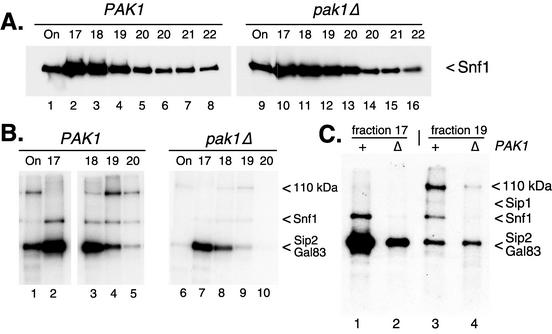
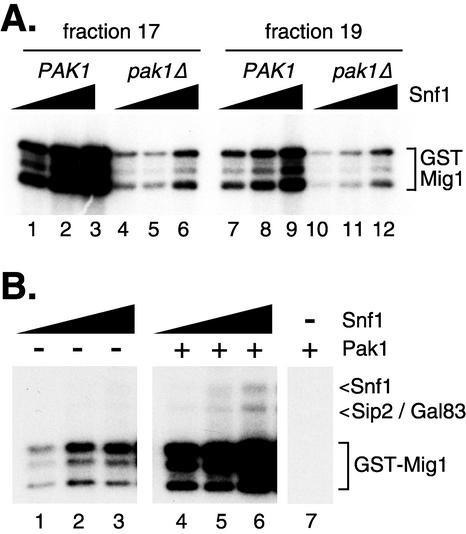
To assess the impact of Pak1 on Snf1 kinase activity, the Snf1 kinase was affinity purified from PAK1 and pak1Δ mutant cells. The pak1Δ mutant strain used in this experiment was a haploid segregant from the homozygous deletion strain set (7). Because of concerns about differences caused by different strain backgrounds, we used a related haploid PAK1 mutant strain as our control (haploids MSY608 and MSY678 were derived from Research Genetics strains 31628 and 34056, respectively). Western blotting established that equivalent amounts of Snf1 protein were recovered from both preparations (Fig. 3A). When Mono Q fractions containing Snf1 kinase complexes were incubated with [γ-32P]ATP, radioactivity was incorporated into three bands (Fig. 3B). On the basis of results reported previously (22), we have identified two of these bands as Sip2/Gal83 and Snf1. The beta subunits Sip2 and Gal83 were prominently labeled with 32P. These proteins comigrated on SDS gels and appeared as a single band. The third beta subunit, Sip1, was much less abundant (22) and was faintly visible in some reaction mixtures (Fig. 1, lane 2).The Snf1 polypeptide was also labeled with 32P; however, label incorporation was greatly reduced when Snf1 complexes were purified from pak1Δ mutant cells (Fig. 3B, compare lanes 2 and 7). Finally, a third band of approximately 110 kDa was also labeled. Putative members of the Snf1 kinase complex that are in this size range are Reg1 and Pak1 (14). The 110-kDa protein cannot be the Pak1 protein itself since it is detected in Snf1 complexes purified from pak1Δ mutant cells. The possibility that the 110-kDa protein is Reg1 is addressed below. Assuming that the abundance of the 110-kDa protein correlates with its 32P incorporation, this protein was present in the Mono Q input (Fig. 3B, lanes 1 and 6) and in the later fractions from the Mono Q column but absent from early Mono Q column fractions (Fig. 3B, lanes 2 and 7). Thus, Snf1 kinase complexes of differing protein compositions may be separated by Mono Q chromatography. Two forms of the Snf1 kinase complex, those with abundant 110-kDa protein (fraction 19) and those lacking the 110-kDa protein (fraction 17), were incubated with [γ-32P]ATP, and the labeled proteins were analyzed side by side (Fig. 3C). First, Snf1 kinase complexes from both fractions showed an almost complete loss of incorporation of 32P into the Snf1 polypeptide when the enzyme was purified from pak1Δ mutant cells. Second, a reduction in overall 32P incorporation was observed in reaction mixtures containing the 110-kDa protein (fraction 17 versus fraction 19).
FIG. 3.
