Abstract
R2 retrotransposons insert into the rRNA-encoding units (rDNA units) that form the nucleoli of insects. We have utilized an R2 integration system in Drosophila melanogaster to study transcription of foreign sequences integrated into the R2 target site of the 28S rRNA genes. The exogenous sequences were cotranscribed at dramatically different levels which closely paralleled the level of transcription of the endogenous R1 and R2 elements. Transcription levels were inversely correlated with the number of uninserted rDNA units, variation in this number having been brought about by the R2 integration system itself. Females with as few as 20 uninserted rDNA units per X chromosome had expression levels of endogenous and exogenous insertion sequences that were 2 orders of magnitude higher than lines that contained over 80 uninserted rDNA units per chromosome. R2 insertions only 167 bp in length exhibited this range of transcriptional regulation. Analysis of transcript levels in males suggested R2 insertions on the Y chromosome are not down-regulated to the same extent as insertions on the X chromosome. These results suggest that transcription of the rDNA units can be tightly regulated, but this regulation gradually breaks down as the cell approaches the minimum number of uninserted genes needed for survival.
In Drosophila melanogaster, and most other insects, variable fractions of the tandemly repeated genes coding for 28S rRNA are interrupted by the non-long terminal repeat (LTR) retrotransposable elements R1 and R2 (13). The R1 insertion site is located 74 bp downstream of the R2 site (Fig. 1A). These elements have been stable components of the rRNA gene loci (rDNA loci) since the origin of the Drosophila genus and possibly since the origin of arthropods (1, 16, 24). Although they share a common location in the genome, the two elements are only distantly related. R1 elements have two overlapping open reading frames (ORFs) and encode an apurinic-like endonuclease (14), while R2 elements have a single ORF and encode an endonuclease with an active site more similar to that of certain restriction enzymes (43).
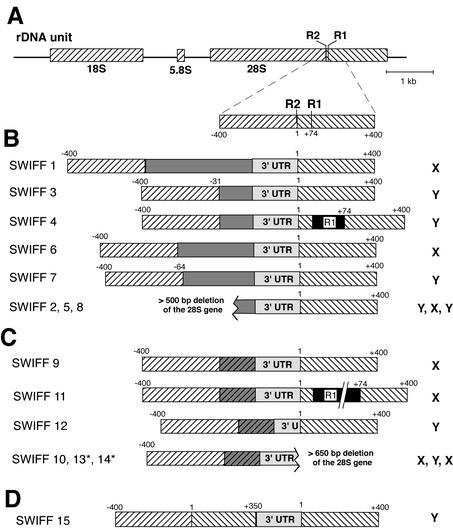
FIG. 1.
D. melanogaster lines containing B. mori sequences within their rDNA arrays. (A) Diagram of the D. melanogaster rDNA unit, with the rRNA genes represented by diagonally hatched boxes. Gene sequences upstream of the R2 site have diagonal hatching in the opposite orientation from sequences downstream of the R2 site. Also shown is an enlarged area spanning the R1Dm and R2Dm insertion sites. Distances in base pairs are relative to the R2 insertion site. (B) Diagrams of eight insertion lines generated by injection of RNA containing the R2Bm 3′ UTR and additional upstream sequences. Dark gray boxes correspond to various lengths of either the R2Bm 5′ UTR, R2Bm ORF, or GFP ORF (see Materials and Methods). If the insertion gave rise to more than a 5-bp deletion of 28S sequences at the 5′ junction, then the length of the deletion is indicated above the junction. A 150-bp R1Dm insertion (black box) is located downstream of the insertion in SWIFF 4. SWIFF 1 and SWIFF 2 were previously referred to as HR4-1 and 10R21, respectively (12). (C) Diagrams of the inserted 28S units in six lines generated by injecting RNA containing the 250-nt R2Bm 3′ UTR and 170 bp of upstream B. mori 28S sequences. The 170-bp B. mori 28S sequences were inserted in most cases resulting in tandem duplications (gray hatched boxes). An asterisk indicates that the entire upstream 170 bp was not duplicated: 23 bp of 28S sequences were duplicated in SWIFF 13, and no duplication occurred in SWIFF 14. A 500-bp R1Dm element is downstream of the SWIFF 11 insertion. Many of the insertions generated with this RNA had deletions of the R2Bm 3′ UTR sequences or downstream 28S sequences at the 3′ junction of the insertion. (D) Diagram of the inserted 28S unit generated by injecting an R2Bm transcript containing 170 bp of upstream and 350 bp of downstream D. melanogaster 28S gene sequences. Integration of this insert involved recombination events that regenerated the region of the 28S gene containing the R1 and R2 sites (see text). The chromosomal location of each insertion on either the X or Y chromosome is shown to the right of each diagram.
Like other non-LTR retrotransposons, one of the hallmarks of R1 and R2 retrotransposition is the presence of elements with truncations of their 5′ ends (2, 16). Because of these 5′ truncations, R1 and R2 elements of D. melanogaster range in length from a few hundred base pairs to 5.3 and 3.5 kb, respectively (18). Using these 5′ truncations as markers for individual R1 and R2 elements, we have studied R1 and R2 turnover both in fruit fly populations and in strains maintained for known periods in the laboratory (35, 36). These studies have suggested that the recombinational processes that give rise to the concerted evolution of the rDNA locus are biased against R1 and R2 and, thus, individual copies of these elements are rapidly eliminated. Only active retrotransposition maintains the presence of R1 and R2 in the rDNA locus.
The relative insertion levels of the R1 and R2 elements vary among D. melanogaster strains, with one study finding 17 to 67% of the 28S units containing R1 and 2 to 28% of the units containing R2 (19). Because such a significant fraction of the ribosomal genes can be interrupted by R1 and R2 sequences, their effect on ribosomal synthesis has long been of interest. Early experiments revealed that these elements (then referred to as type I and type II introns or insertions) have a marked effect on transcription of the inserted units (21, 23, 26, 27). At all developmental stages, R1 and R2 transcripts were determined to be present at levels 2 to 3 orders of magnitude below the level of nascent rRNA chains. This decrease in transcription of R1 or R2 inserted units was also observed by direct electron microscopic observations of actively transcribing rDNA arrays (4, 20). D. melanogaster strains with the highest levels of R1 and R2 transcripts were bobbed strains, strains that contain insufficient numbers of rDNA units to support normal development (25, 40).
Our interest in the transcription of the R1 and R2 insertions includes both how their presence affects the level of rRNA available for ribosome assembly and how their occasional transcription is linked to their retrotransposition cycle. Previous experiments have only monitored the total level of transcription of all endogenous R1 or R2 elements. We have recently developed a procedure by which new R2 copies can be inserted in the rDNA locus of D. melanogaster (12). In this approach, RNA and protein components from the R2 element of the silk moth Bombyx mori are injected into preblastoderm D. melanogaster embryos. B. mori R2 sequences (R2Bm) inserted into the rDNA locus could be recovered from the somatic tissues of the injected flies as well as from their progeny. The latter germ line events provided the means by which to monitor the transcription of individual R2Bm insertions. Because these insertions do not contain promoter sequences, RNA transcripts should be the result of cotranscription with the 28S gene. In addition, because RNA probes can be tailored to the 5′ truncations of particular R2 elements, we have been able to use RNase protection analysis to correlate cotranscription of the new R2Bm insertions with the cotranscription of specific endogenous D. melanogaster R2 (R2Dm) elements present in the same rDNA loci.
MATERIALS AND METHODS
Generation of R2Bm genomic Drosophila lines.
A number of different RNA templates were used in the Drosophila embryo injections (12). All RNAs contained the 250-nt 3′ untranslated region (3′ UTR) of R2Bm at the 3′ end of the RNA and were derived by in vitro transcription of pBluescript plasmids. Described previously are plasmids HR4, which contained 550 bp of the R2Bm ORF, and plasmid micro-R2, which contained 50 bp of the R2Bm 5′ end and 170 bp of upstream 28S rRNA sequences (12). Plasmid GFP (obtained from J. George) contained both an 800-nt cytomegalovirus promoter and the green fluorescent protein (GFP) ORF upstream of the R2Bm 3′ UTR. Plasmid UTR-R2 (obtained from J. Yang) contained the first 375 bp of the 5′ UTR of R2Bm upstream of the 3′ UTR. Plasmid TSR (target site replacement) was constructed by PCR amplification of D. melanogaster genomic DNA by using a primer corresponding to a region 170 bp upstream of the R2 target site (5′-CTAAGTCGACCGTTCTAAGGCGGCACT-3′) and a second primer near the end of the 28S gene (5′-CTGCGTCGACCGAATCATCAAGCAAAGG-3′). The amplified DNA was digested with SalI and KpnI, inserted into puc19, and subjected to site-directed mutagenesis to remove an XmnI site. The clone was then digested with KpnI/HindIII, and the 520-bp fragment was used to replace the upstream sequence of the micro-R2 plasmid. RNAs with 5′ methyl-G caps were synthesized with the T7 mMessage mMachine kit (Ambion) as previously described (12). Prior to transcription, plasmids HR4, UTR-R2, and TSR were linearized with XmnI, plasmid micro-R2 was linearized with EcoRI, and plasmid GFP was linearized with XhoI. Preparation of R2 protein, microinjections into w1118 Drosophila preblastoderm embryos, PCR assays of the progeny, and the generation of homozygous lines were as described previously (12). The 5′ junctions of all insertions were sequenced, as well as those 3′ junctions that appeared atypical in PCR assays.
