Abstract
DNA single-strand breaks (SSB) are one of the most frequent DNA lesions produced by reactive oxygen species and during DNA metabolism, but the analysis of cellular responses to SSB remains difficult due to the lack of an experimental method to produce SSB alone in cells. By using human cells expressing a foreign UV damage endonuclease (UVDE) and irradiating the cells with UV through tiny pores in membrane filters, we created SSB in restricted areas in the nucleus by the immediate action of UVDE on UV-induced DNA lesions. Cellular responses to the SSB were characterized by using antibodies and fluorescence microscopy. Upon UV irradiation, poly(ADP-ribose) synthesis occurred immediately in the irradiated area. Simultaneously, but dependent on poly(ADP-ribosyl)ation, XRCC1 was translocated from throughout the nucleus, including nucleoli, to the SSB. The BRCT1 domain of XRCC1 protein was indispensable for its poly(ADP-ribose)-dependent recruitment to the SSB. Proliferating cell nuclear antigen and the p150 subunit of chromatin assembly factor 1 also accumulated at the SSB in a detergent-resistant form, which was significantly reduced by inhibition of poly(ADP-ribose) synthesis. Our results show the importance of poly(ADP-ribosyl)ation in sequential cellular responses to SSB.
DNA damage is a continual threat to genetic stability, as demonstrated by the cancer-prone phenotype of human diseases in which DNA repair is defective. Among various DNA lesions, single-strand breaks (SSB) are one of the most frequent DNA lesions produced by endogenous reactive oxygen species or generated by ionizing radiation or through base hydrolysis (37). SSB are also intermediate products in various aspects of DNA metabolism, including DNA repair, replication, and recombination. During base excision repair, SSB are produced by DNA glycosylases and apurinic/apyrimidinic (AP) endonucleases at the site of base damage. If SSB are not properly repaired, they may result in double-strand breaks in replicating DNA and may also affect transcription (7, 19). In spite of such harmful effects, however, our knowledge of the cellular responses to and repair of SSB is still incomplete. One reason for this is that no experimental procedure that induces solely SSB in cells has been developed.
In higher eukaryotes, it is known that SSB produced during base excision repair are processed by two alternative repair pathways, a DNA polymerase β (Polβ)-dependent pathway and a proliferating cell nuclear antigen (PCNA)-dependent pathway. Selection of these pathways is considered to be damage and/or dose dependent (12). Poly(ADP-ribosyl)ation, a posttranslational modification of proteins catalyzed by DNA-dependent poly(ADP-ribose) polymerase (PARP), occurs very rapidly in response to SSB. PARP-1 has been proposed to play a key role in SSB repair by its rapid binding to SSB and subsequent activation (3). PARP-2 is also activated by SSB and participates in the repair of SSB (2, 31). XRCC1 is another protein that plays a central role in the repair of SSB. In biochemical studies, XRCC1 was shown to interact with activated PARP-1 (8, 22, 31) and PARP-2 (31), OL β (6, 21), DNA ligase IIIα (4, 5), AP endonuclease (39), and polynucleotide kinase (42). By these interactions, XRCC1 is thought to play a key role in coordinating the DNA OL β-dependent base excision repair pathway and SSB repair (7).
The repair mechanisms for SSB and base excision repair proposed so far have mainly been examined in in vitro cell-free systems reconstituted with purified proteins and/or cell extracts. Since the actions of the DNA repair proteins are performed at the relevant stage (e.g., on complex chromatin substrates) in time-dependent processes, any in vitro assay tends to overlook crucial biological processes and effects. To elucidate the in vivo repair process of SSB, we previously established a nucleotide excision repair-deficient xeroderma pigmentosum group A (XPA) cell line expressing Neurospora crassa UV damage endonuclease (UVDE) (27). UVDE introduces an SSB with a 3′-OH immediately 5′ to UV-induced cyclobutane pyrimidine dimers, 6-4 photoproducts, and Dewar photoproducts (13, 16, 33, 44, 45). We showed that in these cells (XPA-UVDE cells), SSB are produced immediately after UV irradiation by the action of UVDE on various UV-induced DNA lesions, and SSB together with UV-induced lesions are then efficiently repaired (27). A PARP inhibitor, 3-aminobenzamide (3-AB), increases the UV sensitivity of the cells, and XRCC1-deficient Chinese hamster cells harboring UVDE are highly UV sensitive, suggesting the involvement of PARP and XRCC1 in the process of repair of UVDE-induced SSB. We found that the repair synthesis of the SSB is a long-patch repair, with a mean patch size of seven nucleotides (27), suggesting that PARP, XRCC1, and aphidicolin-sensitive DNA polymerase(s) participate in this SSB repair. Since the cells do not possess nucleotide excision repair, UV-induced lesions are processed exclusively by UVDE and subsequent SSB repair. Thus, the XPA-UVDE cell offers a unique experimental system with which to create SSB by UV irradiation.
In order to analyze cellular responses to SSB and dynamic correlations between DNA repair proteins, we made use of a recently developed technique to inflict UV damage in restricted small regions of the nucleus (local UV irradiation) for XPA-UVDE cells. By using antibodies and fluorescence microscopy, the temporal and spatial behavior of the principal proteins involved in the repair of SSB were visualized within the nucleus for the first time.