Effect of PAK1 on the activity of purified Snf1 kinase. (A) Western blot assay of Snf1 protein purified from PAK1 and pak1Δ mutant cells (22). Input and peak fractions across the NaCl gradient of the final Mono Q column were probed with anti-HA antibodies. (B) Input and peak fractions across the NaCl gradient of the final Mono Q column were incubated with [γ-32P]ATP and resolved by SDS-polyacrylamide gel electrophoresis. Incorporation of radioactivity was detected by autoradiography (C) Equivalent aliquots from fractions 17 and 19 prepared from PAK1 and pak1Δ mutant cells were incubated with [γ-32P]ATP and resolved side by side on an SDS-polyacrylamide gel.
The Reg1 protein is present in Snf1 kinase complexes.
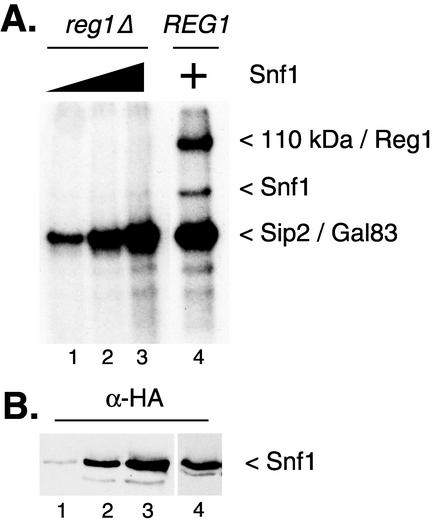
Mass spectrometry analysis of proteins in the Snf1 kinase complex identified Reg1 as a component (6, 14). Furthermore, genetic and biochemical studies have shown that Reg1 associates with Snf1 and is phosphorylated in a Snf1-dependent manner (27). The Reg1 protein has a predicted molecular mass of 112 kDa. In vitro phosphorylation reactions were performed with Snf1 enzyme purified from REG1 and reg1Δ mutant strains. The 110-kDa protein was not detected in reaction mixtures with Snf1 purified from reg1Δ mutant cells (Fig. 4A) even though equivalent levels of Snf1 enzyme were present (Fig. 4B). The simplest explanation for this is that the 110-kDa protein is the Reg1 protein. We have previously shown that the Reg1 protein is required to remove the activating phosphorylation of threonine 210 (21). Further support for the conclusion that Reg1 is the 110-kDa protein is the observation that fractions with an abundance of the 110-kDa band have lower Snf1 kinase specific activity (Fig. 5A, compare titrations of fractions 17 and 19). We noted also that very little 32P was incorporated into the Snf1 protein purified from reg1Δ mutant cells (Fig. 4A). Failure to incorporate 32P into Snf1 may be due to the fact that threonine 210 is already fully phosphorylated. Western blotting experiments demonstrated that the Snf1 protein was present in the complex purified from reg1Δ mutant cells (Fig. 4B) but was not efficiently phosphorylated in vitro.
FIG. 4.
Snf1 kinase complexes contain Reg1 protein. Snf1 kinase complexes prepared from REG1 and reg1Δ mutant cells were assayed for autophosphorylation activity. Increasing concentrations of Snf1 kinase from the reg1Δ mutant cells were used (lanes 1 to 3). Lanes 2 and 4 contain comparable levels of Snf1 protein (approximately 25 ng/ml).
FIG. 5.
Pak1-dependent activation of Snf1 kinase activity. (A) Increasing concentrations of Snf1 kinase complexes (fractions 17 and 19) were incubated with [γ-32P]ATP and recombinant GST-Mig1 protein. Incorporation of radioactivity into the GST-Mig1 protein (and its proteolytic fragments) was detected by autoradiography. Snf1 kinase had been purified from PAK1 (lanes 1 to 3 and 7 to 9) and pak1Δ (lanes 4 to 6 and 10 to 12) mutant cells. (B) Increasing concentrations of Snf1 kinase purified from pak1Δ mutant cells (fraction 17) were incubated with [γ-32P]ATP, GST-Mig1 protein, and either buffer (lanes 1 to 3) or purified Pak1 kinase (lanes 4 to 6). Snf1 kinase was omitted from one reaction mixture (lane 7).
