Abstract
Since the majority of high-grade breast cancers express reduced levels of BRCA1 mRNA, we investigated the factors regulating BRCA1 transcription. Factors with specific affinity for the previously identified positive regulatory region (PRR) in the BRCA1 promoter were purified from whole-cell extracts. Identified proteins included replication protein A and a series of related factors with affinity for the sense strand of PRR. A subset of the identified factors activated the BRCA1 promoter. Identification of these families of proteins regulating the BRCA1 promoter represents an important step in the comprehension of the mechanisms responsible for breast cancer development.
Mutations of BRCA1 are thought to account for 5 to 10% of all cases of breast cancer and approximately 50% of all cases of inherited breast cancer. They are also suggested to be responsible for 80 to 90% of all cases of cancer in families with breast-ovarian cancer syndrome (40). Women inheriting mutations in BRCA1, most of which are truncating mutations, have an 85% chance of developing breast cancer in their lifetimes (32). An interesting aspect of BRCA1 is that, although mutations in the gene are associated with highly invasive inherited breast cancers, few mutations have been detected in the BRCA1 gene in sporadic forms of cancer (17).
Interestingly, BRCA1 expression was observed to be suppressed in most high-grade sporadic breast cancers (47, 50), and it was suggested that the reduction in BRCA1 mRNA (and protein) may be due to dysregulated transcription. Furthermore, hypermethylation of BRCA1 promoters and reduced levels of BRCA1 transcripts in sporadic breast cancers have been reported (3, 6, 10, 13, 15, 16, 26-28, 34, 36-38), suggesting a role for BRCA1 in nonfamilial breast cancers.
Despite the importance of identifying factors regulating BRCA1 transcription, scant information was available to us. Recently, the ID4 factor was proposed to regulate BRCA1 transcription (5). Our laboratory identified a positive regulatory region (PRR) in the BRCA1 promoter (44). Deletion of the PRR resulted in a significant loss of BRCA1 transcriptional activity. In addition, the PRR exhibited a strong and specific affinity for nuclear factors. These results were confirmed by others who demonstrated a regulatory role for the region encompassing the PRR (42). In the present study, we demonstrate by mutational analysis that an intact PRR in the BRCA1 promoter is essential for BRCA1 promoter activity. We purified the three subunits of replication protein A (RPA), a specific factor binding double-stranded as well as single-stranded PRRs. We also purified and identified a family of related factors with a specific affinity for the sense strand of the PRR, and we show evidence for these factors' roles in regulating BRCA1 transcription.
MATERIALS AND METHODS
Mutants of the PRR.
The isolation of the BRCA1 promoter and the conditions used for the generation of BRCA1 promoter mutants have been described previously (44). Systematic deletion mutants of the PRR were constructed by utilizing PCR amplification. Primers with systematic deletions at the 5′ ends of the PRR (−202 to −178) and a 3′ primer described previously were used to amplify 1 ng of the BRCA1 promoter template. The amplified products were digested with MluI and XhoI (restriction enzyme sites incorporated in the primers) and ligated into pGL3 basic vector (purchased from Promega) containing the luciferase reporter gene.
To generate BRCA1 promoter mutants spanning less than 100 bp, respective DNA strands were synthesized, annealed, and ligated directly into pGL3 basic vector. Synthetic primers with progressive 3′ deletions and containing adapters of the MluI and XhoI enzymes at each end were annealed and ligated to the MluI and XhoI sites of the pGL3 basic vector to generate BRCA1 promoters with 3′ deletions. The cyclic AMP response element binding (CREB) site was disrupted in the transcriptionally active BRCA1 promoter fragment spanning positions −202 to −136 by incorporating mutated residues in synthetic primers, which were ligated directly into pGL3 basic vector. Point mutations in the PRR were generated by the ligation of synthetic annealed primers (with MluI and XhoI adapters) in the pGL3 basic vector. All constructs were confirmed by sequencing.
RPA cDNA was a gift from R. Fishel and M. Wold. RPA1- and RPA2-expressing clones were generated by subcloning the respective cDNA clones in the pcDNA 3.1 (−) vector (purchased from Invitrogen) in the EcoRI site. The FBP-1 antibody (24) and cDNA clone of FBP-1 in the pET28a vector were gifts from David Levens. The XbaI/HindIII fragment containing FBP-1 cDNA (obtained from the pET28a construct) was subcloned in the pcDNA 3.1 (−) vector, which contained the cytomegalovirus (CMV) promoter. The cDNAs of the other factors were amplified (with the Gold Taq system purchased from Roche) from human placental, ovarian, skeletal, brain, and lymph node cDNAs. The amplified constructs were cloned in pcDNA3.1/V5 His Topo vector (purchased from Invitrogen) by using the TA cloning technique per the manufacturer's protocol. All the insertions were confirmed by sequencing. The forward (F) and reverse (R) primers from the respective 5′ and 3′ sequences are as follows: DAZAP1 F (5′-ATG AAC AAC TCG GGC GCC GAC GAG A-3′) and DAZAP1 R (5′-CTA GCG TCG GTA GGG GTG GAA CC-3′), ELAVL1 F (5′-ACA ATG TCT AAT GGT TAT GAA GAC C-3′) and ELAVL1 R (5′-GAG CGA GTT ATT TGT GGG ACT TG-3′), HN-RNPK F (5′-GAA TAT GGA AAC TGA ACA GCC AG-3′) and HN-RNPK R (5′-GCA TTA GAA TCC TTC AAC ATC TGC-3′), HN RPA2 F (5′-GAA GCG ACT GAG TCC GCG ATG-3′) and HN RPA2 R (5′-GGA AGA AGC TCA GTA TCG GCT-3′), PCBP1 F (5′-TCG CCA TGG ATG CCG GTG TGA CTG-3′) and PCBP1 R (5′-CTG TTC TAG CTG CAC CCC ATG C-3′), PCBP2 F (5′-TGC TCG ACA TGG ACA CCG GTG TG-3′) andPCBP2 R (5′-ATC TGC ATT GTT CTA GCT GCT C-3′), PTBP1 F (5′-GTG CCA TGG ACG GCA TTG TCC-3′) and PTBP1 R (5′-CCC TAG ATG GTG GAC TTG GAG-3′, and TIA1-F1 (5′-ATG GAG GAC GAG ATG CCC AAG ACT C-3′) and TIA1-R1 (5′-CCT TAT TCA CTG GGT TTC ATA CC-3′.
Transfections, EMSAs, and Northern blot analyses.
Maintenance of MCF-7 cells, transfections, luciferase assays, normalization of transfection efficiencies, preparation of nuclear extracts, and electrophoretic mobility shift assays (EMSAs) were performed essentially as described previously (44, 45). The Ramos B-cell lymphoma line was grown in RPMI medium supplemented with 10% fetal bovine serum. Supershift experiments were performed by adding 1 to 5 μg of the antibodies indicated below to EMSA reaction mixtures. Northern blots were performed by the hybridization of 20 μg of total RNA with probes derived from first 80 bases of published cDNAs.
Purification of the Py/Pu motif and PRR binding proteins.
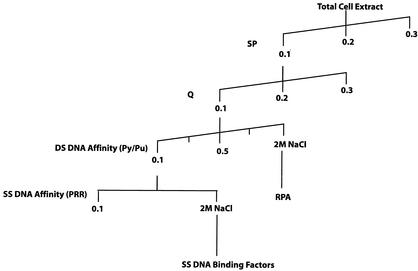
The steps followed in protein purification are outlined in Fig. 4. Total-cell extract was made from 15 liters of the Ramos cell line as described previously (29). The extract was passaged over a cation-exchange column (SP Sepharose purchased from Pharmacia) and collected as unbound Py/Pu binding activity in the flowthrough. The Py/Pu binding activity was pooled and passaged over an anion-exchange column (Q Sepharose purchased from Pharmacia) and once again collected as unbound activity. A third chromatography step involved passage over a DNA affinity column of double-stranded Py/Pu motif DNA as previously described (2), extensive washing in increasing concentrations of potassium chloride in EMSA base buffer up to a concentration of 1 M, and elution of bound proteins with 2 M NaCl. After each enrichment step, eluted proteins were dialyzed in EMSA binding buffer. The eluted proteins were analyzed by standard Coomassie blue staining methods. The protein bands were excised from the gel and subjected to trypsin digestion and matrix-assisted laser desorption ionization (MALDI)-mass spectrometry identification. Immunoblotting was performed with an ECL+Plus kit purchased from Amersham Pharmacia Biotech.
