Abstract
During mitosis, sister kinetochores attach to microtubules that extend to opposite spindle poles (bipolar attachment) and pull the chromatids apart at anaphase (equational segregation). A multisubunit complex called cohesin, including Rad21/Scc1, plays a crucial role in sister chromatid cohesion and equational segregation at mitosis. Meiosis I differs from mitosis in having a reductional pattern of chromosome segregation, in which sister kinetochores are attached to the same spindle (monopolar attachment). During meiosis, Rad21/Scc1 is largely replaced by its meiotic counterpart, Rec8. If Rec8 is inactivated in fission yeast, meiosis I is shifted from reductional to equational division. However, the reason rec8Δ cells undergo equational rather than random division has not been clarified; therefore, it has been unclear whether equational segregation is due to a loss of cohesin in general or to a loss of a specific requirement for Rec8. We report here that the equational segregation at meiosis I depends on substitutive Rad21, which relocates to the centromeres if Rec8 is absent. Moreover, we demonstrate that even if sufficient amounts of Rad21 are transferred to the centromeres at meiosis I, thereby establishing cohesion at the centromeres, rec8Δ cells never recover monopolar attachment but instead secure bipolar attachment. Thus, Rec8 and Rad21 define monopolar and bipolar attachment, respectively, at meiosis I. We conclude that cohesin is a crucial determinant of the attachment manner of kinetochores to the spindle microtubules at meiosis I in fission yeast.
Sister chromatid cohesion is established during S phase and maintained throughout G2 until M phase in eukaryotic cells. This cohesion is mediated by a conserved multisubunit complex, cohesin (4, 5, 11, 16, 29). In fission yeast, the cohesin complex in mitosis comprises at least four subunits: Rad21, Psc3, Psm1, and Psm3 (equivalent to Scc1, Scc3, Smc1, and Smc3, respectively, in budding yeast) (25, 26). Chromosome segregation during mitosis is triggered by the dissolution of sister chromatid cohesion along the whole chromosome length.
During meiosis, cohesin subunit Rad21/Scc1 is largely replaced by its meiotic counterpart, Rec8 (9, 18, 19, 30). There are three crucial features of the chromosome at meiosis I that are not observed in mitosis (13, 26, 32). First, pairing and recombination occurs between homologous chromosomes in meiotic prophase, resulting in the formation of chiasmata. Second, sister kinetochores always attach to microtubules from same spindle pole, which is called monopolar attachment and is known to ensure that sister chromatids segregate to the same pole at meiosis I. Third, cohesion between sister chromatids in the vicinity of centromeres persists until the onset of the second meiotic division. Remarkably, all of these features of meiotic chromosomes are disrupted in Schizosaccharomyces pombe rec8Δ cells, suggesting that a meiosis-specific cohesin complex plays a central role in the assembly of meiotic chromosomes (12, 30). In particular, because Rec8 preferentially locates at centromeres and rec8Δ cells undergo equational chromosome segregation at meiosis I, it has been proposed that Rec8 is required at centromeres to construct kinetochores that orient to the same pole (30, 31). However, it has been shown in Saccharomyces cerevisiae that if the Scc1/Rad21 cohesin subunit, but not Rec8, is expressed by the REC8 promoter, then monopolar attachment is established, although sister chromatid cohesion at the centromeres is disrupted during anaphase I (27). Thus, at least in S. cerevisiae, Scc1/Rad21 can allow proper orientation and monopolar attachment of sister kinetochores at meiosis I. Therefore, there is an argument that the equational segregation observed in S. pombe rec8Δ cells may be due to a loss of cohesin in general rather than a specific requirement for the Rec8 subunit of cohesin (10).
Here we address the open question presented above. First, we examined why S. pombe rec8Δ cells undergo equational rather than random division. We show that if Rec8 is absent during meiosis, Rad21 relocates to the centromeres to back up the cohesion of sister kinetochores. Thus, substitutive Rad21 is the reason for the occurrence of equational division. Moreover, we demonstrate that even though enough Rad21 is transferred to the centromeres by an ectopic expression, rec8Δ cells never undergo reductional division but instead secure equational division. From these results and others we conclude that Rec8 and Rad21 determine the manner of kinetochore attachment to microtubules at meiosis I, at least in fission yeast.
MATERIALS AND METHODS
Media and fission yeast strains.
The complete medium YE or minimal medium (SD, MM, or SSA) was used for the routine culture of S. pombe strains. Sporulation agar plate (SPA) was used to induce meiosis and microscopic examination. MM and MM-N (lacking nitrogen) containing 1% glucose were used for synchronous meiosis. All strains used in the present study are listed in Table 1.