Snf1 kinase requires Pak1 for full activity.
The Snf1 kinase purified from pak1Δ mutant cells showed significantly lower levels of in vitro kinase activity than did an equivalent amount of Snf1 purified from PAK1 cells (Fig. 5A). Snf1 kinase is much more active when assayed with GST-Mig1 as a phosphate acceptor, allowing the use of 10-fold-less Snf1 enzyme. Thus, the autophosphorylation products shown in earlier figures are not observed unless much longer exposures are shown. The Mig1 protein is a known Snf1 target, with four serine residues that are phosphorylated by Snf1 in vivo (33) and in vitro (30). Peak Snf1 complex fractions that lacked Reg1 (fraction 17) or contained abundant Reg1 (fraction 19) were assayed. Deletion of PAK1 led to greatly reduced Snf1 kinase specific activity (compare lanes 1 to 3 with lanes 4 to 6). Equivalent levels of Snf1 protein were included in these reaction mixture, as judged by Western blotting (Fig. 3A). We concluded that Snf1 kinase requires PAK1 for maximal activity.
Purified Pak1 kinase activates Snf1 in vitro.
To determine if the Pak1 kinase could activate Snf1 in vitro, Pak1 kinase was affinity tagged and purified from snf1Δ10 mutant cells (25). The Snf1 kinase was purified from pak1Δ mutant cells and showed a low but detectable level of kinase activity in vitro (Fig. 5B). Addition of purified Pak1 kinase significantly increased Snf1 kinase activity, as judged by the level of GST-Mig1 phosphorylation (compare lanes 4 to 6 with lanes 1 to 3). In the absence of added Snf1 kinase, no phosphorylation of GST-Mig1 was observed, demonstrating that Pak1 itself does not phosphorylate GST-Mig1 (lane 7). Thus, Pak1 kinase is able to stimulate Snf1-dependent phosphorylation of GST-Mig1.
Genetic manipulation of PAK1.
Our data support the idea that the Pak1 kinase is a Snf1-activating kinase. This hypothesis was examined genetically by phenotypic analysis of pak1Δ mutant strains. Our source for the pak1Δ::KAN allele was the homozygous diploid deletion collection (7). Haploid progeny of strain 36128 were crossed into our own strain background, and tetrads were analyzed for Snf phenotypes. None of our pak1Δ mutant strains exhibited even a weak Snf phenotype. All pak1Δ mutant strains grew as well as wild-type cells on raffinose-antimycin medium (Fig. 6A) and on glycerol-ethanol medium at 15, 30, and 37°C. We tested whether deletion of PAK1 could exacerbate the weak Snf phenotype observed in the sip2Δ gal83Δ mutant strain. Deletion of PAK1 did not affect the Snf phenotype of this strain (data not shown). All pak1Δ mutant strains were able to induce invertase expression with normal kinetics and to similar degrees (Fig. 6B). We also crossed our pak1Δ mutant strains with several other strains bearing deletions of other kinases that are candidates for being Snf1-activating kinases (CKA1, CKA2, TOS3, PKH1, PKH2, KIN1, KIN2, NPR1, et al.). None of the double mutants produced a synthetic Snf phenotype. We also examined the effect of an increased PAK1 gene dosage. A wild-type allele of PAK1 and a kinase-dead allele (D277A) were introduced into both wild-type and pak1Δ mutant cells on high-copy-number plasmids. In neither case was any change in growth on alternative carbon sources (data not shown) or invertase expression detected (Fig. 7C).
FIG. 6.
