Abstract
We describe a new computer system, called ARACHNE, for assembling genome sequence using paired-end whole-genome shotgun reads. ARACHNE has several key features, including an efficient and sensitive procedure for finding read overlaps, a procedure for scoring overlaps that achieves high accuracy by correcting errors before assembly, read merger based on forward-reverse links, and detection of repeat contigs by forward-reverse link inconsistency. To test ARACHNE, we created simulated reads providing ∼10-fold coverage of the genomes of H. influenzae, S. cerevisiae, and D. melanogaster, as well as human chromosomes 21 and 22. The assemblies of these simulated reads yielded nearly complete coverage of the respective genomes, with a small number of contigs joined into a smaller number of supercontigs (or scaffolds). For example, analysis of the D. melanogaster genome yielded ∼98% coverage with an N50 contig length of 324 kb and an N50 supercontig length of 5143 kb. The assembly accuracy was high, although not perfect: small errors occurred at a frequency of roughly 1 per 1 Mb (typically, deletion of ∼1 kb in size), with a very small number of other misassemblies. The assembly was rapid: the Drosophila assembly required only 21 hours on a single 667 MHz processor and used 8.4 Gb of memory.
Shotgun sequencing was introduced by Sanger et al. (1977) and has remained the mainstay of genome sequence assembly for nearly 25 years. The method involves obtaining random sequence reads from a genome (which may range from a small plasmid to the complete chromosomal complement of an organism) and assembling them into contigs on the basis of sequence overlap. The method was refined by Ansorge and colleagues (Edwards et al. 1990), who introduced the notion of obtaining paired-end reads by sequencing both ends of a plasmid of known insert size (a technique sometimes referred to as double-barreled shotgun sequencing). The forward and reverse reads from a plasmid can be used to link together distinct sequence contigs.
Shotgun sequencing is straightforward for simple genomes—those with no or few repeat sequences. For such genomes, one can largely assemble the genome simply by merging together reads containing overlapping sequence. Over the years, shotgun sequencing has been applied to simple genomes of increasingly larger size, including plasmids, viruses (Sanger et al. 1982), cosmids and bacterial artificial chromosomes carrying genomic DNA from humans or other organisms, and bacterial genomes (Fleischmann et al. 1995).
Shotgun sequencing is more challenging for complex (repeat-rich) genomes, because false overlaps may occur owing to the repeat sequences. Two primary approaches can be taken to deal with complex genomes. The hierarchical shotgun approach involves generating an overlapping set of intermediate-sized clones [e.g., bacterial artificial chromosomes (BACs) with 200-kb inserts], selecting a tiling path of these clones, and then subjecting each of these clones to shotgun sequencing; this approach thus decomposes a large genome into smaller genomes. The whole-genome shotgun (WGS) approach involves generating sequence reads directly from a whole-genome library and using computational techniques to reassemble the genome sequence in one fell swoop. The WGS approach avoids the laborious steps of generating and mapping a library of BAC clones, but poses a more challenging computational assembly problem and a greater potential for large-scale misassembly. The relative merit of the two approaches is likely to depend on the complexity of the genome under study.
The hierarchical approach has been used for most eukaryotic genomes, including the yeast S. cerevisiae (Goffeau et al. 1996), the nematode C. elegans (C. elegans Seq. Cons. 1998), the mustard weed A. thaliana (Arabidopsis Gen. Init. 2000), and the human (International Human Genome Sequencing Consortium 2001). The WGS approach was used for the fruit fly Drosophila melanogaster (Adams et al. 2000) to generate a draft assembly of the euchromatic portion of the genome (∼3% repeat), although the hierarchical approach is being used to produce a finished sequence. In this paper, we describe a computer system for performing WGS assembly of complex genomes.
Various assembly programs have been previously reported, including SEQAID (Peltola et al. 1984), CAP (Huang 1992), PHRAP (Green 1994), TIGR assembler (Sutton et al. 1995), AMASS (Kim et al. 1999), CAP3 (Huang and Madan 1999), the Celera assembler (Myers et al. 2000; cf. Myers 1995), and EULER (Pevzner et al. 2001). However, only the Celera assembler has been designed and implemented for large and complex eukaryotic genomes.
We present first an overview of the algorithm used for assembly. A description of the generation of simulated data for testing ARACHNE follows, as does a summary of the results of applying ARACHNE to simulated WGS data from H. influenzae, S. cerevisiae, D. melanogaster, human chromosome 21, and human chromosome 22. Technical details of the assembly algorithm are then presented, followed by a general discussion.
Outline of Assembly Algorithm
Input Data
ARACHNE is designed to analyze sequence reads obtained from both ends of plasmid clones—that is, paired forward and reverse reads (although it can also be used with unpaired reads). The mean and standard deviation of the insert length of each plasmid clone can be specified individually. In practice, it is most convenient to describe each clone as belonging to one of several libraries characterized by these parameters.
Each base in each read has an associated quality score, such as that produced by the PHRED computer program (Ewing et al. 1998). A quality score of q corresponds to a probability of 10−q/10 that the base is incorrect; a quality score of 40 thus corresponds to 99.99% accuracy. As an initial step, ARACHNE trims reads to eliminate terminal regions of extremely low quality and to eliminate reads containing very little high-quality sequence (see the later section on technical details, “Details of Assembly Algorithm”). The program also trims known vector sequences and eliminates known contaminants (e.g., sequence from the bacterial host or cloning vector).
Overlap Detection and Alignment: Sort and Extend
ARACHNE starts by detecting and aligning pairs of apparently overlapping reads, referred to here simply as overlaps. Some of these are false overlaps resulting from repeated sequences in the genome and will be eliminated in subsequent steps.
Overlap detection is performed in an efficient manner. Rather than comparing all pairs of reads (which requires N2/2 comparisons, where N is the number of reads), the program uses a sort and extend strategy that scales approximately linearly. This strategy involves producing a sorted table of each k-letter subword (k-mer) together with its source—that is, the read in which it occurs and its position within the read (in practice, we use k = 24). The table is sorted so that identical k-mers appear consecutively (Batzoglou 2000).
The program then excludes k-mers that occur with extremely high frequency, which typically correspond to high-copy, high-fidelity repeated sequences in the genome. We eliminate them to increase the efficiency of the overlap detection process rather than to eliminate repeated sequences, which do not cause a problem per se.
The program then identifies all instances of read pairs that share one or more overlapping k-mer (which can be readily recognized from the sorted k-mer table), and uses a three-step process to align the reads in an efficient manner. The first step merges overlapping shared k-mers, the second step extends the shared k-mers to alignments, and the third step refines the alignments by dynamic programming (details provided in the section below entitled “The Alignment Module”). The overall process bears some resemblance to the FASTA algorithm (Pearson and Lipman 1988).
This three-step process yields the most valid alignments between read pairs. Some valid alignments may be missed if the overlaps are short, of low quality, or consist solely of high-copy, high-fidelity repeats. Some invalid alignments may result from low-copy repeats that produce apparently overlapping sequence.
Error Correction
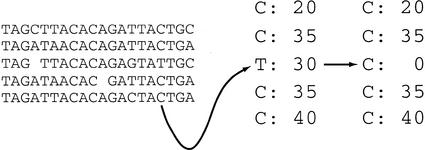
ARACHNE next detects and corrects sequencing errors by generating multiple alignments among overlapping reads. The program then identifies instances in which a base is overwhelmingly outvoted by bases aligned to it and corrects the base (Fig. 1). In practice, this occurs where there is a disagreement at an isolated position involving a single base (or occasionally two bases); the process takes into account the quality scores of the bases. Based on the cases examined, the majority of these instances appear to be sequencing errors rather than slightly different copies of a repeated sequence. In the latter instance, this is due to the fact that there are typically multiple occurrences of each alternative sequence and, thus, there is not an overwhelming vote for one alternative. ARACHNE similarly corrects occasional insertions and deletions resulting from what appear to be sequencing errors. As the reads are corrected, corresponding changes are made to the alignments.