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype |
|---|---|
| JY333 | h−ade6-M216 leu1 |
| JY450 | h90 ade6-M216 leu1 |
| JZ804 | h+mei4::ura4+leu1 ura4-D18 ade6-M210 |
| PY182 | h90 rec8::kanrade6-M216 leu1 |
| PY221 | h+/h+pat1-114/pat1-114 cdc2-L7/cdc2-L7 rec8+-GFP-kanr/rec8+-GFP-kanrade6-M210/ade6-M216 leu2/+ +/lys1 |
| PY322 | h+rec8::kanrlys1+<<lacO his7+<<Pdis1-GFP-lac1-NLS leu 1 ade6-M216 |
| PY445 | h90 mei4::ura4+rec8::kanrrad21-K1<<ura4+lys1+<<lacO his7+<<Pdis1-GFP-lac1-NLS leu1 ade6-M216 |
| PY446 | h90 mei4::ura4+rec8::kanr lys1+<<lacO his7+<<Pdis1-GFP-lac1-NLS leu1 ade6-M216 |
| PY447 | h90 mei4::ura4+lys1+<<lacO his7+<<Pdis1-GFP-lacI-NLS leu1 ade6-M216 |
| PY448 | h90 mei4::ura4+rad21-K1<<ura4+lys1+<<lacO his7+<<Pdis1-GFP-lac1-NLS leu1 ade6-M216 |
| PY545 | h−rec8::kanrleu1 |
| PY594 | h90 mei4::ura4+ade3::lacO-ura4+-kanrhis7+<<Pdis1-GFP-lacI-NLS leu1 lys1 ade6 |
| PY595 | h90 mei4::ura4+ade3::lacO-ura4+-kanr his7+<<Pdis1-GFP-lacI-NLS leu1 lys1 ade6 |
| PY741 | h+/h−mei4::ura4+/mei4::ura4+rec8+-GFP-kanr/rec8+-GFP-kanrade6-M216/ade6-M210 |
| PY748 | h−rec8Δ::Padh1-rad21+-FLAG ade6-M210 |
| PY749 | h−rec8Δ::Prec8-rad21+-FLAG ade6-M210 |
| PY754 | h+/h+pat1-114/pat1-114 cdc2-L7/cdc2-L7 rec8Δ::Padh1-rad21+-GFP/rec8Δ::Padh1-rad21+-GFP leu2/+ +/lys1 ade6-M216/ade6-M210 |
| PY770 | h+/h+pat1-114/pat1-114 cdc2-L7/cdc2-L7 rec8Δ::Prec8-rad21+-GFP/rec8Δ::Prec8-rad21+-GFP leu2/+ +/lys1 ade6-M216/ade6-M210 |
| PY783 | h−rad21-K1<<ura4+rec8::kanrleu1 ade6-M216 |
| PY785 | h+rad21-K1<<ura4+rec8::kanrlys1+<<lacO his7+<<Pdis1-GFP-lacI-NLS ade6-M216 leu1 |
| PY786 | h−rad21-K1<<ura4+ade6-M216 |
| PY787 | h+rad21+-GFP-kanrmei4::ura4+leu1 ura4-D18 ade6-M210 |
| PY801 | h90 rad21+-GFP-kanrleu1 ade6-M210 |
| PY878 | h90 rec8::ura4+rad21+-GFP-kanrade6-M216 |
| PY893 | h+/h+pat1-114/pat1-114 cdc2-L7/cdc2-L7 rec8::kanr/rec8::kanrrad21+-GFP-kanr/rad21+-GFP-kanrleu2/+ +/lys1 ade6-M216/ade6-M210 |
| PZ207 | h90 rec8Δ::Padh1-rec8+-3HA lys1+<<lacO his7+<<Pdis1-GFP-lacI-NLS leu1 ura4-D18 ade6 |
| PZ208 | h90 rec8Δ::Padh1-rec8+-3HA rad21::ura4+lysl+<<lacO his7+<<Pdis1-GFP-lac1-NLS leu1 ura4-D18 ade6 |
Construction of a strain carrying the rad21 allele fused to GFP.
A strain carrying the C-terminally green fluorescent protein (GFP)-tagged rad21+ allele was constructed by direct chromosomal integration of a PCR-generated fragment as described previously (1). The PCR products were purified and transformed into JZ804 cells by using the lithium acetate method. Stable G418-resistant (kanr genotype) transformants were selected and analyzed by PCR to verify their correct integration. The obtained rad21+-GFP cells grew normally, indicating that Rad21-GFP is functional.
Construction of plasmids carrying cyan-GFP variant (CFP)-tagged Gar1 or Mis6 protein.
We constructed pREP81-gar1+-CFP and pREP81-mis6+-CFP. The gar1+ open reading frame was amplified by PCR with wild-type genomic DNA as a template and forward primer 5′-GCCGGGTCGACAATGAGTTTTAGAGGCGGTCGC-3′ (the SalI site is underlined) and reverse primer 5′-GGGAGGGATCCAAGAATCTGCCACGGAAACCAC-3′ (the BamHI site is underlined). The mis6+ open reading frame was amplified with forward primer 5′-CGGCCAGATCTTAATGGAAAGCTTTGAGA-3′ (the BglII site is underlined) and reverse primer 5′-CCCCGCGGCCGCAAACGATTTAATGTTGAAA-3′ (the NotI site is underlined). These PCR products were cloned into pGFT81 (7), and then the C-terminal GFP was replaced with the cloned CFP fragment.
Construction of strains overexpressing Rad21 from the adh1 or rec8 promoter.
To construct pREP-Prec8-rad21+-GFP, the rec8 promoter region (ca. −108 to −1), the rad21+-GFP fusion gene, the ura4+ cassette, and the 3′-untranslated region of rec8 (∼600 bp) were cloned between the PstI and SacI sites of pREP1. To construct pREP-Padh1-rad21+-GFP, an adh1 promoter fragment was additionally inserted into the 5′ site of the rad21 sequence. These plasmid constructs were linearized by SacI and ClaI to cut out the Prec8-rad21+-GFP and Padh1-rad21+-GFP regions and then transformed into rec8::kanr cells to replace the chromosomal rec8::kanr locus with these constructs by homologous recombination at the rec8 promoter region and 3′ untranslated region. For construction of Prec8-rad21+-FLAG and Padh1-rad21+-FLAG alleles on the chromosomes, the GFP sequences of the preceding constructs were replaced by a FLAG fragment.
Preparation of synchronous meiotic cells and observation of chromosomes marked with GFP.