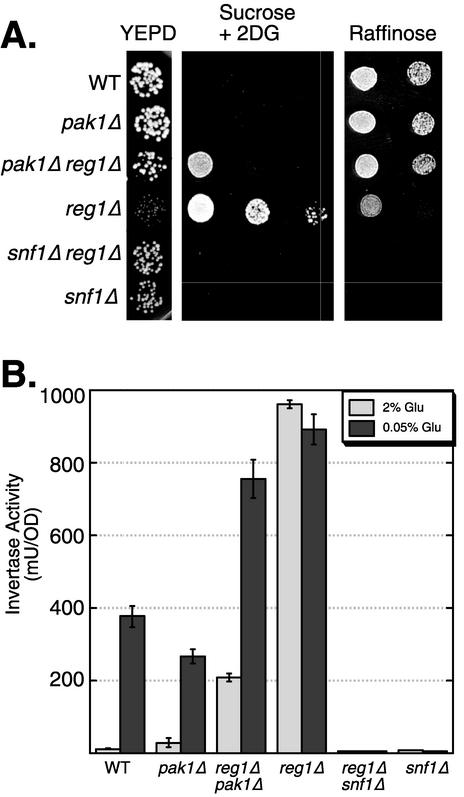
Suppression of reg1Δ by deletion of PAK1. (A) Growth phenotypes were determined by spotting serial dilutions of yeast cultures on YEPD (26), sucrose medium containing 2-deoxyglucose (2DG), and raffinose medium. The relevant genotypes of the yeast strains are shown on the left. (B) Invertase expression of cells grown in 2 or 0.05% glucose (Glu). The relevant genotypes of the yeast strains are shown on the bottom. The mean invertase expression from three independent cultures is plotted. Errors bars represent one standard error. WT, wild type; OD, optical density.
FIG. 7.
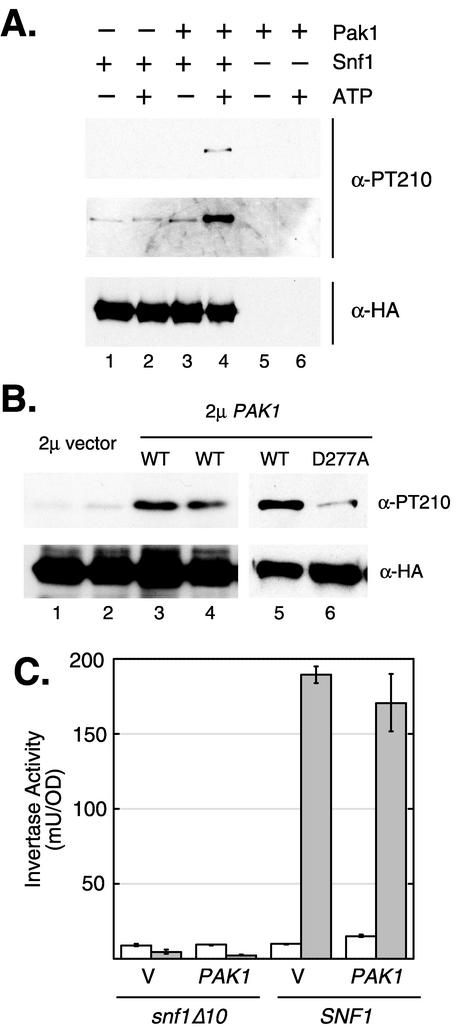
Pak1 phosphorylates Snf1 on activation loop residue threonine 210. (A) Reaction mixtures contained Snf1 kinase purified from pak1Δ mutant cells (lanes 1 to 4), Pak1 kinase purified from snf1Δ10 mutant cells (lanes 3 to 6), and ATP (lanes 2, 4, and 6). Products of the reaction were resolved on an SDS-polyacrylamide gel, transferred to a nylon filter, and probed first with an antibody specific for Snf1 protein containing phosphorylated threonine 210 (21). Both short (top panel) and long (middle panel) exposures of this Western blot are shown (α-PT210). Total Snf1 protein was detected by reprobing the same filter with an antibody directed against the HA epitope (α-HA). (B) Yeast strains expressing HA-tagged Snf1 were transformed with high-copy-number plasmid pRS424 (vector), pPAK1-424 (WT), or pPAK-D277A (D277A), as indicated. Snf1 kinase was immune precipitated from protein extracts prepared from cultures grown in high glucose (2%) and probed by Western blotting with antibodies specific for phosphorylated threonine 210 (α-PT210) or the HA epitope (α-HA). (C) Cells with and without SNF1 were transformed with a high-copy-number plasmid containing either no insert (V) or full-length PAK1. Cells were collected from log-phase cultures grown in 2% (open bars) or 0.05% (filled bars) glucose and assayed for invertase activity. The mean invertase activity (milliunits per unit of optical density [OD]) from three independent transformants is plotted. Error bars represent one standard error.