Figure 1.
Correcting errors in reads. A portion of a multiple alignment between five reads is shown. In the highlighted column of the alignment, a base T of quality 30 is aligned only to bases C, some of which are of quality greater than 30. The base T is changed to a base C of quality 0.
Evaluation of Alignments
ARACHNE assigns a penalty score to each aligned pair of overlapping reads. The program first assigns a penalty score to each discrepant base, based on the sequence quality score at the base and flanking bases on either side. Discrepancies in high-quality sequence are assigned a high penalty, whereas discrepancies in low-quality sequence are penalized less heavily. The penalty scores for the individual discrepancies are then combined to yield an overall penalty score for the alignment; overlaps incurring too high a penalty are discarded (see The “Alignment Module” section for details). This step is stringent, and false overlaps that remain are typically caused by highly conserved repeats. At this point, ARACHNE also detects and discards likely chimeric reads (see the section below entitled “Detection of Chimeric Reads” for details).
Identification of Paired Pairs
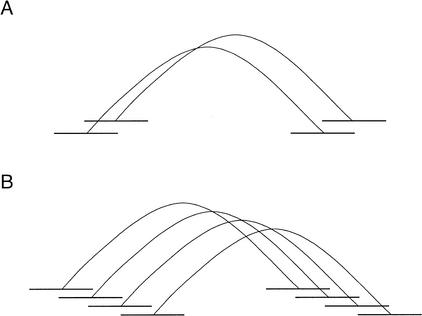
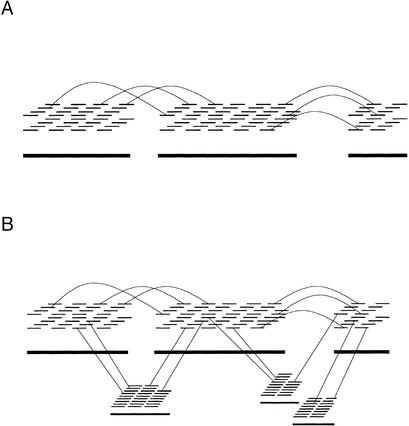
ARACHNE searches for instances of two plasmids of similar insert size with sequence overlaps occurring at both ends (Fig. 2A). Such instances are referred to as paired pairs. These instances can be extended by iteratively building complexes of such paired pairs (Fig. 2B).
Figure 2.
Using paired pairs of overlaps to merge reads. (A) A paired pair of overlaps. The top two reads are end sequences from one insert, and the bottom two reads are end sequences from another. The two overlaps must not imply too large a discrepancy between the insert lengths. (B) Initially, the top two pairs of reads are merged. Then the third pair of reads (from the top) is merged in, based on having an overlap with one of the top two left reads, an overlap with one of the top two right reads, and consistent insert lengths. The bottom pair is similarly merged.
Empirically, we find that these initial assemblies are almost always correct. The only significant exception occurs when the paired pairs come from within a large repeat (larger than the insert size of the plasmid). The program attempts to detect and eliminate such cases by virtue of their having too high a depth of coverage (see below). Such instances are discarded.
The collections of paired pairs (Fig. 2B) are merged together into contigs, and consensus sequences are formed (see below). The resulting contigs are then treated as large reads. In practice, their length varies from slightly more than the size of a typical read to ∼30 kb.
Contig Assembly
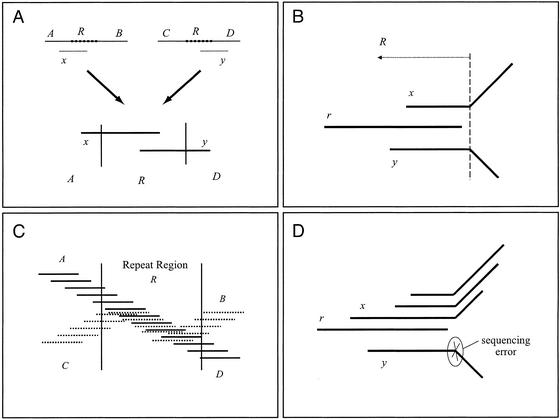
In the absence of repeats, producing the correct layout of reads is straightforward. Any two reads with substantial sequence overlap must truly overlap in the genome, and thus a correct assembly could be obtained simply by merging all overlapping reads. However, false overlaps may arise between reads derived from different copies of a repeat. For example, reads entering different copies of a repeat may have overlapping sequence and could give rise to misassemblies (Fig. 3A).
Figure 3.
Contig assembly. (A) How merging reads across the boundary of a repeat may result in a misassembly. Regions A, B, C, and D are unique regions, and region R is a repeat occurring twice in the genome. Reads x and y overlap in region R. Thus, regions A and D are wrongly joined after merging reads x and y. (B) A potential repeat boundary. Read r overlaps both reads x and y, but reads x and y do not overlap each other; they disagree in their rightmost ends. Here, a repeat R starting inside reads x and y and including the full read r is shown. In practice, sequencing errors rather than repeats often cause such patterns of overlap. (C) Contigs are created by merging reads up to the potential boundaries of repeats. A potential repeat boundary is any place where a read may be extended with two nonoverlapping reads. Two regions of the genome covered with reads are shown here. One region (A-R-D) is covered with solid line reads and the second region (C-R-B) with dotted line reads. The two regions meet in the repeat R creating five contigs: these are the unique contigs corresponding to unique sequences A, B, C, and D, and the repeat contig corresponding to the repeat R, where reads from both copies of R are overcollapsed into one contig. According to the algorithm used to construct contigs, the contig corresponding to R would have exactly the reads that are fully included in the boundaries of R. All the other reads would be assigned to contigs A, B, C, and D. (D) Sequencing errors. Read r dominates read y because the neighbors of y are all neighbors of r. This is caused by a sequencing error on y, which is marked in the figure. Note that if y represented correct sequence, it would likely be extended to the right by some read that did not overlap r, and thus r would not dominate y.
ARACHNE identifies potential repeat boundaries and avoids assembling contigs across such boundaries. The program marks a potential repeat boundary whenever a read r can be extended to the right (or left) by reads x and y, but x and y do not overlap each other (Fig. 3B). Read r is not merged with any read on its right, because it is not clear which read it should be merged with. By contrast, when all neighbors to the right of r overlap each other, then r can be safely merged with its closest neighbor. As in Myers (1995), the program thus merges, at this stage, read pairs that do not cross a marked repeat boundary (Fig. 3C).
This criterion for merging pairs of reads is very conservative, and based on the cases examined, it rarely produces misassemblies. In fact, it is overly conservative because some potential repeat boundaries reflect instances in which reads x and y (Fig. 3D) fail to overlap owing to several sequencing errors. ARACHNE employs a technique to eliminate some of these spurious repeat boundaries (dropping dominated reads, as described in the “Contig Assembly” section below) and performs a second round of contig merger.
In addition to dominated reads, another category of reads that introduces repeat boundaries that can be merged across safely are reads fully included in other reads (the subreads). During the above procedure of constructing contigs, subreads are ignored. They are not used to detect repeat boundaries and they are not included in any contig. After creating the contigs, if read a is fully included in reads b1, …, bk which all belong to contig C, then a is inserted in C. Details are described in the “Contig Assembly” section.
