Abstract
The Athila retroelements of Arabidopsis thaliana encode a putative envelope gene, suggesting that they are infectious retroviruses. Because most insertions are highly degenerate, we undertook a comprehensive analysis of the A. thaliana genome sequence to discern their conserved features. One family (Athila4) was identified whose members are largely intact and share >94% nucleotide identity. As a basis for comparison, related elements (the Calypso elements) were characterized from soybean. Consensus Calypso and Athila4 elements are 12–14 kb in length and have long terminal repeats of 1.3–1.8 kb. Gag and Pol are encoded on a single open reading frame (ORF) of 1801 (Calypso) and 1911 (Athila4) amino acids. Following the Gag-Pol ORF are noncoding regions of ∼0.7 and 2 kb, which, respectively, flank the env-like gene. The env-like ORF begins with a putative splice acceptor site and encodes a protein with a predicted central transmembrane domain, similar to retroviral env genes. RNA of Athila elements was detected in an A. thaliana strain with decreased DNA methylation (ddm1). Additionally, a PCR survey identified related reverse transcriptases in diverse angiosperm genomes. Their ubiquitous nature and the potential for horizontal transfer by infection implicates these endogenous retroviruses as important vehicles for plant genome evolution.
Retrotransposons and retroviruses (collectively referred to as retroelements) replicate by a common mechanism of reverse transcription (for review, see Coffin et al. 1997). Retroelement genomes are delimited by direct long terminal repeats (LTRs), and they encode gag and pol genes, whose products form a particulate replication intermediate wherein reverse transcription takes place. The primary distinguishing feature between the retrotransposons and retroviruses is that the latter have a third gene called envelope (env). env encodes a transmembrane protein that associates with the cell membrane. The replication intermediate buds from the cell as a membrane-bound virion, and Env extends from the virion surface and interacts with cellular receptors to mediate infection.
Phylogenetic relationships based on reverse transcriptase amino acid sequences identify six distinct lineages of retroelements (Xiong and Eickbush 1990; Malik 2000). One of these—the vertebrate retroviruses—encodes env genes and is infectious. The five remaining groups are comprised mostly of retrotransposons and include the well-studied Ty1-copia (Pseudoviridae) and Ty3-gypsy (Metaviridae) elements (van Regenmortel et al. 2000), the so-called DIRS1 and BEL groups, and the caulimoviruses (Malik et al. 2000). With the exception of the caulimoviruses and the sparsely populated DIRS1 group, some members of each lineage encode open reading frames (ORFs) with env-like features—most notably transmembrane domains. These include a large number of invertebrate Ty3-gypsy elements (e.g., gypsy, 17.6, 297, and ZAM from Drosophila melanogaster; TOM from Drosophila ananassae; TED from Trichoplusia ni; Yoyo from Ceratitis capitata; for review, see Lerat and Capy 1999), two Ty1-copia elements from plants (i.e., SIRE-1 from Glycine max [soybean] and Endovir from A. thaliana; Laten et al. 1998; Kapitonov and Jurka 1999; Peterson-Burch et al. 2000), and several BEL group elements (e.g., Tas from Ascaris lumbricoides and Cer7 from Caenorhabditis elegans; Felder et al. 1994; Bowen and McDonald 1999). Analyses of env-like genes from the various retroelement groups suggests that env was independently acquired from viruses multiple times during evolution. The env-like ORFs of several insect Ty3-gypsy elements are closely related to env of the bacculoviruses, and for some Cer elements, the env-like gene is related to env of the phleboviruses (Malik et al. 2000). Despite the widespread presence of env-like ORFs and their similarity to known viral env genes, gypsy of D. melanogaster is the only known retroelement outside of the retroviruses for which Env is known to play a role in infection (Kim et al. 1994; Song et al. 1994).
In our analysis of the A. thaliana genome sequence, we determined that Athila—a degenerate, centromere-associated retroelement (Pelissier et al. 1995, 1996; Copenhaver et al. 1999)—is a Ty3-gypsy group retrotransposon with an env-like ORF (Wright and Voytas 1998). A related element was also described in Pisum sativum (pea) called Cyclops-2 (Chavanne et al. 1998). Because Cyclops-2 was less degenerate than Athila and prevalent in related legumes, we sought potential functional homologs in soybean. The soybean elements, called Calypso, encode an env-like gene that shares 29% amino acid identity to the corresponding gene of Cyclops-2 (Peterson-Burch et al. 2000). This suggests that the env-like ORF has evolved under functional constraint and likely plays a role in the life cycle of these elements. For simplicity, we refer to Athila and related retroelements as endogenous retroviruses, with the understanding that the biological role of their env-like genes remains to be determined. The sequence degeneracy of the endogenous plant retroviruses described to date has frustrated attempts to define their structural features. However, further characterization of the soybean Calypso elements and completion of the A. thaliana genome sequence has enabled us to construct consensus elements that likely approximate functional elements. Here we report a detailed description of these endogenous retroviruses and provide evidence of their widespread distribution in higher plants.
RESULTS
Athila Elements of A. thaliana
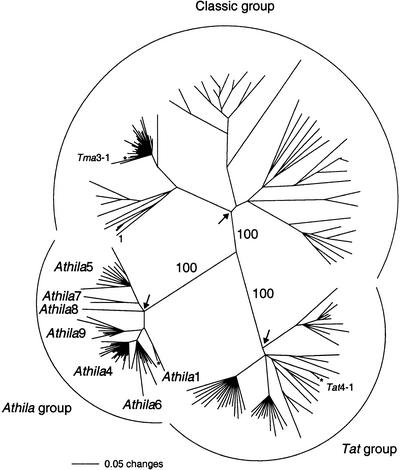
To further characterize the A. thaliana Athila elements, reverse transcriptases from all Ty3-gypsy elements were recovered from the A. thaliana genome sequence (Initiative 2000). BLAST searches (Altschul et al. 1990) were performed with reverse transcriptases from Athila1-1, Tat4-1, and Tma3-1, three divergent A. thaliana Ty3-gypsy elements (Fig. 1; Wright and Voytas 1998). Additional BLAST searches were performed with the most divergent retroelement sequences recovered. A total of 191 unique reverse transcriptases were identified. These were aligned, and when necessary, conservative changes were made to correct frameshift mutations. A phylogenetic tree was generated by the neighbor-joining method (Fig. 1; Saitou and Nei 1987). The elements clustered into three distinct clades designated the classic, Tat, and Athila lineages.