FIG. 4.
Outline of steps used in the purification of PRR binding factors. Washes and elutions, unless otherwise indicated (when a solution of sodium chloride [NaCl] was used), were performed with a molar solution of potassium chloride (KCl). DS, double stranded; SS, single stranded.
For purification of single-stranded PRR binding proteins, an affinity column of sense-strand PRR was generated. Each synthetic primer contained two PRR sites. The eluted fractions were monitored for PRR binding and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and MALDI.
Immunoprecipitation of PRR-protein complex.
We used a method adapted from a chromatin immunoprecipitation (ChIP) assay wherein antibodies recognizing PRR binding proteins were used to immunoprecipitate the prepared DNA-protein complex instead of antibodies recognizing histone proteins. A total of 106 MCF-7 cells were fixed on a petri plate with 1% formaldehyde for 10 min at 37°C and washed with ice-cold phosphate-buffered saline solution containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, and 1 μg of pepstatin A per ml). This treatment resulted in covalent cross-linking of DNA to proteins (including histones). The cells were scraped, pelleted, suspended in 200 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl), incubated on ice for 10 min, and sonicated (the DNA length, kept between 200 bp and 1 kb, was monitored by reverse cross-linking in the presence of 5 M NaCl at 65°C for 4 h with subsequent phenol-chloroform-isoamyl alcohol extraction and visualization with an agarose gel run). The sonicated sample was spun for 10 min, and the supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) supplemented with protease inhibitors. The cell supernatant was precleared by adding 80 μl of salmon sperm DNA-protein A-agarose slurry (400 μg of salmon sperm DNA per ml, 1 mg of bovine serum albumin per ml, and 3 mg of protein A per ml in 10 mM Tris-HCl [pH 8.0]-1 mM EDTA), incubating it for 30 min at 4°C, and spinning to obtain a supernatant ready for immunoprecipitation.
Approximately 5 μg of antibodies recognizing RPA1, RPA2, FBP-1, and DNA-dependent protein kinase (negative control) was added to each preparation and incubated overnight at 4°C. Sixty microliters of salmon sperm DNA-protein A agarose slurry was added and incubated for 1 h at 4°C to collect the antibody-protein-DNA complex, which was pelleted by centrifugation at 4,000 rpm for 1 min in a Microfuge (Eppendorf). The pellet was washed sequentially with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), and lithium chloride [LiCl] immune complex wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]). Finally, the antibody complex was washed twice with Tris-HCl (10 mM, pH 8.0)-EDTA (1 mM) buffer. The complex was eluted twice from the antibody by adding 200 μl of elution buffer (1% SDS, 0.1 M sodium bicarbonate) and incubating it at room temperature for 15 min. The agarose beads were spun down, and the supernatant was collected. Twenty-five microliters of 5 M NaCl was added to the supernatant, and the DNA and proteins were reverse cross-linked by heating the mixture to 65°C for 4 h. The sample was subjected to proteinase K digestion at 45°C for 1 h and then subjected to phenol-chloroform extraction and ethanol precipitation. The precipitated DNA was pelleted, washed with 70% ethanol, air dried, and dissolved in distilled water. The presence of the BRCA1 PRR element was investigated by using PCR amplification (35 cycles; 94, 65, and 72°C) with BRCA1 primers (5′-GCC GCA ACT GGA AGA GTA GAG-3′ and 5′-GAT GGG AGG GAC AGA AAG AGC-3′) encompassing the PRR.
RESULTS
PRR.
The PRR is essential for BRCA1 promoter activity (42, 44). A schematic representation of the sense strand of the BRCA1 PRR is shown in Fig. 1A. Sequence analysis of the PRR suggested the presence of two potential motifs within the PRR. The 5′ motif consists primarily of polypyrimidine and polypurine (Py/Pu) bases, with a single guanine (G) base. The second putative motif is similar to the CREB site. In addition, a previously reported (52) transcription start site is indicated.
FIG. 1.
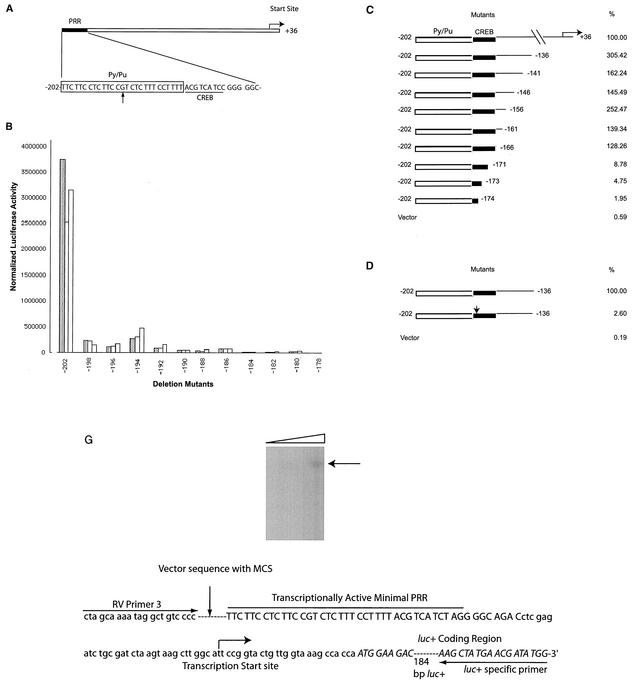
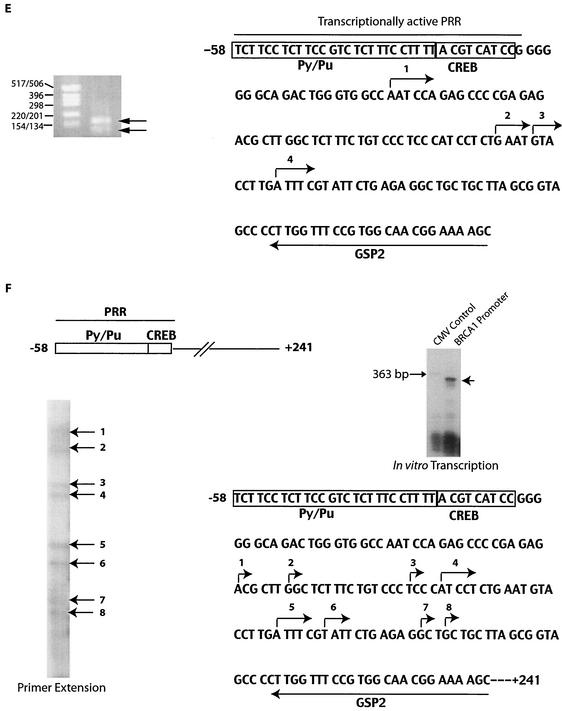
PRR of the BRCA1 promoter, results of mutation studies, and mapping of the start site. (A) Schematic representation and sequence of the PRR. The Py/Pu, CREB, and transcriptional start sites are indicated. The guanine residue in the Py/Pu site is indicated with an arrow. (B) Transcriptional effects of 5′ deletions in the PRR. The 3′ ends of all the mutant sequences correspond to position +36 of the promoter. Transfections were performed in triplicate for each construct and represent results from several repeats. (C) Transcriptional effects of the 3′ promoter deletions. The structures and relative activities (i.e., the percentages of the level of activity of the full-length PRR sequence) are shown. (D) Relative activity of the BRCA1 promoter with the site-directed mutation within the CREB site (indicated by the arrow). The results of panels C and D are from three independent transfection experiments. (E) Mapping of the BRCA1 start site was performed by 5′RACE. RACE-ready cDNA from human testis cells (purchased from AMBION) was amplified with BRCA1 GSP1 (5′-CCC AGT TAT CTG AGA AAC CCC ACA) and outer adapter primer 1 (AP1, 5′-GCT GAT GGC GAT GAA TGA ACA CTG-3′) for 35 cycles. One microliter was reamplified by using the nested primers GSP2 (indicated in the figure) and inner adapter primer 2 (AP2, 5′-CGC GGA TCC GAA CAC TGC GTT TGC TGG CTT TGA TG-3′) for 45 cycles. The amplified product was analyzed on 2% agarose gel, purified, and sequenced. Molecular size markers (in base pairs) are noted at the left of the gel. (F) Start sites used by the BRCA1 promoter in an in vitro system. An amplified fragment of the BRCA1 promoter (positions −58 to +246; primers used were 5′-GCC GCA ACT GGA AGA GTA GA-3′ and 5′-AAG GCC TCC TGA GCG CAG GG-3′) was used as template DNA. The BRCA1 promoter DNA and a control of the CMV promoter (expected to transcribe a 363-bp product) were in vitro transcribed by using HeLa nuclear extract (purchased from Promega) in the presence of radiolabeled UTP and additional ribonucleotides at 37°C for 1 h as suggested by the manufacturer. The transcribed RNA was extracted with phenol-chloroform and isoamyl alcohol, ethanol precipitated, and resolved in 10% Tris-borate-EDTA gel. Reverse transcription of the transcribed RNA was performed with avian myeloblastosis virus reverse transcriptase (purchased from Fisher Scientific) in the presence of radiolabeled cytosine. The products were electrophoresed in a 6% Tris-borate-EDTA-urea gel with dideoxy sequencing reactions (with the fMol sequencing kit from Promega) by using GSP2 with a BRCA1 promoter construct as a template. (G) Mapping of the start site(s) of the minimal BRCA1 promoter within the reporter gene vector. A construct with the BRCA1 PRR (described in the legend to Fig. 3) was used as a template to amplify a segment encompassing the PRR by utilizing vector-based primers (RV Primer 3 and the luciferase-specific primer indicated in the sequence). Start sites were mapped by primer extension analysis of transcribed luciferase RNA with a second luciferase-specific primer (5′-TTA TGC AGT TGC TCT CCA GC-3′). Increasing quantities of primer extension reaction mixtures were loaded in the lanes from left to right. The locations of the multiple cloning site (MCS) of the vector, the BRCA1 PRR, and the transcription start site are indicated. The luciferase coding region is in uppercase and italicized. Vector sequences are in lowercase, and the BRCA1 promoter is in uppercase.