Detection of Repeat Contigs
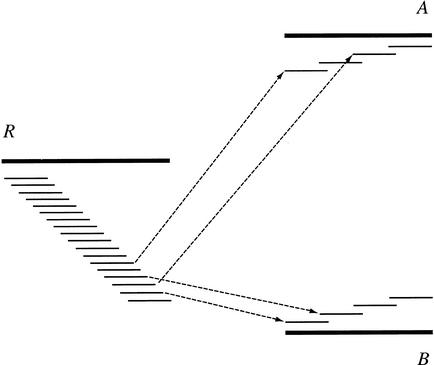
In the previous step, ARACHNE merges reads into contigs up to potential repeat boundaries. Some of these are repeat contigs—defined as contigs in which nearly identical sequence from distinct regions are collapsed together. Such repeat contigs can be identified in two ways. First, repeat contigs will typically have an unusually high depth of coverage. This can be assessed by using the log-odds ratio—that a contig of a given length and density of reads represents unique sequence versus being composed of reads derived from two copies of a repeat (Myers et al. 2000). Second, they will typically have conflicting links to other contigs. Repeat contigs are usually linked to multiple, distinct, nonoverlapping contigs, reflecting the multiple regions that flank the repeat in the genome (Fig. 4).
Figure 4.
Detection of repeat contigs. Contig R is linked to contigs A and B to the right. The distances estimated between R and A and R and B are such that A and B cannot be positioned without substantial overlap between them. If there is no corresponding detected overlap between A and B (if their reads do not overlap), then R is probably a repeat linking to two unique regions to the right.
Creation of Supercontigs
If the repeat contigs have been correctly marked, the remaining contigs should represent correctly assembled sequence. The unmarked contigs are referred to as unique contigs (similar to unitigs, (Myers et al. 2000)), because they usually represent unique sequence in the genome (or genomic repeated sequence that has diverged sufficiently to allow it to assemble as if it were unique).
ARACHNE then uses the forward-reverse links from plasmid reads to order and orient the unique contigs in longer layouts called supercontigs (or scaffolds). Supercontigs are created incrementally. At the beginning, each unique contig is considered a supercontig. Pairs of supercontigs are then merged into a large supercontig if they are joined by at least two forward-reverse links (Fig. 5A). The program requires the presence of at least two links to avoid links due to a single chimeric plasmid. It is unlikely that there will be two independent links supporting the same incorrect merger. Priority is given to those mergers supported by the most links and involving the shortest distances (see the “Supercontig Assembly” section below for details).
Figure 5.
Supercontig creation and gap filling. (A) A supercontig is constructed by successively linking pairs of contigs that share at least two forward-reverse links. Here, three contigs are joined into one supercontig. (B) ARACHNE attempts to fill gaps by using paths of contigs. The first gap in the supercontig shown here is filled with one contig, and the second gap is filled by a path consisting of two contigs.
Filling Gaps in Supercontigs
The layout now consists of a number of supercontigs, each of which is an ordered list of contigs with interleaved gaps. Most of these gaps correspond to regions marked as repeat contigs and are thus omitted from the supercontig construction. Some gaps will also correspond to regions in which there is an insufficient number of shotgun reads to allow assembly.
ARACHNE attempts to fill gaps by using the repeat contigs. For every pair of consecutive contigs with an interleaving gap in a supercontig S, the program tries to find a path of pairwise overlapping contigs that fill the gap. Forward-reverse links from S guide the construction of the path by identifying contigs likely to fall in the gap (Fig. 5B). Further details are given in the section below entitled “Filling Gaps in Supercontigs”.
Consensus Derivation and Postconsensus Merger
The layout of overlapping reads is converted to a consensus sequence with quality scores. This is done by converting pairwise alignments of reads (computed during the overlap detection phase) into multiple alignments. Because multiple sequence alignment is traditionally a time-consuming step, we developed an efficient algorithm, described below under the heading “Consensus Derivation”.
After forming consensus sequence, ARACHNE merges overlapping adjacent contigs in supercontigs. In this final stage, it accepts overlaps that have significant numbers of discrepancies, thereby merging across spurious repeat boundaries that were not detected during layout.
Comparison with Other Assemblers
ARACHNE was developed from an earlier version of the program described in a Ph.D. thesis (Batzoglou 2000). In broad outline, the program follows the overlap-layout-consensus approach that has been used by nearly all assemblers. Many of the algorithmic aspects of ARACHNE are novel, while other aspects are similar to existing assemblers.
Myers and colleagues produced a sophisticated assembly program, called the Celera assembler (Myers et al. 2000), based on various layout algorithms previously developed by Myers (1995). ARACHNE shares some significant similarities with the Celera assembler, most notably in the algorithms for merging reads into contigs up to the boundaries of repeats (Myers 1995). However, the two assemblers also have many significant differences. The Celera assembler screens for predefined repeats; ARACHNE does not and instead uses k-mer frequencies to identify repeats. ARACHNE uses sorting of k-mers to detect overlaps; the initial version of the Celera assembler employed a different approach (although the program has been subsequently revised (G. Myers, pers. comm.). Like PHRAP (Green 1994) and CAP3 (Huang and Madan 1999), ARACHNE creates, refines, and evaluates read alignments using quality scores; the Celera assembler does not use quality scores subsequent to trimming and prior to consensus. ARACHNE uses error correction to more accurately evaluate read overlaps. Both assemblers detect potential repeat boundaries, merge reads up to these boundaries (as in Myers 1995), and use read density to detect repeat contigs. ARACHNE also detects repeat contigs via conflicting links. Moreover, it uses paired pairs and other techniques to produce longer contigs during layout. Both ARACHNE and the Celera assembler require at least two links to order a pair of contigs in a supercontig, but apart from that they use different algorithms to build supercontigs and fill in gaps within supercontigs.
Simulation of WGS Data
We tested ARACHNE on simulated WGS data from known genomes. The data were generated in two steps: (1) selection of random reads from the target genome and (2) assignment of quality scores and introduction of errors. In the first step, plasmids were randomly chosen from the genome by selecting a random location for the start of the insert and a random length from a given length distribution. The corresponding forward and reverse reads were then read from the genome. In the second step, each simulated read was assigned realistic quality scores and errors by pairing it with a real read taken from a finished BAC sequencing project from the Whitehead/MIT Center for Genome Research. For each read, the length, quality scores, and correctness of each base in the read were known (from comparison with the finished sequence). On each simulated read, we imposed the pattern of quality scores and errors seen in its corresponding read.
The simulated reads thus contain fairly realistic error patterns. Nonetheless, they fall short of being completely realistic in several respects (in decreasing order of significance). First, because the plasmids are randomly positioned along the genome, the simulation does not reflect potential cloning bias. In other words, there is a nonuniform distribution of clones across the genome. Second, because the simulated reads are assigned quality scores by randomly pairing them with actual reads, the simulation does not reflect regions of systematically poor sequence quality. For example, sequence following a long poly(A)-stretch may have poor quality, whereas an ARACHNE simulation will pair each such simulated read with an independent real read. Third, because our reads are taken from published genomic sequences, we necessarily omitted regions absent from the published sequence and concatenated sequences flanking small gaps. Consequently, regions that were difficult to clone, sequence, or assemble in the original published sequence will likely also be missing from our results.