MATERIALS AND METHODS
Cell lines and culture conditions and cell synchrony.
Simian virus 40-transformed human fibroblasts were used in all experiments. XPA-UVDE and XPA-Vector cells (27) were grown in Eagle's minimal essential medium (Nissui) supplemented with 10% fetal bovine serum. Synchrony in G1 was achieved by incubation in mimosine (Sigma) according to the reported procedure used for human cells (20): cells were incubated in medium containing 0.5 mM mimosine for 24 h.
DNA transfection.
DNA constructs were made according to standard procedures (30). Briefly, the cDNA of full-length XRCC1 as well as various fragments of XRCC1 were amplified by PCR with 5′ and 3′ primers containing SalI and NotI sites, respectively. The amplified DNA fragments were subcloned into the XhoI and NotI sites of pCY4B-Flag, a modified pCY4B plasmid (26) containing a chicken β-actin promoter which drives the expression of an in-frame N-terminal Flag peptide tag. Two additional pCY4B-Flag constructs of XRCC1 mutants having amino acid replacement mutations in the BRCT1 domain were constructed by PCR with plasmids pcD2EXH W385D and pcD2EXH LI360/361 DD (36) as templates and the same primers used for the full-length XRCC1. The DNA constructs were verified by sequencing. These plasmids were introduced into XPA-UVDE cells with Fugene 6 (Roche) according to the manufacturer's protocol.
Local UV irradiation.
Local UV irradiation was performed essentially as described previously (17). Cell monolayers in 35-mm glass-bottomed culture dishes (poly-d-lysine coated; MatTek, Ashland, Oreg.) were covered with a polycarbonate isopore membrane filter with pores of 3 μm in diameter (Millipore) and UV irradiated with a germicidal lamp (GL-10; Toshiba; predominantly 254-nm UV) at a dose rate of 1.82 J/m2/s. The polycarbonate blocks the 254-nm UV light, and cells are exposed only through the pores of the filter. A filter with a larger pore size results in larger spots of poly(ADP-ribose) synthesis, indicating that SSB are produced only at the UV-exposed areas (see Results).
Immunofluorescence microscopy.
For immunolabeling, XPA-UVDE or XPA-Vector cells were grown for 2 days in glass-bottomed culture dishes, and almost confluent cultures were UV irradiated. To examine the effect of inhibitors of PARP, cells were incubated for 1 h in medium supplemented with 1,5-dihydroxyisoquinoline (DIQ; 100 μM; Sigma) or 3-aminobenzamide (3-AB; 4 mM; Sigma) before irradiation and washed twice with Hanks' solution (Nissui) with DIQ or 3-AB. Then the cells were irradiated with UV at 20 J/m2 as described above. After UV irradiation, cells were incubated in medium with DIQ or 3-AB at 37°C for various periods of time and then fixed with methanol-acetone (1:1) for 10 min at −20°C. The fixed cells were dried, subsequently rinsed once with TNT buffer (0.1 M Tris-HCl, 0.15 M NaCl, 0.05% Tween 20, pH 7.5), and then incubated in TNB buffer (TNT buffer containing blocking reagent [NEN]) at 30°C for 30 min. Cells were then incubated with anti-XRCC1 antibody (ab144; Abcam) at a 1:150 dilution in TNB buffer at 30°C for 1 h.
For observation of the Flag-tagged proteins, XPA-UVDE cells grown in dishes were transfected with the plasmids mentioned above. At 24 h following transfection, cells were treated and UV irradiated as described above. For labeling the Flag-tagged proteins, rabbit anti-Flag (5 μg/ml; Sigma) was used. Cells treated with antibodies against XRCC1 and Flag were then washed three times with TNT buffer and incubated with Alexa Fluor 594 goat anti-rabbit immunoglobulin G conjugate (Molecular Probes) at a 1:400 dilution in TNB buffer for 1 h. After washing with TNT buffer, cells were then incubated with anti-poly(ADP-ribose) (1:200 dilution; Trevigen) or antinucleolin/C23 (1:25 dilution; MS-3; Santa Cruz) or anti-PARP-1 (1:25 dilution; F-2; Santa Cruz) in TNB buffer for 1 h. After washing treatments, cells were incubated with Alexa Fluor 488 goat anti-mouse immunoglobulin G conjugate (Molecular Probes) at a 1:400 dilution in TNB buffer for 1 h. Cell samples were then mounted in drops of PermaFluor (Immunon), and coverslips were added. Confocal imaging was performed with an Olympus FV-500 confocal laser system connected to an Olympus microscope (IX81) with a 60× oil immersion objective lens (PlanApo).
For double staining of PCNA and CAF-1 p150, cells were further treated before fixation with ice-cold detergent solution (0.5% Triton X-100, 0.2 mg of EDTA per ml, and 1% bovine serum albumin in phosphate-buffered saline) for 15 min and washed with phosphate-buffered saline. Then cells were fixed with methanol-acetone and treated as described above. Cells were labeled successively with anti-PCNA (1:25 dilution; FL-261; Santa Cruz), Alexa Fluor 594 goat anti-rabbit immunoglobulin G conjugate, anti-CAF-1 p150 monoclonal antibody (1:250 dilution; SS1; a kind gift from B. Stillman, Cold Spring Harbor Laboratory), and Alexa Fluor 488 goat anti-mouse immunoglobulin G conjugate. Nuclear DNA was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; Wako; 0.5 μg/ml). Cell samples were mounted, and coverslips were added as described above. Fluorescence microscopic images were obtained with a Leica model DMLB30. Images were captured by charge-coupled device camera (DC250; Leica) and colored with Qfluoro software (Leica).