PRR mutants.
In order to study the importance of the PRR in BRCA1 transcription, a series of BRCA1 promoter mutants were constructed. The transcriptional activities of the constructs were measured by reporter gene assays as previously described (44). All transfections were performed in the MCF-7 breast cancer cell line.
5′ promoter deletions.
Mutants with systematic deletions in the PRR site of the BRCA1 promoter were transfected in MCF-7 cells and measured for reporter activity (Fig. 1B). The progressive deletions impaired BRCA1 promoter activities. Deletion of 4 bases from the 5′ end of the PRR resulted in a significant decrease of promoter activity (−202 versus −198). Further deletions correlated with a trend of gradual decline in activity, except in the cases of mutants −194 and −186, for which some increase of activity was noted upon deletion of bases. Mutants −194 and −186 provided the first hint that BRCA1 transcription may be complex. Deletion of the 5′ half of the Py/Pu site (deletions after position −186) resulted in further loss of promoter activity. These results suggest the importance of an intact Py/Pu site for promoter activity.
3′ deletions.
Progressive deletions from the 3′ end of the BRCA1 promoter were also made (Fig. 1C). Surprisingly, constructs lacking the previously reported start site showed strong transcriptional activity comparable to that of the wild-type promoter. Initial deletions resulted in a modest increase of promoter activity, suggesting the presence of inhibitor sequences (mutants −136 to −166). Deletion of more than 6 bases from position −166 caused a significant drop in promoter activity (mutants −171 to −174). Progressive 3′ deletion experiments helped to identify a minimal fragment of 37 bp (positions −202 to −166), which was transcriptionally active and encompassed the PRR.
CREB site mutation.
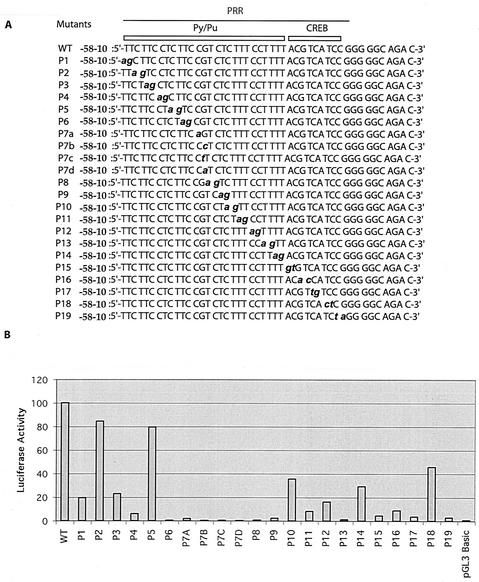
The mutants with 3′ deletions in the PRR (Fig. 1C) indicated the importance of the CREB site in BRCA1 transcription. Deletion of 5 bases within the CREB site (mutant −171) abolished the transcriptional activity of the BRCA1 promoter. In order to further confirm the importance of this site, we generated a BRCA1 promoter construct (positions −202 to −136) in which the CREB site was mutated. A mutation of the first 3 bases (ACG) of the CREB site to thymidines resulted in significantly decreased activity (Fig. 1D). In addition, the introduction of point mutations (see Fig. 3) (mutants P15 to P19) within the CREB site also impaired transcriptional activity.
FIG. 3.
Transcriptional activities of PRR point mutations. (A) The mutated bases in the PRR are in lowercase, italicized, and boldface letters. (B) Transcriptional activities of PRR point mutations. The vector control (pGL3 basic) is indicated. WT, wild type.
The results suggested that the PRR may function as an enhancer for the cryptic promoter(s) present within the reporter gene vectors. However, it was observed that the PRR was orientation and position dependent, that it did not activate a heterologous promoter (data not shown), and therefore that it did not fit the classic definition of an enhancer. Another possibility was that the BRCA1 promoter was encompassed within the PRR and that it used an alternative site(s) (distinct from the ones reported) for transcription initiation.
Transcriptional start sites of the BRCA1 promoter.
The possibility of an alternative start site(s) in close proximity to the PRR was investigated by using rapid amplification of the 5′ cDNA ends (5′RACE). Initially, our attempts to identify the BRCA1 transcription start site were confounded by amplification of the 5′ ends of transcripts derived from the BRCA1 pseudogene (8; data not shown). Therefore, primers were designed by taking into account the expression of transcripts derived from the pseudogene. Template cDNAs derived from human testes were used, as testes express the highest level of BRCA1 transcripts (46). Amplifications resulted in the detection of cDNA moieties which were detected as two bands of 160 and 90 bases (Fig. 1E, left panel). The cDNAs were subcloned and sequenced. Four potential start sites were mapped by this method and are indicated. Since the 160-bp cDNA was predominant and represented the site upstream of all the reported (and detected) sites, it was used for marking the start site in subsequent descriptions. This start site (labeled number 1) is 21 bases downstream of the 3′ end of the transcriptionally active PRR. The 5′end of PRR was accordingly renumbered at −58.
In addition to 5′RACE, primer extension experiments with in vitro-transcribed RNA expressed from the BRCA1 promoter were performed. As a promoter template, an amplified product containing the PRR, the newly mapped start sites, and 241 downstream bases (Fig. 1F, upper-left panel) was transcribed with HeLa nuclear extracts. A control lane expressing a product (363 bp) transcribed by the CMV promoter was included (lane 1, upper-right panel). BRCA1 promoter-transcribed products consistent with approximate sizes of 180 to 200 bp were detected (lane 2). It proved difficult to map the 5′ residues of these RNAs directly by comparison with sequences from sequencing reactions due to RNA degradation in the sequencing gels. Therefore, primer extension experiments (performed by using gene-specific primer 2 [GSP2]) were used to generate more-stable cDNAs (lower-left panel). The in vitro-transcribed RNAs were reverse transcribed in the presence of radiolabeled UTP. The radiolabeled cDNAs were electrophoresed in a sequencing gel with dideoxy sequencing reaction mixtures containing the BRCA1 promoter by using GSP2 (data not shown). Eight start sites were detected and are indicated (lower-right panel). Interestingly, only one start site (site number 5) was identical to the one identified by 5′RACE (site number 4 of Fig. 1E). The transcription start site was not detected within the PRR itself, suggesting that basal transcription machinery assembles within the PRR and initiates transcription 21 to 100 bases downstream of the PRR. Deletion of promoter bases downstream of the PRR does not inhibit promoter activity (Fig. 1C), suggesting that the PRR is utilized for assembly and priming of the basal machinery, which subsequently initiates transcription by using downstream nonspecific vector sequences.