Isolation of total genomic RNA from D. melanogaster.
Twenty-five female or 40 male adults were ground in 200 μl of 10 mM Tris (pH 8.0), 60 mM NaCl, 10 mM EDTA, 0.15 mM spermidine, 0.15 mM spermine, 5% sucrose. An equal volume of 200 mM Tris (pH 9.0), 30 mM EDTA, 2% sodium dodecyl sulfate (SDS), 5% sucrose, 1.25 mg of proteinase K/ml was added, and the solution was incubated at 37°C. After 90 min an additional 250 μg of proteinase K was added, and the incubation continued for another 90 min. The solution was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol. Forty microliters of 3 M Na-acetate and 900 μl of cold ethanol were added, and the solution was centrifuged for 5 min at 13,000 rpm in a Microfuge at 4°C. The pellet was rinsed with 70% ethanol and air dried. The dried pellet was resuspended in 200 μl of DNase I buffer (40 mM Tris, [pH 7.9], 5 mM MgCl2, 5 mM CaCl2), 20 U of DNase I (Ambion) was added, and the solution was incubated at 37°C for 40 min. The solution was then phenol-chloroform-isoamyl alcohol extracted once followed by a chloroform-isoamyl alcohol extraction. After ethanol precipitation, the pellet was rinsed in 70% ethanol, air dried, and resuspended in 25 μl of distilled water. The integrity of the RNAs was checked on a 1% agarose gel, and RNA concentrations were estimated by optical density determinations at 260 nm.
RNase protection assay.
The procedure was essentially as described by Yang et al. (44). Constructs used to generate antisense RNA were as follows. Plasmid pBmR2-249 was used to monitor the R2Bm insertions (28). In the case of the endogenous R2Dm or R1Dm elements, specific segments were PCR amplified, restriction enzyme digested in some cases, and inserted into pBluescript. The following nucleotide positions, indicated in parentheses, are found in accession number X51967 (R2Dm) and X51968 (R1Dm): DmR2-probe 1 (181 to 528); DmR2-probe 2 (3355 to 3693); DmR2-probe 3 (2977 to 3357); R1Dm-3′ UTR (4940 to 5343); and R1Dm-5′ UTR (227 to 480). The constructs were linearized with the appropriate restriction enzyme, and 32P-labeled antisense RNAs were generated using either T7 or T3 polymerase under the conditions specified by the supplier. The full-length runoff transcripts were separated on a 5% polyacrylamide gel containing 8 M urea, and the RNA was eluted by constant shaking for 4 h at room temperature in 300 μl of 300 mM Na-acetate, 1 mM EDTA, 0.1% SDS, and 5 μg of yeast tRNA. The solution was then transferred to a clean tube, and the RNA was purified by phenol-chloroform-isoamyl alcohol extraction followed by ethanol precipitation and resuspended in 30 μl of distilled water. Twenty micrograms of Drosophila RNA was incubated in the presence of the labeled antisense RNA in a 20-μl hybridization reaction mixture containing 50% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.7), 300 mM NaCl, and 1 mM EDTA at 55°C for 18 h. The reaction mixture was diluted by the addition of 300 μl of 500 mM NaCl, 10 mM Tris (pH 7.5), 5 mM EDTA, 12 μg of RNase A, and 0.2 U of RNase T1 (Ambion), and the solution was incubated at 25°C for 30 min to digest unhybridized probe. The solution was then made to 0.6% SDS, 50 μg of proteinase K/ml and incubated for an additional 30 min. After adding 10 μg of yeast tRNA, the RNA hybrids were phenol-chloroform-isoamyl alcohol extracted and ethanol precipitated. The pellets were resuspended in 80% formamide, 0.1 M Tris, 0.1 M boric acid, 2 mM EDTA, 2% bromophenol blue and electrophoresed on a 5% denaturing polyacrylamide gel. Before drying, the gel was fixed in 10% methanol-10% acetic acid for 30 min. Dried gels were exposed in a PhosphorImager cassette and quantitated in a Molecular Dynamics Storm Analyzer using ImageQuant version 1.2.
R2 truncation profiles and copy number determinations.
The PCR primers and amplification procedures used to determine the number and 5′ truncation profiles of the R1 and R2 insertions were as previously described (36). Briefly, the total number of 5′-truncated copies was estimated by counting the bands visible on a 5% native polyacrylamide gel using six different R2 primers, or 10 different R1 primers, each in conjunction with an upstream 28S primer. The number of full-length copies of R2 was estimated using a 32P-end-labeled primer positioned 150 bp downstream from the 5′ end of R2 in conjunction with the 28S primer. In this case, the amplified products were separated on a high-voltage denaturing 8% polyacrylamide gel, and the relative intensities of the bands were quantified using a PhosphorImager cassette and ImageQuant 1.2. The number of R2 elements upstream of R1 elements was determined by repeating the R1 primer amplifications, but substituting for the upstream 28S gene primer with the primer 5′-AAAAAAAAAAAATAGCCAAAT-3′, which anneals to the 3′ junction of the R2Dm element, or the primer 5′-TTGGCAGACCTAGTATCTTTC-3′, which anneals to an internal segment of R2Dm near its 3′ end.
Isolation of genomic DNA and Southern analysis to determine the number of uninserted rDNA units.
The DNA isolation procedure was similar to the RNA isolation procedure, except that the starting material and volumes were doubled and total nucleic acid was originally precipitated with an equal volume of isopropanol. The first pellet was resuspended in 150 μl of TE (10 mM Tris [pH 7.5], 1 mM EDTA) and digested with 35 μg of RNase A at 37°C for 30 min before a second precipitation with ethanol. The spooled DNA was resuspended in 50 μl of TE. Genomic DNA was digested with the appropriate restriction enzymes under the conditions suggested by the supplier. The DNA was separated on a 1% agarose gel, transferred to nitrocellulose, and hybridized to 32P-labeled DNA probes (11). To determine the fraction of the rDNA units corresponding to R1 inserted, R2 inserted, and uninserted units, the genomic DNA was digested with PstI and HindIII and hybridized with a 280-bp fragment from the 28S gene located immediately downstream of the R1 insertion site (11). This digest gives rise to a 3.5-kb fragment corresponding to uninserted rDNA units, a 1.4-kb fragment corresponding to the 3′ end of R1 elements inserted into 28S genes, and a 0.9-kb element corresponding to the 3′ end of R2 elements in the 28S gene (see Fig. 1 and 2 of reference 11 for restriction maps and examples of these blots). Based on the relative fraction of the rDNA units in each of these three bands, the absolute number of rDNA units corresponding to each fraction was calculated based on the number of R2 inserted units, determined from PCR analysis of the R2 5′ truncation profiles (see above). As an independent means to determine the number of uninserted and R2 inserted rDNA units, genomic DNA was digested with PstI, SphI, and HindIII. After transfer of the size-fractionated DNA, the blot was cut into three pieces. The region around 3.0 kb was probed with a fragment of the alcohol dehydrogenase (adh) gene. The 1.0-kb probe was generated from the alcohol dehydrogenase (adh) gene using the primers 5′-GAACTGGAAACCAACAACTA-3′ and 5′-TGGGAGTATCACTTCTTAGA-3′ (see accession number Z00030). The region of the Southern blot around 1.3 kb was probed with an 18S to 5.8S gene probe. This 820-bp probe was generated by digesting the PCR product amplified by the primers 5′-GTAGGTGAACCTGCGGAAGGATC-3′ and 5′-GCATATTAATTAGGGGAGG-3′ with PstI (see accession number M21017). Finally, the region of the Southern blot around 0.6 kb was probed with a fragment from the R2Dm element. This probe was a 375-bp PstI-AluI fragment from near the 3′ end of R2Dm (18). Using the adh blot to standardize the amount of DNA per lane, it was possible to determine both the relative size of the rDNA array on the X chromosomes, using the 18S blot, and the relative number of R2 insertions, using the R2Dm 3′ UTR probe. The probed nitrocellulose films were exposed in a PhosphorImager cassette and quantitated in a Molecular Dynamics Storm Analyzer using ImageQuant 1.2.