Figure 1.
A neighbor-joining tree of reverse transcriptases from Arabidopsis thaliana Ty3/gypsy retroelements. The tree was generated from amino acid sequences encompassing the seven conserved domains that define reverse transcriptase (Xiong and Eickbush 1990). Each major group (classic, Tat, and Athila) is labeled. Numbers along the branches indicate bootstrap support for 100 replicates. Arrows indicate the most recent common ancestor for each of the three lineages. Reverse transcriptases from Tma3-1, Tat4-1, and Athila1-1 were used in BLAST searches and are noted with an asterisk. BAC numbers for reverse transcriptases are in clockwise order, beginning with the branch marked number 1: T5E15, F23C08-2, MTO24, T16I21, T12H3, F23H14-4, T5M2/T17H1, F23H14-3, F9B22, T12K4, T14K23, T26P13-1, T5E15-2, T14A11, F13E17, F3D18, Tma3-1 9D12, MKD10, T1J24, MXE2, F28I8, T13P21/T6B13/F26C24-1-1, F23H14-4-4, F7F23, T5L23, T13H18/F3K12/F14P14-1, F9M13-2, F3H7, MJI6-1, F8D11, T32N15, T32B20, F17L24-2, F28J9-1, F5J5, F5K24, T14K23-2, T7B9-2, T10P12-3, F14I23, F11I2, T1O13, T29A4-3, F17L24, F19C17, T14A11-5, T27D20, T9F8/T4E14, T7B9-3, F15O4-2, MYM9, F7P1-2, F22D12, K11J14-2, F9M13, F6H5-2, T17A11/F15O11, T12K4-3, MTO24-2, T32E20, F12P23/T4D8, T10P12, T21C14, T13H18/F3K12/F14P14-2, F18P9, MJI6-2, F23H6, T8M17, F7K2, MOD1, T13P21/T6B13/F26C24-2, F26G5-2, T5N23, T21B14, K11J14, F1D9, MXC9, MVA11, F16J10/T20G20/T3P4, F28I8/F1O13, T17A11/F15O11-2, F19I11, T5L23, F6H5, F7M19, F1M23-4, F7I20, T24G3, F8N14, F8N14-2, F23M2, T15J14/F15A23, T4P3, T1O16/T13P21-1, Tat4-1 MXA21, T24H24, T12O21, F26H6, F7K15, F4M19, F5M15, T19K21/T17A11, F23H14-1-1, F12P23, T15D5, T19K21/T17A11-2, K11J14, T17A11/F15O11, F28H19, T24H24, F9M13-1, T27D6, F28L22, T7B9-1, F24C20/T27D6, F6H8, F15O11/F14O4/T26C18, F23C08-2, F3D18, F28L21, F1M23-2, F23C08, T13P21, F19N02, T5H22, F1M23-3, F17M07, F14G16, MIL15, F10A2, K2K18, T6L9, T3J11, F15K19/T13H18, Athila1 (Athila1-3 F4I4, Athila1-1 MXI10), Athila6 (F13C19, T32E20-2, F21I2-2, F7I20, F23H6-2, F5K7/F18P14/F28N16, F3D18-2, F26B15, F9M8, Athila6-2 F28N5, Athila6-1 T9E19), Athila4 (T32O2-2, T25N22/T13E11, Athila4-3 F7F22, Athila4-4 F7F22, F28L21, F9M8-2, Athila4-1 F1404, F7P3, Athila4-2 MED5, Athila4-10 T26P13, Athila4-7 F7B19/T15D2, T32O2, F10A2, Athila4-6 F1809, Athila4-9 T15D2 Athila4-5 F7K15, T26P13-2, T29A4, MIF6, T14A16, F21I2, Athila4-8 T6L9, F7B19/T15D9), Athila9 (T13D4,T13H18/F3K12/F14P14, F28H19, Athila9-1 T24G23, T7B9, F14C23, F10A2-2, T27B3), Athila8 (F17L24/F9B22), Athila7 (T3B16, F21A17), Athila5 (F1M23, T22B15, T19K21/T17A11, Athila5-1 F23M2, F23C08, Athila5-1 F14B16, T5E15-3, T32A11, T18O6/F7E22/F23M2, F11B20).
Phylogenetic analysis further resolved the Athila elements into clades, which we designated as distinct families (Fig. 1). These included the previously described Athila1 family (Wright and Voytas 1998) and six additional families, designated Athila4–Athila9. The Athila, Athila2, and Athila3 families are not included in the tree, because they have deletions of reverse transcriptase (Pelissier et al. 1995; Wright and Voytas 1998). Elements in four of the seven families had potential coding regions flanking reverse transcriptase and discernible LTRs (Athila1, Athila4, Athila5, and Athila6). Relatively intact insertions were given species designations (e.g., Athila1-1, Fig. 1). The Athila4 family was the largest and included 22 members. Six of these (designated Athila4-1 to Athila4-6) were ∼14 kb in length and had LTRs of ∼1.8 kb (Fig. 2). Athila4-3 and Athila4-4 were organized in tandem and shared a central LTR. The tandem Athila4-3/Athila4-4 insertion and the individual Athila4 elements were flanked by 5-bp target-site duplications (data not shown). In pairwise comparisons, the six Athila4 elements averaged 94% nucleotide identity across their entirety. Despite this high degree of sequence identity, gag and pol were broken by stop codons and frameshifts.
Figure 2.
Structural organization of Arabidopsis thaliana Athila4 elements. Boxes with filled triangles represent LTRs. Open boxes represent coding sequences, and they are offset to indicate changes in reading frame. Vertical thin lines represent stop codons. Horizontal thin lines represent noncoding sequences. The shaded region identifies the coding region for reverse transcriptase. Shaded boxes indicate the putative envelope (env) gene. The accession number, the BAC designator, and the position within the BAC for each Athila4 element are as follows: Athila4-1: AC007209, F1404, 33315 to 47208; Athila4-2: AB026642, MED5, 3448 to 17452; Athila4-3 and Athila4-4: AC007534, F7F22, 88613 to 114709; Athila4-5: AL353871, F7K15, 86117 to 99436; Athila4-6: AF296831, F1809, 38836 to 52851.