Cells were cultured to log phase, collected by centrifugation, suspended in 20 g of leucine liter−1, and then spotted onto a SPA. When we marked only one chromosome by GFP, we cultured opposite mating-type cells, one marked with GFP and the other not, and mixed them prior to spotting them onto SPA. This allows subsequent synchronous conjugation and meiosis. When cells undergoing meiosis I and II or cells arrested at late prophase I by mei4Δ became abundant in the population, they were observed without fixation or after being fixed with cold methanol; zygotes were monitored for GFP and DAPI (4′,6-diamidino-2-phenylindole). Images were obtained by using a microscope (Axioplan 2; Zeiss) equipped with a cooled charge-coupled device camera (Quantix; Photometrics) and Metamorph software (Universal Imaging Corporation). Seven Z sections for GFP signals were converted into single two-dimensional images by taking the maximum signal at each pixel position in the images.
Synchronous meiosis.
We induced synchronous meiosis with a temperature-sensitive pat1-114 allele as described previously (31). Cells were cultured in MM-N at 25°C for 16 h and then shifted to 32°C, and 0.1 g of NH4Cl liter−1 was added. The cells were harvested and fixed with 1% formaldehyde (Wako) for chromatin immunoprecipitation (ChIP) assay and with methanol for microscopy and fluorescence-activated cell-sorting analysis. For microscopy, fixed cells were washed and suspended in PEMS buffer (100 mM PIPES, pH 6.9; 1 mM EGTA; 1 mM MgSO4; 1.2 M sorbitol) with DAPI.
ChIP analysis.
ChIP assays were carried out essentially as described previously (21). Anti-GFP polyclonal antibodies (Living Colors Full-length A.v. Polyclonal Antibody; Clontech) were used for immunoprecipitation. DNA prepared from whole-cell extracts or immunoprecipitated fractions was analyzed by quantitative PCR with a LightCycler and a LightCycler-DNA Master SYBR Green I kit (Roche). The following primers were used for PCR: forward primer for cnt, 5′-ATCTCATTGCTATTTGGCGAC-3′; reverse primer for cnt, 5′-GCGTTTCTTCGGCGAAATGC-3′; forward primer for imr, 5′-CACATACCAAAAAGTCTGGC-3′; reverse primer for imr, 5′-GCTGAGGCTAAGTATCTGTT-3′; forward primer for dg, 5′-TTTTCAGCGAGACATGTACC-3′; reverse primer for dg, 5′-TCATAAAGCAACACTGGGTG-3′; forward primer for dh, 5′-GTAAGTATGAGCAACTGGCG-3′; reverse primer for dh, 5′-GGAACAAATCAGGAAACCGAG-3′; forward primer for lys1, 5′-ATTTTCGCATCCAACGCTGC-3′; reverse primer for lys1, 5′-ACAACTAAGGCTCTGGGCTT-3′; forward primer for mes1, 5′-CGAAGGCTACTTTCATGCCA-3′; reverse primer for mes1, 5′-CGTACATTCAGACTGTTGAAC-3′; forward primer for rDNA1, 5′-ATGATTCCCTCAGTAACGGCGAG-3′; reverse primer for rDNA1, 5′-CACTCTACTTGTTCGCTATCGGT-3′; forward primer for rDNA2, 5′-CCTCTAACGATAGTTACCTGGTTG-3′; reverse primer for rDNA2, 5′-CAGAAATTTGAATGAACCATCGCC-3′; forward primer for TAS, 5′-CGTTACCCAATTAATTATGAC-3′; and reverse primer for TAS, 5′-CGGTGATGTAGTGTACTATA-3′.
RESULTS
Rad21 is dispensable for normal meiosis, as long as Rec8 function is intact.
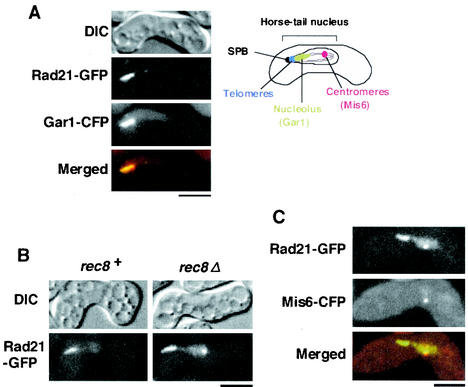
During meiosis, Rad21 is largely replaced by its meiotic counterpart, Rec8, which plays an essential role in establishing reductional chromosome segregation at meiosis I. Because Rad21 has been detected in meiotic nuclei at a residual level in several organisms (9, 20, 30), we first sought to determine whether Rad21 plays some role in meiosis in fission yeast. In normal meiosis, Rad21 is detected near the leading edge of the horsetail nucleus, where the telomeres are clustered (30) (Fig. 1A). ChIP assay indicated that Rad21 associates with ribosomal DNA (rDNA) rather than telomere-associated sequences (see below). Moreover, fluorescence microscopy revealed that Rad21 tagged with GFP colocalizes mostly with a nucleolar protein, Gar1 (3), tagged with CFP in the horsetail nucleus (Fig. 1A). These results suggest that Rad21 is enriched in the nucleolus, rather than telomere-adjacent DNA sequences, although residual association with other chromosomal regions might also occur. Given that Rad21 is essential for cell proliferation but rad21Δ cells become viable by expressing Rec8 with a constitutive adh1 promoter (30), we used rad21Δ Padh1-rec8+ cells to inspect the requirement of Rad21 for meiosis. We first assayed meiotic recombination in rad21-deleted cells but did not detect any decrease in recombination when tested between the lys3 and ura1 loci (located near the telomere of chromosome I) and between the leu1 and mat1 loci (located near the centromere of chromosome II) (data not shown). We next examined the requirement of Rad21 for meiotic chromosome segregation. Chromosome segregation was analyzed by viewing the LacI-GFP fusion protein, bound to LacO DNA repeats that were integrated at the lys1 locus (cen1-GFP dot), ca. 30 kb from the centromere of chromosome I (14). The normal segregation pattern of cen1-GFP, a single GFP dot in every nucleus of the tetranucleated cells, was prevalent in the rad21Δ Padh1-rec8+ cells (PZ208) (94%, n = 543), as well as in its rad21+ version (PZ207) (92%, n = 515). Although we do not know the significance of Rad21 enrichment in the nucleolus, these analyses suggest that Rad21 is virtually dispensable for normal meiotic chromosome segregation and recombination as long as Rec8 function is intact.