We generated full coverage and half coverage in the simulations. Full coverage corresponds to ∼10.3-fold coverage based on Whitehead's definition of trimmed read length and ∼8.3-fold coverage based on the number of bases with a PHRED quality score ≥20. Half coverage is half of this level. We generated data from small plasmids (mean insert size 4 kb) and large plasmids (mean insert size 40 kb) in a ratio of 20:1 in the case of full coverage, and 10:1 in the case of half coverage. In each case, the insert sizes were normally distributed around the mean with a standard deviation of 10%. Only 85% of the reads were successfully paired, the other 15% being single reads (reflecting a realistic sequence failure rate). Moreover, 0.7% of the inserts were chimeric—that is, corresponding forward-reverse linked reads that come from unrelated, random locations on the genome. (This rate was chosen to be higher than the observed rate of chimeric inserts at Whitehead.)
RESULTS
We generated simulated WGS data from the published sequence of H. influenzae (Fleischmann et al. 1995), S. cerevisiae (Goffeau et al. 1996), C. elegans (C. elegans Seq. Cons. 1998), D. melanogaster (Adams et al. 2000), and from human chromosomes 21 (Hattori et al. 2000) and 22 (Hunt et al. 1999).
With the exception of H. influenzae, in each case the published sequence consisted of more than one contig, because of gaps in the published sequence and because the genome had multiple chromosomes. In these cases, we concatenated the contigs to obtain a single contiguous sequence (which we refer to as the genome). This was done to simplify the analysis of the assembly results.
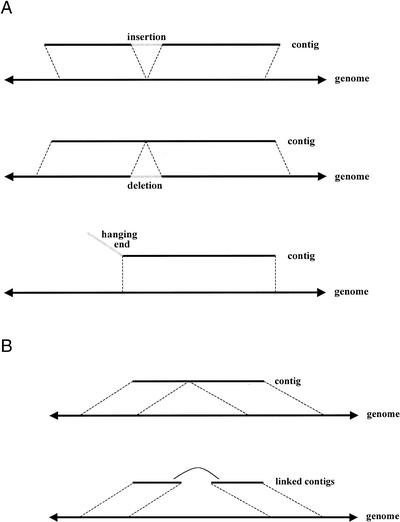
We evaluated the assemblies by attempting to align the entire length of each contig to the genome. If the contig could be aligned along its entire length in more than one place, we chose the best alignment. If the contig could not be aligned along its entire length, we aligned as much as possible and reported the outcome as a misassembly, as described below. Each portion of a contig that could not be aligned to the genome is reported. We also checked each link of each supercontig to see if that link was consistent with the positions of the contigs in their alignments against the genome. If a link did not align, we reported a misassembly. Most of the misassemblies were readily classified into a few types, as described in Figure 6A,B.
Figure 6.
Types of misassemblies. (A) Three types of simple minor misassemblies are shown: insertions, deletions, and hanging ends. In all three cases, a contiguous segment (of a contig or the genome) of length less than 10 kb does not align in the expected location (with the genome or contig). This segment could be aligned at some alternate location in most cases, although we do not do this in practice. Compound minor misassemblies (e.g., contigs having two insertions) are reported as multiple misassembly events. (B) Two types of major misassemblies are shown. In the first type, two pieces of a contig align to distant parts of the genome (if one piece is very short, we instead report a hanging end, as in A). In the second type, adjacent contigs in a supercontig are aligned to distant parts of the genome. In practice, what we typically encounter is a hybrid between these two types: a contig that lies in the middle of a supercontig is split as in the first type. We call this hybrid the standard major misassembly.
The assembly results are described in Tables 1–5. Table 1 reports contig coverage and read usage. The contig coverage is 97%–98% at full coverage and a slightly lower percentage for half coverage. In all cases, at least 92% of the reads were used. Because repeat contigs are occasionally placed in more than one location, a given read may sometimes be used more than once in the assembly if it occurs in a long, perfect repeat in the genome. At full coverage, the rate of multiple usage in H. influenzae is 1% (reflecting the presence of eight exact repeats) and much lower for the other genomes, which lack such long, perfect repeats.
Table 1.
Contig Coverage and Read Usage
| Genome | H. influenzae | S. cerevisiae | D. melanogaster | Human 21 | Human 22 |
|---|---|---|---|---|---|
| Length (Mb) | 1.8 | 12 | 120 | 33.8 | 33.5 |
| Full Coverage (∼10-fold) | |||||
| Genome contained in sequence contigs (%) | 98.8 | 96.1 | 97.9 | 98.2 | 97.4 |
| Bases in undetected overlaps (%) | 0.02 | 0.1 | 0.95 | 0.6 | 0.4 |
| Reads appearing in assembly (%) | 98.7 | 95.1 | 97.3 | 96.7 | 95.3 |
| Reads appearing in multiple locations in assembly, owing to exact repeats (%) | 1.0 | 0.2 | 0.2 | 0.03 | 0.06 |
| Half Coverage (∼5-fold) | |||||
| Genome contained in sequence contigs (%) | 97.1 | 92.4 | 95.4 | 95.0 | 92 |
| Bases in undetected overlaps (%) | 0.9 | 0.2 | 0.2 | 0.3 | 0.5 |
| Reads appearing in assembly (%) | 97.9 | 92.9 | 95.9 | 95.4 | 92.1 |
| Reads appearing in multiple locations in assembly, owing to exact repeats (%) | 1.7 | 0.04 | 0.1 | 0.02 | 0.04 |
For ten assemblies (of five genomes at two levels of coverage), the genomic coverage by contigs and read utilization is given. Bases in undetected overlaps give the fraction of the genome covered multiply by contigs.
Table 5.
Misassemblies
| Genome | H. influenzae | S. cerevisiae | D. melanogaster | Human 21 | Human 22 |
|---|---|---|---|---|---|
| Full Coverage (∼10-fold) | |||||
| Deletions | 2 | 5 | 102 | 13 | 26 |
| Mean length (bp) | 440 | 470 | 1660 | 360 | 430 |
| Insertions | — | 1 | 7 | — | 2 |
| Mean length (bp) | — | 350 | 990 | — | 400 |
| Hanging ends | — | — | 3 | — | 4 |
| Mean length (bp) | — | — | 190 | — | 800 |
| Other | — | — | 3 | 1 | — |
| Misassemblies | — | — | (notea) | (noteb) | — |
| Half Coverage (∼5-fold) | |||||
| Deletions | 3 | 4 | 116 | 26 | 41 |
| Mean length (bp) | 290 | 3790 | 1600 | 220 | 340 |
| Insertions | 2 | — | 12 | 2 | 3 |
| Mean length (bp) | 380 | — | 670 | 90 | 390 |
| Hanging ends | 1 | 2 | 42 | 11 | 14 |
| Mean length (bp) | 78 | 450 | 460 | 1330 | 826 |
| Other | — | — | 5 | 4 | 5 |
| misassemblies | — | — | (notec) | (noted) | (notee) |
See Figure 6A,B for an illustration of the various types of common misassemblies. Other misassemblies are as follows:
Two contigs of lengths 1 kb and 138 kb were exchanged in a supercontig. Two correct contigs of lengths 313 kb and 169 kb, which were separated in the genome by 60 bp, were glued along a 440-bp segment that appeared at the left end of the left contig, and also at the right end of the right contig, yielding a chimeric contig in the final assembly (which we call a slipped join). There was a standard misassembly (Fig. 6B), occurring in a 4.8-Mb supercontig.
At the end of a supercontig, there was a contig of length 10 kb, which aligned at a distant location in the genome relative to the rest of the supercontig.
A supercontig of 467 kb had a standard major misassembly. A supercontig of 1.4 Mb had two stray contigs of 5 kb and 2 kb. A contig had a deletion of 29 kb. Contigs of 27 kb and 37 kb were misassembled.
A 3.4-Mb supercontig had a standard major misassembly. Contigs of 9 kb and 24 kb had slipped joins. A 12-kb contig at the end of a 2.6-Mb supercontig was misassembled.