In order to map the start sites used in reporter constructs by BRCA1 promoters with 3′ deletions, additional 5′ mapping experiments were performed (Fig. 1G). An amplified product containing the BRCA1 PRR, vector sequences, and a section of the luciferase reporter gene (shown in the sequence) was used as template DNA and transcribed. Transcripts of variant sizes were detected (data not shown), and the 5′ ends of these products were mapped by primer extension analysis (upper panel). The experiments suggested the presence of a start site (5′-TTCCG… .) in the vector sequence upstream of the luciferase cDNA. Additional higher-mobility bands reflecting start sites in the coding region of luciferase cDNA were also detected (data not shown), though the derivative RNAs would not be translated into full-length luciferase proteins.
BRCA1 promoter or PRR binding studies.
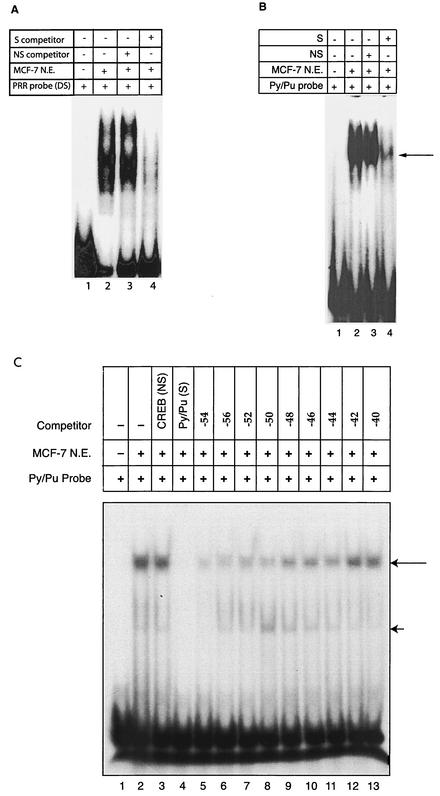
The double-stranded PRR was used as a probe with MCF-7 (breast cancer cell line) nuclear extracts in EMSAs (Fig. 2A). Diffuse DNA-protein complexes were observed (lane 2). Competition with nonspecific oligonucleotides had no significant effect on DNA-protein complexes (lane 3). In contrast, specific competitors abolished the complexes (lane 4).
FIG. 2.
PRR binding studies. (A and B) EMSA was performed with PRR (A) and Py/Pu (B) probes. Specific (S) competitions were performed with a 100-fold excess of double-stranded, annealed, and unlabeled PRR and Py/Pu oligomers. The arrow in panel B indicates the distinct DNA-protein complexes formed by the Py/Pu probe. Nonspecific (NS) competition was performed with a synthetic oligomer (5′-GTC ACT ATG GCT TTC AAT TGG CCC GGC ATA G-3′) annealed to its complementary sequence. (C) DNA binding affinities of mutant constructs with systematic deletions in the PRR. Nonspecific (NS) and specific (S) competitions were performed with the CREB and Py/Pu oligomers, respectively. Additional competitions were performed with the Py/Pu oligomers with systematic 5′ deletions. The long arrow indicates the distinct DNA-protein complex formed by the MCF-7 nuclear extract. The short arrow indicates diffuse and streaked high-mobility DNA-protein complexes. N.E., nuclear extract; DS, double stranded.
EMSAs with DNA probes derived from the Py/Pu and CREB sequences were performed to characterize the factors that bound these sites individually. The Py/Pu probe formed distinct DNA-protein complexes (Fig. 2B, lane 2). These complexes were not affected by nonspecific competition (lane 3). Specific competition disrupted the DNA-protein complexes (lane 4). Thus, Py/Pu protein binding was specific.
Although specific protein binding to the CREB site has been reported previously (28), we did not detect protein binding (data not shown), suggesting that CREB binding may be cell line specific. Significantly, mutations in the CREB site resulted in downregulation of promoter activity (Fig. 1D and 3), indicating the functional importance of the motif. Therefore, the CREB site may function in protein binding in the context of the full-length PRR.
EMSAs with double-stranded Py/Pu domain constructs and competitions with oligonucleotides with progressive deletions matching the deletion promoter constructs in Fig. 1B were performed (Fig. 2C). Incubation of the probe with the MCF-7 nuclear extract resulted in a distinct DNA-protein complex (lane 2). Competition with a nonspecific probe had no effect (lane 3), while specific competition abolished the DNA-protein complex (lane 4). Competitions with oligonucleotides with progressive deletions showed a progressive loss in the ability of these oligonucleotides to compete for protein binding (lanes 5 to 13). The decreases in the binding affinities of competing mutant oligonucleotides were largely concordant with the loss of mutant promoter activities. These experiments reinforced the importance of an intact Py/Pu motif for normal activity of the BRCA1 promoter. Interestingly, we occasionally detected diffuse and streaked high-mobility DNA-protein complexes.
Point mutations of the PRR domain.
Identification of the functional BRCA1 transcriptional unit provided us with a reagent with which to perform a rapid mutation analysis of this region (Fig. 3). The template used for mutagenesis ranged from positions −58 to −10 (extra bases were included to provide flanking sequences). Sequential dinucleotide and mononucleotide point mutations were introduced across the PRR (Fig. 3A). Strikingly, the majority of mutants demonstrated significant loss of activity compared to that of the wild type (Fig. 3B). Particularly, mutations in the center of the Py/Pu domain (P6 to P9) caused a loss of over 95 to 99% of the activity. Mutations in the CREB site (P15 to P19) resulted in a decline of 90 to 95% of promoter activity (except P18). The mutagenesis studies suggested the existence of a complex mechanism influencing BRCA1 transcription in which regulation was dependent upon the recruitment of multiple proteins to the PRR. Thus, efforts to identify PRR binding factors were initiated.
Purification of PRR binding factors.
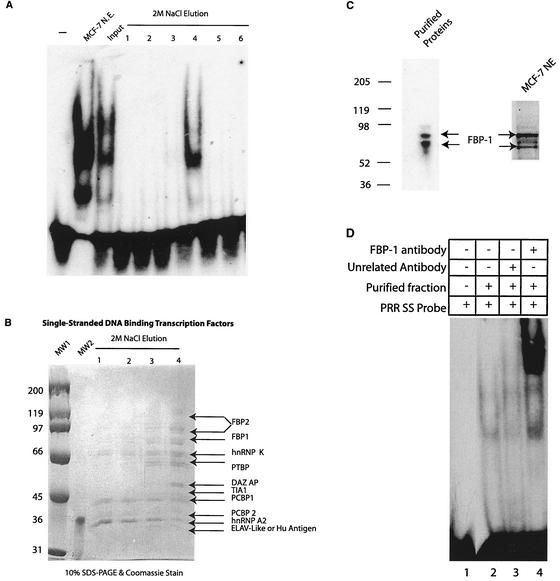
Chromatographic protocols were followed to enrich the PRR binding factors (Fig. 4). Ramos B-cell leukemic cells were used to prepare total-cell extracts, as this cell line could be grown rapidly in large volumes, as opposed to adherent breast cell-derived lines. The approach was adopted with a view to extending the results of these studies to breast cells. As shown below, Ramos and MCF-7 cells exhibited similar PRR binding activities (see Fig. 6A, MCF-7 versus Ramos cell input).
FIG. 6.