Suppression of reg1Δ by pak1Δ.
We did detect genetic interaction between pak1Δ and reg1Δ. The Reg1 protein is one of the regulatory subunits of type 1 protein phosphatase Glc7 (35). The Reg1-Glc7 complex acts in opposition to the Snf1 signaling pathway by promoting the dephosphorylation of Snf1 threonine 210 (21). On glucose medium, reg1Δ mutant strains exhibit derepressed levels of invertase and galactokinase (20, 23) and form small colonies (4). Both phenotypes are Snf1 dependent. If Pak1 kinase is one of the Snf1-activating kinases, then deletion of the PAK1 gene might suppress some or all of the Snf1-dependent phenotypes observed in reg1Δ mutant strains. To test this prediction, we crossed pak1Δ and reg1Δ mutant strains and characterized the phenotypes of the pak1Δ reg1Δ double mutant. We found that deletion of PAK1 effectively suppressed a number of the phenotypes of reg1Δ. First, reg1Δ mutant cells grown on glucose medium formed small colonies and this phenotype was suppressed by deletion of PAK1 (Fig. 6A). Wild-type cells are unable to grow on sucrose medium in the presence of 2-deoxyglucose (23). Deletion of REG1 causes constitutive relief of glucose repression and allows cells to grow on sucrose medium in the presence of 2-deoxyglucose. Deletion of PAK1 severely restricted the ability of reg1Δ mutant cells to grow on 2-deoxyglucose medium (Fig. 6A). In addition, the constitutive invertase expression observed in reg1Δ mutant cells was partially suppressed in reg1Δ pak1Δ mutant cells (Fig. 6B). Therefore, pak1Δ suppresses many of the Snf1-dependent phenotypes observed in reg1Δ mutant cells.
Pak1 phosphorylates Snf1 on threonine 210.
Previous studies have shown that the key regulatory phosphorylation site on the Snf1 protein is threonine 210 within the activation loop (5, 21). To determine whether Pak1 phosphorylates Snf1 on threonine 210, we used a phosphopeptide antibody that specifically recognizes Snf1 that has been phosphorylated on threonine 210 (21). Purified Snf1 kinase was incubated with or without ATP in the presence or absence of Pak1 kinase (Fig. 7A). Increased phosphorylation of threonine 210 was detected only when both Pak1 and ATP were present. Therefore, Pak1 kinase promotes the phosphorylation of threonine 210. We next tested whether manipulation of the PAK1 gene copy number affects the phosphorylation of Snf1 threonine 210 in vivo (Fig. 7B). Strains expressing Snf1-HA were transformed with a high-copy-number plasmid with either no insert (2μm vector) or full-length PAK1 (2μm PAK1). Protein extracts were prepared in duplicate from cells grown under high-glucose conditions (2% glucose), and the Snf1 polypeptide was collected by immune precipitation with anti-HA antibodies. Snf1 protein was then detected by parallel Western blot assays with antibodies directed against Snf1 phosphorylated on threonine 210 or against the HA epitope, as indicated. Increased PAK1 gene dosage resulted in a significant increase in the level of phosphorylated threonine 210. Equivalent levels of total Snf1 polypeptide were present in all four extracts (lanes 1 to 4). To determine whether Pak1 kinase activity is required, a point mutation was introduced into the PAK1 gene such that the catalytic aspartate residue was changed to alanine (D277A). Cells transformed with a high-copy-number PAK1 plasmid with a wild-type (WT) kinase domain and cells transformed with a kinase-dead (D277A) high-copy-number PAK1 plasmid were compared. Pak1-mediated stimulation of Snf1 threonine 210 phosphorylation required an intact Pak1 kinase domain (compare lanes 5 and 6). However, increased phosphorylation of Snf1 threonine 210 was not sufficient to activate Snf1, as judged by invertase assays (Fig. 7C). Taken together, our data indicate that the Pak1 kinase activates the Snf1 kinase by phosphorylation of the Snf1 activation loop in vitro and in vivo.