Calypso Elements of Soybean
In the initial description of the Cyclops-2 element from pea, related DNA sequences (based on Southern hybridizations) were found to be abundant in other legumes, including soybean (Chavanne et al. 1998). Cyclops-2 homologs were recovered from soybean by screening a genomic λ phage library using the Cyclops-2 reverse transcriptase as a hybridization probe. Sixty-three hybridizing phage were characterized, 35 of which were unique based on restriction endonuclease mapping (data not shown). Each of these latter clones was partially sequenced, and 24 had identifiable amino acid sequence similarity to Cyclops-2 and Athila (data not shown). The coding regions of these 24 elements, however, were replete with stop codons, frameshifts, deletions, and insertions. Five of the least degenerate elements (designated Calypso1-1, Calypso2-1, Calypso3-1, Calypso4-1, and Calypso5-1) were sequenced (Fig. 3). Despite being highly degenerate, each had discernable features such as LTRs and coding regions with similarity to gag, pol, and the env-like gene of Cyclops-2. In the case of Calypso2-1, the 5′ LTR depicted in Figure 3 is the 3′ LTR of a second Calypso element that inserted within Calypso2-1. Calypso5-1 contained an insertion within its reverse transcriptase of 1.8 kb, with flanking 5-bp target-site duplications and end sequences suggesting it is a retroelement solo LTR (Fig. 3; data not shown). Despite the high level of sequence degeneracy, the reverse transcriptases of the five Calypso elements shared, on average, 81% amino acid identity.
Figure 3.
Structural organization of the Calypso elements. Elements are depicted as described in the Fig. 2 legend. A Calypso element has inserted into Calypso2-1. The left-most LTR depicted belongs to this element; whereas, the right LTR belongs to Calypso2-1. Calypso5-1 has what appears to be a solo Calypso LTR within reverse transcriptase, which is indicated by the thick horizontal line.
Features of Athila4 and Calypso Elements
For most retroelements, the region adjacent to the 5′ LTR is complementary to a cellular tRNA and serves as the site for priming minus-strand DNA synthesis. The primer binding site (PBS) of Athila4 and Calypso is complementary to the 3′ end of the aspartic acid tRNA for the GAC codon from A. thaliana and soybean (Fig. 4a; Waldron et al. 1985; Wright and Voytas 1998). Complementarity begins at variable positions from the boundary of the 5′ LTR, and extends for 13 bases for the Athila4 elements and for 18 or 19 bases for given Calypso elements. For most retroelements, a stretch of purines adjacent to the 3′ LTR serves as the priming site for plus-strand DNA synthesis. A polypurine tract (PPT) is found at this location in Athila4 and Calypso, and all of the endogenous plant retroviruses share a conserved core consensus sequence (TTTGGGGG), as well as less conserved flanking sequences (Fig. 4B). A second PPT motif (PPT1) is found after the env-like gene. The two PPTs delimit a large noncoding region, which in Athila averages ∼2 kb in length (see Figs. 2, 3). A second noncoding region lies between gag-pol and the env-like gene and approximates 0.7 kb.
Figure 4.
Priming sites for reverse transcription. LTR sequences are underlined and square brackets denote the LTR boundaries. (A) The primer binding site (PBS) is complementary to the 3′ end of the ASP tRNA. Complementary sequences are shaded, including those that form G:U base pairs. (B) Polypurine tracts following the envelope (env)-like ORF (PPT1) and near the 3′ LTR (PPT) are characterized by the shaded, conserved core motif.
Because of the large number of frameshifts and stop codons in Calypso coding sequences, a quasiconsensus Calypso element was generated. Additionally, a strict Athila4 consensus sequence was generated, which was possible because of the high degree of sequence homogeneity. Figure 5A depicts the structural organization of these consensus elements, as well as Cyclops-2 from pea (Chavanne et al. 1998) and three partially sequenced homologs: Diaspora from soybean, BAGY-2 from barley (Shirasu et al. 2000), and a degenerate element from rice that we identified from the rice genome sequence data. The consensus Athila4 and Calypso elements encode Gag and Pol on a single ORF of 1911 and 1801 amino acids, respectively. These coding regions were aligned with Gag-Pol of Cyclops-2, and the percent amino acid identity was plotted along their entirety (Fig. 5A). The first third of the ORFs shares ∼20% amino acid identity, and we define this region as Gag (∼600 amino acid [aa], Fig. 5A). The Calypso and Cyclops-2 Gag proteins encode a conserved finger domain characteristic of retrotransposon and retroviral nucleocapsid proteins (Fig. 5B). This motif is not present in any of the other elements examined. A block of ∼110 amino acid residues is conserved near the N terminus of Gag, suggesting a conserved function. Similarity to this region can be detected in the sequence of Diaspora and the rice element but not in BAGY-2 (data not shown).
Figure 5.
Gag-Pol of the Athila group elements. (A) The structural organization of the Athila4 and Calypso consensus elements are shown along with individual-related elements from pea (Cyclops-2; Chavanne et al. 1998), barley (BAGY-2; Shirasu et al. 2000), rice (accession number AP003054, 36238–53391 bp; an inserted element was removed for the diagram), and soybean (Diaspora, accession number AF095730). Below Cyclops-2 is a graph depicting amino acid identity along the length of Gag-Pol. Light gray depicts protease (PR), medium gray depicts reverse transcriptase (RT), and dark gray depicts integrase (IN). All other aspects of the figure are as described in the Fig. 2 legend. (B ) Amino acid sequence signatures of Gag-Pol. The sequence domains are described in the text. Motifs are shown that define the seven conserved domains of reverse transcriptase (Xiong and Eickbush 1990). For integrase, the zinc-binding domain is shown, as are signatures for the DD35E domain (Fayet et al. 1990) and the GPY/F motif (Malik and Eickbush 1999).