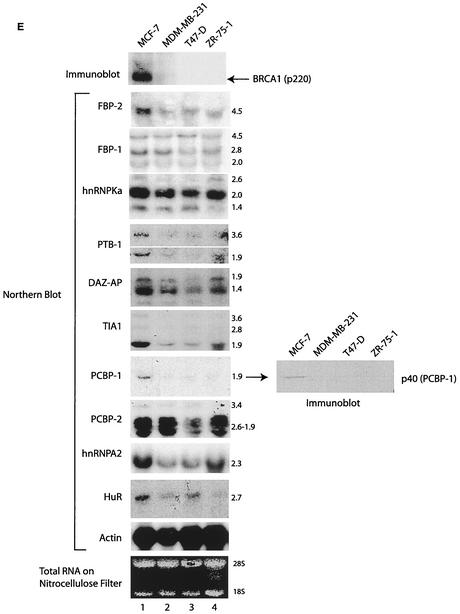
Purification and characterization of factors binding the PRR sense strand. (A) Fractions eluted from the single-stranded PRR affinity column were analyzed by EMSA with a single-stranded PRR probe. An EMSA was also performed with MCF-7 nuclear extracts (N.E.) and partially purified input. (B) The eluted fractions (collected from the single-stranded PRR affinity column) were analyzed by SDS-PAGE and Coomassie blue staining. The identified proteins are indicated. MW1 and MW2, molecular weight markers. (C) Immunoblot of the purified PRR binding fraction and MCF-7 nuclear extract (NE) with FBP-1 antiserum. (D) Supershift experiment with FBP-1 antiserum and purified proteins. Unrelated antibody recognizing DNA-dependent protein kinase (N-20, purchased from Santa Cruz) was also applied as a control. SS, single stranded. (E) Immunoblot and Northern blot analyses of the indicated proteins and genes. Immunoblotting was performed with BRCA1 D-9 and PCBP-1 rabbit polyclonal antibody. Northern blotting was performed by using probes derived from the respective cDNAs. RNA quantity was normalized by Northern blotting with the actin probe and by visualization of ethidium bromide-stained RNA bound to the nitrocellulose filter. Representative panels for actin blots as well as for stained RNA shown. Numbers at the left (B and C) and to the right (E) of the Northern blots indicate molecular size markers (in kilobases).
Initial enrichment of the 300-mg total extract was performed by cation (SP Sepharose)- and anion (Q Sepharose)-exchange chromatography, and the PRR binding activity was monitored by EMSA (data not shown). As the Py/Pu region was observed to be important in BRCA1 promoter function (Fig. 1B) and also demonstrated specific protein binding (Fig. 2B and C), double-stranded Py/Pu binding proteins were purified and identified as the RPA factor. As it was subsequently observed that single-stranded PRR had a specific affinity for proteins, a second round of DNA affinity chromatography with single-stranded PRR was used to purify the binding factors (shown below).
Purification and characterization of double-stranded Py/Pu binding proteins.
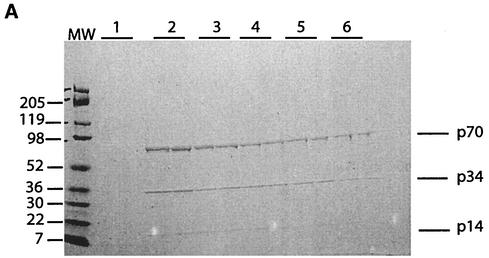
The eluted Py/Pu binding activity was analyzed by SDS-PAGE and Coomassie blue staining. Three proteins with molecular sizes of 70,000, 34,000, and 14,000 (p70, p34, and p14, respectively) were observed (Fig. 5A). These bands were excised and identified by trypsin digestion and MALDI-mass spectrometry. The three proteins constitute a trimer of the RPA factor (with p70 being equivalent to RPA1, p34 being equivalent to RPA2, and p14 being equivalent to RPA3) and have previously been shown to be essential for DNA replication initiation (51).
FIG. 5.
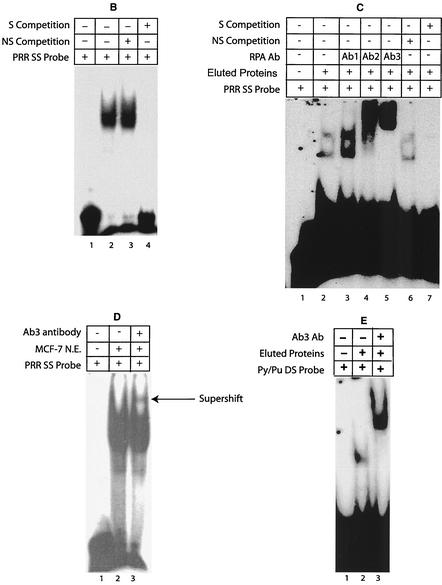
Purification and characterization of Py/Pu binding proteins. (A) Eluted fractions were analyzed by SDS-PAGE and Coomassie blue staining. Each fraction collected from the DNA affinity column was electrophoresed in two lanes. Numbers at the left are molecular weight (MW) markers (in thousands). (B) Single-stranded PRR-specific binding was assessed by using MCF-7 nuclear extract. Nonspecific (NS) (5′-CAA GCG GCC ATC TTG GGT CCA AGC GGC CAT CTT GGG T-3′) and specific (S) competitions were performed. (C) Supershift experiments with RPA antibodies. Monoclonal antibodies (Ab1, Ab2, and Ab3, purchased from Oncogene Research) were used in supershift assays. The specificities of DNA-protein complexes were tested by nonspecific and specific competitions. (D) Supershift experiment with Ab3 antibody and MCF-7 nuclear extract. (E) Binding of purified proteins to the double-stranded Py/Pu probe and supershifting by Ab3. SS, single stranded; DS, double stranded; Ab, antibody; N.E., nuclear extract.
RPA is known to have specific single-stranded DNA binding activity and an apparent affinity for double-stranded DNA (22). These results suggest that single-stranded DNA binding factors such as RPA may bind to the PRR, although earlier studies were all performed with annealed, double-stranded probes. Therefore, EMSAs were performed with a probe made from the sense strand of PRR and MCF-7 nuclear extracts (Fig. 5B). Strong binding of proteins to the single-stranded PRR was observed in the MCF-7 extract (lane 2). Competition studies indicated that the protein binding was specific (lanes 3 and 4). In contrast to the sense-strand PRR probe, the antisense PRR probe did not exhibit specific protein binding (data not shown).
To confirm that purified RPA factor eluted from the double-stranded Py/Pu column bound the single-stranded PRR, additional EMSAs were performed (Fig. 5C), and two DNA-protein bands were detected (lane 2). Monoclonal antibodies recognizing RPA were added to EMSA reaction mixtures (lanes 3 to 5). Ab1 (anti-p70) caused increased DNA-protein binding (lane 3), and the Ab2 and Ab3 (anti-p34) antibodies supershifted the DNA-protein complexes (lanes 4 and 5), thereby confirming RPA's binding to the PRR. The addition of unrelated antibody had no effect on the DNA-protein complex (data not shown). Competition studies with specific and nonspecific oligonucleotides indicated that the binding was specific (lanes 6 and 7).
An additional supershift experiment with MCF-7 nuclear extracts and Ab3 antibody suggested the presence of RPA2 in the single-stranded PRR DNA-protein complex formed (Fig. 5D, lane 3).
Purified RPA bound double-stranded Py/Pu as expected (lane 2), since it was used as bait to purify RPA, and the resultant DNA-protein complex was supershifted by Ab3 antibody (Fig. 5E, lane 3). The affinity of RPA for single- as well as double-stranded DNA suggests a specific role of RPA in the binding and unwinding of double-stranded DNA. Cotransfections with RPA-expressing constructs had no significant effect on BRCA1 promoter activity (data not shown), suggesting that RPA's effects on BRCA1 transcription may involve complex protein interactions and/or modifications which are discussed below.
Though RPA factor bound the PRR, several lines of evidence suggested that RPA factor alone was not involved in PRR binding and the regulation of BRCA1 transcription. Significantly, site-directed mutation studies suggested the existence of multicomponent transcription regulation through the PRR (Fig. 3). In addition, the large PRR-protein complexes formed in EMSAs with a single-stranded PRR probe were not consistent with RPA binding alone (Fig. 5B). RPA2 antibody did not supershift the bulk of the DNA-protein complex, suggesting that the majority of PRR binding factors were not RPA (Fig. 5D). Finally, whereas the full-length PRR was essential for BRCA1 promoter function, RPA bound to a subset of the PRR, the Py/Pu motif. To reconcile these inconsistencies and to further clarify the regulatory process of BRCA1 transcription, we investigated the factors binding the sense strand of full-length PRR.
Purification and characterization of single-stranded PRR binding proteins.
In the purification procedure, single-stranded PRR binding activity eluted as an unbound form from the Py/Pu double-stranded DNA affinity column (Fig. 4). This activity was pooled and passaged through a single-stranded PRR affinity column, washed with 0.1 M KCl, and step eluted with 2 M NaCl. The eluted fractions were analyzed for PRR binding activity (Fig. 6A). In addition, as controls, EMSAs were performed with MCF-7 nuclear extracts and the input fraction. Significantly, the PRR-protein complexes formed by factors from Ramos (input) and MCF-7 lines were similar. This result suggested similarity in the compositions of the PRR binding proteins in Ramos and MCF-7 cells. Specific PRR binding activity, which was similar to that generated by partially purified input, was recovered from the affinity column (lane 4 of eluted fractions).