DISCUSSION
The Snf1 kinase is activated by an upstream kinase (21). However, genetic screens have failed to identify mutations in any protein kinase gene other than SNF1 that produces a Snf− phenotype. We have argued previously that the most likely explanation for these findings is that more than one kinase is capable of activating Snf1 in vivo. In this report, we present the following evidence that the Pak1 kinase is one of the Snf1-activating kinases. First, Pak1 associates with the Snf1 kinase complex in vivo and the association is greatly enhanced under growth conditions that promote Snf1 activation (Fig. 2.). This result puts Pak1 in the right place at the right time. Second, Pak1 kinase activates Snf1 kinase in vitro (Fig. 5B). Third, purified Pak1 is able to phosphorylate Snf1 protein on threonine 210 within the activation loop (Fig. 7A) and an increased PAK1 gene dosage increases the level of threonine 210 phosphorylation in vivo. Finally, Snf1 kinase purified from pak1Δ mutant strains has a lower specific activity than Snf1 kinase purified from PAK1 mutant strains (Fig. 5A). Taken together, these data demonstrate that Pak1 is a Snf1-activating kinase. Our current model for the phosphorylation events occurring in the Snf1 kinase complex is presented in Fig. 8.
FIG. 8.
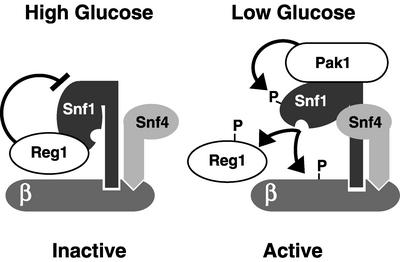
Model of Pak1-dependent activation of Snf1 kinase. In the presence of a high glucose concentration, Snf1 kinase is in a complex with the Reg1 protein, which recruits protein phosphatase Glc7 (not shown). The Reg1/Glc7 complex dephosphorylates Snf1 threonine 210 (21). In the presence of a low glucose concentration, Pak1 associates with the Snf1 kinase complex and phosphorylates Snf1 on threonine 210. Snf4 also participates in activation of the Snf1 kinase through direct interaction with the Snf1 regulatory domain. Once activated, Snf1 kinase phosphorylates its beta subunit (Sip1, Sip2, or Gal83), the Reg1 protein, and additional substrates, such as Mig1 (not shown).
The one troubling aspect of this work is that the pak1Δ allele does not produce any detectable Snf phenotype. We cannot formally rule out the possibility that Pak1 is a kinase whose in vivo function is to activate kinases other than Snf1. However, the findings that Pak1 associates with Snf1 in vivo under conditions in which Snf1 is active and that Pak1 promotes phosphorylation of Snf1 threonine 210 in vivo argue against this interpretation. Furthermore, we show that deletion of PAK1 suppresses the phenotypes associated with reg1Δ. The Reg1-Glc7 PP1 phosphatase is known to act in opposition to Snf1 kinase. Deletion of REG1 results in constitutively activated Snf1. The observation that the pak1Δ mutation suppresses the phenotypes caused by the reg1Δ mutation argues strongly that Pak1 is one of the Snf1-activating kinases in vivo. We favor the interpretation that activation of Snf1 kinase is a redundant function that is shared between Pak1 kinase and one or more other kinases. Hartwell and colleagues have pointed out that redundancy is an important source of buffering genetic variation (10). Many yeast genes have one or more paralogues that may mask phenotypes caused by mutations. In the case of Pak1, the closest relative is Tos3 (16); however, the pak1Δ tos3Δ mutant strain is also Snf+ (data not shown). Identification of the other putative Snf1-activating kinase(s) would settle the question unambiguously and is a high priority for our future studies. An increased PAK1 gene dosage leads to increased phosphorylation of Snf1 threonine 210 under glucose-repressing conditions but does not lead to activation of Snf1 kinase. Previously, we showed that activation of Snf1 kinase is a two-step process (21). We conclude that phosphorylation of Snf1 threonine 210 is required but not sufficient for Snf1 activation.