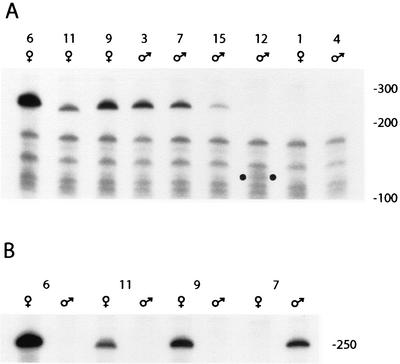
FIG. 2.
Transcription levels of the R2Bm insertions in each SWIFF line. (A) Total RNA from adult females or adult males (as indicated) was subjected to RNase protection. A labeled, antisense probe corresponding to the 250-bp 3′ UTR of R2Bm was hybridized to the RNA, the mixture was digested with RNase T1 and RNase A, and the resistant RNA was run on a 5% polyacrylamide denaturing gel. Numbers above the lanes indicate the SWIFF line used. Due to deletions at the 3′ end of the insertion, the protected fragment for SWIFF 11 is 240 nt and that for SWIFF 12 is 134 nt. The latter is indicated by dots in the lower region of the gel. Locations of single-stranded DNA size standards are indicated to the right. (B) Comparison of transcript levels in males and females for three R2Bm insertions on the X chromosome (SWIFF 6, 11, and 9) and one insertion on the Y chromosome (SWIFF 7).
RESULTS
D. melanogaster lines containing individual R2Bm insertions.
We have previously shown that the injection of purified R2Bm protein and RNA into preblastoderm D. melanogaster embryos resulted in the integration of R2Bm sequences into the 28S gene target site (12). These in vivo integrations show all the features of the target primed reverse transcription (TPRT) reaction characterized in vitro (28-30). While several germ line events were characterized in this previous report, most conclusions were based on the more easily obtained R2Bm insertions in the somatic tissues. Here we describe the characterization of germ line R2Bm insertions obtained by the injection of three classes of RNA templates. We called the lines containing these new R2Bm insertions SWIFF, because they contained a silkworm R2 in a fruit fly.
Each injected RNA contained the 250-nt 3′ UTR of R2Bm at the 3′ end of the RNA, as this sequence is required to initiate the TPRT reaction. The first class of RNA templates injected contained upstream of the R2Bm 3′ UTR either part of the R2Bm ORF (SWIFF 1, 2, 3, and 5), the R2Bm 5′ UTR (SWIFF 4), or a GFP reporter gene (SWIFF 6 to 8) (see Materials and Methods for details of these RNAs). Summary diagrams of the eight germ line events obtained are shown in Fig. 1B. In each case, the insertion was at the correct site of the 28S gene and started at the 3′ end of the injected RNA, suggesting that each integration had been initiated by a TPRT reaction. One insertion occurred in an rDNA unit already containing a 5′-truncated R1 element (SWIFF 4). All eight insertions were 5′ truncated and each resulted in the deletion of upstream 28S sequences. Five of the insertions generated small deletions that are typical of endogenous R2 elements in D. melanogaster (17), while the remaining three resulted in deletions of at least 500 bp upstream of the R2 site. These eight germ line insertions were similar to the previously characterized somatic insertions in which we hypothesized that the 5′ ends were attached by double-strand break repair mechanisms provided by the cell (12).
Because of the 5′ deletions that resulted with the first class of RNAs, the second class of injected RNA contained 170 nt of upstream 28S gene sequences at the 5′ end of the RNA template (micro-R2). The 5′ end of the previously reported somatic insertions obtained with this template resulted from recombination between the newly synthesized cDNA and the upstream 28S sequences, giving rise to insertions with no deletions at their 5′ end (12). However, only one of the germ line insertions resulting from this injected RNA contained a junction similar to that of the somatic events (Fig. 1C, SWIFF 14). The others contained a direct duplication of the 170-bp upstream 28S sequence. Surprisingly, five of the insertions had deletions of R2Bm sequences at the 3′ end of the insertion (10 bp in SWIFF 11, 116 bp in SWIFF 12, >100 bp in SWIFF 13) and/or large (>650-bp) deletions of 28S sequence downstream of the insertion site (SWIFF 10, 13, and 14). The discrepancy between these germ lines and the previously described somatic events could be the result of differences in chromatin structure or in the DNA repair machinery between the rapidly dividing somatic cells and the pole cells (i.e., future germ cells).
Finally, a third RNA template was injected to test a model that the region of the 28S gene containing the R1 and R2 insertion sites is an “enhancer” sequence providing cis transcriptional regulation of the unit. This model is based on the discovery of a number of transposable elements in other organisms that insert near the R1 and R2 insertion sites of the 28S gene (3, 22, 34; W. D. Burke et al., submitted for publication). The third class of injected RNA contained a large region of 28S sequence extending from 170 bp upstream to 350 bp downstream of the insertion site, all located upstream of the R2Bm 3′ UTR sequences on the RNA template. Only a single germ line insertion event was obtained with this injected RNA, despite repeated efforts. This germ line event (SWIFF 15) had a structure that was consistent with a TPRT reaction followed by a recombination of the cDNA with the 28S gene upstream of the insertion site. This R2Bm insertion would appear from the 5′ end of the rDNA unit to be located at a site that was 350 bp downstream of its normal insertion site (Fig. 1D).
Transcription of the R2Bm insertions.
The 15 SWIFF lines shown in Fig. 1 were each tested for the presence of R2Bm transcripts. The insertions did not contain promoter sequences; thus, RNA transcripts from these insertions should be a result of cotranscription with the 28S gene. To determine the best tissue for use in these experiments, total RNA was isolated from different developmental stages of two lines. Equivalent amounts of this RNA were hybridized to a labeled probe (antisense RNA) complementary to the 3′ UTR of R2Bm, the mixture was then digested with RNase T1 and A, and the resistant antisense RNA was run on a denaturing gel. Initial experiments indicated that R2Bm transcript levels were lowest during the late larval instars and during pupation and approximately equal during late embryo-first instar larva and adult stages (data not shown). The adult stage has therefore been used for the following studies, as this stage allowed us to monitor expression separately in males and females.
None of the SWIFF lines that contained large deletions of 28S gene sequences either upstream or downstream of the R2Bm insertion exhibited detectable levels of RNA transcripts (data not shown). Because these insertions cannot be considered part of a typical rDNA transcription unit, they have not been studied further. Results of typical RNase protection assays for the remaining nine R2Bm insertions are shown in Fig. 2A. The RNA was derived from females for the four lines with an R2Bm insertion on the X chromosome and from males for the five lines with an insertion on the Y chromosome. The relative transcript levels from the R2Bm inserts were determined in most lines by the intensity of the 250-nt protected band. It should be noted that the size of the protected RNA was 10 nt shorter in SWIFF 11 and 116 nt shorter in SWIFF 12. These shorter fragments were reproducibly seen and readily explained by the size of deletions found in the integrated R2Bm 3′ UTR sequences of these lines (Fig. 1C).
The level of R2Bm transcripts varied over 140-fold (<0.1 to 14) for the four insertions on the X chromosome and over 40-fold (<0.1 to 3.9) for the five insertions on the Y chromosome (Table 1). The differences in transcript levels for the different R2Bm insertions did not appear to correlate with the size of the insertion or the nature of the sequences upstream of the 3′ UTR. The R2Bm insert in SWIFF 15, which was located in an rDNA unit that contained uninterrupted R1 and R2 target sites upstream of the insertion (Fig. 1D), did not produce higher levels of transcripts than that detected for most of the other R2Bm insertions. Therefore, either the uninterrupted target site region is not sufficient to permit “normal” levels of transcription, or the transcripts derived from this inserted unit are more unstable (see Discussion). The effect on transcription of the R2Bm insertion when an R1 insertion was present in the same rDNA unit was not clear. No transcripts were detected for the R2Bm insertion in SWIFF 4, while transcripts were readily detected for the insert in SWIFF 11.
TABLE 1.
Levels of R2Bm transcripts in the SWIFF lines
| Insertion line | Transcript levela
|
|
|---|---|---|
| Females | Males | |
| SWIFF 1 | <0.1 | <0.1 |
| SWIFF 6 | 14.0 | <0.1 |
| SWIFF 9 | 4.2 | <0.1 |
| SWIFF 11 | 2.1 | <0.1 |
| SWIFF 3 | NAb | 3.9 |
| SWIFF 4 | NA | <0.1 |
| SWIFF 7 | NA | 3.2 |
| SWIFF 12 | NA | 0.7 |
| SWIFF 15 | NA | 1.0 |
Because it represented the lowest level of detectable transcripts, all numbers are relative to the transcript level detected for the SWIFF 15 insertion in male RNA.