Four eluted fractions were analyzed by SDS-PAGE and Coomassie blue staining, followed by identification of stained protein bands by the MALDI procedure (Fig. 6B). The 10 proteins identified by this method included far-upstream element (FUSE) binding proteins 1 and 2 (FBP-1 and -2) (12), heterogeneous nuclear ribonucleoprotein K (hnRNP K) (31), polypyrimidine tract-binding protein (PTBP) (18, 35), deleted- in-azoospermia-associated protein (DAZ AP) (49), T-cell intercellular antigen 1 (TIA-1) (1), poly(ribocytosine) binding proteins 1 and 2 (PCBP-1 and -2) (48), hnRNP A2 (9, 20), and ELAV-like protein (25). Many of these proteins are known to bind RNA and regulate its processing. Interestingly, FBP-1, FBP-2, and hnRNP K have been demonstrated to be DNA binding proteins as well and are known to possess transcription-regulatory activities (14). Significantly, they were reported to regulate the expression of c-Myc, a protein critical in the regulation of human malignancies. The hnRNP C-like protein (which was also detected as a PRR-associated protein [data not shown]) has also been reported to activate the transcription of hepatitis B virus enhancer II (43).
As FBP-1 is known to regulate c-Myc promoter activity and was present in the fractions containing factors with affinity for the BRCA1 transcriptional regulatory site, we investigated its role in BRCA1 promoter activity. The presence of FBP-1 in the fraction eluted from the single-stranded PRR column was confirmed by immunoblot analysis (Fig. 6C), and two major isoforms (60 and 65 kDa) were observed (left panel). Similar results were also obtained by immunoblotting MCF-7 nuclear extracts (right panel). Furthermore, the addition of FBP-1 antibody to EMSA mixtures with a single-stranded PRR probe and eluted fraction (from single-stranded PRR column) resulted in increased protein binding and supershifting (Fig. 6D, lane 3). An unrelated antibody had no effect on the DNA-protein complex (lane 2). These experiments confirmed the presence of FBP-1 in the factors binding the PRR. Initial cotransfection experiments with FBP-1 with short segments of the BRCA1 promoter showed no effects (data not shown). However, when FBP-1 was coexpressed with larger segments of the promoter, transcription was suppressed (see below).
Correlation of expression patterns of BRCA1 and PRR binding factors.
The expression pattern of BRCA1 and transcripts of purified proteins were investigated in a panel of breast cancer cell lines (Fig. 6E). Immunoblot analyses were performed with BRCA1 antibody with the nuclear extracts of the indicated cell lines. A 220-kDa band (consistent with the size of BRCA1 protein) was detected in the MCF-7 cells (lane 1). The BRCA1 protein was expressed at significantly reduced levels in MDM-MB-231, T47D, and ZR-75-1 cells (lanes 2 to 4). The protein was detected upon prolonged exposure (data not shown). The amount of loaded protein was normalized by actin blotting and staining of the filters with Ponsceau S during the immunoblot procedures (data not shown).
The potential coordinate expression of BRCA1 and purified factors was investigated. Northern blotting was performed with probes derived from cDNA sequences of purified proteins. Sizes of the transcripts (in kilobases) are indicated (Fig. 6E). Significantly, all the genes tested were expressed robustly in the BRCA1-positive MCF-7 line and never at levels less than those in lines with reduced BRCA1 expression (Fig. 6E, lane 1 versus lanes 2 to 4). Significant alteration in the expression of FBP-1, hnRNP KA, and DAZ AP transcripts were not apparent in the cell lines studied. Variously reduced expression of FBP-2, PTBP-1, TIA-1, hnRNP A2, PCBP-2, and HuR was noted in the MDM-MB-231, T47D, and ZR-75-1 lines. Strikingly, PCBP-1 transcripts showed reduced expression in all breast cells expressing diminished quantities of the BRCA1 protein. This finding also suggested decreased expression of PCBP-1 protein in the indicated cell lines. Therefore, immunoblotting was performed with PCBP-1 antibody to confirm the possibility (Fig. 6E, right panel). A 40-kDa protein consistent with the size of PCBP-1 was observed in the MCF-7 cells. Reduction of the steady-state level of p40 was observed in the MDM-MB-231, T47-D, and ZR-75-1 cells.
PRR binding factors and BRCA1 transcription.
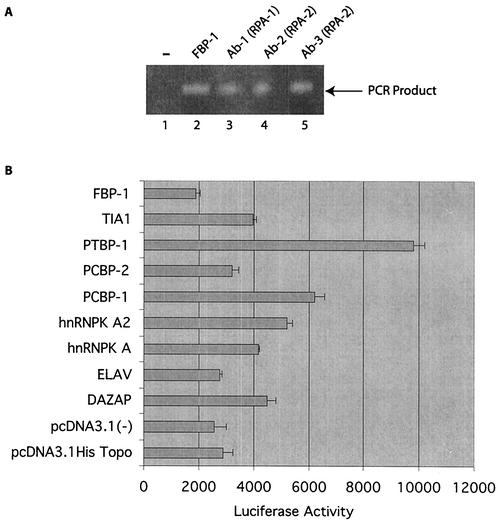
The influence of purified factors on BRCA1 transcription was investigated. In the first series of experiments, immunoprecipitation assays (adapted from the ChIP assay) with extracts from MCF-7 cells were performed with FBP-1 and RPA antibodies (Fig. 7A). The purified DNAs procured after the immunoprecipitation procedures (described in Materials and Methods) were subjected to PCR amplification with primers encompassing the PRR. The negative-control extract (immunoprecipitation with nonspecific DNA-PK antibody) was not amplified (lane 1). In contrast, after immunoprecipitation with FBP-1 and RPA antibodies, an amplified product of the expected size was observed (lanes 2 to 5). The experiment indicates the association of the FBP-1 and RPA antibodies with chromatin-bound PRR.
FIG. 7.
Relationship between single-stranded DNA binding factors and the BRCA1 gene. (A) Immunoprecipitation of the DNA-protein complex was performed with the indicated specific antibodies. The DNA procured from the complex (described in Materials and Methods) was amplified with primers from the PRR. (B) Cotransfection of the BRCA1 promoter with expression constructs of the indicated factors. One microgram of the full-length BRCA1-luciferase construct was cotransfected with 2 μg of the indicated constructs in MCF-7 cells and incubated for 48 h. Standard reporter gene assays were performed. Each transfection was performed in triplicate, the mean values of which were charted with error bars.
Finally, the full-length BRCA1 promoter construct was cotransfected with cDNA expression constructs of PRR binding factors (Fig. 7B). It was observed that a subset of factors (DAZ AP, hnRNP Ka, hnRNP Ka2, PCBP-1, PCBP-2, and TIA-1) exhibited a mild but consistent activation of the promoter. The expression construct of PTBP-1 was noted to have a relatively strong (fourfold increase) positive effect. In contrast, FBP-1 showed a mild, yet consistent, suppressive effect on promoter activity. PCBP-2 and ELAV did not exhibit any effect on BRCA1 transcription. Cotransfected vector controls, pcDNA 3.1 V5 His Topo and pcDNA 3.1, did not elicit any response. The transfection experiment was performed twice independently to corroborate the results.
DISCUSSION
It is accepted that the loss of function of the BRCA1 gene contributes to the development of breast cancer. Significantly reduced levels of BRCA1 mRNA in the majority of high-grade breast cancer cases suggest aberrant transcription. Therefore, studies were carried out to elucidate the factors regulating BRCA1 promoter function. Results of the present study indicate that BRCA1 transcription is regulated by specific factors binding single-stranded DNA. These factors include RPA and a family of proteins which are also known to bind RNA. A subset of these proteins are known to bind specific DNA sequences and to direct transcription.