The PAK1 gene was first isolated in a screen for high-copy suppressors of the temperature sensitivity phenotype caused by the cdc17-1 mutation (15). POL1 (also known as CDC17 or HPR3) encodes one subunit of the alpha DNA polymerase-primase complex. An increased PAK1 copy number partially suppresses the temperature sensitivity phenotype caused by several alleles of POL1. However, a direct connection between Pak1 and DNA polymerase alpha was not established. Furthermore, an increased PAK1 gene dosage did not suppress mutations in other DNA polymerases or even other mutations affecting DNA polymerase alpha. The ability of PAK1 to suppress the cdc17-1 allele required a functional kinase domain. We show here that Pak1 kinase is able to phosphorylate the activation loop threonine of Snf1 kinase (Fig. 7). In addition, the mammalian homologues of Pak1 also activate downstream kinases by phosphorylation of activation loop residues. These observations suggest that Pak1 suppressed the cdc17-1 allele by activating a downstream kinase. It is unlikely that Pak1 is devoted to Snf1 activation. Some other in vivo target of Pak1 might provide an explanation of its ability to suppress mutations affecting the alpha DNA polymerase-primase complex.
Independent experiments demonstrate that Pak1 associates with the Snf1 kinase complex in vivo. In mass spectrometry studies of proteins associated with TAP-tagged Snf1 and TAP-tagged Snf4, both detected Pak1 as a complex member (6). We used myc-tagged Pak1 and HA-tagged Snf1 to demonstrate the association of these proteins by coimmunoprecipitation (Fig. 2). In the course of that experiment, we found that Pak1 accumulation increased under conditions of glucose limitation. A similar increase in Pak1 mRNA was not detected in microarray experiments (3, 18), suggesting that the increase in Pak1 protein abundance may reflect posttranscriptional regulation. Also, a shift in the SDS-polyacrylamide gel mobility of the Pak1 protein in response to glucose limitation was observed, suggesting that the activity or abundance of Pak1 may be regulated by a posttranslational modification.
The mammalian homologue of Snf1 kinase, AMPK, is the subject of intense study, in part because of its potential as a target for treatment of type II diabetes (12). Biochemical studies have shown that AMPK is activated by a distinct protein kinase designated AMPKK (11). The identity of AMPKK has not been determined. Our finding that Pak1 kinase is one of the Snf1-activating kinases in yeast suggests that the mammalian homologue of Pak1, CaMKK-β, may play a similar role in mammalian cells. Biochemical properties of AMPKK and CaMKK-β were used to argue that these enzymes are distinct (13). However, this study based its conclusion on efficiency of activation and relied on partially purified fractions containing AMPKK and CaMKK-β. In some studies, CaMKK-β has been used as a surrogate AMPKK since recombinant CaMKK-β is able to phosphorylate the activation loop threonine of AMPK in vitro (8). Full activation of AMPK may involve more than one phosphorylation event (31). Perhaps efficient activation of AMPK requires the action of more than one kinase enzyme, a possibility that may explain the difficulty with the purification and identification of a single AMPKK. Our data suggest that CaMKK-β may participate in the activation of AMPK in vivo by phosphorylation of the activation loop threonine.
Acknowledgments
This work was supported by grant GM46443 from the National Institutes of Health.
We thank Karen Arndt and Tom Smithgall for thoughtful discussions, Peg Shirra for the reg1Δ mutant strains, Eric Phiziky and Martha Cyert for providing GST-tagged kinases, and Anna Leech for technical assistance.