NA, not applicable because the R2Bm insertion is on the Y chromosome.
The location of four R2Bm insertions on the X chromosome permitted a comparison of transcription of specific units within the nucleolar organizer in females versus males. RNA was isolated from adult males and females from the four X insertion lines as well as the Y insertion line SWIFF 7 (Fig. 2B; Table 1). Consistent with its location on the Y chromosome, R2Bm transcripts in SWIFF 7 were detected in males but not females. Surprisingly, R2Bm transcripts readily detectable in females were not detected in males for the X insertions in SWIFF 6, 9, and 11.
Transcription of endogenous R2Dm elements in the same lines.
Because all nine lines used to study the expression of specific R2Bm insertions were derived from the same D. melanogaster strain, we initially assumed the differences in transcript levels seen in Fig. 2 meant that R2 insertions at different locations within the X and Y rDNA arrays were transcribed at different levels. This interpretation was based on the assumption that total transcription of all insertions in the rDNA loci would be similar across the insertion lines. We tested this assumption by monitoring transcript levels of the endogenous R2Dm elements in these lines.
To study the level of RNA transcripts from the endogenous R2Dm elements, it was first necessary to characterize the R2Dm insertions. The elements were characterized by the same approach used to monitor the turnover of individual R1 and R2 copies in populations and lab strains of Drosophila simulans and D. melanogaster (35, 36). In this approach, the number and length of the R2 elements were determined by PCR amplification by using a series of primers that annealed to different locations along the R2Dm element, each in combination with a primer that annealed to the 28S gene a short distance upstream of the R2 site (see Materials and Methods). This approach indicated that the X chromosome present in the w1118 strain used for the injections contained 20 copies of R2Dm that were 5′ truncated (Fig. 3A). As we have found for other strains of Drosophila (35, 36), each of these 5′ truncations appeared to be at the level of one copy per genome. Finally, the number of full-length R2Dm elements on the X chromosome was also determined by this PCR approach, except that one primer was end labeled, the products were separated on DNA sequencing gels, and the intensity of PCR products corresponding to multiple copies of the same length were quantified with a PhosphorImager (see Fig. 3A in reference 36 for an example of this approach). These results revealed 32 full-length copies of R2, for a total of 52 R2Dm insertions on the X chromosome.
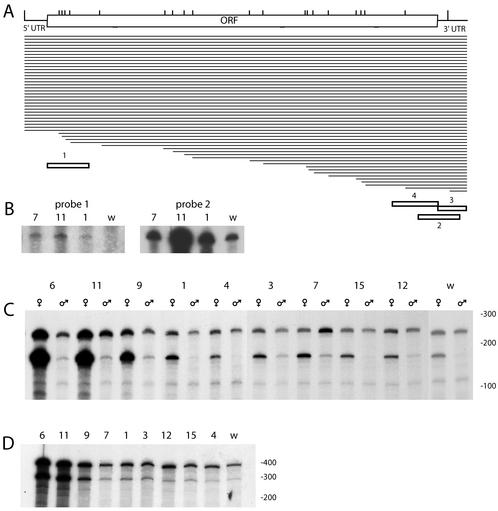
FIG. 3.
Transcription from endogenous R2Dm elements in the SWIFF lines. (A) Schematic representation of the R2Dm element with the 5′ UTR, ORF, and 3′ UTR indicated. Vertical lines represent the extent to which various R2Dm elements in the w1118 line are 5′ truncated. Horizontal lines represent the 32 full-length and 20 5′-truncated R2 copies in w1118. The positions within the element of the R2Dm antisense RNA probes used in the RNase protection assays are shown at the bottom of the diagram. (B) RNase-protected fragment using either antisense probe 1 (left panel) or antisense probe 2 (right panel). RNA was isolated from adult males (SWIFF 7), adult females (SWIFF 1 and 11), and 0- to 6-h embryos (w1118). The panel on the left was exposed 10 times longer than that on the right. (C) RNase-protected fragments from adult females and males of each line, determined by using antisense probe 3. The upper band represents the level of transcripts from all insertions greater than 240 bp. The lower band represents the level of transcripts arising from the smallest truncated element, a 167-bp insertion on the X chromosome. (D) The RNase-protected fragments from adult females generated using antisense probe 4. The upper band corresponds to the level of transcripts derived from all insertions greater than 600 bp in length. The lower band corresponds to the level of transcripts derived from the second-shortest R2Dm element, a 516-bp insertion on the X chromosome. Numbers above the lanes indicate the SWIFF line used, and w indicates the w1118#12 line. Size standards are indicated to the right of each gel.
This same approach was also used to characterize the R2 elements in the various SWIFF lines. In the case of the 5′-truncated copies, the only differences detected between the initial injection line and the SWIFF lines involved single, unique new R2Dm insertions on the X chromosomes in lines SWIFF 1, 9, and 11. In the case of the full-length elements, most lines had approximately 32 copies; however, SWIFF 9 and 11 each had approximately six additional copies (data not shown).
We next determined whether RNA transcripts of the R2Dm elements were derived from both full-length and 5′-truncated elements by using sequences in the RNase protection assays that were derived from either the start (probe 1) or the end (probe 2) of the ORF of the element (Fig. 3A). In both adult and embryonic tissues, RNA transcripts corresponding to the 5′ end of R2Dm were at least 2 orders of magnitude less abundant than transcripts corresponding to the 3′ end (Fig. 3B). To further investigate the 3′ end transcripts, a 3′ UTR-specific probe was generated (Fig. 3A, probe 3). As shown in Fig. 3C, transcript levels from the various SWIFF lines were highly variable. An unexpected result obtained using the 3′ UTR probe was that the predominate protected fragment in many cases was not 240 nt but approximately 165 nt. This shorter-length product was similar to that predicted to arise for the shortest R2Dm element present in these lines (Fig. 3A). This short R2Dm element was cloned from w1118 and sequenced. The insert was found to be 167 bp in length and corresponded to the extreme 3′ end of an R2Dm element. To further confirm that this truncated element was responsible for the 165-nt protected fragment, the RNase protection assay with the 3′ UTR probe was conducted with a D. melanogaster stock (Harwich) that did not contain this short R2Dm insertion. No band corresponding to the 165-bp protected product was observed (data not shown). These results indicate that over 50% of the stable R2Dm transcripts in females from the w1118 and SWIFF lines were derived from a single, highly truncated R2 insertion on the X chromosome.
While the levels of transcripts in females arising from the 167-bp R2Dm insertion can be seen to vary greatly across the lines, the lines can be divided into two groups (Table 2). The first group has relatively low levels of transcripts and includes the original w1118 stock and all lines with an R2Bm insertion on the Y chromosome (SWIFF 3, 4, 7, 12, and 15). The second group has much higher levels of transcripts and includes most of the lines with X chromosome insertions (SWIFF 6, 9, and 11). The only exception to this division was the X insertion line SWIFF 1, which had transcript levels that placed it with the first group.
TABLE 2.
Endogenous R2Dm and R1Dm transcript levels
| Insertion line | R2Dm transcriptsa
|
R1Dm transcriptsa
|
||
|---|---|---|---|---|
| 167 bp
|
516 bp | 3′UTR | ||
| Females | Males | Females | Females | |
| w1118#12 | 1.0 | 0.1 | 1.0 | 1.0 |
| SWIFF 4 | 1.8 | 0.3 | 1.3 | 1.1 |
| SWIFF 12 | 2.4 | 0.5 | 2.2 | 1.6 |
| SWIFF 15 | 2.6 | 0.3 | 2.0 | 1.8 |
| SWIFF 3 | 3.2 | 0.7 | 2.9 | 5.4 |
| SWIFF 1 | 3.6 | 0.3 | 2.9 | 2.6 |
| SWIFF 7 | 4.6 | 1.0 | 1.8 | 1.6 |
| SWIFF 9 | 18.8 | 1.1 | 11.9 | 10.6 |
| SWIFF 11 | 73.8 | 1.1 | 74.0 | 55.5 |
| SWIFF 6 | 101.6 | 0.9 | 77.1 | 60.7 |
For each probe, the transcription levels are given relative to that found in w1118#12 females.
Also shown in Fig. 3C are expression levels of the 167-bp R2Dm insertion in males. While the simplest model would predict a twofold-lower level of transcripts from this X chromosome insertion in males (XY) compared to females (XX), the observed differences were much greater. The lines averaged a 10-fold-lower transcript level in males, with the difference as great as 100-fold in the high expression lines (Table 2). This reduction in expression of a specific endogenous R2Dm insertion on the X chromosome in males compared to females was similar to that observed for the R2Bm insertions on the X chromosome (Fig. 2; Table 1).