A 4.5-Mb supercontig had a standard major misassembly. A 14-kb contig had a slipped join. A 303-kb supercontig was slightly misassembled near one of its ends. A supercontig had a spurious contig of length 5 kb. A 9-kb supercontig consisted of two incorrectly linked contigs.
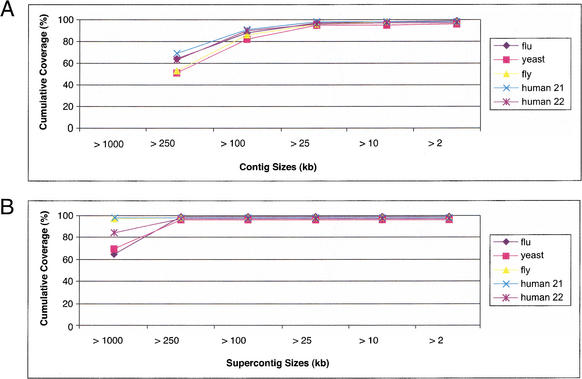
Tables 2 and 3 list size statistics for contigs and supercontigs, respectively. The mean length tends to underestimate the extent of the contigs, because it is unduly influenced by a large number of short contigs that comprise only a small percentage of the genome. The most useful statistic is likely the N50 length, which is the length L such that 50% of the bases are in contigs of size L or greater. The N50 contig length is ∼350 kb for full coverage and ∼17 kb for half coverage. The N50 supercontig length varies considerably among the genomes. The cumulative probability distributions of the contig and supercontig lengths are shown in Figure 7A,B.
Table 2.
Characterization of Contigs
| Genome | H. influenzae | S. cerevisiae | D. melanogaster | Human 21 | Human 22 |
|---|---|---|---|---|---|
| Full Coverage (∼10-fold) | |||||
| Number of contigs | 12 | 88 | 678 | 187 | 230 |
| Mean length (kb) | 150 | 132 | 174 | 178 | 142 |
| N50 length (kb) | 413 | 258 | 324 | 387 | 353 |
| Half Coverage (∼5-fold) | |||||
| Number of contigs | 142 | 865 | 8774 | 2807 | 2917 |
| Mean length (kb) | 13 | 13 | 13 | 11 | 11 |
| N50 length (kb) | 18 | 18 | 19 | 17 | 15 |
The N50 length is the length L such that 50% of the bases are contained in contigs of size at least L.
Table 3.
Characterization of Supercontigs
| Genome | H. influenzae | S. cerevisiae | D. melanogaster | Human 21 | Human 22 |
|---|---|---|---|---|---|
| Full Coverage (∼10-fold) | |||||
| # of supercontigs | 3 | 19 | 65 | 38 | 71 |
| Mean length (kb) | 603 | 6511 | 1811 | 877 | 461 |
| N50 length (kb) | 1192 | 1177 | 5143 | 3986 | 3011 |
| Half Coverage (∼5-fold) | |||||
| # of supercontigs | 10 | 51 | 450 | 168 | 206 |
| Mean length (kb) | 178 | 219 | 254 | 192 | 150 |
| N50 length (kb) | 629 | 1732 | 4258 | 3278 | 3197 |
The lengths include only the known bases actually contained in the assembly and thus exclude the length contained in the gaps (which typically comprise an additional 3–5%). The N50 length is the length L such that 50% of the bases are contained in supercontigs of size at least L.
Figure 7.
Coverage in assemblies of 10-fold simulated reads. (A) Coverage of the genome with contigs. Contigs of sizes >250 kb cover 50%–70% of the genome. (B) Coverage of the genome with supercontigs. Supercontigs of size >1 Mb cover at least 65% of the genome in all test examples.
Table 4 lists base accuracy information. At full coverage, the overall nucleotide accuracy is somewhat better than 99.99% (quality score >40). At half coverage, the overall nucleotide accuracy is somewhat better than 99.9% and ∼95% of the bases have quality scores corresponding to accuracy exceeding 99.99%.
Table 4.
Base Pair Accuracy
| Genome | H. influenzae | S. cerevisiae | D. melanogaster | Human 21 | Human 22 |
|---|---|---|---|---|---|
| Full Coverage (∼10-fold) | |||||
| Average quality | 45.3 | 43.6 | 43.4 | 42.8 | 41.3 |
| Fraction of bases assigned quality 40 | 99.6% | 99.6% | 99.6% | 99.6% | 99.5% |
| Average quality of bases reported to have quality score 40 | (all correct) | 49.5 | 51.5 | 51.3 | 47.6 |
| Half Coverage (∼5-fold) | |||||
| Average quality | 32.3 | 32.6 | 33 | 32.3 | 32.1 |
| Fraction of bases assigned quality 40 | 95.1% | 95.0% | 95.2% | 95.2% | 95.1% |
| Average quality of bases reported to have quality score 40 | 44.0 | 42.6 | 48.2 | 44.2 | 43.6 |
The average quality is defined as: −10 log10 [(# of mismatches, insertions, and deletions)/(# of bases in contigs)]. Thus, bases of quality 30 are 99.9% accurate, bases of quality 40 are 99.99% accurate, and so on. ARACHNE assigns a maximum quality score of 40. The table reports the average quality for the assembly (obtained by comparison with the known genome sequence), the percentage of bases assigned the maximal score, and the actual quality of the bases assigned the maximal score.
Table 5 reports assembly accuracy. With full coverage there is roughly one misassembly per Mb—mostly consisting of small deletions. With half coverage there are slightly more errors, but the assemblies are still quite useful for most purposes. Most are minor misassemblies, as illustrated in Figure 6A. Exceptions are described in the Table 5 footnote.
Table 6 describes ARACHNE's computational performance for the case of full coverage. The largest genome was Drosophila, which used 8.4 Gb of memory and required 21 hours on a single processor machine (667 MHz Compaq Alpha).
Table 6.
Computational Performance at Ten-Fold Coverage
| Genome | Length (Mb) | Time (Hr) | Memory (Gb RAM) |
|---|---|---|---|
| H. influenzae | 1.8 | 0.3 | 0.13 |
| S. cerevisiae | 12 | 1.9 | 1.0 |
| D. melanogaster | 120 | 21 | 8.4 |
| Human 21 | 33.8 | 7.5 | 3.0 |
| Human 22 | 33.5 | 8.7 | 4.9 |
Total CPU times are given, including times for both assembly and evaluation, using a single 667 MHz Alpha processor on a Compaq ES40 machine.
Details of Assembly Algorithm
In this section, we discuss some of the more technical aspects of the assembly algorithm.
Initial Processing of Reads
ARACHNE trims each read, using its base quality scores, as follows. First it finds the longest contiguous sequence of bases in the read such that the expected fraction of errors in the sequence is less than 5%. It trims further to ensure that no base of quality less than 10 is within 12 bases of either end. If after trimming, less than 50 bases remain, the read is discarded.
After trimming, the program uses the alignment module to align the reads with the E. coli host genome, E. coli transposons, the cloning vector and, where appropriate, mitochondrial sequence of the organism under study. Depending on the circumstances, when we find such an alignment we remove the beginning of the read, the end of the read, or the entire read. ARACHNE does not screen reads for matches with repeat sequence.
The Alignment Module
ARACHNE has an alignment module that is used to align reads to reads, reads to contaminants, contigs to each other, and several other purposes. The alignment module accepts as input a collection of DNA sequences of arbitrary lengths, and it produces as output pairwise alignments between them. It proceeds in four phases: sorting of k-mers, coalescence of k-mer hits, alignment generation, and alignment refinement. These phases are designed so that a single k-mer hit (perfect match of length k between two of the sequences) can lead to an alignment between them, and so that k-mer hits that do not lead to alignments can be rapidly processed.