RESULTS
Local and immediate poly(ADP-ribose) synthesis following UV irradiation in XPA-UVDE cells through a membrane filter.
We have previously shown, by Western blotting and alkaline gel analysis, that rapid and transient poly(ADP-ribose) (PAR) synthesis occurs in XPA-UVDE cells after UV irradiation due to the activation of PARP by the SSB produced by UVDE at UV-induced DNA lesions (27). Here, we covered XPA-UVDE cells in a culture dish with an isopore membrane filter and irradiated the cells with UV (20 J/m2) through small pores (pore size, 3 μm) randomly distributed in the filter. This irradiation method was recently used to analyze protein assembly in repair of UV-induced DNA damage (17, 41). This method applied to cells expressing UVDE enabled us to induce SSB only at the UV-irradiated spots by the action of UVDE on UV-induced DNA lesions. In the same way as previously reported (15), we roughly estimated that the number of SSB introduced in the present experimental conditions (20 J/m2) was on the order of 104 per irradiated spot.
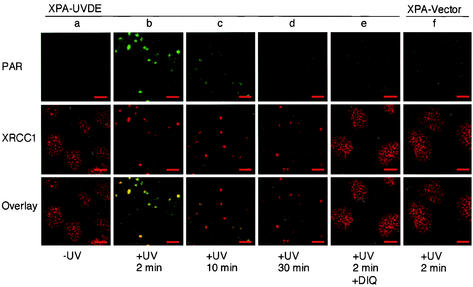
After local UV irradiation, cells were fixed and then immunolabeled with antibodies against PAR. Before irradiation, no evident PAR was observed (Fig. 1a, upper panel). Two minutes after irradiation, PAR synthesis was observed in restricted spots in the nucleus with diameters comparable to the pore size (Fig. 1b, upper panel). Ten minutes after irradiation, the sizable amount of PAR in the spots had faded away (Fig. 1c, upper panel), and 30 min after irradiation, these spots were almost indiscernible (Fig. 1d, upper panel). In irradiated XPA cells harboring empty vector plasmid (XPA-Vector cells), no PAR synthesis was observed (Fig. 1f, upper panel).
FIG. 1.
Fluorescent micrographs of XPA-UVDE and XPA-Vector cells, doubly immunolabeled for PAR and XRCC1. The columns from a to e show XPA-UVDE cells, and f shows XPA-Vector cells, before irradiation (a), 2 min after local UV irradiation with 20 J/m2 (b and f), and 10 and 30 min after local UV irradiation (c and d, respectively). Cells were fixed and costained with anti-PAR antibody (upper row in green) and anti-XRCC1 antibody (middle row in red). Colocalization of PAR and XRCC1 is shown (bottom row in yellow). Column e is DIQ-treated XPA-UVDE cells fixed 2 min after local UV irradiation. Bar, 10 μm.
In the presence of DIQ (100 μM), an inhibitor of PARP which is known to be superior in potency and specificity to monoaryl amide inhibitors such as 3-AB (40), PAR was not detected (Fig. 1e, upper panel). PAR was also not observed in the presence of 3-AB (4 mM; data not shown). These results, together with the previously published data (27), indicate that SSB are produced at the UV-irradiated area through small pores and that activation of PARP is restricted only to the small areas where SSB are induced. The degradation of PAR occurred very rapidly, which may be attributable to the activity of poly(ADP-ribose) glycohydrolase (9).
Poly(ADP ribosyl)ation-dependent accumulation of XRCC1 in SSB.
Before irradiation, XRCC1 was found in many small dots of various sizes in addition to a faint background in the nucleus (Fig. 1a, middle panel, and see the next section). Two minutes after irradiation, XRCC1 had already accumulated at the irradiated spots (Fig. 1b, middle panel), which completely overlapped the sites of synthesis of PAR (Fig. 1b, bottom panel), while the amount of XRCC1 in unirradiated nuclear regions had drastically decreased (Fig. 1b, middle panel). This shows that, upon UV irradiation, XRCC1 translocated from the unirradiated nuclear region to the irradiated spots. In XPA-Vector cells, the recruitment of XRCC1 was not observed after irradiation (Fig. 1f, middle panel). In the presence of DIQ (Fig. 1e, middle panel) or 3-AB (data not shown), no translocation of XRCC1 to the irradiated spots was observed in XPA-UVDE cells. These results indicate that the synthesis of PAR at the sites of SSB is a prerequisite for XRCC1 accumulation at SSB and also that the translocation of XRCC1 from throughout the nucleus to SSB is triggered by the synthesis of PAR. In contrast to PAR, the intensities of the spots of XRCC1 were not drastically changed 10 and 30 min after irradiation (Fig. 1c and 1d, middle panels, respectively).