To determine the relative level of transcripts arising from the second shortest R2Dm element in these lines, another antisense RNA probe was used in the RNase protection assays which targeted the 5′ end of a 516-bp R2Dm insertion (probe 4 in Fig. 3A). In this case, a 288-nt protected RNA fragment would correspond to transcripts from the 516-bp truncated element, while a 380-nt fragment would correspond to transcripts derived from the many R2Dm insertions greater than this length (Fig. 3D). In all lines, the 516-bp R2Dm element accounted for a significant fraction of the R2Dm transcripts observed. The SWIFF lines with the highest levels of transcripts from the 516-bp insertion were the same as those with the highest levels of the 167-bp insertion, with SWIFF 6 and 11 containing over 70-fold-higher levels of transcript than that from the lowest line (Table 2).
Transcription of the endogenous R1Dm insertions.
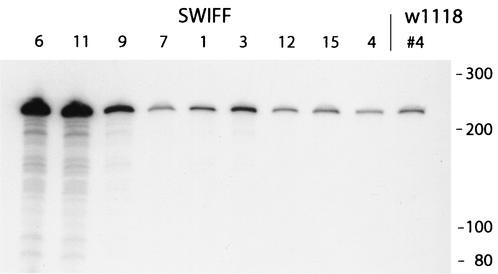
We have also compared transcript levels from the endogenous R1Dm elements of w1118 and the SWIFF lines. There were over 100 R1Dm insertions on the X chromosome in w1118, with 21 of these copies corresponding to 5′-truncated elements (data not shown). An analysis of the 5′-truncated copies in the nine SWIFF lines revealed no new insertions, but several lines had eliminated from one to three old copies (data not shown). RNase protection assays indicated that, similar to the R2Dm elements, transcripts from the 3′ end of the endogenous R1Dm elements were over 2 orders of magnitude more abundant than transcripts from their 5′ end (data not shown). The level of R1Dm 3′ transcripts in females varied widely between strains (Fig. 4), with the highest levels again detected in SWIFF 6, 9, and 11. Indeed, the relative levels of R1Dm transcripts clearly paralleled that of the R2Dm transcripts (Table 2), with only the R1Dm transcript level in SWIFF 3 somewhat higher than that predicted from the R2Dm expression pattern. The shortest R1Dm inserts in the SWIFF lines are around 500 bp in length. Unfortunately, there were more than 10 of these short insertions; therefore, it was not possible to determine the percentage of transcripts due to a specific R1Dm copy.
FIG. 4.
Transcription from endogenous R1Dm elements in the SWIFF lines. Total RNA from adult females of each line was subjected to RNase protection by using a probe from the extreme end of the 3′ UTR of the R1Dm element (see Materials and Methods). All R1Dm elements in these lines are greater than 240 bp in length; thus, the protected bands represent the level of transcripts derived from all R1 insertions. Size standards are indicated to the right of the gel.
Stability of the expression patterns.
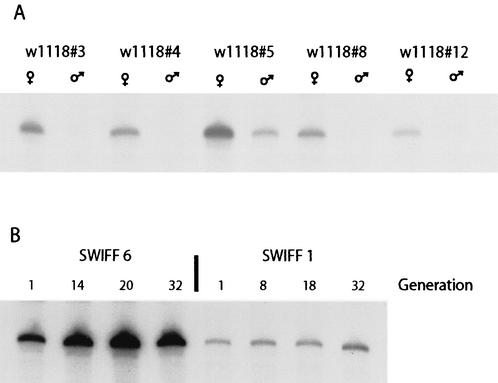
To address whether the differences observed among the SWIFF lines were reflections of the variability within the original w1118 line, the transcript levels of the 167-bp R2Dm insertion were determined in males and females for five stocks of w1118 maintained separately for over 18 months (Fig. 5A). While the level of the 167-nt R2Dm transcripts was 3- to 6-fold higher in subline 5 compared to the other sublines, this is clearly well below the 100-fold difference observed between w1118 and the SWIFF 6 and 11 lines (Fig. 3B). The level of transcripts was on average 10-fold lower in males than in females. These findings indicated that changes in the expression levels of the 167-bp R2Dm insertion could occur in the w1118 stock over time; however, these changes were not great enough to explain the elevated transcript levels found in three of the SWIFF lines with R2Bm insertions on the X chromosome.
FIG. 5.
Stability of endogenous transcript levels over many generations. (A) The relative level of transcripts corresponding to the 167-bp truncated element was examined in both males and females for five stocks of the w1118 line separately maintained as small populations for 18 months. RNase protection assays were carried out with probe 2 (Fig. 3A). (B) The relative level of the 167-bp transcript level was monitored in two of the SWIFF lines over an 18-month period. Total RNA was isolated from adult females at the indicated generation, and RNase protection assays were performed as for panel A.
We next tested whether the very high transcript levels found in SWIFF 6 were stable from generation to generation. In Fig. 5B we show the level of transcripts from the 167-nt R2Dm insertion at four generations over an 18-month period. For comparison, we also show the expression levels over time of the 167-bp R2Dm insert in SWIFF 1, a line that exhibited more typical levels of expression. Over the 30 generations, transcript levels of the 167-bp insertion varied only twofold in SWIFF 1 and fourfold in SWIFF 6. We conclude that the expression of the R2 insertions in the D. melanogaster rDNA locus can vary from generation to generation; however, the higher levels of expression observed in SWIFF 6, 9, and 11 appear to be the result of stable changes in the locus that likely occurred as a result of the R2Bm injections.
Levels of R1 and R2 transcripts are correlated with the number of uninserted rDNA units.
What changes in the structure of the rDNA locus in certain SWIFF lines were induced by the injection process? As described previously, the profile of full-length and 5′-truncated copies of the endogenous R2Dm elements as well as the 5′ truncation profile of the R1Dm elements in each line indicated that minor changes in the number of these elements had occurred between lines. It seemed unlikely, however, that the addition or elimination of a few copies could have caused such a dramatic increase in the transcription from the specific 167-bp and 516-bp R2Dm insertions, as well as from the many R1Dm insertions.
A possible difference in the rDNA loci of the SWIFF lines that would have gone unnoticed in the analysis of the R1 and R2 insertions is in the number of uninserted rDNA units. We therefore determined the number of uninserted units in the rDNA locus on the X chromosome of each line. Because the number of R2 elements in the rDNA locus was directly counted by scoring the 5′ sequence variation of all copies (Fig. 3A), we could calculate the number of uninserted units based on the fraction of the rDNA units that were R2 inserted and the fraction that were uninserted. Genomic DNA was isolated from adult females of the nine SWIFF lines as well as w1118#12. The DNA was digested with the restriction enzymes PstI and HindIII and probed with a short segment of the 28S gene immediately downstream of the R1 insertion site. Three bands were observed in this blot: a 3.5-kb band corresponding to the uninserted units, a 1.4-kb band corresponding to the R1 inserted units, and a 0.9-kb band corresponding to units containing only an R2 insertion (see Fig. 1 and 2 of reference 11 for restriction maps and examples of these blots). The fraction of each unit type for each line is shown in Table 3.
TABLE 3.
Fraction of the rDNA units on the X chromosome that are inserted or uninserted
| Insertion line | Fraction of rDNA units
|
No. of R2b | ||
|---|---|---|---|---|
| Uninserted | R1 inserted | R2 inserteda | ||
| w1118#12 | 0.38 | 0.53 | 0.09 | 28 (1.00) |
| SWIFF 4 | 0.30 | 0.57 | 0.13 | 28 |
| SWIFF 15 | 0.27 | 0.62 | 0.10 | 28 |
| SWIFF 12 | 0.28 | 0.60 | 0.11 | 29 |
| SWIFF 3 | 0.27 | 0.60 | 0.13 | 29 (1.09) |
| SWIFF 1 | 0.32 | 0.55 | 0.13 | 29 |
| SWIFF 7 | 0.28 | 0.59 | 0.13 | 27 (1.07) |
| SWIFF 9 | 0.16 | 0.65 | 0.19 | 36 (1.27) |
| SWIFF 11 | 0.13 | 0.64 | 0.24 | 36 (1.30) |
| SWIFF 6 | 0.15 | 0.65 | 0.21 | 30 (1.09) |
Those rDNA units which have both an R1 and R2 were scored by the Southern approach as R1 inserted.