FIG. 1.
Rad21 relocates to the centromeres in rec8Δ cells. (A) h90 rad21+-GFP cells (PY801) carrying pREP81-gar1+-CFP during the horsetail period were examined by fluorescence microscopy. In the merged figure, Rad21-GFP and Gar1-CFP are represented by green and red, respectively. The positions of the spindle pole body (SPB), nucleolus, telomere, and centromere are shown in the drawing of the horsetail nucleus. (B) h90 rad21+-GFP cells of rec8+ and rec8Δ (PY801 and PY878) were induced to meiosis and examined for GFP fluorescence during the horsetail period. (C) The localization of Rad21-GFP and Mis6-CFP in h90 rad21+-GFP rec8Δ cells (PY878) carrying pREP81-mis6+-CFP. In the merged figure, Rad21-GFP and Mis6-CFP are represented by green and red, respectively. Bars, 5 μm.
Rad21 relocates to the centromeres if Rec8 is absent in meiosis.
We have previously shown that S. pombe rec8Δ cells undergo mostly equational division at meiosis I. If the Rec8 complex is the sole cohesin in meiosis, rec8Δ cells will lose most sister chromatid cohesion during prophase and then undergo random, rather than equational, chromosome segregation. Indeed, the budding yeast rec8 mutant shows such a phenotype in 70% of cells (9). One explanation for the phenotype of fission yeast rec8Δ is that residual Rad21 may substitute for centromeric cohesion, because Rad21 is required at the centromeres for equational segregation in mitosis (2, 17, 28). To investigate this possibility, we determined the localization of Rad21 in rec8Δ cells. Although Rad21 is enriched in the nucleolus of rec8+ cells, an additional distinct dot of Rad21 appears in rec8Δ cells on the opposite side of the leading edge of the horsetail nucleus (Fig. 1B). To determine the position of the Rad21-GFP dot, we coexpressed a kinetochore protein, Mis6 (21), tagged with CFP. Fluorescence microscopy showed the Rad21-GFP dot colocalizing with the Mis6-CFP dot (Fig. 1C), indicating that a significant amount of Rad21 relocates to the centromeres if Rec8 is depleted. These results raise the possibility that substitutive Rad21 may play a role in ensuring equational chromosome segregation at meiosis I in rec8Δ cells.
Rad21 is required for equational division during meiosis I in rec8Δ cells.
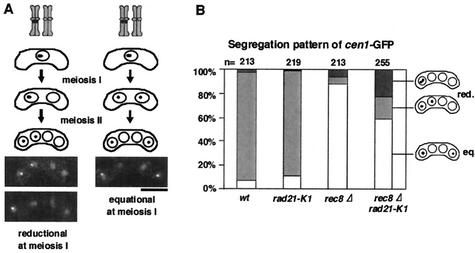
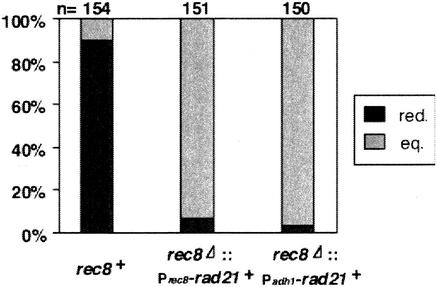
We then decided to introduce a temperature-sensitive mutation, rad21-K1 (24), into rec8Δ and examine chromosome segregation at meiosis I. By crossing the cen1-GFP strain with an untagged strain, we monitored the segregation pattern of one pair of sister chromatids during meiosis. Because the two nuclei on either side of the tetrad are derived from the same meiosis I nucleus in fission yeast, reductional or equational segregation in meiosis I can be determined by examining the distribution of the two cen-GFP dots in the zygotes (Fig. 2A). Wild-type zygotes represented mostly reductional segregation patterns: +/+, −/− (Fig. 2B). The occurrence of some +/−, +/− patterns can be explained by recombination between the GFP-associating sequences and the centromere (appearance of recombinants is expected to be 8%). As reported previously (30), rec8Δ cells mostly displayed equational segregation patterns: +/−, +/− (Fig. 2B). The rad21-K1 mutation by itself showed no meiotic defect. However, when rec8Δ was combined with rad21-K1, the equational segregation at meiosis I was partly disrupted with the reductional population increasing to ∼40% (Fig. 2B). Notably, the rec8Δ rad21-K1 cells with reductional division at meiosis I underwent random second division (Fig. 2B), suggesting that the “reductional” segregation was erroneous. If equational division is disrupted completely, sister cen1-GFP dots will move randomly, producing a complete mixture of equational and reductional patterns (each 50%). Therefore, the observed change would be close to this case. We conclude that Rad21 plays a role in ensuring equational segregation during meiosis I in rec8Δ cells.
FIG. 2.