The demonstration that BRCA1 transcription is regulated by a specific single-stranded sequence is supported by previously reported results of similar studies of a eukaryotic promoter (14, 23). Specifically, it has been shown that FBP-1 and -2 and hnRNP K bind to the positive cis-acting sequence (far-upstream element) on one strand of the human c-Myc gene and promote transcription. The binding to PRR likely involves the displacement of the nonspecific band and the binding of the specific sense-strand sequences. The higher-mobility bands frequently observed in EMSAs with the Py/Pu probe are likely to form due to strand displacement and binding of the probe to single-stranded DNA binding factors (Fig. 2C).
The binding of multiple proteins (including RPA) to the PRR suggests complex regulation of BRCA1, consistent with the upregulation of BRCA1 in developing cells with a rapid rate of proliferation (21, 30). Interestingly, gel filtration experiments indicate that these proteins do not form a single complex but rather form several multimeric complexes (ranging from 40 to 170 kDa), which appear to independently compete (or cooperate) in binding to the PRR (data not shown). Adding complexity to the spectrum of related factors associated with the PRR is the fact that this gamut of proteins is subject to transcriptional, posttranscriptional, and posttranslational alterations. The complex and multicomponent assembly of these single-stranded DNA binding modular proteins on the PRR and alterations in them may be developmentally regulated and tissue specific. The essence of regulation is likely to involve complex interactions of multiple components aggregating on the PRR.
The strong expression of these genes in MCF-7 cells, in relation to expression levels in other breast cell lines, supports the possibility of a role for these proteins in BRCA1 transcription (Fig. 6E, lane 1 versus lanes 2 to 4). One of the proteins, PCBP-1, which has a positive influence on BRCA1 transcription, had reduced expression in lines expressing reduced BRCA1. Interestingly, PCBP-1 and PTBP-1 (which has a greater influence on BRCA1 transcription) are known to possess affinity for pyrimidine-rich nucleotide sequences (11), an observation consistent with their role in associating with the PRR and influencing BRCA1 transcription. PTBP-1 has been suggested to influence the transcription of growth-related genes like Myc and Ras (39); it may also play a role in the development of normal breast cells as well as breast cancer cells. Interestingly PTBP-1 transcripts were observed to be expressed in low quantities in two of the three lines (MDM-MB-231 and T47-D) with reduced BRCA1 transcripts (Fig. 6E).
BRCA1 is known to be cell cycle regulated in cells grown in tissue culture (19) and upregulated in proliferating cells undergoing the process of differentiation and maturation. These reports make the notion that DNA replication may influence BRCA1 transcription an attractive one. Purification of RPA as a PRR binding factor that is essential for replication initiation supports this model. The affinity of RPA for the PRR suggests that replication and BRCA1 transcription may be directly linked. The processes of replication and transcription are known to influence each other (33, 41). RPA and other PRR binding factors may participate and interact in a replication initiation-transcription activation complex, which may be dynamically regulated in proliferating cells undergoing maturation. A specific role of RPA in binding and unwinding double-stranded DNA is also plausible.
It has been suggested that single-stranded factors binding their target site may be coupled with torsional stress due to transcription at nearby promoters (4, 14, 23). The accumulation of supercoils generated by the activity of nearby promoters may be released by generating single strands. Specific protein binding to unwound DNA may stabilize the single strands and contribute to the release of transcriptionally generated supercoiling. Since the BRCA1 promoter is known to be in proximity to the IAI-3B gene (7), transcription of IA1-3B may augment torsional stress on the PRR and generate single strands. In addition to alleviating torsional stress, components of the PRR binding factors are likely to influence basal transcription machinery as well. It is already known that FBP-1 interacts with a transcription repressor, which associates with TFIIH in initiation complex (23, 24). Therefore, FBP-1's suppressive effect may be partially mediated through the function of TFIIH.
In summation, the specific binding of RPA and single-stranded DNA binding factors, coupled with changes in the torsional stress of DNA, may introduce topological changes which could alter and influence interactions between the PRR-protein complexes and the associated basal transcription machinery. These complex interactions would most likely be affected by changes in the states of chromatin during cell cycling and development.
The involvement of single-stranded DNA binding factors in BRCA1 transcription suggests a role for these proteins in breast cell development. It also suggests that aberrant function or expression of these proteins may contribute to the loss of BRCA1 transcription and lead to the development of a neoplastic phenotype. Since PRR binding proteins also bind to RNA, defects in them may also result in aberrant RNA processing and thereby result in abnormal gene expression. Investigation of this functional molecular cascade of single-stranded DNA binding proteins may yield information with regard to the development of breast cancer.
Acknowledgments
This study was supported by program project grants 5P30CA56036 (Translational Research in Cancer) and 5PO1CA76259 (Oncogene Deregulation) from U.S. Public Health Sciences to Carlo M. Croce and by Department of Defense breast cancer fellowship DAMD17-98-1-8170 to Sanjay Thakur.
We thank Arnold B. Rabson for critical reading of the manuscript and helpful suggestions. We also thank Richard Fishel, Marc Wold for RPA cDNA clones, David Levens for FBP-1 reagents, and Maria F. Czyzyk-Krzeska for PCBP-1 antibody. We are also grateful to Devjani Chatterjee at the KCI computing facility for computer-assisted analysis of the sequences of the purified factors.
REFERENCES
- 1.Anderson, P. 1995. TIA-1: structural and functional studies on a new class of cytolytic effector molecule. Curr. Top. Microbiol. Immunol. 198:131-143. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. DNA protein interactions, p. 2.12.1-2.12.11. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 3.Baldwin, R. L., E. Nemeth, H. Tran, H. Shvartsman, I. Cass, S. Narod, and B. Y. Karlan. 2000. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 60:5329-5333. [PubMed] [Google Scholar]
- 4.Bazar, L., D. Meighen, V. Harris, R. Duncan, D. Levens, and M. Avigan. 1995. Targeted melting and binding of a DNA regulatory element by a transactivator of c-Myc. J. Biol. Chem. 270:8241-8248. [DOI] [PubMed] [Google Scholar]
- 5.Beger, C., L. N. Pierce, M. Kruger, E. G. Marcusson, J. M. Robbins, P. Welcsh, P. J. Welch, K. Welte, M. C. King, J. R. Barber, and F. Wong-Staal. 2001. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl. Acad. Sci. USA 98:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianco, T., G. Chenevix-Trench, D. C. Walsh, J. E. Cooper, and A. Dobrovic. 2000. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis 21:147-151. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. A., H. Nicolai, C. F. Xu, B. L. Griffiths, K. A. Jones, E. Solomon, L. Hosking, J. Trowsdale, D. M. Black, and R. McFarlane. 1994. Regulation of BRCA1. Nature 372:733.. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. A., C. F. Xu, H. Nicolai, B. Griffiths, J. A. Chambers, D. Black, and E. Solomon. 1996. The 5′ end of the BRCA1 gene lies within a duplicated region of human chromosome 17q21. Oncogene 12:2507-2513. [PubMed] [Google Scholar]
- 9.Burd, C. G., M. S. Swanson, M. Gorlach, and G. Dreyfuss. 1989. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc. Natl. Acad. Sci. USA 86:9788-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catteau, A., W. H. Harris, C. F. Xu, and E. Solomon. 1999. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18:1957-1965. [DOI] [PubMed] [Google Scholar]
- 11.Czyzyk-Krzeska, M. F., and J. E. Beresh. 1996. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J. Biol. Chem. 271:3293-3299. [DOI] [PubMed] [Google Scholar]