These numbers were based on the PCR analysis of the 5′ sequence variation of all R2 elements. In all lines, 24 R2 insertions were doubly inserted with an R1 element and are subtracted from the total number of R2 insertions. Numbers in parentheses correspond to relative estimates of the number of R2 elements based on the adh Southern analysis, with the R2/adh hybridization ratio in w1118#12 defined as 1.00.
Because the 28S probe used in the Southern blotting was located downstream of the R1 insertion site, any R2Dm insertion located in an rDNA unit also containing an R1Dm insertion (i.e., doubly inserted rDNA units) was not scored by this approach. Therefore, it was necessary to subtract from the total number of R2 elements in the locus those copies located upstream of an R1 insertion. Our PCR assays (see Materials and Methods) indicated that 24 R2Dm insertions (10 corresponding to 5′ truncation and 14 full-length) were part of doubly inserted rDNA units, and that this number remained constant in all lines. The number of rDNA units on the X chromosome of each line inserted with only an R2 element is shown in the last column in Table 3. This number of singly inserted R2 elements was then used in conjunction with the unit percentages to calculate the absolute number of inserted and uninserted rDNA units in each line.
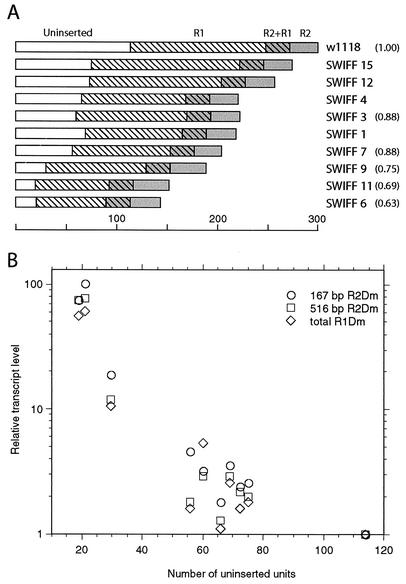
Diagrammed in Fig. 6A is the number of rDNA units on the X chromosome of each line that are uninserted, R1 inserted, R2 inserted, and doubly inserted. The 13 to 38% range in rDNA units that are uninserted corresponds to only 20 copies in SWIFF 6 and 11 and to over 100 units in w1118. Variation in the number of R1 insertions was less extreme and ranged from approximately 70 to 150 units. The loss of R1 inserted units in most of the SWIFF lines, compared to that in w1118, mirrored the loss of uninserted units in these lines.
FIG. 6.
Relationship in females between the transcript levels of various insertions and the number of uninserted units in the X rDNA locus. (A) Schematic diagrams of the number of each type of rDNA unit present in w1118#12 and the various SWIFF lines. The length of each box represents the number of units, based on the scale shown. Unshaded boxes represent uninserted units, hatched boxes represent R1 inserted units, gray boxes represent R2 inserted units, and hatched gray boxes represent doubly inserted units. (B) Plot of the number of uninserted rDNA units per X chromosome (shown in panel A) versus relative levels of transcripts from various R1Dm and R2Dm insertions (Table 2). The transcript level in w1118#12 was defined as 1.0 for each assay. Symbols used to indicate transcript levels for the 167-bp R2Dm insertion, the 516-bp R2Dm insertion, and the 3′ UTR of all R1Dm elements are indicated on the figure.
As an independent estimate of the number of inserted and uninserted rDNA units, genomic DNAs from adult females from six lines (w1118, SWIFF 3, 6, 7, 9, and 11) were digested with the restriction enzymes PstI, SphI, and HindIII. After transfer of the size-fractionated DNA, the blot was cut into three pieces and the individual pieces were probed with either a fragment of the alcohol dehydrogenase (adh) gene, a fragment of the 18S rRNA gene, or a fragment from the 3′ end of R2Dm (see Materials and Methods for a description of the digestions and probes). Using the adh blot to standardize the amount of DNA per lane, it was possible to determine the relative size of the rDNA array on the X chromosomes by using the 18S blot and the relative number of R2 insertions by using the R2Dm 3′ UTR probe. A similar experiment with the R1 elements could not be conducted because the signal from the large number of degenerate R1 sequences located in the centromeric heterochromatin of D. melanogaster complicates that arising from R1 sequences within the rDNA locus (11, 19). The relative number of uninserted and R2 inserted units obtained from the comparisons to adh were consistent with the absolute numbers derived from the first Southern approach. In the case of the R2 insertions, the relative numbers of R2 elements were clearly higher in SWIFF 9 and 11 than in w1118 and intermediate in SWIFF 3, 6, and 7 (Table 3, numbers in parentheses). In the case of the total number of rDNA units, SWIFF 6 was determined to have the lowest number of rDNA units, w1118 had the highest, and the remaining lines had a range of intermediate levels consistent with the previous estimates (Fig. 6A, numbers in parentheses).
Plotted in Fig. 6B is the number of uninserted rDNA units on the X chromosome of each line versus the relative level of transcripts in females corresponding to the 167-bp R2Dm insertion, the 516-bp R2Dm insertion, and total R1Dm 3′ UTR. This graph reveals an inverse correlation between the number of uninserted rDNA units and the level of R1 and R2 transcription. The reduction in the number of uninserted units on the X chromosome to only 20 (or 40 per diploid cell) resulted in a 50- to 100-fold stimulation in the number of transcripts corresponding to R1 and R2 insertions.
The same two Southern approaches used to calculate the number of unit types in females were also conducted with DNA isolated from males of five lines. The numbers of inserted and uninserted units on the Y chromosome were estimated by subtracting the numbers previously calculated for the X chromosome. While the numbers derived for the Y chromosome were inherently less accurate, and are therefore not presented here, this approach has enabled us to estimate that there are approximately 150 uninserted rDNA units on the Y chromosome in lines SWIFF 3, SWIFF 7, and SWIFF 9 and over 200 uninserted units on the Y chromosome in lines w1118#12 and SWIFF 6. Consistent with our PCR analysis of the 5′ junctions, these Southern approaches indicated relatively few R1 and R2 insertions on the Y chromosome. Thus, the size of the rDNA locus is about the same on the X and Y chromosome, but there are significantly more uninserted units on the Y chromosome due to the lower number of R1 and R2 insertions.
Crosses between high and low expression lines give intermediate levels of transcripts.
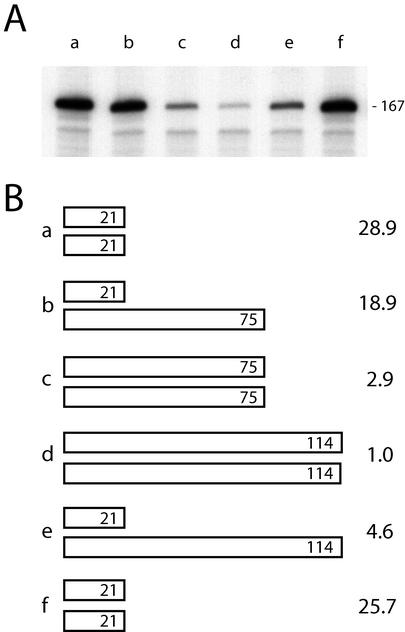
In order to test the model that the level of R1 and R2 transcripts in females was inversely correlated with the total number of uninserted rDNA units, crosses were made between lines that had different levels of R2 transcripts. Males from line w1118#12, which exhibited the lowest level of transcripts (114 uninserted units), were mated to SWIFF 6 females, which had the highest level of transcripts (21 uninserted units). In a similar manner, males from SWIFF 15 (75 uninserted units) were mated to SWIFF 6 females. The heterozygous female progeny from these two crosses would have intermediate numbers of uninserted units (Fig. 7B). RNA was isolated from the female progeny of these crosses as well as from females of the parental lines, and RNase protection assays were conducted to determine the level of transcripts corresponding to the 167-bp R2Dm insertion. As shown in Fig. 7, in both crosses the level of transcripts from the heterozygous females was intermediate to that of females from the two parental lines, consistent with the model that the level of expression is based on the total number of uninserted rDNA units present on the two X chromosomes of females.
FIG. 7.
Transcript levels in heterozygous females. (A) Males from SWIFF 15 and w1118#12 were mated to females from SWIFF 6. Total RNA from the adult female progeny from these crosses as well as from the parental females were subjected to RNase protection assays using probe 2 (Fig. 3A) to monitor transcription of the 167-bp R2Dm insertion. Lane a, SWIFF 6/SWIFF 6; lane b, SWIFF 6/SWIFF 15; lane c, SWIFF 15/SWIFF 15; lane d, w1118#12/w1118#12; lane e, SWIFF 6/w1118#12; lane f, SWIFF 6/SWIFF 6. (B) Schematic diagrams of the number of uninserted units in the parental and heterozygous females for the two crosses assayed in panel A. Lanes a to f refer to the lanes in panel A, and the numbers at right correspond to the relative level of transcripts in each lane, with the transcript level in w1118 (lane d) defined as 1.0.