In phase 1 of this process (sorting), a list is made of all the length k sequences (k-mers) that appear in the sequences and their reverse complements; where k is an adjustable parameter, varying from 8 to 24, depending on the characteristics of the sequences.
To reduce memory usage, phase 1 is usually performed in 100 passes, with each pass processing a subset of the k-mers. Specifically, there are 10 outer passes, in which the first and last bases of the k-mer are varied symmetrically (e.g., one outer pass handles k-mers of the form A…G or its reverse complement C…T) and 10 inner passes, in which the second and second-to-last bases are varied in the same way. In each pass, we scan the input sequences to find all instances of the relevant k-mers and, for each, create a record having four entries: the k-mer, the sequence number, the position of the k-mer in the sequence, and a reverse complementation flag. The role of the latter flag is as follows. For each relevant k-mer in the sequences, we list either the k-mer or its reverse complement depending on which is lexicographically first (with the choice indicated by the flag). In this way, we avoid separate processing of the reverse complements of the given sequences.
The records for each pass are placed in a vector. For each pass of phase 1, a pass of phase 2 is performed. Phase 2 begins by sorting the members of this vector by their first entries, thereby grouping together records having the same k-mer. If we find that the number of records in the vector having the same k-mer as their first entry exceeds a specified (adjustable) threshold, we do not process them. Otherwise, for each two records sharing the same k-mer entry, we consider the perfect k-long partial alignment (or k-mer hit) that it defines.
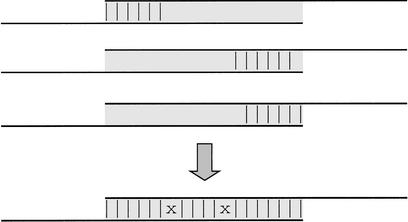
Phase 2 then proceeds by exploiting the inherent redundancy of the data, coalescing the k-mer hits into longer, imperfect, but gap-free partial alignments, as illustrated in Figure 8. These partial alignments are further restricted to have the following properties: no mismatches on either end; any two mismatches are separated by at least two matching bases; and, subject to these restrictions, no extension of the partial alignment is possible. We call these partial alignments extended k-mer hits.
Figure 8.
Partial alignments in the alignment module. Three partial alignments of length k = 6 between a pair of reads coalesce to yield a single full alignment of length k = 19. Vertical bars denote matching bases, whereas x's denote mismatches. This illustrates the commonly occurring situation where an extended k-mer hit is a full alignment between two reads (k = 6 is used in the figure for simplicity).
The alignment module maintains a list of extended k-mer hits, which grows as the passes are processed. For each k-mer hit, phase 2 first checks this list to see if a preexisting extended k-mer hit extends it. This is a rapid operation, because it can be accomplished by checking indices (and without accessing the sequences). If no such extended k-mer hit exists, then the given k-mer hit is extended and appended to the list.
In phase 3, full alignments are created from the extended k-mer hits. For each pair of sequences we form a directed graph, whose vertices are the extended k-mer hits associated to that sequence pair and whose edges correspond to pairs of extended k-mer hits that can likely be combined into a single, good, partial alignment. Then, for each maximal path in this graph, we attempt to construct an alignment. This is done by filling in bases between consecutive extended k-mer hits in the path and extending on both ends to create a full alignment. Several heuristic parameters control this process, which yields, at most, one alignment from each path. Many paths do not yield a full alignment.
In phase 4, each full alignment is refined via dynamic programming (Needleman and Wunsch 1970), under the assumption that the new alignment is close to the old one (using the standard “banded diagonal” approach to improve running time). Once the alignments have been created, they are scored. For each mismatch, we first assess the quality of the aligning bases, assigning a single number, which is approximately the minimum of the quality scores assigned to the two bases, the two bases on their left, and the two bases on their right. Then we assign a penalty score to the mismatch. For example, if we found that the minimum of all six scores was 30, we would assign the penalty score of 1000 (= 1030/10). Penalty scores for insertions and deletions are computed similarly, and the penalty scores for all the discrepancies appearing in the alignment are summed and then normalized by dividing by (overlap length/100)1.5. Alignments whose penalty score exceeds 100 are usually discarded.
Detection of Chimeric Reads
Reads that contain genomic sequence from two disparate locations are termed chimeric. We identify them according to heuristic criteria, which empirically appear to flag all chimeric reads, but also flag a small number of nonchimeric reads. Reads flagged as chimeric are discarded.
Two reads have a good overlap if they overlap with penalty score ≤100. Two reads, for example, a and c, have a transitive good overlap if there exists a read b such that a and b have a good overlap, b and c have a good overlap, and there is an implied overlap of at least 50 bases between a and c.
For a read c to be declared (potentially) chimeric, it must branch, in the sense that it has a transitive good overlap with some read z, yet no good overlap between c and z can be found. To be certain, we align c with z from scratch. Moreover, to declare c chimeric, there must also be evidence that some position x in c can be called the point of chimerism: we must see reads that align with c from the left, up to x, and perhaps slightly beyond, and reads that align with c from the right, back to x, and perhaps slightly before, but no read n that aligns with c, covering bases well to the left and right of x (Fig. 9).
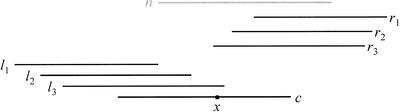
Figure 9.
Detection of chimeric reads. Reads l1, l2, l3, r1, r2, and r3, and the absence of a read n (having long overlaps on both sides of a point x) suggest that read c may be chimeric, consisting of the juxtaposition of two disparate genomic segments: one corresponding to the part of c before x, and one corresponding to the part of c after x. We call x the point of chimerism of c. Note that reads l3 and r3 extend slightly beyond x, as often happens for real chimeric reads.
At the same time that we detect chimeric reads, we also detect and discard dead-end reads: reads a for which no known read overlaps a and extends beyond its left (or right) end, yet for which some read c has a good transitive overlap with a and extends significantly beyond this end. Reads may have dead-ends because they are of low quality at one of their ends, or because they are chimeras, formed by concatenating disjoint genomic segments, one long and one very short. Chimeras of this type are not always detected by the previous test for chimerism, because overlaps with the short segment may be missed.
Contig Assembly
During contig assembly, reads are merged into contigs up to potential repeat boundaries. Reads that are fully included in other reads (the subreads) are excluded from this procedure. The rest of the reads (the full reads) are merged whenever they overlap, except for the following restriction. Suppose read b is the read that extends read a to the right by the least positive amount (denoted by a → b). Suppose there is a read c that overlaps with a by at least 100 bases, and extends a to the right. Suppose b and c do not overlap. Moreover, suppose there is no path of reads x1,…, xk such that (b,x1), (x1,x2), …, (xk,c) are all overlaps and the implied shift between b and c is consistent with the overlaps (a,b) and (a,c) (Fig. 10). Then, merging of reads stops to the right of read a. In other words, the contig where a belongs does not contain any read that extends a to the right.
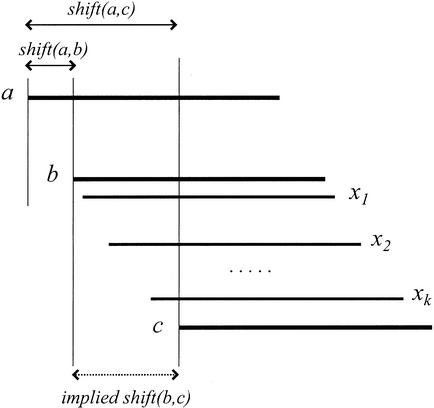
Figure 10.