Distribution of XRCC1 and PARP-1 in nucleoli and nucleoplasm.
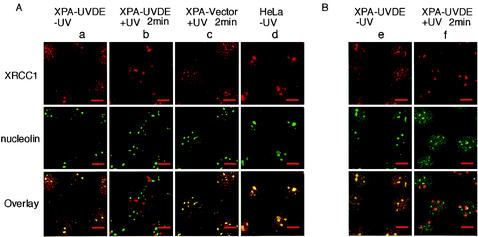
As shown in the previous section, XRCC1 was distributed throughout the nucleus of unirradiated cells. We found that a part of XRCC1 was located in relatively large foci (Fig. 2Aa, top). These foci were identified as the nucleoli by costaining the cells with antibody to the nucleolar protein nucleolin (14) (Fig. 2Aa, middle and bottom panels). In HeLa cells harboring larger nucleoli, the presence of XRCC1 in the nucleoli was much more evident (Fig. 2Ad). Two minutes after irradiation, most of the XRCC1 present in the nucleoli as well as in the nucleoplasm had translocated to the irradiated sites (Fig. 2Ab). In XPA-Vector cells, no discernible translocation of XRCC1 was observed after irradiation (Fig. 2Ac).
FIG. 2.
Nuclear distribution of XRCC1 and PARP-1 before and after local UV irradiation. (A) Double immunolabeling for nucleolin and XRCC1 in XPA-UVDE cells, XPA-Vector cells, and HeLa cells. Panels in column a are unirradiated XPA-UVDE cells, b and c are XPA-UVDE and XPA-Vector cells fixed at 2 min after local UV irradiation (20 J/m2), respectively, and panels in column d are unirradiated HeLa cells. Panels in the upper and middle rows correspond to XRCC1 (red) and nucleolin (green), respectively. Panels in the bottom row represent an overlay of the panels from the upper and middle rows. Colocalization of the two proteins appears yellow. (B) Double immunolabeling of XPA-UVDE cells for XRCC1 (red in the upper-row panels) and PARP-1 (green in middle-row panels); panels in the bottom row are the corresponding overlay; colocalization of the proteins appears yellow. Panels in column e are unirradiated cells, and those in f are cells at 2 min after local UV irradiation (20 J/m2). Bar, 10 μm.
PARP-1 is known to be present in the nucleolus (11). By costaining with antibodies against XRCC1 and PARP-1, both proteins were shown to be colocalized in nucleoli (Fig. 2Be, bottom panel). In contrast to XRCC1, no location change from the nucleoli was observed for PARP-1, nor was any accumulation of PARP-1 found in the irradiated spots (Fig. 2Bf).
In situ determination of domain in XRCC1 responsible for recruitment to SSB.
In order to identify which part of XRCC1 is necessary for the recruitment to SSB sites in situ, we used deletions and site-specific mutagenesis. A series of expression constructs harboring various XRCC1 fragments and two BRCT1 domain mutants with amino acid replacement mutations (W385D and LI360/361DD) were tagged with the Flag epitope at the NH2 terminus and transiently expressed in XPA-UVDE cells. They were then assayed for their ability to be recruited to the PAR-synthesized spots (Fig. 3). As shown in Fig. 3Aa, the full-length Flag-XRCC1 accumulated in nuclear foci in transfected cells, as described previously (35). However, Flag-XRCC1 was not present in nucleoli, nor was XRCC1 which had been tagged with green fluorescent protein (GFP) at the carboxyl terminus located in nucleoli (data not shown), suggesting that the nucleolar distribution of XRCC1 may be very sensitive to its modification.
FIG. 3.
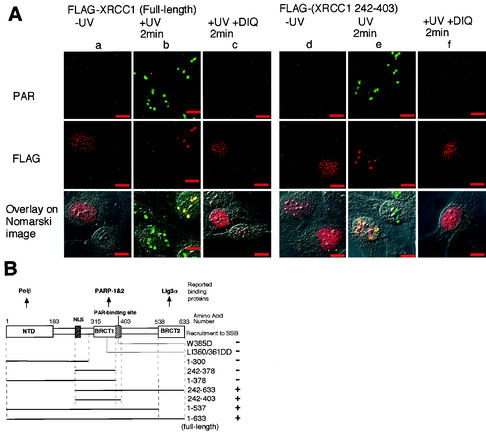
In situ determination of XRCC1 domains necessary for its recruitment to SSB. (A) Double immunolabeling of PAR and Flag-tagged full-length XRCC1 and XRCC1 fragment after local UV irradiation (20 J/m2). Panels in the columns from a to c are f cells transfected with full-length XRCC1 tagged with Flag, and those from d to f are the cells transfected with the Flag-tagged XRCC1 fragment which comprises amino acids 242 to 403 of XRCC1. Columns a and d are unirradiated cells; b and e are cells fixed 2 min after local UV irradiation (20 J/m2); c and f are DIQ-treated cells fixed at 2 min after local UV irradiation (20 J/m2). Not all the cells express the gene. Cells were costained with anti-PAR (panels in the upper row, green) and anti-Flag epitope (panels in the middle row, red). The corresponding fluorescent images were superimposed onto the Nomarski images and are shown in the bottom row. Colocalization appears yellow. Bar, 10 μm. (B) Schematic representation of the ability to be recruited to SSB for deletion and site-specific mutations of XRCC1 polypeptides, all tagged with Flag at the NH2 terminus. Thick lines below the schematic drawing of XRCC1 protein represent the deleted fragments; the corresponding numbers at the right-hand side are the amino acids at both ends of the polypeptides. Two mutants having amino acid replacement mutations are also shown here. The results are given at the extreme right-hand side. The reported binding proteins of XRCC1 are shown uppermost. The N-terminal domain (NTD) contains the binding site for OL β, and the BRCT domains contain the binding site for PARP-1, PARP-2, and ligase IIIα. NLS, bipartite nuclear localization signal.