Following Gag is a motif (LI/CDLGA) that we believe is the active site of an aspartic acid protease (Fig. 5B). We define protease as the region of ∼40% amino acid identity that spans ∼300 amino acid residues between Gag and reverse transcriptase (depicted in light gray, Fig. 5A). Although we do not know the precise boundaries of protease, this region is considerably larger than the proteases of retrotransposons and retroviruses (e.g., 181 aa for Ty1, 99 aa for HIV; Merkulov et al. 1996; Coffin et al. 1997). Following protease is ∼520 amino acids that comprise reverse transcriptase. Reverse transcriptase shares ∼68% amino acid identity among elements. All seven conserved amino acid sequence domains characteristic of retroviral and retrotransposon reverse transcriptases are evident (depicted in gray, Fig. 5A). The remainder of Gag-Pol constitutes an ∼450 amino acid integrase (depicted in dark gray, Fig. 5A). In addition to the conserved N-terminal zinc-binding motif and the DD35E motif of the catalytic domain, integrase has a C-terminal extension with a GPY/F module (Fig 5B; Malik and Eickbush 1999). The GPY/F module is found in some retroviral and Ty3/gypsy element integrases and is thought to bind DNA. Integrase shares ∼64% amino acid identity among Athila4, Calypso, and Cyclops-2.
Features of the env-like Gene
After gag and pol and between the two noncoding regions, the Athila4 and Calypso consensus elements encode ORFs of 619 and 420 amino acids, respectively (Fig. 6A). Recognizable env-like ORFs are also found in members of the Athila, Athila1 thru Athila6, and Athila9 families (data not shown). The env-like ORFs of Athila2, Athila3, Athila4, and Athila6 share an average of 69% amino acid sequence identity in pairwise comparisons (data not shown). The Athila1 and Athila5 elements are divergent (Fig. 1), and their env-like ORFs do not align well with the other Athila families. The consensus Calypso env-like gene shares 29% amino acid sequence identity to the env-like gene of Cyclops-2 (Peterson-Burch et al. 2000). Between the pea/soybean and A. thaliana elements, no significant amino acid sequence similarity was observed.
Figure 6.
Features of the Athila group env-like ORFs. (A) Generalized organization of env-like ORFs from the Arabidopsis thaliana Athila group elements, the soybean Calypso elements, Cyclops-2 of pea, gypsy of Drosophila melanogaster, and HIV1. The open boxes indicate ORFs. Arrowheads indicate signal sequences. Black boxes indicate transmembrane domains. Vertical lines within boxes denote stop codons. The first methionine within each ORF is indicated by a short line. (B) Amino acid sequences of putative signal sequences at the N termini and predicted transmembrane domains. (C) TMpred output for the Athila4 consensus envelope (env)-like ORF with and without a frameshift at the C terminus. Values above 500 (on the X-axis) are significant and indicate likely transmembrane domains (Hofmann and Stoffel 1993). The Y-axis indicates amino acid sequence position. (D) Sequences at the putative splice acceptor site of the Arabidopsis thaliana Athila and Calypso elements. The confidence level indicates the output for NetGene 2 (Brunak et al. 1991; Hebsgaard et al. 1996). The first methionine in the env-like ORF is in bold.
Retroviral Env proteins are typically transported through the endomembrane system, where they are proteolytically cleaved to generate surface (SU) and transmembrane (TM) proteins prior to being released on the cell surface (Coffin et al. 1997). Targeting to the endomembrane system is mediated by a signal sequence at the N terminus of Env. The N termini of the Calypso and Cyclops-2 Env-like proteins are basic in nature (Fig. 6B). Additionally, the N termini of Athila4 and Cyclops-2 are serine-rich. The program PSORT predicts a variety of destinations for the Env-like proteins within the cell (Nakai and Kanehisa 1992). The most confident predictions are for Calypso2-1 and Athila4-1, which suggest targeting to the plasma membrane (70% confidence) and endoplasmic reticulum (85% confidence), respectively.
At the cell surface, the retroviral TM protein spans the plasma membrane. We previously reported a predicted transmembrane domain in the env-like ORFs of several Athila elements (Athila, Athila1, Athila2, and Athila3, Wright and Voytas 1998). The Athila4 consensus env-like ORF also encodes a transmembrane domain (TM1, Fig. 6A–C), to which the program TMpred assigns a score of 2006 (scores above 500 are considered significant; Hofmann and Stoffel 1993). Similarly, a transmembrane domain is predicted near the center of the Calypso env-like ORF (TMpred value of 947; Fig. 6A,B and data not shown). The Cyclops-2 env-like protein has a potential transmembrane domain at a similar location, but at a reduced confidence level relative to the other elements (TMpred value of 650; Fig. 6A,B and data not shown).
In our analysis of the Athila4 env-like gene, we noticed the potential to encode additional transmembrane domains after the stop codon. Strong transmembrane domains were predicted in either the same frame as the env-like ORF (TM2, Fig. 6A–C) or in the +1 frame (TM3, Fig. 6A–C). These potential coding regions extend the env-like ORF to the first polypurine tract (PPT1) and are conserved among some element families (Fig. 6B). Small ORFs with predicted transmembrane domains are also found at the end of the Calypso and Cyclops-2 env-like ORFs. In the consensus Calypso element, the ORF is in a −1 frame, although the degree of degeneracy among Calypso elements reduces confidence in this reading frame assignment. Unfortunately, sequences between Athila families were too divergent to ascertain whether the short ORFs are evolving as coding sequences based on frequencies of synonymous versus nonsynonymous substitutions (data not shown).
Retroviral env genes are typically expressed from a spliced, subgenomic mRNA (Coffin et al. 1997). The Calypso env-like ORF has a predicted splice-site acceptor sequence located at the first methionine, to which the program NetGene2 assigns a confidence level of 100% (Fig. 6D; Brunak et al. 1991; Hebsgaard et al. 1996). Although there are other favorable splice acceptors in the vicinity of the Calypso env-like ORF, only the putative acceptor at the first methionine is conserved (Fig. 6D). For the Athila elements, a number of possible splice acceptors are present near the beginning of the env-like gene, one of which is located just before the first methionine and is consistently predicted with a high level of confidence (>94%, Fig. 6D). In the animal retroviruses, the splice-site donor is typically located near the 5′ LTR or within Gag. Of the several possible donors in these regions, none are well conserved between element families (data not shown).