Contig assembly. If (a,b) and (a,c) overlap, then (b,c) are expected to overlap. Moreover, one can calculate that shift(b,c) ≈ shift(a,c) − shift(a,b). We detect a repeat boundary toward the right of read a, if there is no overlap (b,c), nor any path of reads x1, …, xksuch that (b,x1), (x1,x2), …, (xk,c) are all overlaps, and shift(b,x1) + … + shift(xk,c) ≈ shift(a,c) − shift(a,b).
Missed overlaps between pairs of reads lead to detected repeat boundaries according to the above criterion. The following heuristic is used in order to recover some of the missed overlaps: Let a → b, and c → d. Suppose there are overlaps (a,c) and (b,d), but no overlap (b,c). Then the overlap (b,c) is inserted (we refer to this rule as Rule 1).
A first round of merging reads is performed, with the restriction of not merging any reads across detected repeat boundaries. A second round of merging is performed after repeat boundaries are recomputed under a more stringent definition of what is a repeat boundary. Specifically, only maximal reads are used to detect repeat boundaries in this second round. A read is maximal if no other read dominates it. Read a dominates read b if a and b overlap, and the set of reads extending a both to the left and the right is a strict superset of the set of reads extending b, and all the extension lengths and directions are consistent (e.g., Fig. 3D). For example, if b extends a to the right by 100, a is the only neighbor of b to the left, c is the only neighbor of b to the right and extends b by 50, c extends a to the right by 150 (or, more precisely, within 4% of 150), and d also extends a to the right, then a dominates b.
After the creation of contigs as described above, the subreads (excluded so far from consideration) are inserted into contigs, whenever this can be done unambiguously. Specifically, if read a is fully included in reads b1, …, bk, which all belong to contig C, then a is inserted in C. This insertion takes place if the positions of a in C as implied by its overlaps to b1, …, bk are all similar within three bases.
Detecting Repeat (Super)contigs
We mark repeated contigs using two heuristic methods, namely density of reads and consistency of forward-reverse links. The second of these methods is also used to mark supercontigs as repeated during supercontig assembly.
Density of Reads
We compute the log-odds ratio, that a contig with a given density of reads represents a unique region of the genome versus representing at least two copies of a repeated region (Myers et al. 2000). We use a cutoff of 1 for this ratio (log base-2), so that any contigs with a log-odds ratio less than 1 are marked as repeated.
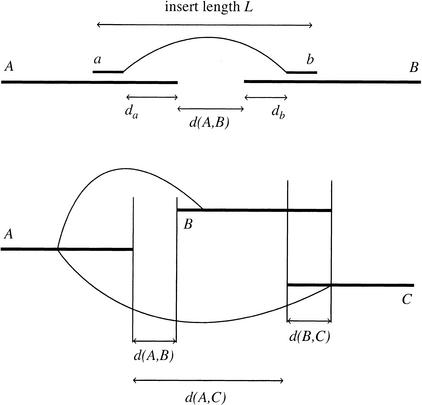
Consistency of Forward-Reverse Links
Contig A is linked with contig B if there are at least two forward-reverse links between reads in A and B. Each forward-reverse link gives an estimated distance between A and B (Fig. 11), which is positive in the case of a gap and negative in the case of an overlap. The mean and standard deviation of the distance between A and B, denoted respectively d(A,B) and ERR(A,B), can be computed from the estimated insert length and standard deviation of each forward-reverse link.
Figure 11.
Consistency of forward-reverse links. (A) The distance d(A,B) (length of gap or negated length of overlap) between two linked contigs A and B can be estimated using the forward-reverse linked reads between them. (B) The distance d(B,C) between two contigs B,C that are linked to the same contig A, can be estimated from their respective distances to the linked contig.
Rule 1. If d(A,B) < −2000 − 4 ERR(A,B) then (super)contigs A and B are marked as repeated. Intuitively, if from the forward-reverse links connecting them, A and B are estimated to overlap by too much, then they are both marked as repeated (and are not subsequently merged with any supercontig).
Rule 2. Similarly, if contig A is linked to contigs B and C, then from d(A,B) and d(B,C) and the lengths of B and C we can compute the estimated gap or overlap length between B and C, denoted d(B,C). We can also compute the standard deviation of d(B,C), denoted ERR(B,C). If d(B,C) < −2000 − 4 ERR(B,C), then we mark contig A as repeated.
The above rules are generalized to apply to supercontigs, so that as supercontigs grow some of them are marked repeated and are not further extended.
Supercontig Assembly
Supercontigs are built incrementally. Initially, every contig is a supercontig, and every subsequent round of merging consists of selecting two supercontigs and merging them, until no more merges can be performed. Specifically, a priority queue (Q) is created, which holds all pairs of supercontigs that could be merged. Each pair of supercontigs has a score that signifies the priority of merging that pair. For two supercontigs, S1, S2, with k ≥ 2 forward-reverse links between them, and estimated distance d(S1,S2), the priority score of merging them is s(S1,S2) = f(k) −| d(S1,S2)|. Intuitively, the first term, f(k), rewards the number of links between S1 and S2, so that supercontigs with more links are merged with higher priority. (In our implementation, f(2), f(3), f(4), … = 50, 875, 1700, 2025, 2350, 2475, 2600, 2625, 2650, 2675. These numbers were derived empirically; however, any scoring scheme that balances between f(k) and |d(S1,S2) | should work. The second term penalizes merging S1 and S2 if doing so would introduce either too much of an overlap or too long a gap.
Initially, Q contains all pairs of contigs that are not marked as repeated and are linked by ≥2 forward-reverse links.
During each round of merging, the following steps are performed:
- 1.
Extract from Q the pair of supercontigs, S1, S2, with the highest priority score.
- 2.
Merge S1, S2, creating supercontig T.
- 3.
Remove all pairs in Q where one of the two supercontigs is either S1 or S2.
- 4.
Find all supercontigs W that share forward-reverse links to T, and insert (T,W) into Q.
- 5.
Apply Rules 1 and 2 for marking repetitive supercontigs, to mark T or any supercontig W linked to T. For any supercontig marked as repetitive, remove all pairs in Q containing it.
The above procedure is first run using only short forward-reverse links (links <10,000 long). It is run again using all the links. In effect, only plasmid links are used in the first round whereas plasmid, cosmid, and BAC links are used in the second round.
Filling Gaps in Supercontigs
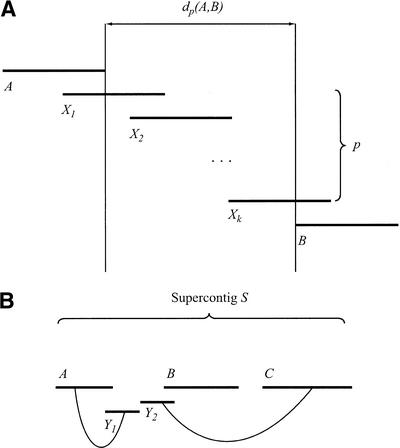
Gaps between two consecutive contigs in supercontigs are filled according to the following algorithm: Graph G = (V,E) is defined, where the set of vertices V is the set of contigs, and an edge connects two contigs if they are known to overlap. A pair of contigs A, B is said to be connected by a path p in G if there are edges (A,X1), …, (Xk,B) in E. This path has an associated distance, dp(A,B), corresponding to the length of sequence between A and B in the path (Fig. 12A).
Figure 12.
Filling gaps in supercontigs. (A) Contigs A and B are connected by a path p of contigs X1, …, Xk. The distance dp(A,B) between A and B (along the path p) is the length of the sequence in the path that does not overlap A or B. (B) Contigs Y1 and Y2 share forward-reverse links with the supercontig S. These links position them in the vicinity of the gap between A and B. Therefore, Y1 and Y2 will be used as possible stepping points in the path closing the gap from A to B.