UV irradiation of the transfected cells initiated accumulation of the Flag-XRCC1 to the irradiated spots (Fig. 3Ab, middle), which coincided with the spots for PAR accumulation (Fig. 3Ab, bottom panel) and was inhibited by DIQ treatment (Fig. 3Ac). Almost the same results as those obtained for the full-length XRCC1 were obtained by transfection of XRCC1 fragment 242 to 403 (Fig. 3Ad, e, and f). In XPA-Vector cells, UV-induced assembly of the full-length XRCC1 and the XRCC1(242-403) to the irradiated spots was not observed (data not shown). XRCC1 fragments containing the BRCT1 domain (1 to 537 and 242 to 633) were found to accumulate at spots after irradiation (Fig. 3B), while XRCC1 fragments devoid of the BRCT1 domain (1 to 300) and XRCC1 fragments having an incomplete BRCT1 domain (1 to 378 and 242 to 378) were not recruited to irradiated spots after irradiation (Fig. 3B).
Transfected XRCC1 fragment containing the BRCT2 domain (538 to 633) without the nuclear localization signal (22) was diffusely distributed throughout the cell, including the nucleus, and was not recruited to the irradiated spots after irradiation (data not shown). Significantly, two BRCT1 domain mutants corresponding to the mutants of the BRCT2 domain shown previously to disrupt its folding (46) or activity (24, 34) were not recruited to irradiated spots (Fig. 3B). These results indicate that the BRCT1 domain, the site for interacting with poly(ADP-ribosyl)ated PARP-1 (22) and PARP-2 (31), plays a crucial role in the recruitment of XRCC1 to SSB.
In situ visualization of assembly of PCNA and CAF-1 at SSB.
PCNA is known to take a detergent-resistant chromatin-bound form during S phase and also after DNA damage induced by UV (18), hydrogen peroxide, or alkylating drugs (12). Chromatin assembly factor 1 (CAF-1) is a histone chaperone that has a role in chromatin assembly during DNA replication and nucleotide excision repair (18). CAF-1 p150 interacts directly with PCNA (23, 32). Involvement of PCNA and the largest subunit (p150) of CAF-1 in the repair of SSB was analyzed in our cell system. Cells were irradiated, permeabilized with Triton X-100, fixed, and then analyzed by immunofluorescence microscopy.
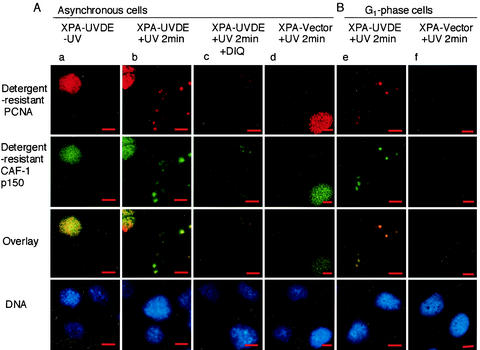
In unirradiated control cells, about 34% of nuclei were found to be in S phase, each having detergent-resistant PCNA and CAF-1 p150 throughout the nucleus (red-stained PCNA and green-stained CAF-1 p150 in Fig. 4Aa, first and second upper panels). Following local irradiation of the cells, detergent-insoluble PCNA and CAF-1 p150 were detected at irradiated spots in cells not in S phase (Fig. 4Ab). The spots overlapped each other (Fig. 4Ab). Cells synchronized in G1 phase by treatment with mimosine were also examined. In these cells, S-phase nuclei disappeared almost completely (data not shown). After irradiation, detergent-resistant spots were observed in every nucleus (Fig. 4Be), while these spots were not observed in either asynchronous or G1-phase cells after irradiation in XPA-Vector cells (Fig. 4Ad and Bf). Thus, the observed detergent-resistant PCNA and CAF-1 p150 do not reflect DNA replication in S-phase cells but do reflect SSB-dependent accumulation and suggest that a PCNA-dependent process is accompanied by chromatin-assembly mediated by CAF-1.
FIG. 4.