Distribution of Endogenous Retroviruses in Plants
To assess the distribution of the endogenous retroviruses, a set of degenerate primers was designed based on conserved sequences flanking the seven domains of the Athila4, Cyclops-2, and Calypso reverse transcriptases. Genomic DNAs were surveyed by PCR from 18 plant species, including several dicots (Gossypium hirsutum, cotton; Platanus occidentalis, sycamore; Lycopersicon esculentum, tomato; Solanum tuberosum, potato; and Nicotiana tabacum, tobacco), old-world monocots (Oryza sativa, rice; Avena sativa, oat; Secale cereale, rye; Hordeum vulgare, barley; Triticum aestivum, wheat; and Sorghum bicolor, sorghum), new-world monocots (Zea mays, corn; Zea mays ssp., Parviglumis, teosinte; a Tripsicum species), and a gymnosperm (Pinus coulteri, pine). A. thaliana, soybean, and pea, served as positive controls. PCR products were cloned and at least three independent clones were sequenced from each species. Most of the PCR products from dicots and old-world monocots encoded reverse transcriptases that shared >60% amino acid identity. In contrast, the new-world monocots and the single gymnosperm surveyed only yielded reverse transcriptases from more distantly related elements (data not shown). The dicot reverse transcriptases had numerous stop codons and insertions/deletions; whereas, sequences from the old-world monocots were considerably less degenerate. The most intact reverse transcriptases were from oat, rye, and barley, which shared >85% nucleotide identity across species. All nucleotide and amino acid sequences were aligned, making it possible to identify and correct frameshifts. A neighbor-joining tree was constructed from these reverse transcriptases and representative Tat elements were used as an outgroup (Fig. 7). The endogenous retroviruses clustered on a single branch, and with few exceptions (e.g., Diaspora from soybean), elements from a single species clustered together.
Figure 7.
Neighbor-joining tree based on amino acid sequences of Tat and Athila group reverse transcriptases. The tree is rooted to six elements from the Tat group (from top to bottom): Tat4-1, F26H6, Cinful, Vulgar, Rire2, and Grande1-4). The numbers on the branches indicate bootstrap support for 100 replicates. Brackets indicate the species from which the elements originated. Branches indicating previously described elements are labeled. Unlabeled branches represent reverse transcriptases from the following species (from top to bottom): sycamore (syc4-3, syc2-3, syc4-2, syc4-7); cotton (cot8-7, cot5-3, cot8-6); lily (TfcII elements AF219190, AF219217, AF219200); rice (rice2-17, rice5-2, rice2-10, rice1, rice); sorghum (sorg4-3, sorg5-6, sorg5-4, sorg5-5, sorg5-2, sorg5-8); tobacco, tomato, and potato (tom4-10, tom10-16, tob1, tob2-2, tob5-3, tob4-1, tom4-4, pot8-8, pot8-3, pot8-4, pot8-5, tom10-4, pot8-10, pot5-1); oat (oat6-7, oat6-1, oat6-8); rye (rye5-4, rye5-2, rye3-4, rye4-4); barley (bar7, bar2-12, bar2-19, bar2-4, BAGY-2); wheat (wheat5-3, wheat8-11, wheat8-2, wheat3-1, wheat8-5); Arabidopsis thaliana (La9-5, T17A11, hau8-3, Athila5-2 F23M2, La9-7, La9-9, Athila1-1 La8-1, La9-6, T16I21, T13D4, Athila9-1 T24G23, F18P14, hau7-2, T13E11, F7B19, hau8-4); pea and favabean (pea1, favabean1, favabean2, pea8-1, pea9-1); soybean (will8-2, hark5, will, L859-3, hark2, will2, hark5-1, L859-6, L859-2, L852will8-3, L858-2).
Athila4 Elements Are Expressed in a Methylation-Deficient Strain
The A. thaliana Athila elements are preferentially located within heterochromatin flanking the centromeres (Pelissier et al. 1996; Initiative 2000). These regions contain repeated sequences that are methylated and likely transcriptionally quiescent (Jeddeloh et al. 1998; Consortium 2000). Some Athila group elements and retrotransposons are expressed in genetic backgrounds, such as ddm1, which have reduced levels of DNA methylation (Hirochika et al. 2000; Steimer et al. 2000; Lindroth et al. 2001). We sought Athila4 mRNAs by RT-PCR in ddm1 backgrounds, using five different Athila4 primers and a poly(T) primer/adaptor. Fifteen separate Athila cDNAs were cloned and sequenced: eight were Athila4 elements, four were Athila6 elements, and three could not be easily assigned to a family because of sequence degeneracy (Fig. 8). No transcripts were recovered from a wild-type strain. All 15 transcripts terminated within a 200-bp window of a consensus Athila LTR. One Athila4 cDNA was primed with a gag primer and was 8.4 kb in length. A portion of this clone (1.8 kb) was sequenced and matched Athila4-6, except for a single base change, which could be the result of a PCR-induced error. No spliced transcripts were detected.
Figure 8.
Transcription termination sites of Arabidopsis thaliana Athila group elements. RNA was isolated from a methylation-deficient strain (ddm1) and amplified by RT-PCR using an Athila4 primer and a poly(T) primer/adaptor. PCR products were cloned and sequenced. At the top of the diagram is a generic Athila LTR with the region denoted wherein the transcripts terminate. Clones are listed that match the Athila6 and Athila4 elements. The numbers next to the arrows indicate the base position for transcription termination sites within the LTR. Clones were recovered with variable termination sites, as indicated by the gaps in the alignment. Three clones (pDW820, pDW825, and pDW780) match highly degenerate, unclassified Athila elements, and the numbers for the BACs that carry these elements are provided.
DISCUSSION
We previously reported that the A. thaliana Athila retroelements have a novel feature—a putative env gene that may enable them to be infectious (Wright and Voytas 1998). Homologs of Athila elements have been described in other plant species (e.g., Cyclops-2 of pea, Chavanne et al. 1998; BAGY-2 of barley, Shirasu et al. 2000), all of which are replete with deletions, rearrangements, or stop codons. To ascertain conserved features of these endogenous plant retroviruses, we analyzed Athila elements in the completed A. thaliana genome sequence. We also recovered Athila homologs from soybean—the so-called Calypso elements. By generating consensus sequences from degenerate insertions, we were able to identify features that likely define a functional element.