DISCUSSION
In this report we utilized an in vivo R2 integration system based on the R2 element from B. mori to study cotranscription of sequences integrated into the R2 target site of the 28S rRNA genes of D. melanogaster. The newly integrated R2 sequences gave rise to dramatically different levels of RNA cotranscript that paralleled the endogenous R1 and R2 elements in each line. The increased level of expression observed in some lines was correlated with the loss of a large fraction of the uninserted rDNA units on the X chromosome. The loss of rDNA units associated with the insertion lines is likely a result of the frequent cleavage within the array made by the injected R2Bm endonuclease, the repair of which results in the elimination of rDNA units. Because rDNA units with an R2 insertion are presumably not subject to cleavage, this model could explain why the number of R2 insertions remained relatively stable in these lines. The R2Bm insertion lines have remained stable and have served as a valuable tool in the study of the expression of the rDNA locus and the ability of R1 and R2 insertions to inhibit that expression.
Many previous studies of D. melanogaster rRNA synthesis have indicated that R1 and/or R2 insertions in an rDNA unit dramatically decrease transcription of that unit (21, 23, 26, 27). Sensitive Northern blot analyses indicated that the R1 and R2 cotranscripts observed in nuclear RNA were at least 3 orders of magnitude lower than that of uninserted rRNA transcripts. While differences in the levels of R1 or R2 transcripts were detected in various lines (23), most studies involved comparisons between D. melanogaster strains with different origins and did not involve a characterization of the size or the R1 and R2 insertion profiles of the rDNA locus. While correlations between insert transcript levels and the number of uninserted genes has been observed in some studies of bobbed lines (25, 31, 40), these studies monitored total levels of transcripts and thus could not determine whether all insertions were behaving in a uniform manner.
Here we have monitored differences in transcript levels in lines that are isolates from the same D. melanogaster strain, w1118. The differences in transcript levels we observed are unlikely to have resulted from variable stability of the RNA transcripts, because we have monitored expression of the same inserted units in all lines. Therefore, these results provide the first direct evidence that cells can differentially regulate transcription of R1 and R2 inserted rDNA units. The range of expression levels was determined to be 100-fold. This could, however, be an underestimate of the total range possible, because transcription in the highest lines, SWIFF 6 and 11, may still be down-regulated relative to that of uninserted units. Indeed, SWIFF 6 and 11 females do not exhibit an overt bobbed phenotype, suggesting rRNA synthesis is not dramatically limiting.
A remarkable aspect of the regulation of transcription of the R1 and R2 inserted units is the uniformity that is exhibited across the locus. The quantitative increase in expression observed in the SWIFF lines as the number of uninserted rDNA units decreased was similar for the 167-bp R2Dm insertion and the 516-bp R2Dm insertion, as well as for the combined R1Dm 3′ UTRs (Table 2; Fig. 6B). Transcription of the exogenous R2Bm insertion was also consistent with the endogenous R1Dm and R2Dm levels. Among the lines with an insertion on the X chromosome, SWIFF 6 (21 uninserted units) had the highest level of R2Bm expression and SWIFF 1 (69 uninserted units) had the lowest level of expression, while SWIFF 9 (30 uninserted units) had an intermediate level of expression. Only the R2Bm element in SWIFF 11 (19 uninserted units) was expressed at levels lower than that predicted from the endogenous transcripts. However, the R2Bm insertion in SWIFF 11 is located in an rDNA unit that also contains a 500-bp R1Dm insertion. It is possible that the presence of both insertions either more severely inhibits transcription of this unit or gives rise to a less stable cotranscript.
The similar response of the different inserted units supports a model in which each unit in the rDNA locus is similarly regulated, independent of its position in the locus. Such a model has been suggested by previous studies of rRNA transcription in various organisms. Direct electron microscopic observation of rRNA transcription in D. melanogaster as well as other species has revealed that rDNA units are activated at different times in development and either are loaded with RNA polymerase and transcribed at near full capacity or completely lack transcription complexes (20, 32, 33). Maximally expressed and inactive units are interspersed in the locus. In a similar manner probes of chromatin structure, including nuclease-sensitive assays and chemical cross-linking probes, have indicated that the rDNA units of eukaryotes are divided into two fractions (7-9, 39, 41). One fraction appears in an accessible conformation for transcription, and a second fraction appears inaccessible for transcription. Based on these findings, we suggest that our observation of the same inserted unit being transcribed at levels that vary over 100-fold is a reflection of the different percentage of time the unit is active (or percentage of cells in which it is active) rather than different transcription rates.
The ability of adjacent rDNA units to exhibit dramatic differences in transcription levels is attributed to the presence of insulator sequences in the nontranscribed, tandemly repeated intergenic region typically found between rDNA transcription units (38). These insulators have been suggested to represent binding sites for the nuclear matrix, and they thereby define individual chromatin loops which are either transcribed or not transcribed. The ability of an R1 or R2 insertion 6 kb downstream of the transcription start site to inhibit transcription of the unit is intriguing. This inhibition is probably not a simple change in the length of the loop, since the length of the intergenic spacer of uninserted rDNA units in D. melanogaster varies over a range of several kilobases (37), while R2 insertions as small as 167 bp are sufficient to repress a unit.
One simple model to explain how R1 or R2 insertions can inactivate a unit is to postulate that the target sites for R1 and R2 are part of cis-acting sequences that promote transcription. Consistent with this model is the discovery of many other transposable elements that insert within 30 bp of the R2 target site of the 28S gene: R3 elements in insects (22), R4 elements in nematodes (3), Pokey elements in crustaceans (34), and R5 elements in flatworms (W. D. Burke et al., unpublished data). Because transcription of an inserted rDNA unit would generate defective rRNA, the insertion element would be under less negative selection if it simply shifted the inserted rDNA unit to the inactive fraction. Thus, insertion into a cis-control sequence of the rDNA unit may be the least harmful location for an insertion. To test this model, we designed an integration strategy that regenerated the R1/R2 insertion region upon insertion (SWIFF 15 [Fig. 1D]). From the perspective of the promoter of this inserted unit, the R2Bm sequence is 350 bp downstream of its normal location. Unfortunately the R2Bm element in SWIFF 15 was not transcribed at a higher level and, therefore, did not provide support for this model. However, only one example of this insertion was obtained, and the relative stability of the transcript was not addressed.
A final conclusion from these experiments is that the level of transcription of the inserted rDNA units in females is based on the number of uninserted units counted across both X chromosomes (Fig. 7). The finding that transcript levels of R2Bm insertions on the X chromosome or the 167-bp R2Dm element on the X chromosome are dramatically reduced in males is also consistent with the 150 to 200 uninserted units found on the Y chromosomes in these lines. Therefore, in males, R2 inserts may also be transcribed at levels that are dependent upon the number of uninserted units on both the X and Y chromosomes. On the other hand, the fact that four of the five R2Bm insertions on the Y chromosome are transcribed at readily detectable levels, with the insertions in SWIFF 3 and 7 transcribed at the very high levels of SWIFF 9 insertions in females, is inconsistent with this simple counting mechanism. (The only R2Bm insertion on the Y that is not transcribed, SWIFF 4, contains a doubly inserted R1Dm/R2Bm unit.) Therefore, transcription of the rDNA units is likely to be more complicated in males, with transcription of the rDNA units on the Y chromosome preferred over those on the X and/or transcription of inserted units less effectively suppressed on the Y chromosome.
It has long been known that the rDNA array on either the X or Y chromosome of D. melanogaster can rescue a deletion of the rDNA locus on the other chromosome. However, it has not been possible to resolve whether, under conditions where both loci contain ample numbers of rDNA units, one chromosome is preferentially transcribed (10). A number of selection and population genetics observations have led to the suggestion that the Y rDNA locus is less likely to be expressed than the X rDNA locus (5, 6, 15, 42). Our studies suggest this may not be the case. Analysis of the expression of individual insertions on the Y chromosome in the context of a variable number of uninserted units will hopefully shed light on the differential expression of inserted and uninserted rDNA units on the X and Y chromosomes of D. melanogaster.
A second question which remains is whether the shortest R1 and R2 insertions are transcribed at higher levels than full-length insertions, or whether rRNA cotranscripts containing longer insertions are simply less stable. Data suggesting that full-length R1 and R2 inserts are not transcribed come from electron microscopic observation of actively transcribing rDNA units (4, 20, 32). Because in these studies the ability to score an rDNA unit as inserted or uninserted was based on length, these studies would not have been able to differentiate an rDNA unit with an insertion only 500 bp in length from an uninserted unit. Greater transcription of shorter R2 elements would also have been missed in previous studies of R2 expression, because the hybridization probes used did not include the 500 bp at the 3′ end of the element (23, 25, 27, 40). We plan to address the question of whether longer R1 and R2 insertions are efficiently transcribed but more unstable, using nuclear run-on experiments. Clearly, many questions remain concerning the regulated transcription of the inserted and uninserted rDNA units of D. melanogaster. Our integration system and our ability to assay the activity of individual inserted units now provide the tools to study these mechanisms.