In situ visualization PCNA and CAF-1 p150 at SSB. (A) Double immunolabeling for PCNA and CAF-1 p150 in asynchronous cells. Panels in column a are unirradiated XPA-UVDE cells; those in c are DIQ-treated XPA-UVDE cells fixed at 2 min after local UV irradiation (20 J/m2). Those in column b are XPA-UVDE cells fixed at 2 min after local UV irradiation (20 J/m2); those in d are XPA-Vector cells fixed at 2 min after local UV irradiation (20 J/m2). Panels in the uppermost row are cells immunolabeled for PCNA (red), whereas those in the second row are cells immunolabeled for CAF-1 p150 (green); both proteins were in the detergent-resistant form, as described in Results. Colocalization of the proteins appears yellow, as seen in the third row for the overlay of the corresponding panels. The nuclei were stained with DAPI and are shown in the bottom row. Bar, 10 μm. (B) Double immunolabeling for PCNA and CAF-1 p150 in G1-phase cells. XPA-UVDE (column e) and XPA-Vector (column f) cells were treated with mimosine as described in Materials and Methods. Cells were exposed to local UV irradiation (20 J/m2) and incubated for 2 min, then doubly immunolabeled for PCNA (uppermost row, red) and for CAF-1 p150 (second row, green); both proteins were in the detergent-resistant form as described in Results. Colocalization of the proteins appears yellow, as seen in the third row for the overlay of the corresponding panels. The nuclei were stained with DAPI (bottom row). Bar, 10 μm.
To our surprise, in the presence of DIQ, the accumulation of PCNA and CAF-1 was significantly reduced. This indicates that both proteins accumulate at PARP-activated SSB sites. However, in contrast to XRCC1, the assembly of PCNA and CAF-1 p150 was not completely blocked by DIQ, and there was residual accumulation of both proteins at irradiated spots (Fig. 4Ac). These data suggest that the accumulation of PCNA and CAF-1 p150 at SSB depends partially on PARP activation.
DISCUSSION
SSB are very frequent DNA lesions, but their actual repair processes in living cells and the physiological significance of poly(ADP-ribosyl)ation associated with SSB repair are not well understood. To address these questions, we applied local UV irradiation to the previously established human XPA-UVDE cells, which are completely deficient in nucleotide excision repair but express UVDE, which introduces nicks immediately 5′ to various UV-induced lesions, leaving 3′-OH and 5′-P UV damage. This experimental system enabled us to produce SSB with a 5′ block and 3′-OH when and where the cells are irradiated with UV (27). Poly(ADP-ribosyl)ation of proteins by PARP and its automodification have been shown to be produced by SSB and are involved in various processes associated with physiology and pathophysiology (3). Our present in situ results directly demonstrate that a physiological significance of poly(ADP-ribosyl)ation is the recruitment of DNA repair proteins to the sites of SSB, as has been presumed from in vitro studies (29, 42).
First of all, by production of SSB at the UV-irradiated areas in the nucleus, we demonstrated that the transient synthesis of PAR occurs at the restricted area of SSB in the actual context of the nucleus (Fig. 1). This may be the first in situ visual demonstration of the widely accepted model for PARP activation based on in vitro results. Second, we showed that before UV irradiation, XRCC1 protein was present throughout the nucleus, including nucleoli (Fig. 2). Park et al. reported that XRCC1 was not observed in the nucleolus (28). This discrepancy may be due to the difference in the fixation procedures and/or antibodies used in the experiments. The binding of XRCC1 to the structural component of the nucleolus may be weak, as tagged constructs of XRCC1 were not observed in the nucleolus. XRCC1 has been proposed to serve as the molecular scaffold or docking platform for other repair proteins (7, 21). Nucleoli may simply be a storage place for XRCC1. However, it is tempting to consider that this nucleolar localization may be responsible for some other unidentified functions of XRCC1 in transcription or recombination, as proposed previously (37, 39).
Depending on the presence of PAR at SSB, XRCC1 is recruited very rapidly and efficiently from throughout the nucleus distant from the sites of SSB (Fig. 1 and 2). In accordance with these results, after treatment of HeLa cells with H2O2 the distinct nucleolar XRCC1 disappeared, suggesting that H2O2-induced SSB also recruit XRCC1 (data not shown). Our results suggest that XRCC1-mediated repair is considerably impaired in PARP-deficient cells compared with wild-type cells. In agreement with this, the high sensitivity of PARP-1 knockout mice and the derived mouse embryonic fibroblasts to alkylating agents and gamma irradiation (38) and prolonged delays in SSB repair in PARP-1- and PARP-2-deficient cells following treatment with alkylating agents (31, 38) have been reported.
The region near the N terminus of XRCC1 binds to OL β (21), and the BRCT2 domain at the C terminus of XRCC1 binds to DNA ligase IIIα (25). By deletion analysis along with site-specific mutagenesis, we demonstrated that the BRCT1 domain of XRCC1 protein, the interaction site with poly(ADP-ribosyl)ated PARP-1 (22) and PARP-2 (31), is responsible for its recruitment to SSB (Fig. 3). Thus, the ability of XRCC1 to bind SSB, OL β, and DNA ligase IIIα was not necessarily required for its recruitment to SSB (Fig. 3). The C-terminal portion of the BRCT1 domain of XRCC1 (379 to 400; PAR-binding site) has recently been reported to interact noncovalently with PAR (29). Our present results showed that XRCC1 fragments 1 to 378 and 242 to 378 devoid of the PAR-binding site were not recruited after irradiation (Fig. 3). Thus, the site for interacting with PAR may play an essential role in the recruitment of XRCC1 to SSB.