Shared Features Among Plant Endogenous Retroviruses
The characterized plant endogenous retroviruses range from 12 to 14 kb in length and have LTRs ranging from 1.3 to 1.8 kb, among the largest LTRs described to date for Ty3/gypsy elements. Like many plant retroelements, Gag and Pol are encoded on a single ORF. One striking feature among Athila4, Calypso, and Cyclops-2 is the high degree of sequence conservation of pol. Between these elements, reverse transcriptase and integrase, respectively, share ∼68% and 64% amino acid identity. Because reverse transcription is error prone and often leads to accelerated rates of sequence evolution (Gabriel and Mules 1999), this suggests that either Pol is under very tight functional constraints or that the elements have invaded their plant hosts relatively recently. The phylogenetic tree of A. thaliana Ty3/gypsy elements provides some support for the recent acquisition of Athila elements. The short branch lengths supporting the Athila and Tat element groups suggest they share a more recent common ancestor relative to classic Ty3/gypsy element families (see arrows in Fig. 1). Because the Athila elements encode an env-like ORF, horizontal transfer by infection is one possibility for the apparent difference in their evolutionary history.
In contrast to pol, gag shows higher levels of sequence divergence. This is typical of retroelement gag genes, whose products carry out structural roles. Nonetheless, Calypso and Cyclops-2 Gag have conserved finger motifs characteristic of nucleocapsid proteins, and all three elements have a conserved domain near the Gag N terminus. Gag averages 675 amino acid residues (measured from the first methionine to the active site of protease), which is larger than most classic plant Ty3/gypsy element Gag proteins (e.g., Reina, 482 aa; Avramova et al. 1996). If the endogenous retroviruses are infectious, Gag may carry out functions related to transmission. Many plant viruses encode movement proteins that transport viral nucleic acids from cell to cell (Ghoshroy et al. 1997) or factors that facilitate spread by insect vectors (Woolston et al. 1983). These proteins are typically not well-conserved, and no similarity to the Gag proteins of the endogenous retroviruses is evident.
Another characteristic feature of the endogenous retroviruses is the presence of two large noncoding regions that flank the env-like ORF. The upstream region approximates 0.7 kb, and the downstream region approximates 2 kb. In most retroelements, noncoding sequences are very small, and it is generally thought that extraneous sequences are lost to maximize the amount of genetic information that can be encoded within an element. The conservation of noncoding domains among the endogenous retroviruses suggests they play a role in replication. Possibilities include regulating gene expression (either transcription or translation) or facilitating expression of the env-like ORF (e.g., in splicing or in enabling internal ribosome entry). Of the two noncoding regions, the 3′ region is flanked by conserved polypurine tracts (PPTs), which might serve as priming sites for plus-strand DNA synthesis during reverse transcription. Multiple PPTs are found in other retroelements such as Ty1 and HIV, although in these elements, the upstream PPT resides within pol (Hungnes et al. 1993; Heyman et al. 1995). A third, small noncoding region is also found between the 5′ LTR and the start of the gag-pol ORF. This region carries the putative primer binding site (PBS) for minus-strand DNA synthesis, which is complementary to an Asp tRNA. This is a distinguishing feature of the endogenous retroviruses, for the classic Ty3/gypsy elements and the Ty1/copia group elements have PBSs complementary to initiator Met tRNAs, and the Tat elements have PBSs complementary to Asn, Lys, and Arg tRNAs (Wright and Voytas 1998; D.A. Wright, unpublished observation).
The env-Like ORF and Its Potential Role in Infection
We previously concluded that the env-like genes of the endogenous retroviruses likely play a functional role in replication, based on sequence conservation between the Cyclops-2 and Calypso env-like genes (Peterson-Burch et al. 2000). With the availability of the A. thaliana genome sequence, additional Athila env-like genes made it possible to discern conserved features. Computer models predict the Env-like proteins are expressed from a spliced subgenomic mRNA. The Env-like proteins are also predicted to encode a central transmembrane domain. Env-like proteins of animal retroviruses often have both central and C-terminal transmembrane domains, the latter of which anchors Env within the endoplasmic reticulum. In most endogenous plant retroviruses, there is a short ORF after the env-like gene that is predicted to encode a transmembrane domain and could serve an anchoring role. Expression of the short ORF as part of Env would require read-through of a stop codon. Alternatively, because a transmembrane domain is also encoded in adjacent reading frames, ribosomal frameshifting may be employed. Attempts to determine if this region is evolving as a coding sequence were not productive because of the high degree of sequence divergence between element families. As other endogenous plant retroviruses are identified, it will be of interest to determine whether they too have this short transmembrane domain-encoding ORF. A functional element will be required to determine experimentally whether it has a biological role.
If the endogenous retroviruses are infectious, then the Env-like protein is likely important in this process. During infection by retroviruses, Env facilitates the merging of the membrane-bound virion with the target cell. The plant cell wall poses an obstacle to membrane-mediated infection. Nonetheless, enveloped plant viruses do exist, including members of Bunyaviridae and the Rhabdoviridae (van Regenmortel et al. 2000). These viruses bud from the endomembrane system and accumulate in the cell until a feeding invertebrate ingests them and carries them to another plant. Recent work has shown that some animal retrotransposons have acquired env genes from viruses (Malik 2000). For example, the env gene of the D. melanogaster gypsy element is related to env of the baculoviruses and was likely acquired by gypsy through transduction. To date, however, we have not identified similarity between the env-like ORFs of the endogenous plant retroviruses and those of viruses or other genes in the databases. It should be mentioned that some plant Ty1/copia group retrotransposons have env-like ORFs (Laten et al. 1998, 1999; Kapitonov and Jurka 1999; Peterson-Burch et al. 2000). These genes are unrelated to the env-like genes of Athila and its homologs, but they are predicted to be transmembrane proteins. It is tempting to speculate that env-like genes play a similar role in both groups of elements.