REFERENCES
- 1.Anderson, K. A., R. L. Means, Q. H. Huang, B. E. Kemp, E. G. Goldstein, M. A. Selbert, A. M. Edelman, R. T. Fremeau, and A. R. Means. 1998. Components of a calmodulin-dependent protein kinase cascade: molecular cloning, functional characterization and cellular localization of Ca2+/calmodulin-dependent protein kinase kinase beta. J. Biol. Chem. 273:31880-31889. [DOI] [PubMed] [Google Scholar]
- 2.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 4.Dombek, K. M., V. Voronkova, A. Raney, and E. T. Young. 1999. Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 19:6029-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton, S. R., J. B. O'Donnell, Jr., A. Hammet, D. Stapleton, S. A. Habinowski, A. R. Means, B. E. Kemp, and L. A. Witters. 2002. AMP-activated protein kinase kinase: detection with recombinant AMPK α1 subunit. Biochem. Biophys. Res. Commun. 293:892-898. [DOI] [PubMed] [Google Scholar]
- 9.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 10.Hartman, J. L., B. Garvik, and L. Hartwell. 2001. Principles for the buffering of genetic variation. Science 291:1001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Hawley, S. A., M. Davison, A. Woods, S. P. Davies, R. K. Beri, D. Carling, and D. G. Hardie. 1996. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 271:27879-27887. [DOI] [PubMed] [Google Scholar]
- 12.Hawley, S. A., A. E. Gadalla, G. S. Olsen, and D. G. Hardie. 2002. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes 51:2420-2425. [DOI] [PubMed] [Google Scholar]
- 13.Hawley, S. A., M. A. Selbert, E. G. Goldstein, A. M. Edelman, D. Carling, and D. G. Hardie. 1995. 5′-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin activates the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J. Biol. Chem. 270:27186-27191. [DOI] [PubMed] [Google Scholar]
- 14.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 15.Hovland, P. G., M. Tecklenberg, and R. A. Sclafani. 1997. Overexpression of the protein kinase Pak1 suppresses yeast DNA polymerase mutations. Mol. Gen. Genet. 256:45-53. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, T., and G. D. Plowman. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22:18-22. [DOI] [PubMed] [Google Scholar]
- 17.Jaffer, Z. M., and J. Chernoff. 2002. p21-activated kinases: three more join the PAK. Int. J. Biochem. Cell Biol. 34:713-717. [DOI] [PubMed] [Google Scholar]
- 18.Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez, V. C. Culotta, G. R. Fink, and L. Guarente. 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344-348. [DOI] [PubMed] [Google Scholar]
- 19.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153-1155. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto, K., T. Yoshimatsu, and Y. Oshima. 1983. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J. Bacteriol. 153:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase: activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 22.Nath, N., R. R. McCartney, and M. C. Schmidt. 2002. Purification and characterization of Snf1 kinase complexes containing a defined beta subunit composition. J. Biol. Chem. 277:50403-50408. [DOI] [PubMed] [Google Scholar]
- 23.Neigeborn, L., and M. Carlson. 1987. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics 115:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phizicky, E. M., M. R. Martzen, S. M. McCraith, S. L. Spinelli, F. Xing, N. P. Shull, C. Van Slyke, R. K. Montagne, F. M. Torres, S. Fields, and E. J. Grayhack. 2002. Biochemical genomics approach to map activities to genes. Methods Enzymol. 350:546-559. [DOI] [PubMed] [Google Scholar]
- 25.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 26.Rose, M. D., F. Winston, and P. Hieter (ed.). 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Sanz, P., G. R. Alms, T. A. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schillberg, S., D. Schumann, and R. Fischer. 1997. PCR-based multiplex method for rapid screening of recombinant bacteria. BioTechniques 23:212-214, 216. [DOI] [PubMed]
- 29.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, F. C., S. P. Davies, W. A. Wilson, D. Carling, and D. G. Hardie. 1999. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 453:219-223. [DOI] [PubMed] [Google Scholar]
- 31.Stein, S. C., A. Woods, N. A. Jones, M. D. Davison, and D. Carling. 2000. The regulation of AMP-activated protein kinase by phosphorylation. Biochem. J. 345:437-443. [PMC free article] [PubMed] [Google Scholar]
- 32.Thiagalingam, S., K. W. Kinzler, and B. Vogelstein. 1995. PAK1, a gene that can regulate p53 activity in yeast. Proc. Natl. Acad. Sci. USA 92:6062-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu, H., and M. Wigler. 1999. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu, J., and M. Carlson. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 38.Winder, W. W., and D. G. Hardie. 1999. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am. J. Physiol. 277:E1-E10. [DOI] [PubMed] [Google Scholar]
- 39.Woods, A., M. R. Munday, J. Scott, X. Yang, M. Carlson, and D. Carling. 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269:19509-19515. [PubMed] [Google Scholar]