Two XRCC1 mutants (XRCC1W385D and LI360/361DD) could not repair SSB in G1 or S/G2 phase (36), while the XRCC1 W385D mutant still bound to OL β and DNA ligase IIIα in vitro (36). We showed that these mutant XRCC1 forms were not recruited to SSB (Fig. 3). These results, together with the deletion data, strongly suggest that the ability of XRCC1 to bind to activated PARP plays an indispensable role in both G1- and S/G2-specific repair. In striking contrast to XRCC1, no discernible translocation of PARP-1 to the SSB was observed after local irradiation (Fig. 2B). This may be due to the amount of PARP-1, which is normally present in considerable molar excess (threefold) compared with XRCC1 (37), or may be due to the accumulation of many XRCC1 molecules in one poly(ADP-ribosyl)ated PARP at SSB.
We showed that a PCNA-dependent process is involved in the repair of the SSB introduced by UVDE (Fig. 4). This is consistent with our previous result showing that after whole-cell UV irradiation with 20 J/m2, repair synthesis was decreased to about 30% by aphidicoline, suggesting polymerase δ/ε-dependent long-patch repair for UVDE-induced SSB (27). Our present results suggest that this pathway is accompanied by chromatin assembly mediated by CAF-1 (Fig. 4). This is the first demonstration that the chromatin assembly mediated by CAF-1 is involved in the process of SSB repair in vivo. The recruitment of both PCNA and CAF-1 p150 was reduced in the absence of PAR at the sites of SSB. In contrast to XRCC1, the assembly of PCNA with SSB does not fully depend on the synthesis of PAR (Fig. 4). These results are in harmony with a model to explain the function at the molecular level: by PARP activation at SSB, the relaxation or opening-up of chromatin superstructure by covalent or noncovalent modification of histones by PAR occurs, allowing access for the DNA repair machinery to the SSB (1, 10). In the presence of PARP inhibitors, PARP-1 and probably histones would remain bound to SSB and may partially block the access of the PCNA-loading machinery and CAF-1, thereby reducing the assembly of these proteins at SSB. Our results indicate the involvement of PAR synthesis in the PCNA-dependent repair of SSB.
Sequential assembly of repair proteins at the sites of DNA damage has been considered to enable various DNA repair systems to work quite effectively. So far, in vitro studies with purified proteins and cell extracts along with structural studies have clarified many features and provided various models for base excision repair and SSB repair processes (7, 42, 43). Our in situ results show that SSB repair is a sequential process in which the order of protein assembly is regulated by PAR. Although many questions concerning the processes by which SSB are repaired remain open, our experimental system provides a powerful tool for understanding the spatial and temporal aspects of the cellular responses to SSB in situ, as exemplified here.
Acknowledgments
We thank B. Stillman (Cold Spring Harbor Laboratory) for providing us with the anti-CAF-1 p150 antibody. We also thank Y. Watanabe (Olympus) and D. Suzuki (Hamamatsu) for technical support in microscopy. We thank S. McCready for editing the text.
This work was supported by Grants-in-Aid for Scientific Research (no. 12143201 and no. 13480162) from the Ministry of Education, Science, Sports and Culture of Japan and by the Mitsubishi Foundation to A.Y.
REFERENCES
- 1.Althaus, F. R. 1992. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J. Cell Sci. 102:663-670. [DOI] [PubMed] [Google Scholar]
- 2.Ame, J. C., V. Rolli, V. Schreiber, C. Niedergang, F. Apiou, P. Decker, S. Muller, T. Hoger, J. Menissier-de Murcia, and G. de Murcia. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860-17868. [DOI] [PubMed] [Google Scholar]
- 3.Burkle, A. 2001. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays 23:795-806. [DOI] [PubMed] [Google Scholar]
- 4.Caldecott, K. W., C. K. McKeown, J. D. Tucker, S. Ljungquist, and L. H. Thompson. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldecott, K. W., J. D. Tucker, L. H. Stanker, and L. H. Thompson. 1995. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldecott, K. W., S. Aoufouchi, P. Johnson, and S. Shall. 1996. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ′nick-sensor' in vitro. Nucleic Acids Res. 24:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldecott, K. W. 2001. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays 23:447-455. [DOI] [PubMed] [Google Scholar]
- 8.Dantzer, F., G. de la Rubia, J. Menissier-de Murcia, Z. Hostomsky, G. de Murcia, and V. Schreibr. 2000. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39:7559-7569. [DOI] [PubMed] [Google Scholar]
- 9.Davidovic, L., M. Vodenicharov, E. B. Affar, and G. G. Poirier. 2001. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 268:7-13. [DOI] [PubMed] [Google Scholar]
- 10.de Murcia, G., A. Huletsky, and G. G. Poirier. 1988. Modulation of chromatin structure by poly(ADP-ribosyl)ation. Biochem. Cell Biol. 66:626-635. [DOI] [PubMed] [Google Scholar]
- 11.Desnoyers, S., S. H. Kaufmann, and G. G. Poirier. 1996. Alteration of the nucleolar localization of poly(ADP-ribose) polymerase upon treatment with transcription inhibitors. Exp. Cell Res. 227:146-153. [DOI] [PubMed] [Google Scholar]
- 12.Dogliotti, E., P. Fortini, B. Pascucci, and E. Parlanti. 2001. The mechanism of switching among multiple base excision repair pathways. Prog. Nucleic Acid Res. Mol. Biol. 68:3-27. [DOI] [PubMed] [Google Scholar]