Acknowledgments
This work was supported by National Institutes of Health grant GM42790.
We thank Bill Burke for discussions and comments on the manuscript.
REFERENCES
- 1.Burke, W. D., H. S. Malik, W. C. Lathe, and T. H. Eickbush. 1998. Are retrotransposons long term hitchhikers? Nature 239:141-142. [DOI] [PubMed] [Google Scholar]
- 2.Burke, W. D., H. S. Malik, J. P. Jones, and T. H. Eickbush. 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16:502-511. [DOI] [PubMed] [Google Scholar]
- 3.Burke, W. D., F. Müller, and T. H. Eickbush. 1995. R4, a non-LTR retrotransposon specific to the large subunit rRNA gene of nematodes. Nucleic Acids Res. 23:4628-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chooi, W. Y. 1979. The occurrence of long transcription units among the X and Y ribosomal genes of Drosophila melanogaster: transcription of insertion sequences. Chromosoma 74:57-74. [DOI] [PubMed] [Google Scholar]
- 5.Clark, A. G., F. M. Szumski, and E. S. M. Lyckegaard. 1990. Population genetics of the Y chromosome of Drosophila melanogaster: rDNA variation and phenotypic correlates. Genet. Res. 58:7-13. [DOI] [PubMed] [Google Scholar]
- 6.Cluster, P. D., D. Marinkovic, R. W. Allard, and F. J. Ayala. 1987. Correlations between developmental rates, enzyme activities, ribosomal DNA spacer-length phenotypes, and adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conconi, A., J. M. Sogo, and C. A. Ryan. 1992. Ribosomal gene clusters are uniquely proportioned between open and closed chromatin structures in both tomato leaf cells and exponentially growing suspension cultures. Proc. Natl. Acad. Sci. USA 89:5256-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conconi, A., R. M. Widmer, T. Koller, and J. M. Sogo. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753-761. [DOI] [PubMed] [Google Scholar]
- 9.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1995. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15:5294-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durica, D. S., and H. M. Krider. 1977. Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids. Dev. Biol. 59:62-74. [DOI] [PubMed] [Google Scholar]
- 11.Eickbush, D. G., and T. H. Eickbush. 1995. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics 139:671-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eickbush, D. G., D. D. Luan, and T. H. Eickbush. 2000. Integration of Bombyx mori R2 sequences into the 28S ribosomal RNA genes of Drosophila melanogaster. Mol. Cell. Biol. 20:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eickbush, T. H. 2002. R2 and related site-specific non-long terminal repeat retrotransposons, p. 813-835. In N. L. Craig, R. Craigie, M. Gellart, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 14.Feng, Q., J. V. Moran, H. H. Kazazian, and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 15.Frankham, R., D. A. Briscoe, and R. K. Nurthen. 1980. Unequal crossing over at the rDNA tandon as a source of quantitative genetic variation in Drosophila. Genetics 95:727-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentile, K. L., W. D. Burke, and T. H. Eickbush. 2001. Multiple lineages of R1 retrotransposable elements can coexist in the rDNA loci of Drosophila. Mol. Biol. Evol. 18:235-245. [DOI] [PubMed] [Google Scholar]
- 17.George, J. A., W. D. Burke, and T. H. Eickbush. 1996. Analysis of the 5′ junctions of R2 insertions with the 28S gene: implications for non-LTR retrotransposition. Genetics 142:853-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakubczak, J. L., Y. Xiong, and T. H. Eickbush. 1990. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J. Mol. Biol. 212:37-52. [DOI] [PubMed] [Google Scholar]
- 19.Jakubczak, J. L., M. K. Zenni, R. C. Woodruff, and T. H. Eickbush. 1992. Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics 131:129-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamrich, M., and O. L. Miller, Jr. 1984. The rare transcripts of interrupted rRNA genes in Drosophila melanogaster are processed or degraded during synthesis. EMBO J. 3:1541-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolly, D. J., and C. A. Thomas. 1980. Nuclear RNA transcripts from Drosophila melanogaster ribosomal RNA genes containing introns. Nucleic Acids Res. 8:67-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerrebrock, A. W., R. Srivastava, and S. A. Gerbi. 1989. Isolation and characterization of ribosomal DNA variants from Sciara coprophila. J. Mol. Biol. 210:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Kidd, S. J., and D. M. Glover. 1981. Drosophila melanogaster ribosomal DNA containing type II insertions is variably transcribed in different strains and tissues. J. Mol. Biol. 151:645-662. [DOI] [PubMed] [Google Scholar]
- 24.Lathe, W. C., III, and T. H. Eickbush. 1997. A single lineage of R2 retrotransposable elements is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 14:1232-1241. [DOI] [PubMed] [Google Scholar]
- 25.Long, E. O., M. Collins, B. I. Kiefer, and I. B. Dawid. 1981. Expression of the ribosomal DNA insertions in bobbed mutants of Drosophila melanogaster. Mol. Gen. Genet. 182:377-384. [DOI] [PubMed] [Google Scholar]
- 26.Long, E. O., and I. B. Dawid. 1979. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell 18:1185-1196. [DOI] [PubMed] [Google Scholar]
- 27.Long, E. O., M. L. Rebbert, and I. B. Dawid. 1980. Structure and expression of ribosomal RNA genes of Drosophila melanogaster interrupted by type 2 insertions. Cold Spring Harbor Symp. Quant. Biol. 45:667-672. [DOI] [PubMed] [Google Scholar]
- 28.Luan, D. D., and T. H. Eickbush. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15:3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan, D. D., and T. H. Eickbush. 1996. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell. Biol. 16:4726-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 31.Makni, M., M. Marrakchi, and N. Prud'homme. 1989. The occurrence of long ribosomal transcripts homologous to type I insertions in bobbed mutants of Drosophila melanogaster. Genet. Res. 54:127-135. [DOI] [PubMed] [Google Scholar]
- 32.McKnight, S. L., and O. L. Miller, Jr. 1976. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8:305-319. [DOI] [PubMed] [Google Scholar]
- 33.Meyer, G. F., and W. Henning. 1974. The nucleolus in primary spermatocytes of Drosophila hydei. Chromosoma 46:121-144. [DOI] [PubMed] [Google Scholar]
- 34.Penton, E. H., B. W. Sullender, and T. J. Crease. 2002. Pokey, a new DNA transposon in Daphnia (Cladocera: Crustacea). J. Mol. Evol. 55:664-673. [DOI] [PubMed] [Google Scholar]
- 35.Perez-Gonzalez, C. E., and T. H. Eickbush. 2001. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics 158:1557-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Gonzalez, C. E., and T. H. Eickbush. 2002. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162:799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polanco, C., A. I. González, Á. de la Fuente, and G. A. Dover. 1998. Multigene family of ribosomal DNA in Drosophila melanogaster reveals contrasting patterns of homogenization for IGS and ITS spacer regions: a possible mechanism to resolve this paradox. Genetics 149:243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinett, C. C., A. O'Connor, and M. Dunaway. 1997. The repeat organizer, a specialized insulator element within the intergenic spacer of the Xenopus rRNA genes. Mol. Cell. Biol. 17:2866-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stancheva, I., R. Lucchini, T. Koller, and J. M. Sogo. 1997. Chromatin structure and methylation of rat rRNA genes studied by formaldehyde fixation and psoralen cross-linking. Nucleic Acids Res. 25:1727-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terracol, R. 1986. Transcription of rDNA insertions in bobbed mutants of Drosophila melanogaster. Genet. Res. 48:167-174. [DOI] [PubMed] [Google Scholar]
- 41.Wayne, R. L., Z. D. Sharp, and J. D. Procunier. 1985. Preferential DNase I sensitivity of insert-free ribosomal RNA of Drosophila melanogaster. Nucleic Acids Res. 13:2869-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, S. M., G. R. Furnier, E. Fuog, and C. Strobeck. 1987. Evolution of the ribosomal DNA spacers of Drosophila melanogaster: different patterns of variation on the X and Y chromosomes. Genetics 116:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Q., L. M. Angerer, and R. C. Angerer. 1989. Structure and tissue-specific developmental expression of a sea urchin arylsulfatase gene. Dev. Biol. 135:53-65. [DOI] [PubMed] [Google Scholar]