A mutation in rad21 impairs equational division at meiosis I in rec8Δ cells. (A) The segregation patterns of cen1-GFP during meiosis are illustrated with examples. Bar, 5 μm. (B) Segregation of cen1-GFP during meiosis was examined in the indicated cells. A rec8Δ rad21-K1 strain tagged with cen1-GFP (PY785) was crossed with the indicated strain without the tag (wt, JY333; rad21-K1, PY786; rec8Δ, PY545; rec8Δ rad21-K1, PY783) and induced to meiosis at a semipermissive temperature (32°C). Reductional (red.) and equational (eq.) segregation patterns at meiosis I are shown. Note that nondisjunction at meiosis II could be measured only in cells that underwent reductional meiosis I.
Rad21 plays a role in cohesion at centromeres during meiosis I in rec8Δ cells.
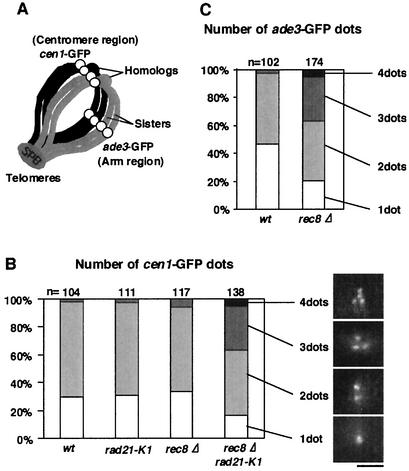
For the faithful processing of equational chromosome segregation, centromere cohesion must be preserved at least until the spindle microtubules capture kinetochores. Therefore, centromeric cohesion of rec8Δ cells would be intact. This was examined by monitoring cen1-GFP marking near the centromere of chromosome I. We utilized a mutation of mei4Δ to synchronize the meiotic cells at late prophase I (6). In wild-type cells, the cen1-GFP dots were mostly doublet or single at this stage, indicating that sister chromatid cohesion at the centromere was intact and that there was some pairing between homologs (Fig. 3A). The rad21-K1 cells also displayed no defect in sister chromatid cohesion during meiosis, although there were severe defects during mitosis (25; data not shown). Interestingly, rec8Δ cells preserved centromeric cohesion at the mei4Δ arrest point in meiotic prophase, although they showed substantially impaired arm cohesion (Fig. 3). Remarkably, the residual centromeric cohesion of rec8Δ cells was substantially disrupted by the introduction of the rad21-K1 mutation. Taken together, these results strongly suggest that if Rec8 is depleted, Rad21 has a concealed role in preserving sister centromere cohesion at meiosis I, thereby ensuring subsequent equational division.
FIG. 3.
Sister chromatid cohesion at the centromere is intact in rec8Δ cells but impaired by the introduction of a rad21-K1 mutation. (A) Chromosomes at the horsetail period are depicted. (B) h90 cen1-GFP mei4Δ cells carrying the indicated mutations (wt, PY447; rad21-K1, PY448; rec8Δ, PY446; rec8Δ rad21-K1, PY445) were induced to meiosis at the permissive temperature of 25°C for 8 h and then shifted to 32°C for 4 h. The cells arrested at prophase I (horsetail period) were observed for GFP dots. The number of dots per nucleus is shown with photos of representative nuclei. Bar, 2 μm. (C) Arm cohesion was similarly assayed in wild-type (wt) PY594 and rec8Δ PY595 cells by observing ade3-GFP, which locates in the middle of the left arm of chromosome I.
Sufficient amounts of Rad21 are transferred to the centromeres in meiosis by ectopic expression.
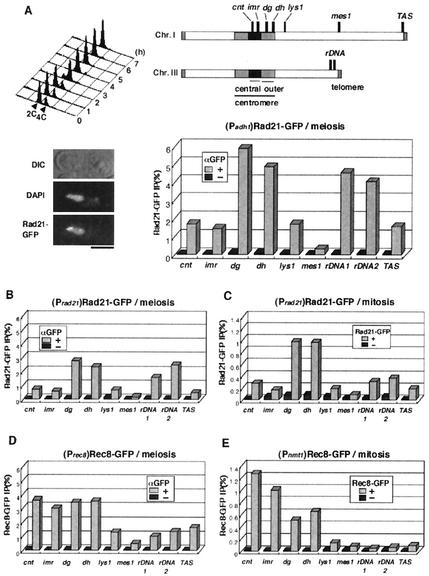
In budding yeast, endogenous Scc1 (Rad21 homolog) likely plays little part in centromeric cohesion in rec8Δ cells, which undergo rather random chromosome segregation at meiosis I (9). The fact that the S. pombe Rad21 complex has an affinity for heterochromatin, which covers a vast region of the centromeres (2, 17), whereas S. cerevisiae has no centromeric heterochromatin, might explain why substantial amounts of Rad21/Scc1 relocate to centromeres in fission yeast but not in budding yeast. When Scc1, but not Rec8, is expressed by the REC8 promoter, however, monopolar attachment at meiosis I is mostly established in budding yeast (27). This observation contradicts our current findings that substitutive Rad21 ensures equational division at meiosis I in fission yeast rec8Δ cells. One may argue that the amount of substitutive Rad21 at the centromeres in fission yeast is not sufficient to provide a full level of “cohesin function.” To address this issue, we expressed Rad21-GFP (carrying full function) from the rec8 promoter or the more robust adh1 promoter in rec8Δ cells and monitored their meioses. Fluorescence microscopy of Rad21-GFP indicated that the centromere signal is indeed stronger in these strains, especially in rec8Δ::Padh1-rad21+-GFP cells (Fig. 4A). To demonstrate further the association of Rad21 with centromere DNA, we performed a ChIP assay. Extracts were prepared from rec8Δ::Padh1-rad21+-GFP cells under synchronous meiosis and mostly within prophase I. In accordance with the cytological observations, the ChIP assay indicated that Rad21-GFP associates mainly with the centromere and rDNA sequences (Fig. 4A and B). S. pombe centromeres are organized into two distinct domains: a heterochromatic outer repeat region and a central core region associated with histone H3 variant CENP-A (Fig. 4A). Endogenous Rad21 associates with the outer centromere regions but less with the central core during mitosis (31) (Fig. 4C) and also during meiosis (Fig. 4B), as has been similarly observed in meiotic rec8Δ::Padh1-rad21+-GFP cells, but the association with chromatin was several times greater in the latter cells (compare Fig. 4A with C). Notably, the increased amount of Rad21 association with the outer centromeric repeat sequences (dg or dh) exceeds that of endogenous Rec8 association during meiosis (Fig. 4A and D). Similar results were obtained with meiotic rec8Δ::Prec8-rad21+-GFP cells, although the amount of Rad21 associated with the outer centromere was similar to that of endogenous Rec8 (data not shown).