- 13.Freyer, G. A., S. Davey, J. V. Ferrer, A. M. Martin, D., Beachand, and P. W. Doetsch. 1995. An alternative eukaryotic DNA excision repair pathway. Mol. Cell. Biol. 15:4572-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginisty, H., H. Sicard, B. Rogerand, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 15.Imoto, K., N. Kobayashi, S. Katsumi, Y. Nishiwaki, T. A. Iwamoto, A. Yamamoto, Y. Yamashina, T. Shirai, S. Miyagawa, Y. Dohi, S. Sugiura, and T. Mori. 2002. The total amount of DNA damage determines ultraviolet-radiation-induced cytotoxicity after uniformor localized irradiation of human cells. J. Investig. Dermatol. 119:1177-1182. [DOI] [PubMed] [Google Scholar]
- 16.Kanno, S., S. Iwai, M. Takao, and A. Yasui. 1999. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res. 27:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsumi, S., N. Kobayashi, K. Imoto, A. Nakagawa, Y. Yamashina, T. Muramatsu, T. Shirai, S. Miyagawa, S. Sugiura, F. Hanaoka, T. Matsunaga, O. Nikaido, and T. Mori. 2001. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Investig. Dermatol. 117:1156-1161. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman, P. D., and G. Almouzni. 2000. DNA replication, nucleotide excision repair, and nucleosome assembly, p. 24-48. In S. C. R. Elgin and J. L.Workman (ed.) Chromatin structure and gene expression, 2nd ed. Oxford University Press, New York, N.Y.
- 19.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 20.Krude, T. 1999. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247:148-159. [DOI] [PubMed] [Google Scholar]
- 21.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 22.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murciaand, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moggs, J. G., P. Grandi, J. P. Quivy, Z. O. Jonsson, U. Hubscher, P. B. Becker, and G. Almouzni. 2000. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20:1206-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, D. J., R. M. Taylor, P. Clements, and K. W. Caldecott. 2000. Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA 97:13649-13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nash, R. A., K. W. Caldecott, D. E. Barnes, and T. Lindahl. 1997. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry 36:5207-5211. [DOI] [PubMed] [Google Scholar]
- 26.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 27.Okano, S., S. Kanno, S. Nakajima, and A. Yasui. 2000. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J. Biol. Chem. 275:32635-32641. [DOI] [PubMed] [Google Scholar]
- 28.Park, S. Y., W. Lam, and Y. C. Cheng. 2002. X-ray repair cross-complementing gene I protein plays an important role in camptothecin resistance. Cancer Res. 62:459-465. [PubMed] [Google Scholar]
- 29.Pleschke, J. M., H. E. Kleczkowska, M. Strohmand, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275: 40974-40980. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028-23036. [DOI] [PubMed] [Google Scholar]
- 32.Shibahara K.-I., and B. Stillman. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96:575-585. [DOI] [PubMed] [Google Scholar]
- 33.Takao, M., R. Yonemasu, K. Yamamoto, and A. Yasui. 1996. Characterization of a UV endonuclease gene from the fission yeast Schizosaccharomyces pombe and its bacterial homolog. Nucleic Acids Res. 24:1267-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, R. M., B. Wickstead, S. Cronin, and K. W. Caldecott. 1998. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr. Biol. 16:877-880. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, R. M., D. J. Moore, J. Whitehouse, P. Johnson, and K. W. Caldecott. 2000. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol. 20:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, R. M., A. Thistlethwaite, and K. W. Caldecott. 2002. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol. 22:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, L. H., and M. G. West. 2000. XRCC1 keeps DNA from getting stranded. Mutat. Res. 459:1-18. [DOI] [PubMed] [Google Scholar]
- 38.Trucco, C., F. J. Oliver, G. de Murcia, and J. Menissier-de Murcia. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal, A. E., S. Boiteux, I. D. Hickson, and J. P. Radicella. 2001. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 20:6530-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virag, L., and C. Szabo. 2002. The therapeutic potential of poly(ADP-Ribose) polymerase inhibitors. Pharmacol. Rev. 54:375-429. [DOI] [PubMed] [Google Scholar]
- 41.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 42.Whitehouse, C. J., R. M. Taylor, A. Thistlethwaite, H. Zhang, F. Karimi-Busheri, D. D. Lasko, M. Weinfeld, and K. W. Caldecott. 2001. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104:107-117. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, S. H., and T. A. Kunkel. 2000. Passing the baton in base excision repair. Nat. Struct. Biol. 7:176-178. [DOI] [PubMed] [Google Scholar]
- 44.Yajima, H., M. Takao, S. Yasuhira, J. H. Zhao, C. Ishii, H. Inoue, and A. Yasui. 1995. A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J. 14:2393-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasui, A., and S. J. McCready. 1998. Alternative repair pathways for UV-induced DNA damage. Bioessays 20:291-297. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X., S. Morera, P. A. Bates, P. C. Whitehead, A. I. Coffer, K. Hainbucher, R. A. Nash, M. J. Sternberg, T. Lindahl, and P. S. Freemont. 1998. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 17:6404-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]