Distribution of Endogenous Retroviruses in Plants
Using a PCR-based assay, we found that endogenous retroviruses are widely distributed among angiosperms. The recovered reverse transcriptases were strikingly similar and shared >60% amino acid identity. This high degree of sequence conservation belied the fact that most carried mutations, the exception being elements from cereals, namely oat, rye, and barley. The integrity of the cereal reverse transcriptases implies that these elements have undergone more recent episodes of replication, and to date, they are the best candidates for functional endogenous retroviruses. Elements were not recovered from a gymnosperm (pine) and the three new-world monocot species tested (corn, teosinte, and tripsicum). It may be that the endogenous retroviruses are not present in the genomes of these plants or that they are divergent and cannot be amplified by the primers. Phylogenetic analyses of the reverse transcriptases indicated that, with few exceptions, the relationships among the elements reflected relationships among their hosts. This suggests that either the endogenous retroviruses are inherited vertically or if they are viruses, they have a limited host range. As more plant genomes are characterized in greater detail, it will be of interest to determine whether high levels of sequence conservation is a general feature of the endogenous plant retroviruses. This will help address the question as to whether or not they are young retroelements relative to the classic Ty3/gypsy elements.
Expression and Activity of A. thaliana Athila Group Elements
Most A. thaliana Athila elements are located within centromeric heterochromatin, which is typically highly methylated (Vongs et al. 1993; Pelissier et al. 1996; Copenhaver et al. 1999). Methylation is thought to control transposable element activity (Yoder et al. 1997; Martienssen 1998), and several recent studies in plants have shown that decreases in DNA methylation are associated with increased transposable element activity (Hirochika et al. 2000; Lindroth et al. 2001; Miura et al. 2001; Singer et al. 2001). Of particular relevance to this study, truncated Athila transcripts have been reported in strains with mom1 mutations, which derepress transcriptionally silent loci (Amedeo et al. 2000; Steimer et al. 2000).
We performed RT-PCR on RNAs isolated from ddm1 plants and were able to amplify cDNA from Athila4 and Athila6 elements, two of the most intact Athila families. Transcripts terminated at a similar position within the LTR, thereby defining the LTR R/U5 boundary. cDNA as large as 8.4 kb was recovered; however, no spliced messages were identified. Although Athila elements are expressed in ddm1 backgrounds, they are probably not replicating because of sequence degeneracy. For future studies, it will be important to identify a functional Athila group element. We envision two approaches for how this might be accomplished: 1) a consensus Athila4 element could be constructed or 2) elements could be further characterized from species such as the small grains that appear to have structurally intact elements. The identification of a replication-competent Athila group element will be necessary to test the hypothesis that these elements are infectious plant retroviruses. If this proves to be the case, the Athila group elements may be useful as vectors for gene transfer and the genetic modification of plants.
METHODS
DNA Manipulations and Filter Hybridizations
A soybean genomic λ phage library (Chen et al. 1998) was screened with a reverse transcriptase probe under low stringency conditions (50°C with a 1% SDS wash; Ausubel et al. 1987). The probe was obtained by PCR amplification of Pisum sativum DNA using primers based on the Cyclops-2 reverse transcriptase (DVO701 5′-CCG-TCA-TCC-GGA-ATG-ACA-AGG-ATG and DVO702 5′-ACG-GAT-GAG-CCT-TTG-CTT-CGA-ATC). Phage subclones were sequenced by primer walking. Genomic DNAs from 18 plant species (see Results) were surveyed by PCR to identify Athila-group reverse transcriptases. DNAs were prepared using genomic tips and protocols supplied by Qiagen. Degenerate primers were designed based on two conserved amino acid sequence motifs flanking the seven core domains of reverse transcriptase (Xiong and Eickbush 1990; VRKEVLKL, DVO1197 5′-GTG-CGN-AAR-GAR-GTN-NTN-AAR-YT, and FIKDFSKV, DVO1198 5′-AAC-YTT-NGW-RAA-RTC-YTT-DAT-RAA). PCR was performed in 50 μL reactions with ∼100 ng genomic DNA, 3 μmole of each primer, 2.5 units Taq DNA polymerase, 1× Taq buffer (Promega), and 2.5 mM MgCl2. PCR was performed for 30 cycles under the following conditions: 92°C for 20 sec, 50°C for 30 sec, and 72°C for 90 sec. The PCR products were purified on low-melting agarose gels and cloned into T-vector prepared from pBluescript II KS- (Hadjeb and Berkowitz 1996). Athila-group reverse transcriptases were sequenced in their entirety from vector-based primers.
Sequence Analysis
DNA Sequence analysis was performed using the GCG software package (Devereux et al. 1984), DNA Strider 1.2 (Marck 1991), and the BLAST search tool (Altschul et al. 1990). Phylogenetic relationships were determined by the neighbor-joining distance algorithm using PAUP v4.0 beta 4a (Saitou and Nei 1987; Swofford 1991) and were based on reverse transcriptase amino acid sequences that had been aligned with CLUSTALX v1.63b (Thompson et al. 1994). Transmembrane helices were identified using the PHDhtm program and TMpred (Hofmann and Stoffel 1993; Rost et al. 1995). Splice-site analysis was performed with NetGene2 (Brunak et al. 1991; Hebsgaard et al. 1996). All DNA sequences have been submitted to the DDBS/EMBL/GenBank databases. The Calypso elements are under accession numbers AF186182, AF186183, AF186184, AF186185, and AF186186. BAC or P1 clone numbers for the Ty3/gypsy reverse transcriptases are listed in the Figure 1 legend. Accession numbers for the Athila4 elements are listed in the Figure 2 legend. The accession numbers of the Athila-group reverse transcriptases from various species are AF378012 to AF378081. Additional details regarding these sequences can also be found at our Web site (http://www.public.iastate.edu/∼voytas/).
RT-PCR
Total RNA was isolated from A. thaliana ddm1 plants using the PUREscript RNA isolation kit (Gentra Systems, Inc.). RNA was annealed to the primer DVO1247, which is a poly(T) oligo with a specific tail (5′-GGA-CTT-CAG-GAC-TGC-TTG-ACA-AAG-T30). First-strand DNA synthesis was performed at 42°C for 2 h using Superscript II reverse transcriptase and the manufacturer's protocol (GIBCO BRL). RNase activity was inhibited by the addition of Super RNase IN per the manufacturer's instructions (Ambion). PCR was carried out using the Expand Long Template PCR System (Roche Molecular Biochemicals) with Athila-element-specific primers, along with DVO1248, which is specific to the tail of DVO1247.