We first compute, for every pair of contigs A,B in V, the shortest path p in G that connects the contigs (shortest in terms of the distance dp(A,B)). In order to perform this step efficiently, we restrict the computation to finding the shortest path p for which dp(A,B) ≤ 2000. We store the results in a new graph, GPATHS(V,EPATHS), where edges in EPATHS correspond to the computed shortest paths. Assuming that on average a contig has a path of sequence distance ≤ 2000 to a constant number of other contigs, this computation takes time linear in the size of V. In practice, we did not experience bottlenecks in this step.
Then, for every pair of contigs A,B that are consecutive in a supercontig S we attempt to fill the gap between A and B as follows:
- 1.
If (A,B) is in EPATHS, that is, a path has already been computed for A,B, then simply fill the gap with the contigs in the path.
- 2.
Otherwise, first find every contig T that shares forward-reverse links with W, and such that from these links T is positioned between A and B in W (Fig. 12B). The set of all such contigs is called the set of targets, VT.
- 3.
Using breadth-first search, look in GPATHS for a path from A to B that uses only nodes in VT. If a path is found in GPATHS from A to B, this corresponds to a path p in G from A to B. Use the ordered contigs in p to fill the gap between A and B.
Consensus Derivation
Once reads have been laid out, we create consensus sequence efficiently by converting pairwise read alignments (computed during the overlap detection phase; see the section above entitled “Overlap Detection and Alignment: Sort and Extend”) into multiple-read alignments. This process begins with a list of oriented reads (for a given contig), approximate placements for them measured from the beginning of the contig, and all the preexisting pairwise alignments between the reads.
From the pairwise alignments, we select those that are consistent with the placements and with each other. If the placements imply the existence of alignments that we don't already have (which might, for example, correspond to alignments not extending a perfect 24-long match), then we attempt to recover them.
We then find an initial read that (according to the consistent alignments) has no bases to its left. This is usually the leftmost read of the contig. Starting at the left end of the initial read (which we will call the current read), we move base-by-base to the right. At each base, we use the consistent alignments to determine which bases are aligned with it in a multiple alignment. A quality-weighted vote determines the base placed in the consensus and the score assigned to it, which we cap at 40. As we move to the right, at each base, we also reconsider the choice of current read. Switching is necessary at the right end of a read, but also to correctly handle indels and low-quality regions. This process continues until (according to the consistent alignments) there are no bases to the right. Normally, all reads in the contig have been used, yielding a single consensus sequence for the contig.
DISCUSSION
ARACHNE is a whole-genome shotgun assembler designed to exploit sequence data from paired-end reads. The program has been tested on simulated data generated by decomposing genomic sequence from bacterial, yeast, fly, and human chromosomes. The assemblies produced are very good, although not perfect. The vast majority of these genomes are covered in large contigs and supercontigs, with less than one misassembly per megabase and excellent sequence accuracy. The algorithm is also reasonably efficient, being subquadratic in time and linear in space, allowing for efficient scaling to larger genomes.
These results provide confidence that ARACHNE can be used for the assembly of genomes of intermediate size (a few hundred megabases)—such as flies, worms, tunicates, and pufferfish. Indeed, we have used ARACHNE in a project to sequence the 40-Mb genome of the fungus Neurospora crassa (B. Birren et al., in prep.). The assembly can be viewed at www.genome.wi.mit.edu/annotation/fungi/neurospora. In an experimental assembly using a portion of the Neurospora reads provided ∼10X coverage of the genome, we obtained contigs with an N50 size of 52 kb which covered ∼99% of the genome. The contigs were assembled into 323 supercontigs with an N50 size of 256 kb. The accuracy of the assembly was checked by comparing it to 4 Mb of independently generated finished sequence. There were only two discrepancies (one involved exchanging the order of two short contigs within a supercontig; the other involved the incorrect addition of a single sequence read to the end of a contig). ARACHNE reported quality scores of at least 40 (that is, 99.99% accuracy) for 98% of the sequence in the assembly; comparison with finished sequence showed that this sequence was 99.996% accurate. We also used ARACHNE to generate initial assemblies of the genomes of Tetraodon nigroviridans (400 Mb) and Ciona savignyi (180 Mb) (manuscripts in preparation).
We are optimistic that ARACHNE can be used to produce reasonable initial WGS assemblies of large, complex mammalian genomes, such as the human, mouse, and rat. Such initial WGS assemblies are likely to be useful for many applications (although producing high-quality finished sequence of such genomes will require at least some clone-based sequencing).
The tests above show that it is possible to produce reasonable WGS assemblies of human chromosomes 21 and 22. These results are encouraging, although they are based on simulated data that ignore the crucial issue of cloning bias, which may result in underrepresentation of some sequences. Recently, we used ARACHNE (with appropriate memory optimization) to produce an initial WGS assembly from 4X coverage of the mouse genome (available at http://mouse.ensemble.org). The analysis required 8 days on a single Compaq Alpha processor running at 833 MHz and used less than 24-Gb RAM. This demonstrates the feasibility of the program for mammalian-sized genomes.
The current version of ARACHNE (version 1.0) is freely available as both source code and executable for Compaq Alpha machines (www.genome.wi.mit.edu). We are continuing to further develop the program to improve its accuracy and its performance.
Acknowledgments
We thank Bruce Birren, Matt Endrizzi, James Galagan, John Klein, Pen Macdonald, Jerome Naylor, Chad Nusbaum, Ralph Santos, and Mike Zody for useful discussions. We thank Dan Brown for comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lander@wi.mit.edu; FAX (617) 252-1933.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.208902.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Batzoglou S. “Computational genomics: Mapping, comparison, and annotation of genomes.” Ph.D. dissertion. Massachusetts Institute of Technology, Department of Electrical Engineering and Computer Science; 2000. [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chen, T. and Skiena, S.S. 1997. Trie-based data structures for sequence assembly, In: Proceedings of The Eighth Symposium on Combinatorial Pattern Matching, pp. 206–223. Springer-Verlag, New York.
- Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwager C, Zimmermann J, Erfle H, Caskey CT, Ansorge W. Automated DNA sequencing of the human HPRT locus. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using PHRED. I: Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- Green P. PHRAP documentation. 1994. http://www.phrap.org http://www.phrap.org. . [Google Scholar]
- Hattori M, Fujiyama A, Taylor T D, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, Ainscough R, Almeida JP, Babbage A, Bagguley C, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Kim S, Segre AM. AMASS: A structured pattern matching approach to shotgun sequence assembly. J Comp Biol. 1999;6:163–186. doi: 10.1089/cmb.1999.6.163. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–941. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Myers EW. Toward simplifying and accurately formulating fragment assembly. J Comp Biol. 1995;2:275–290. doi: 10.1089/cmb.1995.2.275. [DOI] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KHJ, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequences of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;4:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola H, Sunderland H, Ukkonen E. SEQAID: A DNA sequence assembling program based on a mathematical model. Nucleic Acids Res. 1984;12:307–321. doi: 10.1093/nar/12.1part1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner, P., Tang, H., and Waterman, M.S. 2001. A new approach to fragment assembly in DNA sequencing. Proceedings of the Fifth Annual International Conference in Computational Molecular Biology (RECOMB), April 22–25, 2001, Montreal, pp. 256–267. ACM Press, New York.
- Sanger F, Coulson AR, Hong GF, Hill DF, Peterson GB. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulsen AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, White O, Adams M, Kerlavage A. TIGR assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]