FIG. 4.
Rad21 overexpressed in rec8Δ cells associates fully with the outer centromere regions in meiosis. (A) Diploid h+/h+ pat1-114 cdc2-L7 rec8Δ::Padh1-rad21+-GFP cells (PY754) were induced to synchronous meiosis and examined for the association of Rad21-GFP at the indicated regions by ChIP assay. Flow cytometric determination of the cellular DNA content indicates that DNA replication was complete at 3 h (full-level association of cohesin was expected to occur at around this time). Therefore, we used this time point for ChIP assay. Microscopic images of the cell at 3 h are shown. Bar, 5 μm. Schematic representation of S. pombe chromosomes I and III, and the primers (cnt, imr, dg, dh, lys1, mes1, rDNA1, rDNA2, and TAS) used for ChIP assay. DNA prepared from whole-cell extracts or immunoprecipitated fractions was analyzed by quantitative PCR. +, sample immunoprecipitated with anti-GFP antibodies; −, negative control without antibodies. (B) ChIP assay was performed in rec8Δ cells expressing Rad21-GFP from the native rad21 promoter (PY893) as described in panel A. (C) Proliferating rad21+-GFP cells (PY787) were analyzed by ChIP assay with anti-GFP antibodies. We used untagged rad21+ cells (JY333) as a negative control, in this case with anti-GFP antibodies. (D) Meiotic rec8+-GFP cells (PY741) arrested at prophase by mei4Δ were analyzed by ChIP assay with or without anti-GFP antibodies. (E) Proliferating rec8Δ cells (PY182) expressing Rec8-GFP from the nmt1 promoter (pREP81 plasmid) were analyzed by ChIP assay. We used wild-type cells (JY450) as a negative control.
Rad21 ectopically localized at the centromeres secures bipolar attachment at meiosis I.
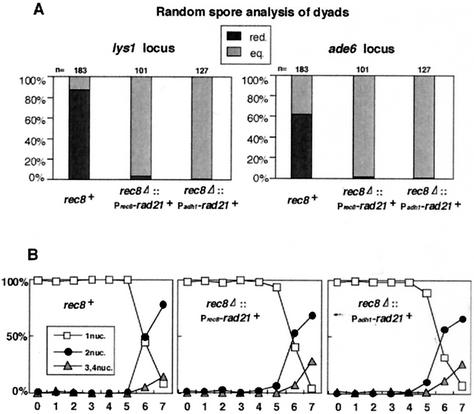
To examine chromosome segregation in rad21+ overproducers at meiosis I, we monitored the segregation patterns of two centromere-linked markers, lys1 and ade6 (16 centimorgans from the centromere of chromosome III), both of which are carried as heterozygous (+/−) in diploids. For this assay, we used a cdc2 temperature-sensitive mutation that blocks meiosis II so that cells form two spored dyads after the completion of meiosis I. Therefore, cells undergoing meiotic division in a reductional or equational manner will generate dyads with +/+, −/− or +/−, +/− genotypes, respectively. By performing random spore analysis, we calculated the percentages of reductional and equational segregation occurring in the cells (Fig. 5A [see the legend for the calculation]). Although wild-type cells showed a reductional segregation pattern (some occurrence of equational segregation can be explained by recombination between centromeres and GFP-associated sequences), both rec8Δ::Prec8-rad21+-GFP and rec8Δ::Padh1-rad21+-GFP cells predominantly displayed an equational pattern (Fig. 5A). Moreover, we monitored meiotic chromosome segregation more directly by observing the segregation pattern of cen1-GFP marked on one pair of sister chromatids. The results were consistent; both rec8Δ::Prec8-rad21+-FLAG and rec8Δ::Padh1-rad21+-FLAG cells underwent equational division at meiosis I (Fig. 6), as in the case of rec8Δ cells (Fig. 2B). These results indicate that even if sufficient amounts of Rad21 are loaded into centromeres in meiosis I, rec8Δ cells never reestablish monopolar attachment but instead establish bipolar attachment. Finally, we examined the kinetics of nuclear division of rec8+, rec8Δ::Prec8-rad21+-FLAG, and rec8Δ::Padh1-rad21+-FLAG cells in synchronous meiosis. We found that the first and second meiotic divisions progressed with similar kinetics in all strains (Fig. 5B). Given that the rec8Δ rad21+ overproducers undergo equational segregation at the first meiotic division, these results imply that the cohesion between sister kinetochores established by Rad21 is not protected during meiosis I, in contrast to the cohesion mediated by Rec8. Taken together, these findings led us to conclude that Rec8 bears specific roles at meiosis I in establishing monopolar attachment and also in maintaining cohesion between sister centromeres and that Rad21 cannot substitute for either of these functions even if it is fully localized to the centromeres during meiosis.