Acknowledgments
We thank Jim Keck for assistance with the figures and members of the Voytas lab for helpful comments on the manuscript. This work was supported by a grant from Phytodyne, Inc., the Center for Advanced Technology Development at Iowa State University, and NIH grant number R41 GM61420. This is journal paper No. J-19446 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, project No. 3383 and was supported by Hatch Act and state of Iowa funds.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL voytas@iastate.edu; FAX 515-294-7155.
Article published on-line before print in December 2001: Genome Res., 10.1101/gr.196002.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.196001.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amedeo P, Habu Y, Afsar K, Scheid OM, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene/Wiley Interscience; 1987. [Google Scholar]
- Avramova Z, Tikhonov A, SanMiguel P, Jin YK, Liu C, Woo SS, Wing RA, Bennetzen JL. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, McDonald JF. Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res. 1999;9:924–935. doi: 10.1101/gr.9.10.924. [DOI] [PubMed] [Google Scholar]
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Chavanne F, Zhang DX, Liaud MF, Cerff R. Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol Biol. 1998;37:363–375. doi: 10.1023/a:1005969626142. [DOI] [PubMed] [Google Scholar]
- Chen W, Jie C, Atherly G. Construction of a soybean genomic and root cDNA library from Phyophthora resistant L85–3044. Soybean Genetics Newsletter. 1998;25:132–133. [Google Scholar]
- Coffin JM, Hughes SH, Varmus H. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Consortium. The complete sequence of a heterochromatic island from a higher eukaryote. The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. Cell. 2000;100:377–386. [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O, Ramond P, Polard P, Prere MF, Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990;4:1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Felder H, Herzceg A, de Chastonay Y, Aeby P, Tobler H, Muller F. Tas, a retrotransposon from the parasitic nematode Ascaris lumbricoides. Gene. 1994;149:219–225. doi: 10.1016/0378-1119(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Gabriel A, Mules EH. Fidelity of retrotransposon replication. Ann NY Acad Sci. 1999;870:108–118. doi: 10.1111/j.1749-6632.1999.tb08871.x. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S, Lartey R, Sheng J, Citovsky V. Transport of proteins and nucleic acids through plasmodesmata. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:27–50. doi: 10.1146/annurev.arplant.48.1.27. [DOI] [PubMed] [Google Scholar]
- Hadjeb N, Berkowitz GA. Preparation of T-over-hang vectors with high PCR product cloning efficiency. Biotechniques. 1996;20:20–22. doi: 10.2144/96201bm02. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman T, Agoutin B, Friant S, Wilhelm FX, Wilhelm ML. Plus-strand DNA synthesis of the yeast retrotransposon Ty1 is initiated at two sites, PPT1 next to the 3′ LTR and PPT2 within the pol gene. PPT1 is sufficient for Ty1 transposition. J Mol Biol. 1995;253:291–303. doi: 10.1006/jmbi.1995.0553. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- Hungnes O, Jonsrud K, Tjotta E, Grinde B. Sequence comparison and mutational analysis of elements that may be involved in the regulation of DNA synthesis in HIV-1. J Mol Evol. 1993;37:198–203. doi: 10.1007/BF02407356. [DOI] [PubMed] [Google Scholar]
- Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes & Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- Kim A, Terzian C, Santamaria P, Pelisson A, Purd'homme N, Bucheton A. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten HM. Phylogenetic evidence for Ty1-copia-like endogenous retroviruses in plant genomes. Genetica. 1999;107:87–93. [PubMed] [Google Scholar]
- Laten HM, Majumdar A, Gaucher EA. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci. 1998;95:6897–6902. doi: 10.1073/pnas.95.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Capy P. Retrotransposons and retroviruses: Analysis of the envelope gene. Mol Biol Evol. 1999;16:1198–1207. doi: 10.1093/oxfordjournals.molbev.a026210. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S, Eickbush TH. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- Marck C. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R. Transposons, DNA methylation, and gene control. Trends Genet. 1998;14:263–264. doi: 10.1016/s0168-9525(98)01518-2. [DOI] [PubMed] [Google Scholar]
- Merkulov GV, Swiderek KM, Brachmann CB, Boeke JD. A critical proteolytic cleavage site near the C-terminus of the yeast retrotransposon Ty1 Gag protein. J Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier T, Tutois S, Deragon JM, Tourmente S, Genestier S, Picard G. Athila, a new retroelement from Arabidopsis thaliana. Plant Mol Biol. 1995;29:441–452. doi: 10.1007/BF00020976. [DOI] [PubMed] [Google Scholar]
- Pelissier T, Tutois S, Tourmente S, Deragon JM, Picard G. DNA regions flanking the major Arabidopsis thaliana satellite are principally enriched in Athila retroelement sequences. Genetica. 1996;97:141–151. doi: 10.1007/BF00054621. [DOI] [PubMed] [Google Scholar]
- Peterson-Burch BD, Wright DA, Laten HM, Voytas DF. Retroviruses in plants? Trends Genet. 2000;16:151–152. doi: 10.1016/s0168-9525(00)01981-8. [DOI] [PubMed] [Google Scholar]
- Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, Yordan C, Martienssen RA. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1) Genes & Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes & Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- Steimer A, Amedeo P, Afsar K, Fransz P, Scheid OM, Paszkowski J. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell. 2000;12:1165–1178. doi: 10.1105/tpc.12.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony and other methods. Laboratory of Molecular Systematics, Smithsonian Institute; 1991. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Regenmortel MHV, Fauquet CM, Bishop DHL, Carsten EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, et al. Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press; 2000. [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Waldron C, Wills N, Gesteland RF. Plant tRNA genes: Putative soybean genes for tRNAasp and tRNAmet. J Mol Appl Genet. 1985;3:7–17. [PubMed] [Google Scholar]
- Woolston CJ, Covey SN, Penswick JR, Davies JW. Aphid transmission and a polypeptide are specified by a defined region of the cauliflower mosaic virus genome. Gene. 1983;23:15–23. doi: 10.1016/0378-1119(83)90212-3. [DOI] [PubMed] [Google Scholar]
- Wright DA, Voytas DF. Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics. 1998;149:703–715. doi: 10.1093/genetics/149.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]