- 12.Davis-Smyth, T., R. C. Duncan, T. Zheng, G. Michelotti, and D. Levens. 1996. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 271:31679-31687. [DOI] [PubMed] [Google Scholar]
- 13.Dobrovic, A., and D. Simpfendorfer. 1997. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 57:3347-3350. [PubMed] [Google Scholar]
- 14.Duncan, R., L. Bazar, G. Michelotti, T. Tomonaga, H. Krutzsch, M. Avigan, and D. Levens. 1994. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8:465-480. [DOI] [PubMed] [Google Scholar]
- 15.Esteller, M., J. M. Silva, G. Dominguez, F. Bonilla, X. Matias-Guiu, E. Lerma, E. Bussaglia, J. Prat, I. C. Harkes, E. A. Repasky, E. Gabrielson, M. Schutte, S. B. Baylin, and J. G. Herman. 2000. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J. Natl. Cancer Inst. 92:564-569. [DOI] [PubMed] [Google Scholar]
- 16.Fearon, E. R. 2000. BRCA1 and E-cadherin promoter hypermethylation and gene inactivation in cancer-association or mechanism? J. Natl. Cancer Inst. 92:515-517. [DOI] [PubMed] [Google Scholar]
- 17.Futreal, P. A., Q. Liu, D. Shattuck-Eidens, C. Cochran, K. Harshman, S. Tavtigian, L. M. Bennett, A. Haugen-Strano, J. Swensen, Y. Miki, et al. 1994. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266:120-122. [DOI] [PubMed] [Google Scholar]
- 18.Gil, A., P. A. Sharp, S. F. Jamison, and M. A. Garcia-Blanco. 1991. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 5:1224-1236. [DOI] [PubMed] [Google Scholar]
- 19.Gudas, J. M., T. Li, H. Nguyen, D. Jensen, F. J. Rauscher, III, and K. H. Cowan. 1996. Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ. 7:717-723. [PubMed] [Google Scholar]
- 20.Kumar, A., K. R. Williams, and W. Szer. 1986. Purification and domain structure of core hnRNP proteins A1 and A2 and their relationship to single-stranded DNA-binding proteins. J. Biol. Chem. 261:11266-11273. [PubMed] [Google Scholar]
- 21.Lane, T. F., C. Deng, A. Elson, M. S. Lyu, C. A. Kozak, and P. Leder. 1995. Expression of Brca1 is associated with terminal differentiation of ectodermally and mesodermally derived tissues in mice. Genes Dev. 9:2712-2722. [DOI] [PubMed] [Google Scholar]
- 22.Lao, Y., C. G. Lee, and M. S. Wold. 1999. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry 38:3974-3984. [DOI] [PubMed] [Google Scholar]
- 23.Liu, J., S. Akoulitchev, A. Weber, H. Ge, S. Chuikov, D. Libutti, X. W. Wang, J. W. Conaway, C. C. Harris, R. C. Conaway, D. Reinberg, and D. Levens. 2001. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104:353-363. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., L. He, I. Collins, H. Ge, D. Libutti, J. Li, J. M. Egly, and D. Levens. 2000. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 5:331-341. [DOI] [PubMed] [Google Scholar]
- 25.Ma, W. J., S. Cheng, C. Campbell, A. Wright, and H. Furneaux. 1996. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271:8144-8151. [DOI] [PubMed] [Google Scholar]
- 26.Magdinier, F., L. M. Billard, G. Wittmann, L. Frappart, M. Benchaib, G. M. Lenoir, J. F. Guerin, and R. Dante. 2000. Regional methylation of the 5′ end CpG island of BRCA1 is associated with reduced gene expression in human somatic cells. FASEB J. 14: 1585-1594. [DOI] [PubMed] [Google Scholar]
- 27.Magdinier, F., S. Ribieras, G. M. Lenoir, L. Frappart, and R. Dante. 1998. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene 17:3169-3176. [DOI] [PubMed] [Google Scholar]
- 28.Mancini, D. N., D. I. Rodenhiser, P. J. Ainsworth, F. P. O'Malley, S. M. Singh, W. Xing, and T. K. Archer. 1998. CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene 16:1161-1169. [DOI] [PubMed] [Google Scholar]
- 29.Manley, J. L., A. Fire, A. Cano, P. A. Sharp, and M. L. Gefter. 1980. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl. Acad. Sci. USA 77:3855-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquis, S. T., J. V. Rajan, A. Wynshaw-Boris, J. Xu, G. Y. Yin, K. J. Abel, B. L. Weber, and L. A. Chodosh. 1995. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat. Genet. 11:17-26. [DOI] [PubMed] [Google Scholar]
- 31.Matunis, M. J., W. M. Michael, and G. Dreyfuss. 1992. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 33.Murakami, Y., and Y. Ito. 1999. Transcription factors in DNA replication. Front. Biosci. 4:D824-D833. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, Y., T. Oyama, and T. Nakajima. 2000. BRCA1 expression status in relation to DNA methylation of the BRCA1 promoter region in sporadic breast cancers. Jpn. J. Cancer Res. 91:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patton, J. G., S. A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237-1251. [DOI] [PubMed] [Google Scholar]
- 36.Rice, J. C., and B. W. Futscher. 2000. Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation and chromatin condensation of the BRCA1 promoter. Nucleic Acids Res. 28:3233-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, J. C., K. S. Massey-Brown, and B. W. Futscher. 1998. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene 17:1807-1812. [DOI] [PubMed] [Google Scholar]
- 38.Rice, J. C., H. Ozcelik, P. Maxeiner, I. Andrulis, and B. W. Futscher. 2000. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis 21:1761-1765. [DOI] [PubMed] [Google Scholar]
- 39.Rustighi, A., M. A. Tessari, F. Vascotto, R. Sgarra, V. Giancotti, and G. Manfioletti. 2002. A polypyrimidine/polypurine tract within the Hmga2 minimal promoter: a common feature of many growth-related genes. Biochemistry 41:1229-1240. [DOI] [PubMed] [Google Scholar]
- 40.Sattin, R. W., G. L. Rubin, L. A. Webster, C. M. Huezo, P. A. Wingo, H. W. Ory, and P. M. Layde. 1985. Family history and the risk of breast cancer. JAMA 253:1908-1913. [PubMed] [Google Scholar]
- 41.Stucki, M., I. Stagljar, Z. O. Jonsson, and U. Hubscher. 2000. A coordinated interplay: proteins with multiple functions in DNA replication, DNA repair, cell cycle/checkpoint control, and transcription. Prog. Nucleic Acid Res. Mol. Biol. 65:261-298. [DOI] [PubMed] [Google Scholar]
- 42.Suen, T. C., and P. E. Goss. 1999. Transcription of BRCA1 is dependent on the formation of a specific protein-DNA complex on the minimal BRCA1 bi-directional promoter. J. Biol. Chem. 274:31297-31304. [DOI] [PubMed] [Google Scholar]
- 43.Tay, N., S. H. Chan, and E. C. Ren. 1992. Identification and cloning of a novel heterogeneous nuclear ribonucleoprotein C-like protein that functions as a transcriptional activator of the hepatitis B virus enhancer II. J. Virol. 66:6841-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thakur, S., and C. M. Croce. 1999. Positive regulation of the BRCA1 promoter. J. Biol. Chem. 274:8837-8843. [DOI] [PubMed] [Google Scholar]
- 45.Thakur, S., H. C. Lin, W. T. Tseng, S. Kumar, R. Bravo, F. Foss, C. Gelinas, and A. B. Rabson. 1994. Rearrangement and altered expression of the NFKB-2 gene in human cutaneous T-lymphoma cells. Oncogene 9:2335-2344. [PubMed] [Google Scholar]
- 46.Thakur, S., H. B. Zhang, Y. Peng, H. Le, B. Carroll, T. Ward, J. Yao, L. M. Farid, F. J. Couch, R. B. Wilson, and B. L. Weber. 1997. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 17:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, M. E., R. A. Jensen, P. S. Obermiller, D. L. Page, and J. T. Holt. 1995. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 9:444-450. [DOI] [PubMed] [Google Scholar]
- 48.Tommerup, N., and H. Leffers. 1996. Assignment of human KH-box-containing genes by in situ hybridization: HNRNPK maps to 9q21.32-q21.33, PCBP1 to 2p12-p13, and PCBP2 to 12q13.12-q13.13, distal to FRA12A. Genomics 32:297-298. [DOI] [PubMed] [Google Scholar]
- 49.Tsui, S., T. Dai, S. Roettger, W. Schempp, E. C. Salido, and P. H. Yen. 2000. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics 65:266-273. [DOI] [PubMed] [Google Scholar]
- 50.Wilson, C. A., L. Ramos, M. R. Villasenor, K. H. Anders, M. F. Press, K. Clarke, B. Karlan, J. J. Chen, R. Scully, D. Livingston, R. H. Zuch, M. H. Kanter, S. Cohen, F. J. Calzone, and D. J. Slamon. 1999. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat. Genet. 21:236-240. [DOI] [PubMed] [Google Scholar]
- 51.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 52.Xu, C. F., M. A. Brown, J. A. Chambers, B. Griffiths, H. Nicolai, and E. Solomon. 1995. Distinct transcription start sites generate two forms of BRCA1 mRNA. Hum. Mol. Genet. 4:2259-2264. [DOI] [PubMed] [Google Scholar]