FIG. 5.
rec8Δ cells ectopically expressing rad21+ undergo equational chromosome segregation at meiosis I. (A) Diploid h+/h+ pat1-114 cdc2-L7 rec8+, h+/h+ pat1-114 cdc2-L7 rec8Δ::Prec8-rad21+-GFP, and h+/h+ pat1-114 cdc2-L7 rec8Δ::Padh1-rad21+-GFP cells carrying heterozygous centromere-linked markers (lys1, ade6) (PY221, PY754, and PY770) were induced to synchronous meiosis. Because of the cdc2-L7 mutation, the cells mostly produced dyads. These were treated with glusulase to obtain free spores, which were plated on YE media containing Magdala Red at 26.5°C for 7 days. Diploid colonies were selected as dark red staining and tested for auxotrophy. Cells undergoing reductional division generate dyads with +/+, −/− genotypes ([+], [−] phenotypes) whereas cells undergoing equational division generate dyads with +/−, +/− genotypes ([+], [+] phenotypes). The number of reductional divisions was calculated by n[red.] = n[−] and that of equational divisions was calculated by n[eq.]= (n[+] − n[−])/2. (B) The nuclear divisions during synchronous meiosis in panel A were monitored by DAPI staining. Note that only some cells underwent meiosis II because of the cdc2-L7 mutation.
FIG. 6.
The segregation pattern of cen1-GFP during meiosis I was examined in rec8+, rec8Δ::Prec8-rad21+-FLAG, and rec8Δ::Padh1-rad21+-FLAG cells. A rec8Δ strain tagged with cen1-GFP (PY322) was crossed with the indicated strain without tag (rec8+; rec8Δ::Prec8-rad21+-FLAG, PY749; rec8Δ::Padh1-rad21+-FLAG, PY748), and the zygotes that formed were analyzed as in Fig. 2A.
DISCUSSION
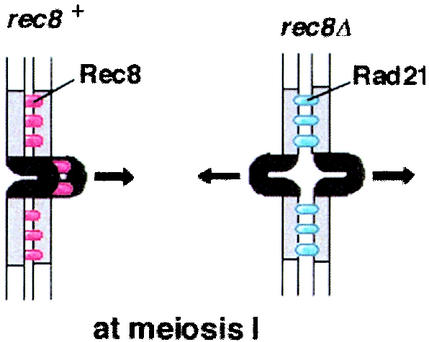
In the present study, we illuminated the inherent difference of the kinetochore roles of two cohesin subunits, Rad21 and Rec8, the former mitotic and the latter meiotic. Regarding the centromere location of cohesin, large amounts of Rec8 locate at both the outer and the central regions of the centromere in meiosis; however, whereas much Rad21 is located at the outer region, less is present at the central core in mitosis (31) (Fig. 4C and D). We found here that ectopically expressed Rad21 in meiosis displays a location pattern similar to that seen in mitosis, especially at the centromere (Fig. 4A). In excellent contrast, Rec8 ectopically expressed in mitosis displays a location pattern similar to that seen in meiosis (Fig. 4D and E). Thus, the centromeric location pattern of Rec8 or Rad21 does not depend on the developmental state, whether mitotic or meiotic, but is determined by intrinsic properties of the Rec8 and Rad21 molecules. Central core regions of the centromere are likely to assume an inverted configuration, protruding from other regions of the centromeres, and would provide the site for microtubule attachment (15, 23). Based on the distinct localization pattern of Rec8 and Rad21 at the centromeres, we speculate that only Rec8 may possess the competency to establish cohesion of the sequences along the central region, which would lead to the mono-oriented configuration of sister kinetochores (31). This model fits well with the present finding that, even though much Rad21 locates at the centromeres (especially at the outer regions) and establishes centromeric cohesion, it cannot establish the monopolar orientation of sister kinetochores but instead establishes bipolar attachment (Fig. 7). Thus, Rec8 and Rad21 are apparently the determinants for the orientation of sister kinetochores during meiosis I in fission yeast. The forced expression of Rec8 during mitosis, however, does not lead to the full occurrence of reductional chromosome segregation by itself (30). This inability to establish monopolar attachment in mitosis is probably not due to the lack of Rec8 at the central core regions of the centromere (Fig. 4E), although we do not exclude the possibility that the amount of Rec8 at centromere during mitosis is not sufficient to fulfill its function. Moreover, even in the normal meiotic cell cycle, residual Rec8 at the centromere after meiosis I does not promote monopolar attachment any more at meiosis II. These facts rationalize the assumption of a meiosis I-specific factor that must cooperate with Rec8, but not Rad21, at the kinetochore to establish monopolar attachment.
FIG. 7.
Model for the regulation of kinetochore orientation by cohesin Rec8 and Rad21 during meiosis I. Rec8 interacts with both the central core and outer repeat regions of the centromere, leading to the monopolar orientation of sister kinetochores at meiosis I. Rad21 associates less with the central core but more with outer repeat regions, thereby ensuring sister kinetochore cohesion and bipolar attachment at meiosis I in rec8Δ cells.
That Rec8 can support monopolar attachment during meiosis I is not the only meiosis-specific centromere feature that Rad21 cannot support. Rec8 is removed from chromosome in a two-step manner—from chromosome arms in meiosis I but from centromeres only in meiosis II—thus preventing the sister chromatids separating in meiosis I. Rad21, however, is apparently removed from all along the chromosome arms at meiosis I because rec8Δ::Prec8-rad21+ cells undergo the first meiotic divisions with the same kinetics as wild-type cells. In budding yeast rec8Δ::Prec8-Scc1+/rad21+ cells, cohesion between sister centromeres mediated by Scc1/Rad21 is also destroyed at the onset of the first meiotic division (27). Thus, the instability of Rad21/Scc1 regarding this aspect of meiosis I is conserved between budding and fission yeast.
Earlier work in budding yeast identified a meiosis-specific kinetochore factor, Mam1, that functions in establishing monopolar attachment, and this role can be executed only in the presence of cohesin (either Rec8 or Scc1 complexes) and only during meiosis (27). Another meiotic factor, Spo13, is also involved in the regulation of monopolar attachment in budding yeast (8, 22). So far, Mam1 or Spo13 homologs have not been found in other organisms, including fission yeast, but it may be a conserved feature that a meiosis I-specific factor(s) cooperates with cohesin to establish monopolar attachment at meiosis I. The definitive difference between the two yeasts is that cohesin specificity plays a crucial role in establishing monopolar attachment in fission yeast but not in budding yeast. Studies of the distribution of Rec8 at centromeres in comparison with that of Rad21/Scc1 in other organisms will be interesting, as will further examinations to determine whether cohesins are determinants of monopolar or bipolar attachment at meiosis I in other eukaryotes.
Acknowledgments
We thank all of the members of our laboratory for their help and discussion, especially T. Shimada for assisting in the observation of Gar1-CFP. We appreciate Y. Hiraoka and A. Yamamoto for providing the ade3-GFP strain.
This work was supported in part by grants from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Bähler, J., J. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 3.Girard, J. P., M. Caizergues-Ferrer, and B. Lapeyre. 1993. The SpGAR1 gene of Schizosaccharomyces pombe encodes the functional homologue of the snoRNP protein GAR1 of Saccharomyces cerevisiae. Nucleic Acids Res. 21:2149-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in Saccharomyces cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 6.Horie, S., Y. Watanabe, K. Tanaka, S. Nishiwaki, H. Fujioka, H. Abe, M. Yamamoto, and C. Shimoda. 1998. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 18:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitayama, C., A. Sugimoto, and M. Yamamoto. 1997. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 137:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klapholz, S., and R. E. Esposito. 1980. Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics 96:589-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein, F., P. Mahr, M. Galova, S. B. C. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91-103. [DOI] [PubMed] [Google Scholar]
- 10.Lee, J. Y., and T. L. Orr-Weaver. 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell Dev. Biol. 17:753-777. [DOI] [PubMed] [Google Scholar]
- 11.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 12.Molnar, M., J. Bahler, M. Sipiczki, and J. Kohli. 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141:61-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore, D. P., and T. L. Orr-Weaver. 1998. Chromosome segregation during meiosis: building an unambivalent, p. 263-299. In M. A. Hamdel (ed.), Meiosis and gametogenesis. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed]
- 14.Nabeshima, K., T. Nakagawa, A. F. Straight, A. Murray, Y. Chikashige, Y. M. Yamashita, Y. Hiraoka, and M. Yanagida. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9:3211-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakaseko, Y., G. Goshima, J. Morishita, and M. Yanagida. 2001. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 11:537-549. [DOI] [PubMed] [Google Scholar]
- 16.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 17.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 18.Parisi, S., M. J. McKay, M. Molnar, M. A. Thompson, P. J. van der Spek, E. van Drunen-Schoenmaker, R. Kannar, E. Lehmann, J. H. J. Hoeijmakers, and J. Kohli. 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family, conserved from fission yeast to humans. Mol. Cell. Biol. 19:3515-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer, and J. Loidl. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto, I., N. Pezzi, J. M. Buesa, L. Kremer, I. Barthelemy, C. Carreiro, F. Roncal, A. Martinez, L. Gomez, R. Fernandez, A. C. Martinez, and J. L. Barbero. 2002. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3:543-550. [DOI] [PMC free article] [PubMed]
- 21.Saitoh, S., K. Takahashi, and M. Yanagida. 1997. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell 90:131-143. [DOI] [PubMed] [Google Scholar]
- 22.Shonn, M. A., R. McCarroll, and A. W. Murray. 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16:1659-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi, K., S. Murakami, Y. Chikashige, H. Funabiki, O. Niwa, and M. Yanagida. 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3:819-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatebayashi, K., J. Kato, and H. Ikeda. 1998. Isolation of a Schizosaccharomyces pombe rad21 ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics 148:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S. E. Kearsey, F. Uhlmann, K. Nasmyth, and M. Yanagida. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toth, A., K. P. Rabitsch, M. Galova, A. Schleiffer, S. B. Buonomo, and K. Nasmyth. 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103:1155-1168. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda, Y., K. Furuya, G. Goshima, K. Nagao, K. Takahashi, and M. Yanagida. 2002. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12:347-358. [DOI] [PubMed] [Google Scholar]
- 29.Uhlmann, F. 2001. Chromosome cohesion and segregation in mitosis and meiosis. Curr. Opin. Cell Biol. 13:754-761. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400:461-464. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe, Y., S. Yokobayashi, M. Yamamoto, and P. Nurse. 2001. Premeiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409:359-363. [DOI] [PubMed] [Google Scholar]
- 32.Zickler, D., and N. Kleckner. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619-697. [DOI] [PubMed] [Google Scholar]